Abstract
Influenza viruses cause significant morbidity and mortality in adults each winter. At the same time, other respiratory viruses circulate and cause respiratory illness with influenza-like symptoms. Human respiratory syncytial virus (HRSV), human parainfluenza viruses (HPIV) and human metapneumovirus have all been associated with morbidity and mortality in adults, including nosocomial infections. This study evaluated 154 respiratory specimens collected from adults with influenza-like/acute respiratory illness (ILI) seen at the Edward Hines Jr VA Hospital, Hines, IL, USA, during two successive winters, 1998–1999 and 1999–2000. The samples were tested for ten viruses in two nested multiplex RT-PCRs. One to three respiratory viruses were detected in 68 % of the samples. As expected, influenza A virus (FLU-A) infections were most common (50 % of the samples), followed by HRSV-A (16 %). Surprisingly, HPIV-4 infections (5.8 %) were the third most prevalent. Mixed infections were also relatively common (11 %). When present, HPIV infections were approximately three times more likely to be included in a mixed infection than FLU-A or HRSV. Mixed infections and HPIV-4 are likely to be missed using rapid diagnostic tests. This study confirms that ILI in adults and the elderly can be caused by HRSV and HPIVs, including HPIV-4, which co-circulate with FLU-A.
INTRODUCTION
Yearly cases of influenza-like/acute respiratory illness (ILI) in acute-care settings throughout the USA are reported to public health authorities and attributed to influenza virus, mostly influenza A virus (FLU-A), with some influenza B virus (FLU-B). The information that characterizes the seasonal epidemics of respiratory viruses has been obtained from sentinel acute-care clinics and emergency-room visits from mixed populations of patients. More information is needed to characterize the viruses that cause ILI in ambulatory adults and the elderly.
In this study, we focused on other negative-strand RNA respiratory viruses that circulate before, after and concurrently with influenza viruses in the ambulatory elderly adult population, including human respiratory syncytial virus (HRSV), human parainfluenza viruses (HPIV) and human metapneumovirus (HMPV). HRSV infection has been suggested to cause significant disease in the elderly, perhaps rivalling non-pandemic influenza (Gonzalez et al., 2000). HRSV infection has been identified as the aetiological agent for pneumonia and death in adults with chronic lung disease, and in elderly adults living in nursing homes (Lee et al., 2005). Current information on the incidence and severity of HRSV in ambulatory adults still contains large gaps in knowledge about the disease (Walsh et al., 2007).
HMPV can cause severe respiratory disease in children and the elderly (Boivin et al., 2002, 2003), but the disease that this virus causes is not clinically distinct. The prevalence of HMPV in ILI has not been well studied.
There are four types of HPIV, estimated to cause 10 % of acute respiratory infections during the winter (Boivin et al., 2002; Henderson, 1987; Sharova et al., 1989). HPIVs also cause severe disease, including pneumonia and death in transplant recipients (Whimbey et al., 1996), as well as nosocomial infections and nursing-home outbreaks (Glasgow et al., 1995), similar to influenza virus.
Nested multiplex RT-PCR enables sensitive, simultaneous detection of multiple viruses from patient specimens and can be completed in a time frame that enables clinical decision-making (Atmar et al., 1996; Claas et al., 1993; Osiowy, 1998). To test clinical respiratory samples for multiple viruses, we improved two nested multiplex RT-PCR assays (Aguilar et al., 2000; Coiras et al., 2003), enabling simple and relatively rapid diagnosis of ten of the most important causative agents of respiratory illness.
METHODS
Clinical specimens.
A total of 154 specimens was collected from adult patients who were hospitalized or received medical care in the emergency department of Edward Hines Jr VA Hospital during the 1998–1999 and 1999–2000 autumn/winter seasons. Patients had influenza symptoms or respiratory symptoms and most had fevers of at least 10 °F/37.8 °C. All specimens were obtained from the nose, throat or nasopharynx using sterile cotton swabs and placed in virus transport medium tubes. Following routine testing by the Clinical Microbiology Laboratory, Edward Hines Jr VA Hospital, the remaining portion of each specimen was stored at −80 °C until used in this analysis. This study was approved by the Institutional Review Boards of Edward Hines Jr VA Hospital, Rush University Medical Center and Nationwide Children's Hospital.
