Abstract
Microscopy is an essential tool for analysis of cellular structures and function. With the advent of new fluorescent probes and super-resolution light microscopy techniques, the study of dynamic processes in living cells has been greatly facilitated. Fluorescence light microscopy provides analytical, quantitative, and three-dimensional (3D) data with emphasis on analysis of live cells using fluorescent markers. Sample preparation is easy and relatively inexpensive, and the use of appropriate tags provides the ability to track specific proteins of interest. Of course, only electron microscopy (EM) achieves the highest definition in terms of ultrastructure and protein labeling. To fill the gap between light microscopy and EM, correlative light and electron microscopy (CLEM) strategies have been developed. In particular, hybrid techniques based upon immuno-EM provide sensitive protein detection combined with high-resolution information on cell structures and protein localization. By adding the third dimension to EM with electron tomography (ET) combined with rapid freezing, CLEM techniques now provide additional tools for quantitative 3D analysis. Here, we overview the major methods applied and highlight the latest advances in the field of CLEM. We then focus on two selected techniques that use cryosections as substrate for combined biomolecular imaging. Finally, we provide a perspective of future developments in the field. (J Histochem Cytochem 57:1103–1112, 2009)
Keywords: correlative light and electron microscopy, electron microscopy, EM tomography, morphometry, confocal microscopy, cryosection, immunofluorescence, immunogold, tetracysteine biarsenical system, quantum dots
According to the resolution equation theorized by Emile Verdet, Ernst Abbe, and Lord Rayleigh (Verdet 1869; Abbe 1873; Lord Rayleigh 1896), lens-based light microscopes can resolve two objects that are separated by ∼200 nm. As a consequence, images of objects that are closer than a quarter of a micron are blurred into one single diffracted spot, so that fluorescent signals cannot be unequivocally assigned to a specific structure. Mathematically, Abbe's expression states that the resolution of a light microscope is limited to λ/[2nsinα] in the focal plane (xy) and 2/[nsin2α] along the optical axis (z), where the wavelength of the light is used, n is the refractive index, and [nsinα] is the numerical aperture of the lens. Because the majority of lenses have a numerical aperture of <1.5 (α is <70°), the common resolution limit for cell imaging is 150 nm laterally and 500 nm in the axial plane. Although many textbooks still report this paradigm, an increasing number of studies demonstrate that fluorescence light microscopy (FLM) has developed into “nanoscopy,” revolutionizing the view of sub-cellular life. We are now able to analyze large data sets of digital images and produce high–fidelity, three-dimensional (3D) models with spatial resolution on the order of 50–100 nm (Hell and Stelzer 1992; Schrader and Hell 1996; Betzig et al. 2006; Gustafsson 2008; Gustafsson et al. 2008; Huang et al. 2008; Lippincott-Schwartz and Manley 2009). Overcoming the diffraction limit has made it possible to observe and zoom in on dynamic events as they occur and even visualize protein–protein interactions and single molecules in living cells. At the extreme of the resolution spectrum, electron microscopy (EM) provides near-atomic-level spatial resolution, but prevents live-cell imaging. Despite this limitation, EM provides the structural basis for the understanding of molecular and cellular functions. Indeed, cell imaging in a nearly native state with high-pressure freezing (HPF) and 3D tomography are two recent breakthrough innovations in electron microscopy (Studer et al. 2008; Hoenger and McIntosh 2009). These implementations have greatly facilitated the accumulation of new insights into the realm of the 3D structural organization of cells. Indeed, the giant leaps forward taken by both fields (FLM and EM) can be considered essentially the results of synergistic innovations in light and electron microscopy equipment, protocols, and fluorescence probes. Correlative light and electron microscopy (CLEM) methods integrate such synergistic innovations, bridging the gap between FLM and EM (Table 1). Although CLEM refers to light microscopy in general, in our review, the term is mostly used for describing CLEM methods involving FLM.
Table 1.
Imaging features of fluorescence and electron microscopy
| Fluorescence microscopy | Electron microscopy | |
|---|---|---|
| Resolution power | + | +++ |
| Observation of living cells | + | — |
| Identification of labeled structures | + | + |
| Identification of unlabeled structures |
— |
+ |
CLEM: Is Two Better Than One?
During its history, CLEM has proven to be a powerful hybrid interface aimed at combining the best of the light and electron microscopy worlds. In fact, no single microscopy technique is capable of providing all of the desired information. Although light microscopy offers the rapid screening of a large area of the sample at relatively low resolution, fast determination of labeling specificity, and the simultaneous detection of multiple antigens by live-cell imaging, EM provides the unique “reference space” where all objects (labeled and unlabeled) can be visually explored at the highest resolution. The most-informative approach would be the convergence of these two worlds, allowing direct correlation of the data sets obtained from one sample (Figures 1 and 2). However, CLEM is disadvantaged by the complexity of the protocols, which are often time-consuming and laborious (Hayat 1987; Schwarz and Humbel 2007). The requirement of expensive equipment and specific expertise has also prevented the expansion of the CLEM field. Other drawbacks include the lack of efficient multimodal labeling strategies and the limited quantitative interpretability of EM. These factors have motivated a flood of efforts whose major goal is to combine existing/new probes and advanced optical equipment in novel easy-to-apply procedures.
Figure 1.
