Abstract
The signaling pathways orchestrating both the evolution and development of language in the human brain remain unknown. To date, the transcription factor FOXP2 (forkhead box P2) is the only gene implicated in Mendelian forms of human speech and language dysfunction1,2,3. It has been proposed, that the amino acid composition in the human variant of FOXP2 has undergone accelerated evolution, and this change occurred around the time of language emergence in humans4,5. However, this remains controversial, and whether the acquisition of these amino acids in human FOXP2 has any functional consequence in human neurons remains untested. Here, we demonstrate that these two amino acids alter FOXP2 function by conferring differential transcriptional regulation in vitro. We extend these observations in vivo to human and chimpanzee brain, and use network analysis to identify novel relationships among the differentially expressed genes. These data provide experimental support for the functional relevance of changes in FOXP2 that occur on the human lineage, highlighting specific pathways with direct consequences for human brain development and disease. Since FOXP2 has an important role in speech and language in humans, the identified targets may have a critical function in the development and evolution of language circuitry in humans.
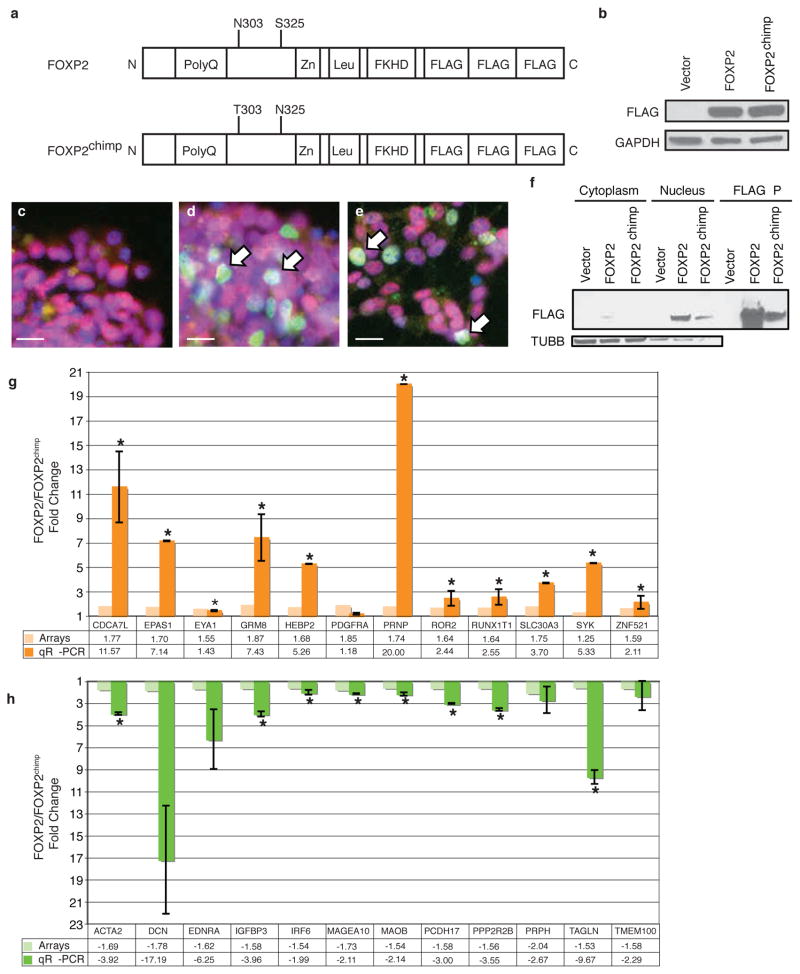
The amino acid structure of FOXP2 had been highly conserved along the mammalian lineage until the common ancestor of humans and chimpanzees, when the human variant of FOXP2 acquired two different amino acids under positive selection, which has been interpreted as evidence for accelerated evolution4,5. To test whether the amino acids under positive selection in human FOXP2 have a distinct biological function, which would support the role of these changes in evolution, we expressed either human FOXP2 or the same construct mutated at two sites to yield the chimpanzee amino acid content, FOXP2chimp, in human neuronal cells without endogenous FOXP2 (Fig. 1a–f). Exogenous FOXP2 protein expressed from both constructs was localized in the nucleus as determined by immunocytochemistry (Fig. 1c–e) and subcellular fractionation (Fig. 1f), consistent with its endogenous expression. To determine if modifying two amino acids leads to changes in gene expression, we conducted whole genome microarray analysis. We identified 61 genes significantly upregulated and 55 genes downregulated by FOXP2 compared to FOXP2chimp (Supplementary Table 1), as well as genes regulated by both FOXP2 and FOXP2chimp (Supplementary Table 2). Interestingly, FOXP2chimp overexpression resulted in more changes in gene regulation than FOXP2 (Supplementary Table 3). In replicate experiments in a different human neuronal cell line, FOXP2chimp again regulated more genes than FOXP2 even though its expression was higher than FOXP2 in these cells (data not shown). To control for any potential confounding effects of FOXP2 levels, we performed correlations of the levels of every gene on the array to either FOXP2 or FOXP2chimp levels, as well as performed random permutation testing, and found no significant differences between other genes’ correlations to either FOXP2 or FOXP2chimp. These data indicate that the differentially expressed genes are not due to different levels of FOXP2 or FOXP2chimp, and are a true indication of differential transcriptional regulation by these two proteins.