Control viruses and cells.
Egg-grown FLU-A (strain Udorn), FLU-B (strain Lee/40) or tissue-culture-grown FLU-C, HPIV1–3, -4A and -4B, HRSV-A and -B, and HMPV-A (CAN97-83) and -B (CAN97-75), were used to standardize and confirm the specificity of the RT-PCRs. LLC-MK2 cells were grown in minimum essential medium with 10 % fetal bovine serum (FBS), MDCK cells were grown in RPMI 1640 with 5 % FBS, and HEp-2 cells were grown in Opti-MEM with 2 % FBS.
RNA isolation.
RNA was extracted from 250 μl clinical sample using RNA-Bee (Tel-Test) according to the manufacturer's protocol. GlycoBlue (15 μg; Ambion) was added to the aqueous phase before precipitation to increase recovery of the viral RNA and to enable visualization of the RNA pellet. The pellet was resuspended in 30 μl RNase-free water. RNA samples were frozen on dry ice and stored at −80 °C.
Nested multiplex RT-PCR assays.
Two nested multiplex RT-PCR assays to detect the common pathogenic negative-strand RNA respiratory viruses, based on published protocols (Aguilar et al., 2000; Boivin et al., 2002, 2003; Coiras et al., 2003), were optimized and HMPV detection was added. FLU-A, -B and -C, HRSV-A and –B, and HMPV, were all assessed in one reaction. HPIV-1, -2, -3 and -4 were assessed in a separate reaction.
Primer improvements were made with the help of Primer Designer software (Scientific & Educational Software) to avoid 3′-end hybridization with any of the other primers in the reaction while maintaining the ability to detect any sequence in GenBank. Primary and nested amplification primers specific for FLU-A, -B and -C, HRSV-A and –B, and the internal control (Coiras et al., 2003) were modified as indicated in Table 1. To the primer mix for FLU-A, -B, -C, and HRSV-A and -B, were added HMPV primers. These HMPV primers were designed from sequences in GenBank to amplify the N gene from both the A and B strains of HMPV.
Table 1.
Primers used for the detection of HRSV-A and -B, HMPV, FLU-A, -B and -C, and HPIV1, -2, -3 and -4 by nested multiplex RT-PCR
| Primer | Sequence (5′→3′) | Gene | ORF position (nt) | Amplicon size (bp) | ||||
|---|---|---|---|---|---|---|---|---|
| HRSV/HMPV/FLU RT-PCR | ||||||||
| HRSVAB1 | ATGGAGYTGCYRATCCWCARRRCAARTGCAAT | F | 1–31 | 738 (HRSV-A); 738 (HRSV-B) | ||||
| HRSVAB2 | AGGTGTWGTTACACCTGCATTRACACTRAATTC | F | 737–705 | |||||
| FluAC1 | GAACTCRTYCYWWATSWCAAWGRRGAAAT | NP | 319–347 (A); 346–374 (C) | 722 (Flu-A); 992 (Flu-B); 739 (Flu-C) | ||||
| FluB1 | ACAGAGATAAAGAAGAGCGTCTACAA | NP | 217–242 | |||||
| FluABC2 | ATKGCGCWYRAYAMWCTYARRTCTTCAWAIGC | NP | 1040–1009 (A); 1208–1177 (B); 1084–1053 (C) | |||||
| HMPV 8426* | CCYTCAGCACCAGACACACC | NP | 508–527 | 444 | ||||