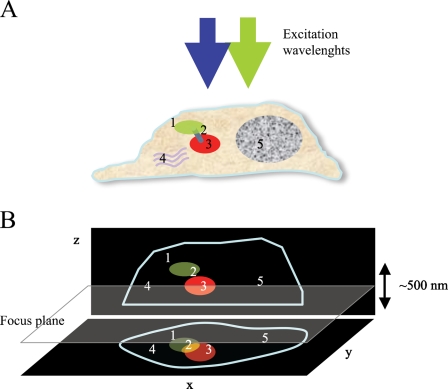
Advantages of correlative light and electron microscopy (CLEM). (A) Five features of a cell are highlighted (1–5). Blue and green arrows indicate different excitation wavelengths used for light microscopy imaging. Whereas fluorescence imaging allows for the identification only of labeled items (1 and 3), electron microscopy (EM) provides the reference space where all objects are visible (1–5). Due to the limited axial resolution of fluorescence light microscopy (FLM), objects that are closer than 200 nm are blurred into a single spot in the x-y projection (B). CLEM combines two techniques to obtain the highest resolution and information about structures or events occurring within cells.
Figure 2.
Example of CLEM using cryosections and the high-data-output CLEM (HDO-CLEM) method. Smooth Russell bodies (SRBs, tubular shape) (A–C) and rough Russell bodies (RRBs, round shape) (D–K)–expressing HeLa cells (Mattioli et al. 2006) have been analyzed by EM and light microscopy. SRBs represent enlarged endoplasmic reticulum–Golgi intermediate compartments, whereas RRBs represent enlarged rough endoplasmic reticulum (RER) compartments. Immunofluorescence staining identifies μ-ΔCH1 (Cy3), a recombinantly expressed mutant immunoglobulin chain accumulated in both SRBs and RRBs. Green labeling refers to calreticulin, a marker for RER. (A) Immunofluorescence labeling for μ-ΔCH1 (Cy3). Confocal laser scanning microscopy (CLSM) optical sectioning poorly resolves the complex tubular architecture of an SRB imaged through the xy middle plane of a three-dimensional (3D) stack; xz axial views along the dotted lines are shown. (B) Transparent surface rendering of the 3D stack shown in A, obtained by MICROSCOBIOJ software. The 3D organization of the SRB tubular structures is not identifiable. The white dot in B corresponds to the cross-point of the dotted lines in A. (C) Immunogold labeling for μ-ΔCH1. Transmission electron microscopy (TEM) image of a 200-nm-thick cryosection of an SRB showing the intricate tubular structure. (D–G) Double immunolabeling on 60-nm or 200-nm (H–K) cryosections of RRBs. (D,H) Immunofluorescence colocalization of calreticulin (Cy3, red) and μ-ΔCH1 (Cy2, green). Nuclei labeled with 4′,6-diamidino-2-phenylindole (blue). Images were collected either by CLSM (D) or by wide-field microscopy (WFM) (H). (G–K) Immunogold labeling colocalization of calreticulin (10 nm) and μ-ΔCH1 (15 nm) of the same sections shown in D and H. (G,K) Higher magnification of the squared areas in E,F and I,J, respectively. Bars: A = 2 μm; C,F,J = 1 μm; D,E,H,I = 5 μm; G,K = 0.5 μm. With permission, Vicidomini et al. Traffic 9:1828–1838, 2008.
Hybrid Labeling Strategies for CLEM
With the emergence of the green fluorescent protein (GFP) and its variants, in the early 1990s, and novel hybrid probes, e.g., fluorescence derivatives of nanogold, such as FluoroNanoGold (Takizawa and Robinson 2000), there has been increased interest in fluorescent proteins for CLEM techniques. The major advantage of FluoroNanoGold is that smaller reagents yield enhanced labeling efficiency. However, these reagents rely on silver enhancement to amplify the nanogold signal, which is otherwise extremely difficult to detect on ultrathin sections. Although promising, the use of FluoroNanoGold has been limited to a few applications in fixed cells. Recently, two captivating labeling options have been developed, the tetracysteine system and quantum dots (Qdots). The tetracysteine-ReAsH system, originally designed by the Roger Tsien group (Griffin et al. 1998; Giepmans et al. 2006; Sosinsky et al. 2007; Adams and Tsien 2008; Giepmans 2008), is an EM-visible tag in which proteins can be genetically fused to express a 12-residue peptide sequence containing tetracysteine additions, which binds membrane-permeable, bi-arsenical compounds with high affinity. Besides other applications, this system represents an important hybrid labeling technique for CLEM studies. After exposing the cells to a membrane-permeant, non-fluorescent, biarsenical derivative of fluorescein, FlAsH–1,2-ethanedithiol (EDT)2, the presence of the tetracysteine motif causes the dye to fluoresce strongly at green/yellow wavelengths. Another biarsenical derivative of FlAsH is the red fluorophore ReAsH-EDT2. REAsH emission peaks strongly at red wavelengths, so that the use of both FlAsH and ReAsH in a time-sequential fashion enables pulse–chase experiments to be performed. Dithiols are added to minimize binding and toxicity to endogenous proteins. Importantly, ReAsH can be applied for CLEM studies using photoconversion of diaminobenzidine (DAB), which is added to the cells. This photooxidation reaction polymerizes the DAB, which then reacts readily with the osmium fixative and becomes easily visible by EM. This approach has been successfully used to study the dynamics of different pools of connexin43 within gap junctions (Gaietta et al. 2002) and more recently, in the visualization of Golgi twins in late mitosis by live-cell imaging (Gaietta et al. 2006). Although representing an excellent alternative to large fluorescence proteins (e.g., GFP) for live cells, limitations in the FlAsH–ReAsH method are the possibility of performing multiple labeling only at the light microscopy level and the requirement for ectopic expression of protein, with the consequent potential for perturbation of the molecule of interest. Nonspecific labeling in light microscopy is another drawback that must be taken into consideration (Martin et al. 2005). Therefore, this method should be used carefully and in parallel with other approaches.