Figure 1.
FOXP2 and FOXP2chimp differentially regulate genes in SH-SY5Y cells. a) Schematic of human FOXP2 showing its major functional protein domains and the two amino acid changes in the mutant FOXP2chimp. b) Representative immunoblot for FLAG-tagged FOXP2 and FOXP2chimp stable overexpression in SH-SY5Y cells. c–e) Immunofluorescent staining of antibodies against FLAG epitope (green) and FOXP1 (red), and DAPI (blue) for nuclei. c) Vector cells demonstrate no FLAG expression, while both FOXP2 (d) and FOXP2chimp (e) expressing cells have FLAG-tagged FOXP2 in the cell nucleus. Scale bars are 5 microns. f) Subcellular fractionation followed by immunoblotting. g–h) Quantitative RT-PCR of genes that were differentially expressed in cells expressing FOXP2 compared to FOXP2chimp. Asterisks indicate P≤0.05 and error bars are ± s.e.m. (two-tailed Student’s t-test, n=3 or 4).
To confirm the validity of differentially expressed FOXP2 target genes, we conducted qRT-PCR using independent RNA samples. We confirmed 93% of the FOXP2 upregulated genes and 75% of the downregulated genes examined (Fig. 1g–h and Supplementary Figure 1). Five genes confirmed by qRT-PCR (COL9A1, ROR2, SLIT1, SYK, and TAGLN; Fig. 1g–h and Supplementary Figure 1) were previously identified as direct FOXP2 targets using ChIP-chip6,7. Sixty percent of promoters of the identified differentially expressed genes have at least one canonical FOXP2 binding site, 92% have at least one forkhead domain binding site, and 99% have at least one “core” FOXP2 binding site (Supplementary Table 4). The canonical FOXP2 binding site CAAATT, as well as the core site AAAT, is significantly enriched in the downregulated genes (P=3.3e-04 and P=8.6e-03, respectively) compared to randomly permuting the same number of promoters from the genome. Genes with promoters containing a canonical FOXP2 binding site are likely to be direct FOXP2 or FOXP2chimp targets.
To confirm that these findings were not an artifact of the cell lines used, we further assessed whether a different primary neural cell, human neural progenitors (NHNPs), would exhibit similar differential regulation by FOXP2 and FOXP2chimp. We confirmed one-third of the genes examined in these human cells using both a different method of gene transduction, and populations of cells with greater levels of FOXP2chimp compared to human FOXP2 over-expression, which complements the SH-SY5Y data to further show that the observed relationships are not due to FOXP2 levels (Supplementary Figure 2). As an additional level of validation and to extend the findings to the level of protein, we confirmed two genes, CACNB2 and ENPP2, by immunoblotting in additional SH-SY5Y cell lines (Supplementary Figure 3).
To explore the potential function of the differential FOXP2 targets, we determined enrichment of gene ontology (GO) categories. GO categories enriched for genes upregulated by FOXP2 compared to FOXP2chimp are involved in transcriptional regulation of gene expression and cell-cell signaling. Those GO categories enriched for genes downregulated by FOXP2 compared to FOXP2chimp are important for protein and cell regulation (Supplementary Table 5). These data support the idea that FOXP2 and FOXP2chimp have distinguishable downstream effects as reflected by their differences in gene regulation.
