Abstract
The putative protein C7orf24 is encoded by Homo sapiens chromosome 7 open reading frame 24. C7orf24 was first identified as a 21-kDa cytochrome c–releasing factor detected in the cytosolic fraction of human leukemia U937 cells after treatment with geranylgeraniol. C7orf24 protein was recently identified as a γ-glutamyl cyclotransferase, an enzyme in the γ-glutamyl cycle. However, the exact localization of C7orf24 mRNA in normal tissues remains unknown. The present study examined the distribution pattern of C7orf24 mRNA in rat tissues using reverse transcription-polymerase chain reaction (RT-PCR) and in situ hybridization histochemistry. The RT-PCR experiments demonstrated that C7orf24 and a variant C7orf24 mRNA were expressed in various tissues. Quantitative RT-PCR analysis revealed significantly high levels of both C7orf24 mRNAs in the liver and kidney, compared with other tissues examined. In situ hybridization histochemistry localized C7orf24 mRNA to hepatocytes in the liver and renal tubules in the kidney. The present results thus implicated an important role for C7orf24 in liver and kidney. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials. (J Histochem Cytochem 57:1121–1126, 2009)
Keywords: glutathione, detoxification, drug metabolism, in situ hybridization
The putative protein C7orf24 is encoded by Homo sapiens chromosome 7 open reading frame 24 (GenBank accession number: NM_024051). The C7orf24 protein was first identified as a 21-kDa cytochrome c–releasing factor in the cytosolic fraction of human leukemia U937 cells after treatment with geranylgeraniol (Masuda et al. 2006). Geranylgeraniol has potent apoptosis-inducing activity in various tumor cell lines, implicating the C7orf24 protein in apoptotic pathways of cancer cells (Masuda et al. 2006).
Independently, Kageyama et al. (2007) identified C7orf24 as a highly expressed protein in cancer cells in a proteomic analysis of bladder cancer. The expression was markedly increased in several cancer cell lines, and the knockdown of C7orf24 expression using small interfering RNAs induced significant anti-proliferative effects (Kageyama et al. 2007). DNA microarray analysis also flagged the C7orf24 gene as upregulated in cancer cells (Zhang et al. 2006; Xu et al. 2007). On the basis of these findings, C7orf24 was proposed as a novel cancer marker protein, although the functional role of C7orf24 in normal tissues has not been elucidated.
Recently, Oakley et al. (2008) reported C7orf24 as a γ-glutamyl cyclotransferase, an enzyme in the γ-glutamyl cycle, that mediates the degradation and synthesis of glutathione (Orlowski and Meister 1973; Meister and Anderson 1983). Although the distribution of γ-glutamyl cyclotransferase expression has been characterized in rat and mouse tissues at the protein level (Orlowski and Meister 1973; Tulchin and Taylor 1981), little information is available about its mRNA distribution in normal tissues. This study therefore investigated the distribution and expression levels of C7orf24 mRNA in rat tissues using quantitative reverse transcription-polymerase chain reaction (RT-PCR) and in situ hybridization histochemistry.
Materials and Methods
Animals
Adult male Wistar rats weighing 220–360 g were used in this study, in accordance with the Public Health Service policy on the humane care and use of laboratory animals, the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the Animal Welfare Act (7 U.S.C. et seq.). The institutional animal care and use committee of the Shiga University of Medical Science approved all experimental protocols.
Tissue Preparation
Three rats were used for the RT-PCR analysis. Under deep anesthesia with sodium pentobarbital (80 mg/kg body weight), rats were transcardially perfused with 10 mM phosphate-buffered saline (PBS), pH 7.4, which was pretreated with diethyl pyrocarbonate (DEPC) (Nacalai Tesque; Osaka, Japan) to protect against RNase action. The rat tissues were dissected out for mRNA analysis.