Qdots
A promising alternative labeling approach for correlative microscopy comes from the use of Qdots. Qdots are a class of non–genetically encoded probes commonly used in single-molecule imaging. With respect to other dyes, Qdots have intriguing optical properties: superior brightness, minimal photobleaching, and size-dependent simultaneous excitation of multiple fluorescence colors. Qdots are typically synthesized from various semiconductor materials that can provide a wide range of potential emission wavelengths, and are used as either core-only or core-shell structures. Core nanoparticles containing CdSe or CdTe (cadmium selenide or telluride, respectively) coated by a ZnS (zinc sulfide) shell and by a polymer coat that allows linking of biomolecules are more suitable for biological applications, inasmuch as the shell passivates the cores and improves their optical properties. (Delehanty et al. 2009). Moreover, the electron-dense crystal is directly visible by EM, allowing straightforward switching between FLM and EM. Several sizes and shapes of Qdots have been developed to gain a variety of fluorescent properties that allow multiple detection of different targets. The different sizes and shapes of Qdot cores make them also suitable for multiple labeling at the EM level. In some recent applications in cellular imaging, Qdots have been conjugated with streptavidin, which was used as a tag to recognize a biotinylated antibody binding connexin43 in gap junctions (Giepmans et al. 2005). Other studies report their use in identifying the distribution of proteins in the nucleus and other cellular compartments (Wu et al. 2003). With the dawn of the nano era, Qdot applications are at the cutting edge of the nanomedicine field. With other nanomaterials, Qdots are widely used as a platform to study and manipulate stem cells (Ferreira et al. 2008). Qdots also offer a unique platform for studying the dynamics of biomolecules, as demonstrated for surface trafficking of neurotransmitter receptors (Groc et al. 2007). However, the use of Qdots in live-cell imaging is still challenging because of their poor biocompatibility. A large fraction of Qdot size comes from the passivating layer, often a polyacrylic acid polymer or phospholipid micelle. However, it is difficult to reduce the passivating layer without increasing nonspecific interactions between Qdots and cells, self-aggregation, and reduction of the quantum emission. Indeed, despite the fact that the size of the core as seen in EM is in the 5–10 nm range, Qdot nanoparticles conjugated to biomolecules (antibodies, peptides, DNAs), are typically 20–30 nm in diameter for red-emitting Qdots, which restricts their use to permeabilized cells or extracellular or endocytosed proteins. Qdot multivalency is also a concern, inasmuch as surface cross-linking might dramatically modify receptor mobility and signaling pathways (Saxton and Jacobson 1997). Recently, Ting and coworkers successfully engineered smaller Qdots, directly coupled with the detection probe (e.g., monovalent streptavidin) (Howarth et al. 2008). This improvement resulted in more-efficient labeling of synapses, compared with the larger, older generation of Qdots. However, whether monovalent Qdots are more useful new tools applicable for CLEM than the first generation will have to be tested. Another possibility is the combination of Qdot labeling with ET, to localize specific targets in three dimensions. Although challenging, this approach will help in elucidating the in situ topology of proteins.
Peroxidase- and Photoconversion-based CLEM Protocols
The peroxidase substrate DAB was originally introduced as an EM labeling method by Graham and Karnovsky (1966) for the localization of horseradish peroxidase (HRP) used as a fluid-phase tracer. First attempts to use the photooxidation of DAB for CLEM studies were obtained upon strong illumination of a fluorescent molecule in conventional EM, i.e., Lucifer Yellow (Maranto 1982). Other initial CLEM studies were performed by conjugating a secondary antibody used for immunofluorescence with a fluorochrome such as eosin 5-isothiocyanate (BODIPY), which is, again, a substrate sensitive for photooxidation of DAB (Deerinck et al. 1994). DAB photooxidation, a reaction similar to the classic HRP, generates an electron-dense, insoluble polymer clearly visible by conventional EM. As a result, the excited fluorophore becomes detectable by the electron microscope. Because eosin is smaller in size than other conventional markers, this method results in improved penetration of labeling reagents, compared with gold- or enzyme-based procedures. This strategy has been successfully applied to label specific lipids in their target organelles, for example, BODIPY ceramide staining in the trans-Golgi network (Pagano et al. 1989). Attempts to use GFP to catalyze photoconversion of DAB have been generally unsuccessful, probably owing to the intrinsic shielding properties of GFP. However, a recent study successfully showed the development of a correlative method for direct photooxidation of GFP. This method was used to visualize the intra-Golgi stack and intracisternal distribution of a human Golgi resident glycosylation enzyme, nacetylgalactosaminyltransferase-2 fused either to enhanced GFP or cyan fluorescent protein in plastic-embedded cells (Grabenbauer et al. 2005). In addition, the photoconversion of different fluorochromes [GFP, BODIPY, and yellow fluorescent protein (YFP)] and different overexpressed target proteins (PDI, golgin 84, SR-B1, and GAT1) has been recently optimized by methodological adaptations of the DAB photooxidation step, providing high-quality ultrastructural localization, 3D tomographic analysis of complex cellular architectures, and correlative microscopy of consecutive ultrathin sections (Meiblitzer-Ruppitsch et al. 2008). In another set of studies, researchers added the temporal dimension to CLEM, matching real-time imaging on single cells of GFP-vesicular stomatitis virus G protein (VSVG) with DAB immunoelectron microscopy (IEM) (Mironov et al. 2000,2003; Polishchuk et al. 2000). This technique was successfully applied for the analysis of transport intermediates from the Golgi apparatus to the plasma membrane and between endoplasmic reticulum (ER) and Golgi, revealing an unexpected complexity of the VSVG transport carriers. The immunolabeling was directed against the lumenal part of the transport structures, and so the precipitate formed by the subsequent oxidation of DAB delineated these structures and allowed their detection in EM. In spite of the optimal preservation of samples, the limitation with photooxidation, and generally of methods using the substrate DAB, is that they work best for ectopically expressed proteins concentrated within a specific organelle. Early in 2009, the transport of cargo VSVG (VSVGpEGFP) across individual Golgi stacks (visualized by GalT-venusYFP) was monitored by super-resolution 4Pi-microscopy and segmented with a zero-crossing procedure. In the same report, the distribution of procollagen-3 within isolated Golgi stacks was mapped by 4Pi microscopy and correlated by nanogold labeling instead of DAB oxidation, providing the first comparative analysis of their imaging capabilities (4Pi-CLEM) (Perinetti et al. 2009).