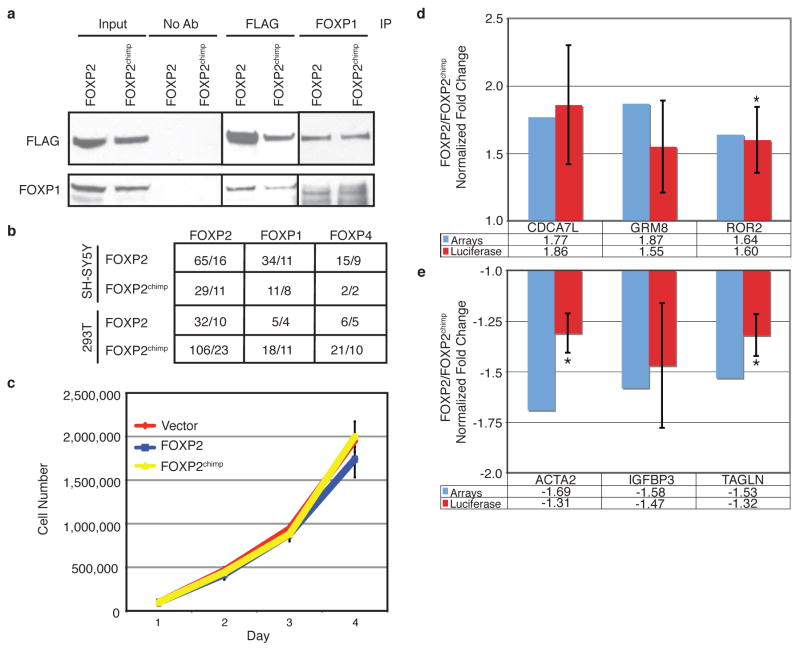
To determine the potential mechanisms by which FOXP2 or FOXP2chimp might differentially regulate gene expression, we first examined whether either protein preferentially interacts with FOXP1 or FOXP4, two proteins known to heterodimerize with FOXP28. Both FOXP2 and FOXP2chimp co-localized with FOXP1 in the cell nucleus, co-immunoprecipitated with FOXP1 as evidenced by immunoblotting, and co-immunoprecipitated with both FOXP1 or FOXP4 when assayed by mass spectrometry (Fig. 1c–e, 2a–b, and Supplementary Fig. 4b–g), ruling out a major difference in FOXP1 or FOXP4 interaction causing differential gene expression. Mass spectrometry showed no significant difference in either co-immunoprecipitation experiment, indicating that differences in hetero- or homo-dimerization did not underlie the observed differences in gene expression between the chimpanzee and human FOXP2. We also tested whether changes in cell proliferation could account for gene expression differences, but did not find significant changes in growth with either FOXP2 construct (Fig. 2c).
Figure 2.
FOXP2 and FOXP2chimp differentially transactivate target promoters independent of FOXP1 or FOXP4 interaction. a) Immunoblotting for FLAG or FOXP1 following immunoprecipitation with either FLAG or FOXP1 Abs. b) Mass spectrometry results from SH-SY5Y or 293T cells overexpressing FOXP2 or FOXP2chimp. The first number indicates the number of spectra and the second is the number of unique peptides. c) Cell growth analysis does not show a significant difference in proliferation between cells expressing FOXP2 or FOXP2chimp over time (P≤0.05). Error bars are ± s.e.m. (two-tailed Student’s t-test, n=3). d–e) Dual luciferase assays in 293T cells transiently transfected with promoter fragments driving luciferase and either FOXP2 or FOXP2chimp. Asterisks indicate P≤0.05 and error bars are ± s.e.m. (two-tailed Student’s t-test, n=3–6).
We next assessed whether FOXP2 and FOXP2chimp expression led to differential promoter transactivation of target genes. We selected eight genes confirmed by qRT-PCR that also contained at least one forkhead binding site (Supplementary Table 6). Six of the promoters tested showed differential regulation by FOXP2 compared to FOXP2chimp in the same direction as the microarrays (Fig. 2d–e), while two did not demonstrate significant transactivation in either direction (data not shown). In contrast, a canonical FOXP2 binding site in triplicate alone, outside of a genomic context, was regulated equally by both FOXP2 and FOXP2chimp (Supplementary Figure 5). Given the complexity of cis-acting gene transactivation elements, these data are particularly compelling considering our use of simplified 5′ promoter regions. These data demonstrate that at least a subset of differentially regulated genes is also differentially transactivated by FOXP2 and FOXP2chimp, indicating they are likely direct FOXP2 targets.