For in situ hybridization, three rats were put under deep anesthesia with sodium pentobarbital (80 mg/kg body weight) and covered with ice. The animals were then transcardially perfused with ∼200 ml of DEPC-treated 10 mM PBS, followed by 300 ml of 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB). The animals were kept in ice for 30 min and subsequently perfused with 1 liter of 15% sucrose containing 0.1% (v/v) DEPC. The kidney and liver were dissected out and snap frozen using CO2 gas. Sections were cut at 20-μm thickness in a cryostat and picked up on RNase-free silane-coated glass microscope slides (Dako Japan; Tokyo, Japan). Slides were stored at −80C until use.
RT-PCR Analysis
Total RNA was isolated from the cortex, choroid plexus, eye, lung, heart, stomach, liver, spleen, kidney, adrenal gland, small intestine, large intestine, and urinary bladder using an RNeasy Mini Kit (QIAGEN; Tokyo, Japan) according to the manufacturer's instructions. The total RNA was incubated for 1 hr with 10 U RNase-free DNase (Roche Applied Science; Mannheim, Germany) and 20 U recombinant RNase inhibitor (Wako Pure Chemicals; Osaka, Japan) at 37C to eliminate DNA contamination. Ten micrograms of each total RNA sample was reverse transcribed for the first-strand cDNA synthesis for 1 hr at 42C using SuperScript III (Life Technologies; Gaithersburg, MD) and 500 pmol of oligo dT12-18 (Amersham Biosciences; Piscataway, NJ). The samples were then heated for 5 min at 65C to inactivate the reverse transcriptase, before a further incubation for 20 min with 100 U/ml RNase H (Invitrogen; Carlsbad, CA).
The C7orf24 mRNA was amplified using a set of primers designed to amplify the entire coding region: sense primer, 5′-ATGGCGAGCTCCGACTGTGAGGAC-3′; antisense primer, 5′-TTATAACAGTTTTGCCTCTCCCTT-3′. The PCR reaction mixture consisted of 2 ng/μl of the template cDNA, 0.8 μM of each primer, 0.2 mM of each of the four deoxynucleotide triphosphates, and 2.0 U Taq polymerase (AmpliTaqGold, Perkin Elmer Japan; Tokyo, Japan) dissolved in 1× PCR buffer containing 1.5 mM MgCl2. After heat activation for 10 min at 95C, products were amplified using the following thermal cycle profile: (1) denaturation at 95C for 30 sec, (2) annealing at 60C for 30 sec, and (3) extension at 72C for 60 sec. The PCR was run for 36 cycles, and obtained products were subcloned into the pCR2.1 vector using a TA cloning kit (Invitrogen; San Diego, CA). Plasmid DNA was prepared by the alkaline lysis method, and the target inserts were sequenced using an ABI 3100 DNA sequencer (Applied Biosystems; Foster City, CA).
Real-time PCR on a LightCycler (Roche Applied Science) was used to detect C7orf24 mRNA using the sense primer, 5′-AAGTCCTGGCGATGAAGTGTGG-3′, and the antisense primer, 5′-CCAGGACTTTGAAAAATGGTAG-3′. Real-time PCR analysis for a variant mRNA of C7orf24 was employed using the sense primer, 5′-AGGTGGCACGGAGACAAGAAGGAG-3′, and the antisense primer, 5′-CTCCTTTTGTCTCCGTGCCACCT-3′. Real-time PCR was also run for β-actin mRNA to assess the variability of mRNA content. The sense and antisense primers for β-actin were 5′-TGGTGGGTATGGGTCAGAAGGACTC-3′ and 5′-CATGGCTGGGGTGTTGAAGGTCTCA-3′, respectively. A plasmid containing C7orf24, its variant, or β-actin cDNA was also analyzed to construct a standard curve for quantitating the real-time PCR.
cDNA was amplified using a real-time PCR procedure with the LC Faststart DNA Master SYBR Green I Kit (Takara Bio; Otsu, Japan) in a Light Cycler instrument. Each reaction volume of 20 μl contained 4 μl of cDNA and 16 μl of a reaction mixture comprising 10 μl of SYBR Premix Ex Taq II (2×), 0.4 μl of each primer (20 pmol/μl), and 5.2 μl of autoclaved distilled water. Cycling conditions typically included an initial denaturation step of 30 sec at 95C, followed by 60 cycles of 15 sec at 95C, 5 sec at 60C, and 15 sec at 72C. The detection of amplicons was verified using the melting curve analysis feature of the LightCycler instrument.