CLEM and Cryofixation: The Leading Edge
To reduce the artifacts associated with conventional EM preparations (Murk et al. 2003), investigators have started to use HPF and cryo-electron tomography (cryo-ET) in CLEM experiments. The native state of a structure is best captured using rapid freezing techniques, and frozen specimens can either be examined directly, avoiding dehydration artifacts, or they can be freeze-substituted, replacing the vitreous ice with a solvent, usually acetone, at low temperature. For example, the development of Caenorhabditis elegans embryos was monitored in FLM and stopped at specific stages of early mitosis using HPF (Muller-Reichert et al. 2007). Other progress with fast-frozen material comes from the Parton group (Nixon et al. 2009). Their work shows a simple and robust CLEM method suitable for whole animals (e.g., zebrafish). This method allows clear visualization of expressed (cav3-GFP) in the notochord and muscle fibers, and provides excellent ultrastructure and immunolabeling for EM. The procedure is also compatible with EM tomography, providing an important tool for researchers who need to perform CLEM with frozen materials. Cryo-ET is an emerging technique that has already produced some impressive results; however, the main limitations are the high sensitivity of the biological samples to electron irradiation, the sample quality, and ice thickness. Recently, two independent groups have developed strategies for correlative light-electron microscopy and cryo-tomography (Lucic et al. 2007; Sartori et al. 2007; Schwartz et al. 2007; Plitzko et al. 2009). Sartori and coworkers successfully overcame most of the aforementioned limitations and obtained correlated light-electron cryo-tomography of mature neurons and keratinocytes. This method is suitable for investigating complex cellular patterns and has the advantage that correlation is obtained without exposing the sample to extra irradiation and does not rely on visual recognition of cellular features. They introduced a novel, software-based system that allows for direct correlation of positions in two coordinate systems, such as pixels on FLM images with goniometer positions at which EM micrographs were recorded. Schwartz and coworkers have built a cryo-light microscope stage that allows for the generation of light microscopy images of samples prepared for cryo-EM, using vital dyes or fluorescent proteins as markers. Extremely low-temperature imaging (−140C) also provided the additional advantage of limited photobleaching. In collaboration with Leica Microsystems, Paul Verkade created the Rapid Transfer System (RTS) for HPF, a device attached to the Leica EMPACT2 (Verkade 2008). The RTS is an automated system that allows for very fast processing of samples, making possible the use of fast freezing for correlative studies. Indeed, with the RTS system, it is possible to capture specific events occurring in living cells as observed by light microscopy, cryofix the sample within 4 sec, and analyze that event at high resolution by EM. Other investigators developed an integrated confocal and electron microscope for CLEM. This electron microscope provides a proof-of-principle setup for correlative microscopy carried out at room temperature (Agronskaia et al. 2008). All these approaches are of great interest in both life science and material science, and represent significant improvements for the routine use of CLEM, increased throughput, and success rate of experiments.
CLEM and Tokuyasu Cryosections: Exploiting the Advantages of a Powerful Technique
Although producing some artifacts, high-resolution IEM methods have been profoundly successful. Among them, cryo-immunogold on ultrathin cryosections (referred to as the Tokuyasu method) (Tokuyasu 1973; Koster and Klumperman 2003; Slot and Geuze 2007), recently optimized for immunoreactions on fast-frozen sections (Van Donselaar et al. 2007), is an elegantly simple protocol that guarantees almost-perfect visualization of cell membranes and excellent labeling of single or multiple antigens by the use of small gold particles. Recently, the Tokuyasu method, in combination with EM tomography (immunoelectron tomography), has been successfully demonstrated by Zeuschner et al. (2006). In this study, the authors developed approaches to combine quantitative immunogold labeling (to identify COPII vesicles) and 3D ET of ER exit sites, establishing the existence of free COPII-coated transport carriers at the ER–Golgi interface. A limitation of immunolabeling of sectioned material is that it underestimates numbers of a particular protein in a 3D volume because high-affinity binding with gold-conjugated antibodies or protein A relies on accessibility of epitopes. Therefore, immunolabeling of cryosections is essentially considered a 2D (surface) method, and cannot be regarded as quantitative for 3D labeling. In addition, the Tokuyasu method does not allow proper visualization of cytoskeleton elements. In early CLEM procedures based upon cryo-immunolabeling, immunofluorescence on cryosections, using either standard fluorochromes and/or GFP-tagged proteins, was successfully combined with immunogold to label endogenous or ectopically expressed proteins (Robinson et al. 2001; Takizawa and Robinson 2003a,b). These studies demonstrated that EM correlation of immunofluorescence performed on ultrathin cryosections is relatively simple and provides several advantages. First, it works equally well for detection of single and multiple molecules, endogenous or ectopically expressed. Second, antigenicity is preserved, owing to mild fixation. Third, fluorescence images are exceptionally sharp, z-axis resolution is higher (only limited by the section thickness), signal-to-noise ratio is lower, and the chromatic aberration can be reduced. Hence, although new preparation techniques are emerging, we think that the Tokuyasu method stands out beyond other procedures for its simplicity, efficiency, and rapidity of observable results. The potential of immuno-EM is illustrated below in two CLEM protocols that have been recently released: the software-based high-data-output CLEM (HDO-CLEM) and the live-cell CLEM (van Rijnsoever et al. 2008; Vicidomini et al. 2008).