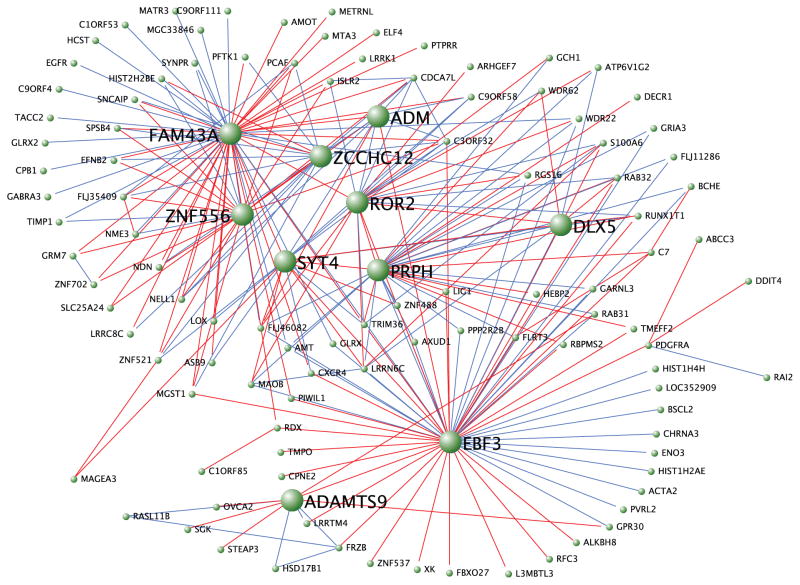
To place these gene expression changes within a more systematic context, we applied weighted gene co-expression network analysis9,10 to the entire SH-SY5Y microarray data set to examine co-regulation of gene expression across all genes. We uncovered two networks where the module eigengene was driven by differences in FOXP2 and FOXP2chimp, and one network driven by similar gene regulation (Figure 3 and Supplementary Figure 6). Using this unsupervised analysis, we found additional genes of interest that do not meet the criteria for differential expression, but that are co-regulated with differences in FOXP2 and FOXP2chimp expression (Supplementary Table 7). Strikingly, two of the genes with the most connections, so-called “hub” genes, in one of the differential networks are DLX5 and SYT4, two genes important for brain development and function11,12.
Figure 3.
Visualization of one of the modules containing FOXP2 and FOXP2chimp differentially expressed genes. Five hundred pairs of genes with the greatest topological overlap are shown. Positive correlations are depicted in red and negative correlations are depicted in blue. The gene symbols for hub genes are accentuated in large bold text.
To extrapolate these findings to true in vivo expression and provide external validation, we compared the differentially expressed genes in SH-SY5Y cells to differentially expressed genes from adult human and chimpanzee brain tissue. We performed microarray analysis on tissue from three brain regions where FOXP2 is expressed in developing brain: caudate nucleus, frontal pole, and hippocampus. We examined gene expression in human compared to chimpanzee for each brain region separately as well as for all brain regions combined, for a total of eight comparisons. There was a significant overlap in seven out of eight of these comparisons, a remarkable convergence with the in vitro data (Table 1). These data are particularly notable, since the tissue was from adult brain. We surmise that a subset of the overlapping differentially expressed genes found in adult brain is the result of differential functions by FOXP2 in the developing brain, and may lead to increased vulnerability to disease. For example, mutations in both FGF14 and PPP2R2B lead to spinocerebellar ataxia (SCA27 and SCA12, respectively), which involves motor-related speech defects13,14. Since both of these genes play a critical role in cerebellar function, it is of note that patients with FOXP2 mutations have decreased gray matter in the cerebellum15, and Foxp2 knockout mice have their most pronounced morphological phenotype in the cerebellum16. Mutations in COL9A1 lead to Stickler syndrome in which patients have craniofacial abnormalities17, and patients with mutations in GJA12 present with ataxia, nystagmus, other motor impairments, and often mental retardation18.
Table 1.