| HMPV 8427* | AGATTCAGGRCCCATTTCTC | NP | 951–932 | |||||
| N1 | CTTGGGCGTGTCTCAAAATCT | Control | 10–30 | 1129 | ||||
| N2 | GTCGCCACGGTTGATGAGAGCT | Control | 1137–1116 | |||||
| HRSV/HMPV/FLU nested PCR | ||||||||
| HRSVA3 | TTATACACTCAACAATRCCAAAAAWACC | F | 347–374 | 364 (HRSV-A) | ||||
| HRSVA4 | AAATTCCCTGGTAATCTCTAGTAGTCTGT | F | 710–682 | |||||
| HRSVB3 | ATCTTCCTAACTCTTGCTRTTAATGCATTG | F | 30–59 | 612 (HRSV-B) | ||||
| HRSVB4 | GATGCGACAGCTCTGTTGATTTACTATG | F | 641–614 | |||||
| FluAB3 | GATCAAGTGAKMGRRAGYMGRAAYCCAGG | NP | 718–746 (A); 892–920 (B) | 302 (Flu-A); 227 (Flu-B); 114 (Flu-C) | ||||
| FluC3 8425† | AAATTGGAATTTGTTCCTTT | NP | 950–969 | |||||
| FluAC4 | TCTTCAWATGCARSWSMAWKGCATGCCATC | NP | 1019–990 (A); 1063–1034 (C) | |||||
| FluB4 | CTTAATATGGAAACAGGTGTTGCCATATT | NP | 1118–1090 | |||||
| HMPV 8422* | CACCHATAATYTTATTATGTGTHGGTG | NP | 524–550 | 235 | ||||
| HMPV 8423* | GADGATGAGCCTAAHGCTTTBC | NP | 737–757 | |||||
| N3 | GGGGTGTTATGAGCCATATTCAACGG | Control | 108–133 | 888 | ||||
| N4 | AGCCGCCGTCCCGTCAAGTCAG | Control | 995–974 | |||||
| HPIV RT-PCR | ||||||||
| HPIV1:PIP1+ | CCTTAAATTCAGATATGTAT | HN | 748–767 | 478 | ||||
| HPIV1:PIP1− | GATAAATAATTATTGATACG | HN | 1225–1206 | |||||
| HPIV1:PIP2+ | AACAATCTGCTGCAGCATTT | HN | 803–822 | 508 | ||||
| HPIV1:PIP2− | ATGTCAGACAATGGGCAAAT | HN | 1310–1291 | |||||
| HPIV1:PIP3+ | CTGTAAACTCAGACTTGGTA | HN | 689–708 | 488 | ||||
| HPIV1:PIP3.1− † | TGGAATTGAGTTTAAGCC | HN | 1176–1159 | |||||
| HPIV1:PIP4+ | CTGAACGGTTGCATTCAGGT | P | Non-coding 5′ | 414 (A) | ||||
| HPIV1:PIP4− | TTGCATCAAGAATGAGTCCT | P | 433–452 | 414 (B) | ||||
| CAT gene I.C.+ | CAGTCAGTTGCTCAATGTA | CAT | 76–94 | 528 | ||||
| CAT gene I.C.− | GACATGGAAGCCATCAC | CAT | 587–603 | |||||
| HPIV nested PCR | ||||||||
| HPIV1:PIS1.1+ | CCGGTAATTTCTCATACCTATG | HN | 780–801 | 317 | ||||
| HPIV1:PIS1.1− † | CTTTGGAGCGGAGTTGTTA | HN | 1096–1078 | |||||
| HPIV1:PIS2.1+† | CCATTTACCTAAGTGATGGA | HN | 847–866 | 202 | ||||
| HPIV1:PIS2.1− † | GCCCTGTTGTATTTGGAAG | HN | 1048–1030 | |||||
| HPIV1:PIS3.1+† | ACTCCCAAAGTTGATGAA | HN | 811–828 | 356 | ||||
| HPIV1:PIS3.1−* | TTTAAGCCCTTGTCAACAAC | HN | 1166–1147 | |||||
| HPIV1:PIS4.1+† | AAAGAATTAGGTGCAACCAG | P | 76–95 | 245 | ||||
| HPIV1:PIS4−* | GTGTCTGATCCCATAAGCAGC | P | 320–300 | |||||
| CAT gene N+† | CTGGAGTGAATACCACGA | CAT | 315–332 | 149 | ||||
| CAT gene N− † | TGGTGAAACTCACCCAG | CAT | 463–447 | |||||
| HPIV-4 subtyping | ||||||||
| HPIV4A:PIS4A+ | ATGATGGTGGAACCAAGATT | P | 146–165 | 175 (A); 82 (B) | ||||
| HPIV4B:PIS4B+ | AACCAGGGAAACAGAGCTC | P | 239–257 | |||||
| HPIV4:PIS4−* | GTGTCTGATCCCATAAGCAGC | P | 320–300 | |||||
*Newly created primers.