HDO-CLEM: Integrating Still-cell Imaging With Immuno-EM
Our group recently set up the HDO-CLEM with the aim of defining a method offering maximal labeling efficiency and statistically relevant data for correlative studies (Vicidomini et al. 2008). The protocol is suitable for (serial) cryosections, allowing both immunofluorescence and immunolabeling of endogenous and ectopically expressed proteins. The first advantage of HDO-CLEM relies on the ability to obtain statistically significant data. This is obtained by correlation of large-scale or synchronized events occurring in a population of living cells, visualized after EM processing in single or serial cryosections. The second advantage is the provision of new tools to perform 3D analysis and correlation. We designed the software MicroSCoBioJ, a collection of three distinct plug-ins able to create 3D multicolor volume rendering from 2D images, at a resolution intermediate between FLM and EM tomography (http://imagejdocu.tudor.lu/doku.php?id=plugin:stacks:microscobioj:start) (Frosi et al. 2008). Any study of cellular structures with details below the limit of confocal axial resolution can benefit from this method. The use of MicroSCoBioJ has been fundamental to highlighting previously unresolved tubular interconnections of rough and smooth Russell bodies, used as a model system (Mattioli et al. 2006). With HDO-CLEM, the fine architecture of Russell bodies was better interpreted and analyzed on a large number of organelles. The third improvement addresses the possibility of performing hybrid morphometry analysis based on the evaluation of a systematic error, mainly due to image blurring, occurring between FLM and EM samples. Correcting large-scale measurements obtained by FLM with the systematic error would improve these measurements to the accuracy of EM. For this reason, HDO-CLEM applications might include the comparison of labeling density of a cell compartment by correcting the fluorescence pixel intensity with the measurements obtained by HDO-CLEM morphometry. In Figure 3, we report the flowchart of the HDO-CLEM protocol.
Figure 3.
HDO-CLEM: Schematic workflow. Pellets from cell cultures are prepared according to the Tokuyasu technique (trimmed block). Ribbons of serial sections are placed on “finder” grids and immunolabeled for light and electron microscopy. A thin layer of methylcellulose is added to the sections to protect them from damage and photobleaching. Grids are next mounted on glass coverslips with glycerol and imaged by WFM and/or CLSM. Once imaging has been completed, grids are extensively washed and stained for EM with uranyl acetate and methylcellulose. Single or serial sections are scanned for areas of interest using grid references and analyzed by standard EM or electron tomography (ET). FLM and TEM-ET images are processed.
Live-cell CLEM: Integrating Live-cell Imaging With Immuno-EM
The Klumperman group in 2008 published a method conceived as a variant of their previous flat-embedding/CLEM protocol (Oorschot et al. 2002). Live-cell CLEM is aimed at studying the dynamics of organelles, shape remodeling events, and a wide range of dynamic processes within a limited number of cells. The protocol combines live-cell imaging of fluorescently tagged proteins with immunogold labeling of ultrathin cryosections (van Rijnsoever et al. 2008). Briefly, gridded coverslips are coated with an optimized mixture that allows the efficient release of cells destined for CLEM after in vivo imaging. Once imaging is completed, oriented cells are easily detached from the coverslip and processed for flat-embedding immuno-EM. Using this method, the authors successfully monitored the kinetics and localization of the lysosomal membrane protein LAMP-1 in endosomes loaded with dextran and/or transferrin in live cells. This approach is very useful and provides all the benefits intrinsic to the immunogold technique. Because fixation is gentle compared with conventional EM, there is only limited cross-linking, generally resulting in well-preserved epitope antigenicity. The HDO-CLEM and live-cell CLEM approaches add to the growing number of CLEM techniques, and both are especially suited for those research subjects that require fine structural information and the quantitative analysis of multiple proteins.
Innovative applications of CLEM based on immuno-EM are emerging in the complex field of neurobiology. The nervous system displays an intricate architecture and complex synapses that have hampered its full exploration with the current imaging techniques. The array tomography method (Micheva and Smith 2007) is an automated imaging tool designed to image complex patterns of neural circuits. The design of array tomography is in principle similar to the HDO-CLEM method, but differs in the sample preparation technique and electron microscopy analysis. In particular, array tomography is based upon repeated cycles of immunolabeling of serial ultrathin conventional sections, followed by 3D reconstruction and scanning electron microscope analysis. Array tomography promises new ways to address many upcoming questions in neurobiology, such as neuron classification and relationships in situ, distribution of receptors, and ion channels, within the tissue context and circuit connectivity.
We have so far reported several CLEM methods and labeling approaches. Each of them appears to have a favorite field of application and specific advantages and disadvantages. In Tables 2 and 3 we have schematically illustrated and summarized the principal labeling strategies available for CLEM studies, their applicability, and the major advantages and limitations of each technique selected.
Table 2.