In Situ Hybridization Histochemistry
Digoxigenin (DIG)-labeled riboprobes for detecting C7orf24 mRNA were prepared as follows. The entire coding region of rat C7orf24 mRNA was subcloned into the pGEM-T Easy vector (Promega; Madison, WI) using the TA cloning kit (Invitrogen; San Diego). Sense and antisense riboprobes were prepared by transcribing the pGEM-T Easy vector with T7 and Sp6 RNA polymerase after linearization with SalI and NcoI, respectively. Transcription reactions were done in the presence of DIG–uridine triphosphate using a DIG RNA labeling kit (Roche Diagnostics; Basel, Switzerland) according to the manufacturer's instructions. DIG-labeled riboprobes were purified by ethanol precipitation.
The expression of C7orf24 mRNA in rat kidney and liver was examined by in situ hybridization histochemistry. Sections mounted on RNase-free silane-coated glass slides were immersed for 10 min in DEPC-treated 0.1 M PBS, pH 7.4. Sections were then treated for 10 min at room temperature with 10 μg/ml of proteinase K in 10 mM Tris-HCl buffer, pH 8.0, containing 150 mM NaCl, before postfixing in 4% PFA in 0.1 M PB for 10 min at room temperature. The sections were washed three times for 5 min each with 0.1 M PBS. The sections were prehybridized for 1 hr at 37C in hybridization buffer containing 50% formamide, 5× Denhardt's solution, 1× saline/sodium citrate (SSC) (1× SSC = 150 mM NaCl and 15 mM sodium citrate), 0.5 mg/ml yeast tRNA (Invitrogen; San Diego), and 0.5 mg/ml of heat-denatured salmon sperm DNA (Wako Pure Chemicals). The probes were diluted in hybridization buffer to a final concentration of 2 μg/ml, and hybridization was performed overnight at 60C.
After hybridization, the sections were washed briefly in prewarmed 1× SSC at 60C, and then rinsed for 2 hr in prewarmed 0.1× SSC buffer at 60C, before a final rinse for 5 min at room temperature in 0.1 M Tris-HCl, pH 7.5, containing 150 mM NaCl (NT buffer). Subsequently, the sections were treated for 60 min with 1% skim milk in NT buffer to block nonspecific protein binding, and then reacted overnight at 4C with alkaline phosphatase–labeled anti-DIG antibody (Roche Diagnostics; Mannheim, Germany) diluted 1:200, in NT buffer containing 1% skim milk. After washing with NT buffer, positive signals were detected by incubating in 0.1 M Tris-HCl buffer, pH 9.5, containing 100 mM NaCl, 50 mM MgCl2, 500 μg/ml of nitroblue tetrazolium chloride, and 187 μg/ml of 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt.
Results
RT-PCR Analysis
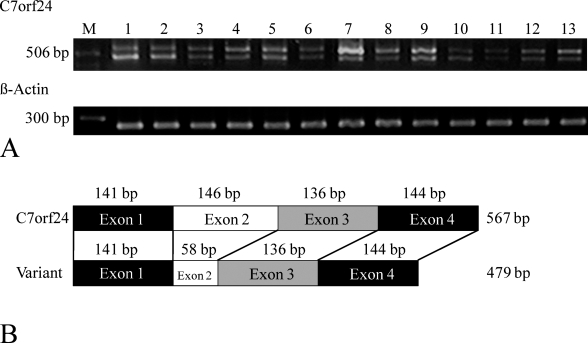
Figure 1A shows a typical RT-PCR experiment using primer sets for the entire coding region of C7orf24 (upper) and β-actin (lower). Two bands were detected for the C7orf24 PCR reaction in all tissues examined: a band of 567 bp, corresponding to the expected size of C7orf24 mRNA (arrow in Figure 1A), and a second, slightly smaller band (arrowhead in Figure 1A). The nucleotide sequence of the longer PCR product was identical to that reported for rat C7orf24 cDNA (GenBank accession number NM_024051), encoding 188 amino acids at a molecular weight of 21,200. The sequence of the shorter PCR product revealed a variant C7orf24 cDNA with 88 nucleotides deleted from exon 2 (Figure 1B). The variant cDNA encoded a frameshift and encodes an open reading frame of 77 amino acids at a molecular weight of 8800.