†Modified from the original primers.
The initial reverse transcription and PCR were performed in a one-tube process using an Access RT-PCR kit (Promega) with conditions similar to those described by Coiras et al. (2003) except that the nested reaction cycle number was increased to 45. Each RT-PCR included an internal positive-control target sequence, and each set of reactions included negative controls using water instead of sample. All reactions were performed in a GeneAmp PCR system 2700 thermocycler (Applied Biosystems).
For HPIV-1, -2, -3 and -4 detection, primary and nested primers (Aguilar et al., 2000; Echevarría et al., 1998) were shortened to avoid 3′-end interactions with other primers in the same mix (Table 1). Conditions used for the single-step RT-PCR were as follows: the primary RT-PCR was incubated at 48 °C for 45 min and 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 1.5 min and 72 °C for 1 min, with extension at 72 °C for 10 min, and storage at 4 °C until collected. The nested PCR was carried out at 94 °C for 4 min, followed by 45 cycles of 94 °C for 30 s, 55 °C for 1 min and 72 °C for 30 s, with extension at 72 °C for 5 min, and storage at 4 °C until collected. To subtype HPIV-4, an additional nested reaction was performed on the first PCR product with primers PIS4A+ and PIS4B+, paired with PIS4− (Aguilar et al., 2000).
Internal control plasmid (100 copies) was added to the RT-PCR at the same time as the specimen RNA. The FLU/HRSV/HMPV control plasmid has been described by Coiras et al. (2003). For the HPIV reactions, we designed a different internal control, from the chloramphenicol acetyl transferase (CAT)-encoding gene, to reduce 3′-end interactions.
Each reaction tube contained AmpliWax (Perkin Elmer) to reduce contamination. Samples were separated into workable groups for running the reactions. Each group included a set of positive-control virus RNAs for each of the viruses being tested and negative controls to ensure that primers were working and that there was no contamination. Positive results in nested multiplex RT-PCR were confirmed by repeating with a new aliquot of RNA, using only the primers specific for that virus. Samples with multiple viruses were retested with each of the individual primer pairs and with the multiplex primer mixture.
All PCR products were analysed in 2 % agarose gels by electrophoresis, stained with ethidium bromide and visualized on a UV light box. Each nested PCR product was designed to migrate at a unique position, enabling virus identification. Samples were considered negative if: (i) no PCR product corresponding to a virus was detected and (ii) the internal control product was present. In many reactions, viral PCR products were generated without the internal control PCR product, indicating that production of the control did not compete with viral product generation. All samples produced one or more PCR products that corresponded to a virus or the control or both, indicating that no samples contained inhibitors. In a few samples, minor PCR products that did not correspond to a virus product appeared, suggesting non-specific amplification. In these cases, the procedure was repeated. If no viral PCR product was detected in the repeat reaction, the sample was reported as negative.
RESULTS AND DISCUSSION
Multiplex RT-PCRs were used to identify respiratory viruses from an adult population by modifying earlier protocols (Aguilar et al., 2000; Coiras et al., 2003) and adding detection primers for HMPV. The primers (Table 1) were designed to produce a uniquely sized PCR product for each virus. These products were separated by agarose gel electrophoresis and stained with ethidium bromide, allowing clear identification of ten respiratory viruses or subtypes.
One or more viruses were detected in 104 (68 %) of the 154 patient specimens. The most frequently detected viruses were FLU-A in 77 cases (50 % of the total specimens), HRSV-A in 25 cases (16 %), and HPIV-4A and 4B in 9 cases (5.8 %). All other viruses were present at less than 3 % frequency: HPIV-2 in four cases (2.6 %) and HPIV-1, FLU-B and HRSV-B in three cases each (1.9 %). HMPV, HPIV-3 and FLU-C were not detected, even thought the primers were designed to detect strains present in GenBank at the time they were designed. It is possible that HMPV, HPIV-3 and FLU-C may not have caused ILI symptoms severe enough to seek medical attention or they may be less likely to occur as mixed infections with a virus that does cause ILI. It is also possible that these viruses were not circulating in this geographical area during these two autumn/winter seasons.