Five CLEM selected techniques
| Live cells | LM, fixed cells | Endogenous antigens | Ectopic-expressed antigens | Multi-labeling LM | Multi-labeling EM | |
|---|---|---|---|---|---|---|
| Cryo-embedding | ||||||
| HDO-CLEM | + | +++ | +++ | +++ | Yes | Yes |
| Live-cell CLEM | +++ | − | − | +++ | Yes | Yes |
| Plastic embedding | ||||||
| QDots | + | ++ | ++ | ++ | Yes | Yes |
| FlASH/ReAsH | +++ | +++ | − | +++ | Yes | No |
| GFP photooxidation |
+++ |
+++ |
− |
+++ |
Yes |
No |
LM, light microscopy; CLEM, correlative light and electron microscopy; QDot, quantum dot; GFP, green fluorescent protein; EM, electron microscopy. The applicability of each method is indicated on a scale ranging from negative (−) to most-optimal (+++). Key characteristics of five CLEM techniques are compared. To determine localization of endogenous proteins in cells, immuno-based methods are preferred (HDO-CLEM, immunogold methods, and QDots). GFP and FlAsH/ReAsH genetic labeling are suitable for live-cell imaging, including pulse–chase experiments with the tetracysteine tag. EM detection can be performed by immuno-based methods or direct photooxidation for GFP (live-cell CLEM and GFP photooxidation) and DAB photooxidation for ReAsH labeling. For light microscopy imaging requiring photostability, Qdots are the best choice. For correlative studies, Qdots of different size can be used in fixed cells to discriminate multiple endogenous or overexpressed proteins simultaneously.
Table 3.
Advantages and disadvantages of each selected technique
| Advantages | Disadvantages | |
|---|---|---|
| Cryo-embedding | ||
| HDO-CLEM | High-efficiency, single and multiple events | Antibody penetration (except nanogold) |
| Live-cell CLEM | High-efficiency, single and multiple events; in vivo imaging | Antibody penetration (except nanogold); limited number of cells |
| QDots | Brightness, photostability, hybrid, multiple events | Background, large size |
| Plastic embedding | ||
| FlASH/ReAsH | In vivo, tag small size | Overexpression; background LM; single labeling for EM |
| GFP photooxidation |
In vivo specificity |
Overexpression; single labeling |
Main advantages and disadvantages of five CLEM techniques are illustrated. CLEM techniques that use cryo-embedded material show the highest potential for in vivo imaging (live-cell CLEM), high efficiency, and possibility of performing single and multiple labeling (HDO-CLEM and live-cell CLEM). However, these methods are both limited in their quantitative aspects, owing to the limited penetration of an antibody within the section thickness. For statistical purposes, a limited number of cells can be analyzed with live-cell CLEM, whereas HDO-CLEM allows for potential analysis of an entire ribbon of cryosections. For techniques that use plastic-embedded material, QDots are an excellent choice for direct hybrid labeling of single or multiple antigens as well as for light microscopy imaging that requires high illumination doses. Further improvements in QDot technology are expected, especially the production of smaller and more-specific QDots. Genetic labeling with both GFP and the tetracysteine system allows for in vivo imaging, and both are highly specific for their target; however, they rely on ectopic expression of the protein of interest. Although GFP-targeted proteins can be detected at the EM level by direct photooxidation or indirectly by immunogold, only ReAsH is visible at the electron microscope upon DAB photooxidation.
CLEM: The Future
Forecasting the future of a rapidly evolving subject is potentially misleading, but we offer some indication of the directions toward which the field of CLEM is moving. EM has undergone enormous evolution over recent years with innovations like tomography, HPF, and cryo-tomography. Since super-resolution fluorescence techniques came out, many groups have been working intensively on improvements and applications of nanoscopy, a novel domain for optical microscopy. The emergence of collaborative research institutions, where biologists and physicists work together to develop new methods to approach biological questions, is one future path that advanced microscopy will follow. We think that future CLEM will be deeply integrated and complementary to these novel super-resolution techniques (e.g., STED, PALM, STORM, 4Pi, RESOLFT, FPALM). Some progress in this direction has already been made with the integration of 4Pi microscopy and CLEM (Perinetti et al. 2009). As stated by Lippincott-Schwartz and Manley (2009): “Unorthodox super-resolution microscopy discoveries will also need support from … electron microscopy. The latter is especially important as it provides the needed nanometer-scale resolution of cell ultrastructure to correlate with super resolution images. Hence, as the focus of SR microscopy shifts from novelty to biological application, careful controls using correlative electron microscopy and other strategies must be adopted to ensure the results are accurate.” Although super-resolution light microscopy provides more-precise information about subcellular structures, these methods do not reveal ultrastructural components and typically require high illumination doses, which may limit their use to intact cells and make the correlative approach even more important. In addition, many open questions regarding the molecular architectures of molecular players involved in cellular processes still have to be resolved. CLEM methods, in particular those involving the emerging cryo-tomography techniques, have great potential in imaging cellular features at the molecular level in many functional states. Integration of super-resolution light microscopy techniques with cryo-tomography may, in principle, allow detection of individual fluorescently labeled molecules in correlated cryo-tomograms. In the future, many improvements are expected to make CLEM and cryo-ET more attractive. In particular, we expect that software designed for the correlative procedure will be further developed to develop high-throughput and easy computer routines suitable for different correlative approaches with minimal interference by users (e.g., automated pattern recognition software). Moreover, the sample preparation and optimization aspect remains one of the most important tasks, especially for correlative methods based upon HPF and cryo-tomography, likely representing a key aspect for further developments of these procedures. We forecast that CLEM, integrated with the different options to study functional cell architecture and protein organization, will be placed among the best tools to decipher the complexity of cellular events. With the 3D and live-cell current capabilities, we expect that the expansion of CLEM will greatly accelerate in the next few years.
Acknowledgments
We thank Mike J. Waters (Institute Molecular Bioscience, University of Queensland, Brisbane, Australia) for useful comments. We also acknowledge all the members of MicroScoBio Center, in particular Giuseppe Vicidomini and Maria Cristina Gagliani for helpful discussions. K.C. is grateful to Davide Pagnotta (London Business School, UK), Maura Porta (Midwestern University, Downers Grove, IL), and Gabriele Garzoglio (Fermi National Accelerator Laboratory, Batavia, IL) for support and useful comments.