Overlap of cell and in vivo microarray data.
| Upregulated | ||||||
|---|---|---|---|---|---|---|
| Cells | All Brain Areas | Hippocampus | Caudate | Frontal Pole | ||
| 7.36E-04 | 4.49E-02 | 4.04E-02 | 7.21E-02 | Overlap P value | ||
| ADAMTS9 | 1.38 | 1.91 | 1.72 | 2.35 | 1.73 | |
| BCAN | 1.29 | 1.64 | ||||
| COL9A1 | 1.26 | 1.23 | 1.26 | |||
| EXPH5 | 1.27 | 1.41 | 1.29 | 1.43 | 1.53 | |
| FRZB | 1.32 | 1.65 | 1.39 | 2.04 | 1.57 | |
| IGFBP4 | 1.27 | 1.35 | 1.59 | |||
| ISLR2 | 1.30 | 1.27 | 1.53 | |||
| MGST1 | 1.26 | 2.33 | 1.79 | 3.34 | 2.13 | |
| NPTX2 | 1.24 | 1.28 | 1.46 | |||
| PDGFRA | 1.84 | 1.27 | 1.35 | |||
| PRICKLE1 | 1.45 | 1.43 | ||||
| RUNX1T1 | 1.64 | 1.24 | 1.33 | 1.24 | ||
| SLC30A3 | 1.75 | 1.92 | 2.20 | 1.83 | 1.74 | |
| Downregulated | ||||||
| Cells | All Brain Areas | Hippocampus | Caudate | Frontal Pole | ||
| 1.10E-06 | 4.07E-08 | 2.86E-02 | 1.86E-04 | Overlap P value | ||
| ACCN2 | −1.27 | −1.33 | −1.31 | −1.28 | −1.39 | |
| B3GNT1 | −1.24 | −1.73 | −1.47 | −2.55 | −1.39 | |
| C6orf48 | −1.23 | −1.56 | −1.53 | −1.65 | −1.51 | |
| C8orf13 | −1.27 | −1.36 | −1.64 | −1.29 | ||
| CACNB2 | −1.23 | −1.69 | −1.4 | −2.58 | −1.35 | |
| DCN | −1.78 | −1.39 | −1.68 | −1.33 | ||
| ELMO1 | −1.28 | −1.34 | −1.64 | −1.32 | ||
| ENPP2 | −1.39 | −1.43 | −1.72 | |||
| FAM43A | −1.40 | −1.32 | ||||
| FAM43B | −1.40 | −1.41 | ||||
| FGF14 | −1.23 | −1.34 | −1.24 | −1.57 | ||
| FLJ11286 | −1.27 | −1.38 | −1.32 | −1.31 | −1.51 | |
| GJA12 | −1.25 | −1.35 | ||||
| GLRX | −1.29 | −1.30 | ||||
| HIST2H2BE | −1.30 | −1.39 | −1.61 | −1.39 | ||
| IFIT2 | −1.24 | −1.34 | −1.49 | |||
| IGFBP3 | −1.58 | −1.30 | ||||
| MAOB | −1.54 | −1.26 | ||||
| PPP2R2B | −1.56 | −1.23 | −1.66 | −1.35 | −1.41 |
While comparisons of developing brain between human and chimpanzees are challenged by a lack of tissue, a recent study examined gene expression in many regions of human fetal brain19. Comparing the list of 116 differentially expressed genes with those focally expressed during human fetal development, we find 14 genes specifically expressed in one brain region, including FOXP2 (Supplementary Table 8). Two regions of the human fetal brain with high FOXP2 expression19, perisylvian cortex and cerebellum, have a significant number of enriched genes that overlap with the differentially expressed FOXP2 and FOXP2chimp genes (P=1.1e-04 and P=1.3e-04, respectively; Supplementary Table 8). A significant number of the differentially expressed genes are also associated with human-specific accelerated highly conserved noncoding sequences (haCNS), but not with chimpanzee highly conserved noncoding sequences (P=1.2e-06 and P=0.04; Supplementary Table 8)19,20. We confirmed a number of these genes, such as GRM8, MAOB, PPP2R2B, PRICKLE1, RUNX1T1 either by qRT-PCR and/or with the adult in vivo dataset (Figure 1 and Table 1). Together, these data suggest that the FOXP2 differentially expressed genes identified here may have important roles in brain development and patterning, and may also have evolved cis-regulatory elements important for their expression specifically in human brain.
Previously, we identified ChIP-chip targets of FOXP2 that themselves were also under positive selection6. We hypothesized that networks of genes important for language circuitry had been positively selected through selective pressure on human brain evolution. Thus, we also examined whether any differential FOXP2 targets were themselves under positive selection. Five genes (AMT, C6orf48, MAGEA10, PHACTR2, and SH3PXD2B) met the standard criteria of Ka/Ks ≥ 1.0 for positive selection on the human lineage (Supplementary Table 9)21. These data, along with the haCNS and expression data mentioned above, suggest that a subset of differential FOXP2 targets may have co-evolved to regulate pathways involved in higher cognitive functions.