The frequency of FLU-positive specimens was high, as expected, as collection was biased towards patients exhibiting ILI. Most of these FLU-positive specimens contained FLU-A (77 cases), compared with FLU-B (3 cases). Over the 1998–1999 season, 11 % of FLU-positive specimens were FLU-B, which was lower than the 36 % rate in the Centers for Disease Control and Prevention (CDC) influenza surveillance data for this region. During the 1999–2000 season, the FLU-B-positive rate of 1.6 % was higher than the rate in the CDC data (0.5 %). Over the same two seasons, other causes of ILI that would have been attributed to FLU by clinical presentation alone were identified.
Multiplex RT-PCR identified an unexpectedly high number of HPIV-4 isolates in the study specimens collected from adults with ILI. Virus identification in cell culture was attempted on several of the specimens without success; in retrospect, these were identified as HPIV-4. The optimal cell line for culturing HPIV-4 varies from isolate to isolate and tissue culture isolation is generally inefficient (Lau et al., 2005). RT-PCR screening has been found to be much more sensitive and consistent for detecting HPIV-4 in both culture-positive and culture-negative specimens from children (Aguilar et al., 2000; Bellau-Pujol et al., 2005; Lau et al., 2005; Templeton et al., 2004, 2005). Of the 154 patient samples, 17 (11 %) contained more than one virus. The identity of each virus in these multiple infections was confirmed by amplification with primers specific for each virus. The most commonly detected virus in single and multiple infections was FLU-A, detected in all but 1 of the 17 mixed infections, followed by HRSV-A in 9 cases and HPIV-4A in six cases. Of the FLU-A specimens, 21 % (16/77 isolates) contained another virus. Among the HRSV-A specimens, 32 % (8/25) contained another virus. Strikingly, HPIVs were all found more frequently in mixed infections: 67 % (2/3) of HPIV-1 infections, 75 % (3/4) of HPIV-2 infections, 86 % (6/7) of HPIV-4A infections and 50 % (1/2) of HPIV-4B infections. The appearance of HPIV types in mixed infections at a much higher proportion than FLU-A or HRSV-A may indicate that FLU-A or HRSV-A enhances the infectivity of HPIVs. Alternatively, solitary HPIV infections may be less likely to cause patients to seek medical attention.
Some of these specimens were collected in order to identify the agent causing nosocomial ILI cases. These specimens were screened with a rapid antigen test, Directigen Flu A (Pachucki et al., 2004). Although this assay is specific for FLU-A, it missed 58 % of the FLU-A samples that were subsequently identified by nested multiplex RT-PCR, and it cannot detect other viruses. Another quick antigen test, such as a direct fluorescent antibody test, could have been used, but can suffer from similar insensitivity (Casiano-Colón et al., 2003). In addition, pooled respiratory direct fluorescent antibody tests can detect common respiratory viruses, but they do not include an antibody against HPIV-4.
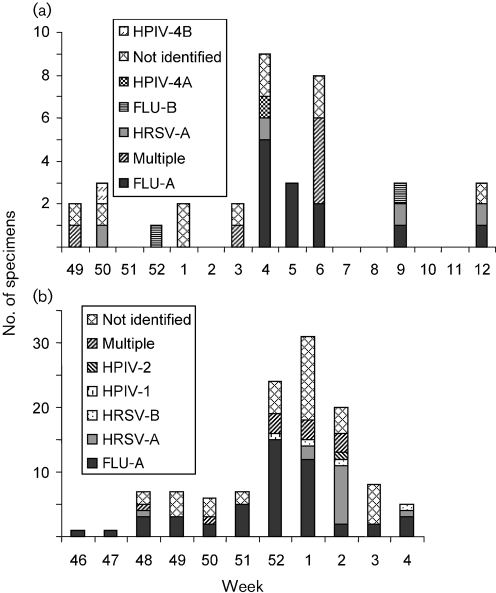
The viruses identified during the first season of surveillance and their times of appearance are shown in Fig. 1(a). In weeks 4 and 5, each identified ILI was caused by a single virus: five FLU-A, one HRSV-A and one PIV-4A in week 4, and three FLU-A in week 5. In contrast, during week 6, four of the six identified ILI samples contained multiple viruses. The HPIV-4A from week 4 was detected in a specimen from a hospitalized patient with ILI. Eight days later, HPIV-4A was detected in specimens from three other hospitalized patients, suggesting that HPIV-4A had probably spread nosocomially. All three of the second-round HPIV-4 specimens also contained another virus: FLU-A in two cases and HRSV-A in the third, suggesting that the second virus had come from a different source.