This work was financially supported by the Italian Cancer Research Foundation, Italian Minister of Research and University (Programmi di ricerca di Rilevante Interesse Nazionale), and San Paolo Foundation (to CT).
References
- Abbe E (1873) Beitrage zur Theorie des Mikroskops und der Mikroskopischen Wahrnehmung. Arch Mikr Anat 9:413–468 [Google Scholar]
- Adams S, Tsien R (2008) Preparation of the membrane-permeant biarsenicals FlAsH-EDT2 and ReAsH-EDT2 for fluorescent labeling of tetracysteine-tagged proteins. Nat Protoc 3:1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agronskaia A, Valentijn J, Van Driel L, Schneijdenberg C, Humbel B, van Bergen en Henegouwen P, Verkleij A, et al. (2008) Integrated fluorescence and trasmission electron microscopy. J Struct Biol 164:183–189 [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645 [DOI] [PubMed] [Google Scholar]
- Deerinck T, Martone M, Lev-Ram V, Green D, Tsien R, Spector D, Huang S, et al. (1994) Fluorescence photooxidation with eosin: a method for high-resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol 126:901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delehanty JB, Mattoussi H, Medintz IL (2009) Delivering quantum dots into cells: strategies, progress and remaining issues. Anal Bioanal Chem 393:1091–1105 [DOI] [PubMed] [Google Scholar]
- Ferreira L, Karp J, Nobre L, Langer R (2008) New opportunities: the use of nanotechnologies to manipulate and track stem cells. Cell Stem Cell 3:136–146 [DOI] [PubMed] [Google Scholar]
- Frosi F, Vicidomini G, Diaspro A, Tacchetti C, Boccacci P (2008) MicroSCoBioJ (ImageJ documentation Wiki) home page. http://imagejdocu.tudor.lu/doku.php?id=plugin:stacks:microscobioj:start (last modified October 10, 2008)
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, et al. (2002) Multicolor and electron microscopic imaging of connexin trafficking. Science 296:503–507 [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, et al. (2006) Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci USA 103:17777–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans B (2008) Bridging fluorescence microscopy with electron microscopy. Histochem Cell Biol 130:211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans B, Adams S, Ellisman M, Tsien R (2006) The fluorescence toolbox for assessing protein location and function. Science 312:217–223 [DOI] [PubMed] [Google Scholar]
- Giepmans B, Deerinck T, Smarr B, Jones Y, Ellisman M (2005) Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat Methods 2:743–749 [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts W, Fernadez-Rodriguez J, Hoenger A, Koster A, Nilsson T (2005) Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods 11:857–862 [DOI] [PubMed] [Google Scholar]
- Graham R, Karnovsky M (1966) The early stages of absorption of injected horseradish peroxidase in the proximal tubules of mouse kidney: ultrastructural cytochemistry by a new technique. J Histochem Cytochem 14:291–302 [DOI] [PubMed] [Google Scholar]
- Griffin B, Adams S, Tsien R (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269–272 [DOI] [PubMed] [Google Scholar]
- Groc L, Lafourcade M, Heine M, Renner M, Racine V, Sibarita J, Lounis B, et al. (2007) Surface trafficking of neurotransmitter receptor: comparison between single-molecule/quantum dots strategies. J Neurosci 27:12433–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG (2008) Super-resolution light microscopy goes live. Nat Methods 5:385–387 [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, et al. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J 94:4957–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat M (1987) Correlative microscopy in biology. In Hayat MA, ed. Instrumentation and Methods. London, Academic Press.
- Hell S, Stelzer E (1992) Fundamental improvement of resolution with a 4Pi-confocal fluorescence microscope using two-photon excitation. Opt Commun 93:277–282 [Google Scholar]
- Hoenger A, McIntosh J (2009) Probing the macromolecular organization of cells by electron tomography. Curr Opin Cell Biol 21:89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall L, Schmidt M, Wittrup K, et al. (2008) Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat Methods 5:397–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X (2008) Three-dimensional super resolution imaging by stochastic optical reconstruction microscopy. Science 319:810–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster AJ, Klumperman J (2003) Electron microscopy in cell biology: integrating structure and function. Nat Rev Mol Cell Biol 4(suppl):SS6–10 [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Manley S (2009) Putting super-resolution fluorescence microscopy to work. Nat Methods 6:21–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord Rayleigh (1896) On the theory of optical images, with special reference to the microscope. Philos Mag XLII:167–195
- Lucic V, Kossel A, Yang T, Bonhoeffer T, Baumeister W, Sartori A (2007) Multiscale imaging of neurons grown in culture: from light microscopy to cryo-electron tomography. J Struct Biol 160:146–156 [DOI] [PubMed] [Google Scholar]
- Maranto A (1982) Neuronal mapping: a photooxidation reaction makes Lucifer yellow useful for electron microscopy. Science 217:953–955 [DOI] [PubMed] [Google Scholar]
- Martin B, Giepmans B, Adams S, Tsien R (2005) Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol 23:1308–1314 [DOI] [PubMed] [Google Scholar]
- Mattioli L, Anelli T, Fagioli C, Tacchetti C, Sitia R, Valetti C (2006) ER storage diseases: a role for ERGIC-53 in controlling the formation and shape of Russell bodies. J Cell Sci 119:2532–2541 [DOI] [PubMed] [Google Scholar]