Figure 1.
RT-PCR analysis (A) and structures of C7orf24 and the variant cDNAs (B). (A) Primers for the entire coding regions of C7orf24 (upper) and β-actin (lower), respectively, were used for RT-PCR to generate products as typified here. A band of 567 bp, corresponding to the expected size of C7orf24 mRNA, and a slightly smaller, second band were detected. M, bp marker; 1, cortex; 2, choroid plexus; 3, eye; 4, lung; 5, heart; 6, stomach; 7, liver; 8, spleen; 9, kidney; 10, adrenal gland; 11, small intestine; 12, large intestine; and 13, urinary bladder. (B) The sequence of the variant C7orf24 cDNA deletes 88 nucleotides from exon 2.
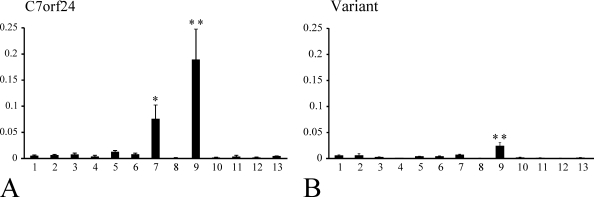
The real-time PCR demonstrated that the distribution patterns of the C7orf24 (Figure 2A) and the variant mRNAs (Figure 2B) were very similar. The expression level of the C7orf24 mRNA is higher than that of the variant mRNAs. Both C7orf24 and the variant cDNA showed significantly higher expression in the kidney compared with the levels in other tissues (p<0.001). The liver also expresses high levels of C7orf24 mRNA (p<0.05).
Figure 2.
Real-time PCR analysis of C7orf24 (A) and the variant cDNAs (B). The values are represented as a ratio of the cDNA content to that of β-actin cDNA. 1, cortex; 2, choroid plexus; 3, eye; 4, lung; 5, heart; 6, stomach; 7, liver; 8, spleen; 9, kidney; 10, adrenal gland; 11, small intestine; 12, large intestine; and 13, urinary bladder. Both C7orf24 and the variant cDNA showed significantly higher expression in the kidney (9) compared with the levels in other tissues (**p<0.001). The liver (7) also expresses high levels of C7orf24 mRNA (*p<0.05).
In Situ Hybridization Histochemistry
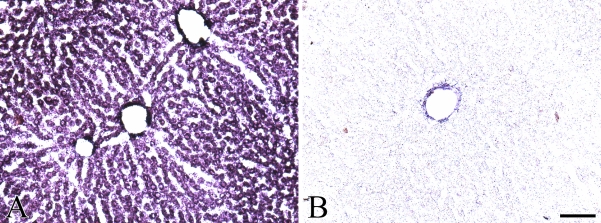
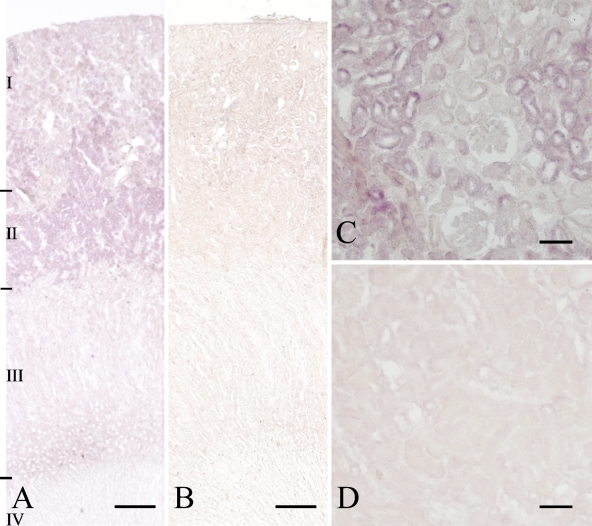
Based on the high liver and kidney expression, the C7orf24 mRNA was next localized in those tissues by in situ hybridization. Transcripts of C7orf24 mRNA were detected in the cytoplasm of hepatocytes in the liver sections (Figure 3A). The sense probe showed only nonspecific binding to blood vessels (Figure 3B). In the kidney, positive signals for the C7orf24 mRNA were relatively intense in the outer layer of the outer zone of the medulla (II in Figure 4A), whereas weak signals were localized in the cortex (I in Figure 4A) and in the inner layer of the outer zone of the medulla (III in Figure 4A). The inner zone of the medulla showed no hybridization signal (IV in Figure 4A). At higher magnification, (Figure 4C), cells in the renal tubules were positive for C7orf24, but the Bowman's capsule showed only weak positivity (Figure 4C). Binding of the sense probe was not detected (Figures 4B and 4D).