Fig. 1.
Viruses identified during the 1998–1999 (a) and 1999–2000 (b) autumn/winter seasons. (a) During the 1998–1999 season, the peak of ILI occurred in weeks 4–6. FLU-A was the predominant virus identified in all 3 weeks. In weeks 4 and 5, all infections were caused by single viruses, but in week 6, multiple infections predominated: two FLU-A/HPIV-4A, one HRSV-A/HPIV-4A and one FLU-A/HRSV-A/HPIV-2. HPIV-4A appeared as the only detected virus in one patient during week 4, 8 days before HPIV-4A was found in three multiple infections in week 6. Additional multiple infections included: FLU-A/HPIV-4A (week 49) and FLU-A/HPIV-4B (week 3). (b) During the 1999–2000 winter season, the ILI peak occurred in weeks 52 to 2. FLU-A predominated in weeks 52 and 1, and HRSV-A in week 2. Multiple infections included: FLU-A/HRSV-A/HPIV-4A (week 48); FLU-A/HPIV-1 (week 50); FLU-A/HPIV-1 and two FLU-A/HPIV-2 (week 52); FLU-A/HPIV-4A, FLU-A/FLU-B/HRSV-A and FLU-A/HRSV-A (week 1); and three FLU-A/HRSV-A (week 2).
During the second season of surveillance (Fig. 1b), FLU-A was detected at increased levels in weeks 52 and 1. The following week, most ILI cases were found to be negative for FLU by rapid testing (Directigen). In what appeared to be the third week of this FLU outbreak, only two specimens contained FLU-A alone. Ten others contained HRSV-A alone, whilst three others contained both HRSV and FLU-A. Clearly, this was a second, overlapping outbreak caused primarily by HRSV instead of FLU-A.
Three of the cases involved in this HRSV cluster required hospitalization. The initial patient had spinal cord injury, developed viral pneumonia and died from respiratory insufficiency. Another was hospitalized for dyspnoea, wheezing and pneumonia. The third was transferred to the hospital for exacerbation of chronic obstructive lung disease. In addition to these nosocomial cases, 12 patients with cases of HRSV-A infection presented to the emergency department in week 2, and 5 of these patients required admission to the hospital. Only two cases of HPIV-4A were detected over this second season.
No viral aetiology was identified in 32 % of the specimens. As only the negative-strand viruses known to cause respiratory symptoms were tested here, the remainder may have been caused by other respiratory viruses, such as rhinovirus, adenovirus or coronavirus, or by other micro-organisms. These other viruses or micro-organisms could also have been additional pathogens in the positive specimens.
The use of nested multiplex RT-PCR as a diagnostic tool at the time of diagnosis would have changed the reporting of a large number of these ILIs. HRSV and HPIV infections were attributed to FLU-A because they were presented during FLU-A outbreaks. Such misattributions can have wide-reaching public health implications, as influenza morbidity and mortality in adults is based on ILI cases without substantial information about the presence of other respiratory viruses.
Whilst nested multiplex RT-PCR is a useful method for detecting respiratory viruses, real-time PCR is being used with increasing frequency. It is sensitive, rapid and reduces the potential for contamination. However, real-time PCR requires expensive equipment and probes, and is limited in its ability to support large-scale multiplexing desired for screening samples for multiple viruses at a reasonable cost. Facilities that do not have the equipment or resources for real-time PCR may find nested, multiplex RT-PCR useful, as long as they maintain good laboratory practices to reduce contamination.