The positive selection of two amino acids in human FOXP2 was previously hypothesized as a mechanism by which human FOXP2 might assume a novel biological function with implications for speech and language evolution4,5. A recent study made an elegant attempt to examine the role of these two amino acids by generating a transgenic mouse with the human version of FOXP222. These mice exhibit a number of interesting phenotypic alterations including increases in dendritic length in striatal neurons and changes in ultrasonic vocalizations, as well as some modest changes in gene expression. Although the mouse is an experimentally tractable model system, from a strictly evolutionary standpoint, the interpretation of data obtained in the mouse specifically for the study of human evolution is challenged by the vast differences in human and mouse brain and the amount of time since the human and mouse common ancestor diverged (70 million years23). Here, we demonstrate that these two amino acid changes have a functional consequence in human cells, validate these differences in vivo in tissue, and elucidate some of the downstream pathways affected by this adaptive evolutionary change.
Using whole genome microarrays, we uncovered genes that are differentially regulated upon mutation of these two amino acids, including some with functions critical to the development of the human CNS. Moreover, this study reveals enrichment of differential FOXP2 targets with known involvement in cerebellar motor function, craniofacial formation, and cartilage and connective tissue formation, suggesting an important role for human FOXP2 in establishing both the neural circuitry and physical structures needed for spoken language. The significant overlap of human FOXP2 targets in cell lines with genes enriched in human compared to chimpanzee brain tissue presents the possibility that human and chimpanzee FOXP2 have differentially regulated targets during brain development. As suggested by King and Wilson over 30 years ago24, and reaffirmed by the sequencing of both the human and chimpanzee genomes, the phenotypic differences exhibited by humans and chimpanzees cannot be explained by differences in DNA sequence alone, and are likely due to differences in gene expression and regulation. Previous microarray studies identified differences in gene expression between human and chimpanzee brains25,26. Here, we link new whole genome expression microarray data from human and chimpanzee brain to direct differences in gene regulation by the human and chimpanzee version of the transcription factor FOXP2. Since normal FOXP2 function is critical for speech in humans, these differentially regulated targets may be relevant to the evolution and establishment or function of pathways necessary for speech and language in humans.
Methods Summary
Cell Culture and Stable Line Generation
SH-SY5Y cells (ATCC) and human fetal neuronal progenitors (Lonza) were grown according to the manufacturer’s instructions, with some modifications (see Methods).
Microarrays
Total RNA was extracted using Qiagen’s RNeasy kit. Illumina HumanRef-8 v2 (SH-SY5Y samples) or v3 (tissue samples) were used and analyzed as described27. Sample information is in Methods.
Full Methods accompany this paper.
Methods
Antibodies
The following antibodies were either used for immunoblotting (IB) or immunofluorescence (IF): anti-FLAG (mouse monoclonal, Sigma; 1:10,000 (IB), 1:10,000 (IF)); anti-GAPDH (mouse monoclonal, Chemicon; 1:2500 (IB)); anti-beta-tubulin (rabbit polyclonal, Abcam; 1:1000 (IB); anti-FOXP1 (6; 1:5000 (IB), 1:1000 (IF); anti-CACNB2 (mouse monoclonal, Abcam; 1:100 (IB)); anti-ENPP2 (rabbit polyclonal, Cayman Chemical; 1:400 (IB)); goat anti-rabbit horseradish peroxidase (Cell Signaling, 1:2500); goat anti-mouse horseradish peroxidase (Chemicon, 1:5000); goat anti-mouse Alexa Fluor 488 (Invitrogen, 1:1500); goat anti-rabbit Alexa Fluor 594 (Invitrogen, 1:1500).
Cell Culture and Stable Line Generation
Stable SH-SY5Y cell lines were generated by transfecting cells with pCMV-Tag4a expression constructs using FuGENE (Roche Applied Science) according to the manufacturer’s instructions. Populations of stable cells were selected using 1mg/ml Geneticin (Invitrogen). Multiple independent lines were generated from independent transfections. Stable human fetal neuronal progenitors cell lines were generated by transducing cells with lentiviruses as previously described28. FOXP2-producing lentiviral vectors were generated by replacing the eGFP in pLUGIP (ATCC) with FOXP2.