- Meiblitzer-Ruppitsch C, Vetterlein M, Stangl H, Maier S, Neumuller J, Freissmuth M, Pavelka M, et al. (2008) Electron microscopic visualization of fluorescent signals in cellular compartments and organelles by means of DAB-photoconversion. Histochem Cell Biol 130:407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva K, Smith S (2007) Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 55:25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A, Mironov AJ, Beznoussenko G, Trucco A, Lupetti P, Smith J, Geerts W, et al. (2003) ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev Cell 5:583–594 [DOI] [PubMed] [Google Scholar]
- Mironov A, Polishchuk R, Luini A (2000) Visualizing membrane traffic in vivo by combined video fluorescence and 3D electron microscopy. Trends Cell Biol 10:349–353 [DOI] [PubMed] [Google Scholar]
- Muller-Reichert T, Srayko M, Hyman A, O'Toole E, McDonald K (2007) Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol 79:101–119 [DOI] [PubMed] [Google Scholar]
- Murk J, Posthuma G, Koster A, Geuze H, Verkleij A, Kleijmeer M, Humbel B (2003) Influence of aldehyde fixation on the morphology of endosomes and lysosomes: quantitative analysis and electron tomography. J Microsc 212:81–90 [DOI] [PubMed] [Google Scholar]
- Nixon S, Webb R, Floetenmeyer M, Schieber N, Lo H, Parton R (2009) A single method for cryofixation and correlative light, electron microscopy and tomography of zebrafish embryos. Traffic 10:131–136 [DOI] [PubMed] [Google Scholar]
- Oorschot V, de Wit H, Annaert W, Klumperman J (2002) A novel flat-embedding method to prepare ultrathin cryosections from cultured cells in their in situ orientation. J Histochem Cytochem 50:1067–1080 [DOI] [PubMed] [Google Scholar]
- Pagano R, Sepanski M, Martin O (1989) Molecular trapping of fluorescent ceramide analogue at the Golgi apparatus of fixed cells: interaction with endogenous lipids provides a trans-Golgi marker for both light and electron microscopy. J Cell Biol 109:2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perinetti G, Muller T, Spaar A, Polishchuk R, Luini A, Egner A (2009) Correlation of 4Pi- and electron microscopy to study transport through single Golgi stacks in living cells with super resolution. Traffic 10:379–391 [DOI] [PubMed] [Google Scholar]
- Plitzko J, Rigort A, Leis A (2009) Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes. Curr Opin Biotechnol 20:83–89 [DOI] [PubMed] [Google Scholar]
- Polishchuk R, Polishchuk E, Marra P, Alberti S, Buccione R, Luini A, Mironov A (2000) Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. J Cell Biol 148:45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Takizawa T, Pombo A, Cook P (2001) Correlative fluorescence and electron microscopy on ultrathin cryosections: bridging the resolution gap. J Histochem Cytochem 49:803–808 [DOI] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko J (2007) Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol 160:135–145 [DOI] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K (1997) Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct 26:373–399 [DOI] [PubMed] [Google Scholar]
- Schrader M, Hell S (1996) 4Pi-confocal images with axial super-resolution. J Microsc 183:189–193 [Google Scholar]
- Schwartz C, Sarbash V, Ataullakhanov F, McIntosh J, Nicastro D (2007) Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J Microsc 227:98–109 [DOI] [PubMed] [Google Scholar]
- Schwarz H, Humbel B (2007) Correlative light and electron microscopy using immunolabeled resin sections. In Electron Microscopy: Methods and Protocols. 2nd ed. Totowa, NJ, Humana Press, Inc. [DOI] [PubMed]
- Slot J, Geuze H (2007) Cryosectioning and immunolabeling. Nat Protoc 2:2480–2491 [DOI] [PubMed] [Google Scholar]
- Sosinsky G, Giepmans G, Deerinck T, Gaietta G, Ellisman M (2007) Markers for correlated light and electron microscopy. Methods Cell Biol 79:575–591 [DOI] [PubMed] [Google Scholar]
- Studer D, Humbel B, Chiquet M (2008) Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem Cell Biol 130:877–889 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Robinson J (2000) Fluoronanogold is a bifunctional immunoprobe for correlative fluorescence and electron microscopy. J Histochem Cytochem 48:481–485 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Robinson J (2003a) Correlative microscopy of ultrathin cryosections is a powerful tool for placental research. Placenta 24:557–565 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Robinson J (2003b) Ultrathin cryosections: an important tool for immunofluorescence and correlative microscopy. J Histochem Cytochem 51:707–714 [DOI] [PubMed] [Google Scholar]
- Tokuyasu K (1973) A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol 57:551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Donselaar E, Posthuma G, Zeuschner D, Humbel B, Slot J (2007) Immunogold labeling of cryosections of high-pressure frozen cells. Traffic 8:471–485 [DOI] [PubMed] [Google Scholar]
- van Rijnsoever C, Oorschot V, Klumperman J (2008) Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat Methods 11:973–980 [DOI] [PubMed] [Google Scholar]
- Verdet E (1869) Leçons d'optique physique. Paris, Victor Masson et fils
- Verkade P (2008) Moving EM: the rapid transfer system as a new tool for correlative light and electron microscopy and high throughput for high-pressure freezing. J Microsc 230:317–328 [DOI] [PubMed] [Google Scholar]
- Vicidomini G, Gagliani M, Canfora M, Cortese K, Frosi F, Santangelo C, Di Fiore P, et al. (2008) High data output and automated 3D correlative light-electron microscopy method. Traffic 9:1828–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu H, Liu W, Haley K, Treadway J, Larson J, Ge N, et al. (2003) Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol 21:2410–2491 [DOI] [PubMed] [Google Scholar]
- Zeuschner D, Geerts W, Van Donselaar E, Humbel B, Slot J, Koster A, Klumperman J (2006) Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat Cell Biol 8:377–383 [DOI] [PubMed] [Google Scholar]