As far as we are aware, this is the first report of a high frequency of HPIV-4 infection (5.8 %) in adults with ILI. This virus was circulating among adult patients during both seasons at the same time that FLU-A was present and causing ILI in adult patients. It also appeared to have spread nosocomially during the first year of surveillance. Most clinical laboratories do not include HPIV-4 in their detection schemes because it is not considered to be a major disease-causing virus (Chanock et al., 2001; Henrickson, 2003). However, HPIV-4 was also recently identified as the cause of an outbreak of respiratory disease in institutionalized children, and has also been shown to cause pneumonia, bronchiolitis and aseptic meningitis (Aguilar et al., 2000; Lau et al., 2005; Lindquist et al., 1997; Rubin et al., 1993; Slavin et al., 2000). HPIV-4 detection should be included in the analysis of respiratory specimens.
This study confirms that multiple respiratory viruses may circulate concurrently with FLU-A among adults, and that HPIVs may cause nosocomial infection. In addition, adults seek acute medical care when they are infected with HRSV or an HPIV.
Acknowledgments
We thank the Clinical Microbiology Laboratory at the Rush University Medical Center; Richard Compans, Peter Collins, Robert Lamb, Guy Boivin and Dean Erdman for the standard viruses; Barbara Newton, Stephanie Downing and Anumeha Gupta for their excellent technical help; Doug Salamon and Amy Leber for helpful discussions; and Mayte Coiras for helpful discussions and reagents. This work was supported by National Institutes of Health (NIH), USA, grant AI047213.
Abbreviations
FBS, fetal bovine serum
FLU, influenza virus
HMPV, human metapneumovirus
HPIV, human parainfluenza virus
HRSV, human respiratory syncytial virus
ILI, influenza-like illness
References
- Aguilar, J. C., Pérez-Breña, M. P., García, M. L., Cruz, N., Erdman, D. D. & Echevarría, J. E. (2000). Detection and identification of human parainfluenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription-PCR. J Clin Microbiol 38, 1191–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar, R. L., Baxter, B. D., Dominguez, E. A. & Taber, L. H. (1996). Comparison of reverse transcription-PCR with tissue culture and other rapid diagnostic assays for detection of type A influenza virus. J Clin Microbiol 34, 2604–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellau-Pujol, S., Vabret, A., Legrand, L., Dina, J., Gouarin, S., Petitjean-Lecherbonnier, J., Pozzetto, B., Ginevra, C. & Freymuth, F. (2005). Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods 126, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin, G., Abed, Y., Pelletier, G., Ruel, L., Moisan, D., Côté, S., Peret, T. C., Erdman, D. D. & Anderson, L. J. (2002). Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis 186, 1330–1334. [DOI] [PubMed] [Google Scholar]
- Boivin, G., De Serres, G., Côté, S., Gilca, R., Abed, Y., Rochette, L., Bergeron, M. G. & Déry, P. (2003). Human metapneumovirus infections in hospitalized children. Emerg Infect Dis 9, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiano-Colón, A. E., Hulbert, B. B., Mayer, T. K., Walsh, E. E. & Falsey, A. R. (2003). Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol 28, 169–174. [DOI] [PubMed] [Google Scholar]
- Chanock, R. M., Murphy, B. R. & Collins, P. L. (2001). Parainfluenza viruses. In Fields Virology, 4th edn, pp. 1341–1379. Edited by D. M. Knipe & P. M. Howley. Philadelphia: Lippincott Williams & Wilkins.