Immunoprecipitation
Nuclear extract were incubated with either 1 μg of FLAG antibody (Sigma) or a polyclonal FOXP1 antibody6.
Cell Proliferation Assay
Equal numbers of cells (2.0E+04) were plated on time zero and counted every subsequent day after trypsinization using a hemacytometer.
Dual luciferase assays
293T cells (ATCC) were transfected with 50ng of reporter construct expressing Photinus pyralis (firefly) luciferase, 1ng of Renilla luciferase plasmid (pRL-EF), and 50ng of pCMV-Tag4a FOXP2 expression plasmid using FuGENE (Roche Applied Science) according to the manufacturer’s instructions. Forty-eight hours later, cells were lysed and analyzed using the dual luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Co-transfection of Renilla was used for transfection normalization, and values were additionally normalized to cells transfected with a promoter-less luciferase construct. Promoter information is in Supplementary Table 6. The canonical FOXP2 binding site driving luciferase was generated by cloning AATTTG in triplicate into pGL4 (Promega).
Gene Ontology Analysis
GO analysis was performed as described6 using DAVID (http://david.abcc.ncifcrf.gov). The differentially expressed genes were compared to all of the genes on the microarrays and a P value computed using a Fisher’s Exact Test.
Immunoblotting
Whole cell protein lysates were generated and immunoblotted as described28.
Immunofluorescence
Cells were grown on glass coverslips, fixed in 2% paraformaldehyde, and permeabilized in 0.2% Triton-X. TBST containing 10% milk, and 10% normal goat serum was used as blocking solution at room temperature for one hour. Antibodies were diluted in TBS with 0.25% BSA, 0.25% normal goat serum and 0.1% Triton-X and applied to cells overnight at 4°C. Secondary antibodies were diluted in blocking solution and added at room temperature for one hour. Coverslips were mounted to glass slides and images taken using a Zeiss Axio Imager D1.
Mass Spectrometry
FOXP2 immunoprecipitates were precipitated by the addition of trichloroacetic acid and proteolyzed by the sequential addition of Lys-C and trypsin proteases29. Digested peptide samples were then analyzed by mass spectrometry as described29. Proteins were considered to be present in a sample if at least two peptides per protein were identified using a false positive rate of less than 5% per peptide as determined using a decoy database strategy30.
Microarrays
For the SH-SY5Y data, we analyzed four biological replicates of each genotype from three independently generated cell lines for a total of 12 microarrays per genotype. Each of these cell lines was created from populations of cells rather than single clones, and as such, the expression data represents changes from hundreds of independent integrations throughout the cells’ genomes. Further, as the endogenous FOXP2 expression is very low in SH-SY5Y cells, the potential confound of heterodimerization with endogenous human FOXP2 is mitigated in these cells. For the tissue data, we analyzed three to six independent samples for each brain region in each species.
Permutation Testing
For FOXP2 correlations, we computed the average correlation for each gene on the microarray to either the level of the human or the chimpanzee FOXP2. We then derived the absolute difference in correlation for each gene between the human and chimpanzee FOXP2 arrays. The average of these differences was not statistically different from performing the same test, but randomizing the correlation values for all of the genes on the arrays or using the values from only the differentially expressed genes. For promoter binding site calculations, we calculated the number of promoters from differentially expressed genes with a given motif and compared them to the average number from a random selection of the same number of promoters from the genome. We assumed a normal distribution and a Z-score less than 0.05 was called significant. Similar analysis was done for comparing genes with a haCNS and expression in human fetal brain. For microarray overlap comparisons, we included the number of differentially expressed genes as well as the total number of probesets on the microarrays for each comparison. We used a hypergeometric distribution test with 10,000 permutations to calculate the mean and standard deviation of the overlap. We assumed a normal distribution and a Z-score less than 0.05 was called significant.
Real-time PCR
RNA extraction and RT-PCR was performed as described6. Primer sequences are in Supplementary Table 10.
Site-directed mutagenesis
Mutagenesis of pCMV-Tag4a/FOXP26 was carried out using GeneTailor Site-Directed Mutagenesis System (Invitrogen) according the manufacturer’s instructions using the following primers: site 1 (asparagine to threonine): F-5′-CCTCCTCGACTACCTCCTCCACAACTTCCAAAGC-3′; R-5′-GGAGGAGGTAGTCGAGGAGGAATTGTTAGTA-3′; site 2 (serine to asparagine): F-5′-ATGGACAGTCTTCAGTTCTAAACGCAAGACGAGA-3′; R-5′-TAGAACTGAAGACTGTCCATTCACTATGGAA-3′. Mutagenesis was confirmed by both sequencing and mass spectrometry.