Figure 3.
In situ hybridization histochemistry of rat liver using antisense (A) and sense (B) probes to C7orf24. Transcripts for C7orf24 were distributed in the cytoplasm of hepatocytes. Bar = 100 μm.
Figure 4.
In situ hybridization histochemistry of the rat kidney using antisense (A,C) and sense (B,D) probes to C7orf24. I, II, III, and IV indicate the cortex, the outer layer of the outer zone, the inner layer of the outer zone, and the inner zone of the medulla, respectively. Bars: A,B = 500 μm; C,D = 50 μm.
Discussion
The present study demonstrated the first reported distribution pattern of C7orf24 mRNA in rat tissues using RT-PCR analysis and in situ hybridization histochemistry. RT-PCR revealed that both the full-length and the variant forms of C7orf24 mRNA were expressed in all tissues examined. This result is consistent with the reported protein expression of variant forms of γ-glutamyl cyclotransferase (Tulchin and Taylor 1981). The variant C7orf24 cDNA lacks 80 bp from exon 2, resulting in a frameshift and truncated protein comprising 77 amino acids. The variant protein lacks the active site pocket that contains Glu98 (Oakley et al. 2008) and thus should have no γ-glutamyl cyclotransferase activity. We investigated a product of C7orf24 alternate splicing by Western blotting of tissues and cell homogenates transfected with the specified expression vectors (see supplementary Figure 1). We detected both the full-length and variant proteins purified from Escherichia coli cells transfected with the appropriate vector. In tissue homogenates, we detected the full form of the protein but failed to observe any bands of the variant protein. The results indicated that the variant protein was not translated or was at least very unstable in tissues. Further studies will be needed to clarify the expression of the variant protein.
The real-time PCR demonstrated very similar expression patterns for the full-length and variant C7orf24 mRNA, although the full-length transcripts were significantly more abundant than the variants. Both forms showed significantly higher expression in liver and kidney than in the other tissues. The results accord well with a previous study showing that the liver and kidney had the highest γ-glutamyl cyclotransferase protein expression in rats (Orlowski and Meister 1973). However, there are some discrepancies between our results and the human expressed sequence tags (EST) database analysis for this protein (Oakley et al. 2008), which showed high expression in the bladder. The reason for this discrepancy could not be determined from this study, although there are several possible explanations. One possibility is the effects of difference among species. Another possibility is that the expression level of C7orf24 in bladder may be affected by the physiological and pathological status, particularly inasmuch as C7orf24 expression is increased in bladder cancer (Kageyama et al. 2007). As pointed out by Oakley et al. (2008), there were no transcripts of γ-glutamyl transpeptidase, another enzyme of the γ-glutamyl cycle, in the EST database from bladder. The role of C7orf24 in the bladder thus remains unknown, and further studies are needed to clarify these findings.