- Claas, E. C., van Milaan, A. J., Sprenger, M. J., Ruiten-Stuiver, M., Arron, G. I., Rothbarth, P. H. & Masurel, N. (1993). Prospective application of reverse transcriptase polymerase chain reaction for diagnosing influenza infections in respiratory samples from a children's hospital. J Clin Microbiol 31, 2218–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiras, M. T., Pérez-Breña, P., García, M. L. & Casas, I. (2003). Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested-PCR assay. J Med Virol 69, 132–144. [DOI] [PubMed] [Google Scholar]
- Echevarría, J. E., Erdman, D. D., Swierkosz, E. M., Holloway, B. P. & Anderson, L. J. (1998). Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR. J Clin Microbiol 36, 1388–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow, K. W., Tamblyn, S. E. & Blair, G. (1995). A respiratory outbreak due to parainfluenza virus type 3 in a home for the aged – Ontario. Can Commun Dis Rep 21, 57–61. [PubMed] [Google Scholar]
- Gonzalez, I. M., Karron, R. A., Eichelberger, M., Walsh, E. E., Delagarza, V. W., Bennett, R., Chanock, R. M., Murphy, B. R., Clements-Mann, M. L. & Falsey, A. R. (2000). Evaluation of the live attenuated cpts 248/404 RSV vaccine in combination with a subunit RSV vaccine (PFP-2) in healthy young and older adults. Vaccine 18, 1763–1772. [DOI] [PubMed] [Google Scholar]
- Henderson, F. W. (1987). Pulmonary infections with respiratory syncytial virus and the parainfluenza viruses. Semin Respir Infect 2, 112–121. [PubMed] [Google Scholar]
- Henrickson, K. J. (2003). Parainfluenza viruses. Clin Microbiol Rev 16, 242–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S. K., To, W. K., Tse, P. W., Chan, A. K., Woo, P. C., Tsoi, H. W., Leung, A. F., Li, K. S., Chan, P. K. & other authors (2005). Human parainfluenza virus 4 outbreak and the role of diagnostic tests. J Clin Microbiol 43, 4515–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, F. E., Walsh, E. E., Falsey, A. R., Liu, N., Liu, D., Divekar, A., Snyder-Cappione, J. E. & Mosmann, T. R. (2005). The balance between influenza- and RSV-specific CD4 T cells secreting IL-10 or IFNγ in young and healthy-elderly subjects. Mech Ageing Dev 126, 1223–1229. [DOI] [PubMed] [Google Scholar]
- Lindquist, S. W., Darnule, A., Istas, A. & Demmler, G. J. (1997). Parainfluenza virus type 4 infections in pediatric patients. Pediatr Infect Dis J 16, 34–38. [DOI] [PubMed] [Google Scholar]
- Osiowy, C. (1998). Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol 36, 3149–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachucki, C. T., Khurshid, M. A. & Nawrocki, J. (2004). Utility of reverse transcriptase PCR for rapid diagnosis of influenza A virus infection and detection of amantadine-resistant influenza A virus isolates. J Clin Microbiol 42, 2796–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, E. E., Quennec, P. & McDonald, J. C. (1993). Infections due to parainfluenza virus type 4 in children. Clin Infect Dis 17, 998–1002. [DOI] [PubMed] [Google Scholar]
- Sharova, N. K., Kubinova, I., Vorkunova, G. K., Tumova, B. & Bukrinskaia, A. G. (1989). Conservativism of the matrix protein of paramyxoviruses. Vopr Virusol 34, 573–575 (in Russian). [PubMed] [Google Scholar]
- Slavin, K. A., Passaro, D. J., Hacker, J. K., Hendry, R. M. & Kohl, S. (2000). Parainfluenza virus type 4: case report and review of the literature. Pediatr Infect Dis J 19, 893–896. [DOI] [PubMed] [Google Scholar]
- Templeton, K. E., Bredius, R. G., Scheltinga, S. A., Claas, E. C., Vossen, J. M. & Kroes, A. C. (2004). Parainfluenza virus 3 infection pre- and post-haematopoietic stem cell transplantation: re-infection or persistence? J Clin Virol 29, 320–322. [DOI] [PubMed] [Google Scholar]
- Templeton, K. E., Bredius, R. G., Claas, E. C., Kroes, A. C. & Walther, F. J. (2005). Parainfluenza virus 4 detection in infants. Eur J Pediatr 164, 528–529. [DOI] [PubMed] [Google Scholar]
- Walsh, E. E., Peterson, D. R. & Falsey, A. R. (2007). Is clinical recognition of respiratory syncytial virus infection in hospitalized elderly and high-risk adults possible? J Infect Dis 195, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Whimbey, E., Champlin, R. E., Couch, R. B., Englund, J. A., Goodrich, J. M., Raad, I., Przepiorka, D., Lewis, V. A., Mirza, N. & other authors (1996). Community respiratory virus infections among hospitalized adult bone marrow transplant recipients. Clin Infect Dis 22, 778–782. [DOI] [PubMed] [Google Scholar]