Weighted Gene Coexpression Network Analysis (WGCNA)
WGCNA was performed as previously described9,10. Briefly, genes were chosen for inclusion into the network based on their consistent presence on the array and high coefficient of variation, and they were clustered based on their topological overlap. For each module, singular value decomposition (X = UDV′) was performed, and the expression was re-calculated without the first principal component because it corresponded to cell line differences. The modules reported in this study were created using expression data with the first principal component removed, as it represented an experimental batch effect.
Supplementary Material
Acknowledgments
We thank Michael Oldham for generating the Illumina microarray mask file, Jing Ou and Elizabeth Spiteri for performing site-directed mutagenesis, Leslie Chen for technical assistance, and Lauren Kawaguchi for lab management. Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD (NICHD Contract No. N01-HD-4-3368 and N01-HD-4-3383). The role of the NICHD Brain and Tissue Bank is to distribute tissue, and therefore, cannot endorse the studies performed or the interpretation of results. This work was supported by grant R21MH075028, R37MH60233-06A1 (D.H.G.), T32HD007032, an A.P. Giannini Foundation Medical Research Fellowship, and a NARSAD Young Investigator Award (G.K.), T32MH073526 (K.W.), and a James S. McDonnell Foundation grant, JSMF#21002093 (T.M.P).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions G.K. and D.H.G. designed the study, analyzed the data, and wrote the paper; G.K. performed all of the experiments; J.M.B. made contributions to an earlier phase of the project including generating cell lines, immunoblotting and qRT-PCR; K.W. performed statistical analysis and WGCNA analysis; G.C. conducted promoter analysis, and G.C. and F.G. analyzed the microarray data; Z.O.J. and J.A.W. performed mass spectrometry; S.P. performed some of the qRT-PCR; T.M.P. performed tissue dissections and provided non-human primate samples; all authors discussed the results and commented on the manuscript.
Author Information Gene expression data have been deposited into NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo), and are accessible using GEO series accession number GSE18142.
References
- 1.Feuk L, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 3.MacDermot KD, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiteri E, et al. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vernes SC, et al. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81:1232–1250. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol CellBiol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
- 11.Acampora D, et al. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 12.Yoshihara M, Adolfsen B, Galle KT, Littleton JT. Retrograde signaling by Syt 4 induces presynaptic release and synapse-specific growth. Science. 2005;310:858–863. doi: 10.1126/science.1117541. [DOI] [PubMed] [Google Scholar]
- 13.Brusse E, et al. Spinocerebellar ataxia associated with a mutation in the fibroblast growth factor 14 gene (SCA27): A new phenotype. Mov Disord. 2006;21:396–401. doi: 10.1002/mds.20708. [DOI] [PubMed] [Google Scholar]
- 14.Holmes SE, et al. Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat Genet. 1999;23:391–392. doi: 10.1038/70493. [DOI] [PubMed] [Google Scholar]
- 15.Belton E, Salmond CH, Watkins KE, Vargha-Khadem F, Gadian DG. Bilateral brain abnormalities associated with dominantly inherited verbal and orofacial dyspraxia. Hum Brain Mapp. 2003;18:194–200. doi: 10.1002/hbm.10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu W, et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci U S A. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Camp G, et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am J Hum Genet. 2006;79:449–457. doi: 10.1086/506478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlenberg B, et al. Mutations in the gene encoding gap junction protein alpha 12 (connexin 46.6) cause Pelizaeus-Merzbacher-like disease. Am J Hum Genet. 2004;75:251–260. doi: 10.1086/422763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 21.Dorus S, et al. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 22.Enard W, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 24.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 25.Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 26.Caceres M, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coppola G, et al. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol. 2008;64:92–96. doi: 10.1002/ana.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konopka G, Tekiela J, Iverson M, Wells C, Duncan SA. Junctional adhesion molecule-A is critical for the formation of pseudocanaliculi and modulates E-cadherin expression in hepatic cells. J Biol Chem. 2007;282:28137–28148. doi: 10.1074/jbc.M703592200. [DOI] [PubMed] [Google Scholar]
- 29.Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 30.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.