We further confirmed the localization of C7orf24 mRNA in the liver and kidney using in situ hybridization histochemistry. In the liver, transcripts were detected in the cytoplasm of hepatocytes. It is well known that hepatocytes function in detoxification, drug metabolism, and anti-oxidatation mechanisms, where glutathione produced in the γ-glutamyl cycle plays an important role (Lu 1999; Yuan and Kaplowitz 2009). In the kidney, C7orf24 mRNA was observed in the nephron system, particularly in the renal tubules, the cortex, and the outer layer of the outer zone. These findings agree with previous studies showing that glutathione is more abundant in the renal cortex than in the medulla (Mohandas et al. 1984), and an earlier study suggested that γ-glutamyl cyclotransferase is important for amino acid transport in renal tubules (Meister 1973). In addition, the renal tubules are a major site for glutathione metabolism (Meister and Anderson 1983), with 80–90% of renal tubular glutathione arising from the kidney cells. The remaining glutathione is then filtered from the plasma and reabsorbed in the proximal tubules (Meister and Anderson 1983). Conjugation of potentially toxic electrophilic compounds with glutathione and excretion into urine is also an important detoxification mechanism (Lohr et al. 1998).
In conclusion, this study demonstrated the distribution of C7orf24 mRNA in rat tissues for the first time. The strong expression of C7orf24 mRNA in hepatocytes and renal tubule cells indicated that C7orf24 might play an important role in liver and kidney.
References
- Kageyama S, Iwaki H, Inoue H, Isono T, Yuasa T, Nogawa M, Maekawa T, et al. (2007) A novel tumor-related protein, C7orf24, identified by proteome differential display of bladder urothelial carcinoma. Proteomics Clin Appl 1:192–199 [DOI] [PubMed] [Google Scholar]
- Lohr JW, Willsky GR, Acara MA (1998) Renal drug metabolism. Pharmacol Rev 50:107–141 [PubMed] [Google Scholar]
- Lu SC (1999) Regulation of hepatic glutathione synthesis: current concepts and controversies. FASEB J 13:1169–1183 [PubMed] [Google Scholar]
- Masuda Y, Maeda S, Watanabe A, Sano Y, Aiuchi T, Nakajo S, Itabe H, et al. (2006) A novel 21-kDa cytochrome c-releasing factor is generated upon treatment of human leukemia U937 cells with geranylgeraniol. Biochem Biophys Res Commun 346:454–460 [DOI] [PubMed] [Google Scholar]
- Meister A (1973) On the enzymology of amino acid transport. Transport in kidney and probably other tissues is mediated by a cycle of enzyme reactions involving glutathione. Science 80:33–39 [DOI] [PubMed] [Google Scholar]
- Meister A, Anderson ME (1983) Glutathione. Annu Rev Biochem 52:711–760 [DOI] [PubMed] [Google Scholar]
- Mohandas J, Marshall JJ, Duggin GG, Horvath JS, Tiller DJ (1984) Differential distribution of glutathione and glutathione-related enzymes in rabbit kidney. Possible implications in analgesic nephropathy. Biochem Pharmacol 33:1801–1807 [DOI] [PubMed] [Google Scholar]
- Oakley AJ, Yamada T, Liu D, Coggan M, Clark AG, Board PG (2008) The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle. J Biol Chem 283:22031–22042 [DOI] [PubMed] [Google Scholar]
- Orlowski M, Meister A (1973) γ-Glutamyl cyclotransferase. J Biol Chem 248:2836–2844 [PubMed] [Google Scholar]
- Tulchin N, Taylor BA (1981) γ-Glutamyl cyclotransferase: a new genetic polymorphism in the mouse (mus musculus) linked to LYT-2. Genetics 99:109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Geman D, Winslow RL (2007) Large-scale integration of cancer microarray data identifies a robust common cancer signature. BMC Bioinformatics 8:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Kaplowitz N (2009) Glutathione in liver diseases and hepatotoxicity. Mol Aspects Med 30:29–41 [DOI] [PubMed] [Google Scholar]
- Zhang C, Li HR, Fan JB, Wang-Rodriguez J, Downs T, Fu XD, Zhang MQ (2006) Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics 7:202. [DOI] [PMC free article] [PubMed] [Google Scholar]