Abstract
LcrF, a Multiple Adaptational Response (MAR) transcription factor, regulates virulence in Yersinia pestis and Yersinia pseudotuberculosis. In a search for small molecule inhibitors of LcrF, an acrylic amide series of N-hydroxybenzimidazoles was synthesized, and the SAR (structure-activity relationship) was examined. Selected test compounds demonstrated inhibitory activity in a primary cell-free LcrF-DNA binding assay as well as in a secondary whole cell assay (Type III secretion system dependent Y. pseudotuberculosis cytotoxicity assay). The inhibitors exhibited no measurable antibacterial activity in vitro, confirming that they do not target bacterial growth. These results demonstrate that N-hydroxybenzimidazole inhibitors, exemplified by 14, 22 and 36, are effective anti-virulence agents, and have the potential to prevent infections caused by Yersinia spp.
Introduction
Yersinia pestis (Y. pestis), a Gram-negative pathogen, is the causative agent of plague,1 and poses a serious concern for its potential use as a biological weapon.2 Clinical isolates of single- and multi-drug resistant (MDR) Y. pestis have been reported,3 which undermines the effectiveness of current therapeutics. Therefore, with the ever-growing antimicrobial resistance problems world-wide,4 efforts to identify new therapies for MDR Y. pestis infections are viewed as a pressing public health need.
Most antibiotics that have entered the market, or advanced to late-stage clinical development in recent years are improved derivatives of existing chemical classes, with very few exceptions.5,6 Such antibiotics will certainly benefit patients infected with drug resistant bacteria. However, with increasing use of new drugs, emergence of resistant strains will probably be a matter of time. The effort to identify new chemical classes of antibiotics with novel mechanisms of action through target based high-throughput screening has not been very fruitful.6 Clearly, circumventing bacterial resistance has proven to be a challenging task, and will require more innovative research approaches.
As an alternative to traditional antimicrobial chemotherapy, strategies for targeting pathogen virulence have been reported in the recent literature.7–9 In this regard we have been developing small molecule anti-infection drugs that target the ability of the bacteria to cause infection (virulence), rather than growth.10 Such anti-virulence agents could be used to prevent infection of individuals in a high-risk environment such as in a bioterrorism event. The bacterial virulence targets of interest are Multiple Adaptational Response (MAR)11,12 transcription factors that are regulators of virulence expression.13
MAR proteins are characterized by two highly conserved helix-turn-helix (HTH) DNA-binding domains.14 They are present in many clinically important Gram-negative and Gram-positive bacteria.15 They control the ability of bacteria to cause infections, resist antibiotics, and adapt to hostile environments. Inactivation of MAR proteins by mutation attenuates the virulence of bacteria in animal models of infection, but does not affect bacterial growth in vitro. 13,16–18 Thus, by design, inhibitors of MAR proteins lack inherent antimicrobial activity, and are far less likely than antibiotics to apply selective pressure for the development of resistance. MAR proteins have not been described in eukaryotic cells, making this family of proteins a desirable target in antimicrobial drug discovery.
LcrF is a MAR transcription factor associated with virulence in Y. pestis and Y. pseudotuberculosis. It regulates expression of a type III secretion system (T3SS), a major virulence determinant of many Gram-negative bacteria (Figure 1). Upon contact with a host cell or following a change in temperature (e.g., from 26 °C in the flea to 37 °C in a human host), LcrF is expressed, and then activates expression of Yersinia outer proteins (Yops) and the T3SS.19 The Yops (i.e., cytotoxins) are secreted into host cells through the T3SS, and result in cellular apoptosis.20,21 Mutants that do not express the T3SS show dramatic attenuation of virulence in a whole cell and animal models of infection.22–24 Flashner et al. have recently investigated the effects of lcrF deletion (ΔlcrF) on the pathogenicity of Y. pestis in a mouse model of septic infection.16 The LD50 (50% lethal dose) of wild-type Y. pestis in this model is approximately 1 colony forming unit (CFU), whereas the LD50 of ΔlcrF Y. pestis is >100 CFUs. This result demonstrates that LcrF is a valid anti-virulence target in Yersinia spp.25
Figure 1.
Diagramatic representation of Yop expression and T3SS (type III secretion system) expression in Yersinia spp.
Small molecule compounds that inhibit the virulence of Yersinia are described in the literature.26–28 These compounds are known to inhibit the type III secretion of Yersinia; however, their specific bacterial targets are not defined. In contrast, we aim to inhibit Yersinia virulence at the transcription level by targeting the LcrF protein, which in turn regulates the expression of the T3SS. Small molecule inhibitors that directly target LcrF have not been reported in the literature to date.29
Previously we reported N-hydroxybenzimidazole compounds (1) (Figure 2) that demonstrated inhibitory activity against the MAR proteins MarA, SoxS, and Rob in E. coli.10 The N-hydroxybenzimidazole scaffold was initially identified from small molecule docking experiments using MarA-DNA and Rob-DNA co-crystal structures.30,31 While the X-ray crystal structure of LcrF-DNA complex or LcrF protein has not been resolved to date, the amino acid sequence of LcrF in the DNA binding domain is known to be homologous to that of MarA, SoxS, and Rob with 31% identity and 53% similarity over a 92 amino acid stretch.32 Therefore, initial screening of the previously identified inhibitors was viewed as a reasonable starting point in a search for LcrF inhibitors. Our goal was to identify, and to develop inhibitors for LcrF-DNA binding using in vitro cell-free and whole cell assays as well as in vivo mouse models of infection caused by Yersinia spp.
Figure 2.
General structure of N-hydroxybenzimidazoles
Chemistry
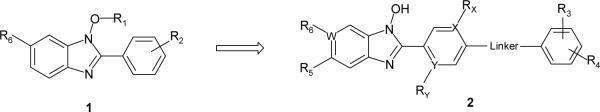
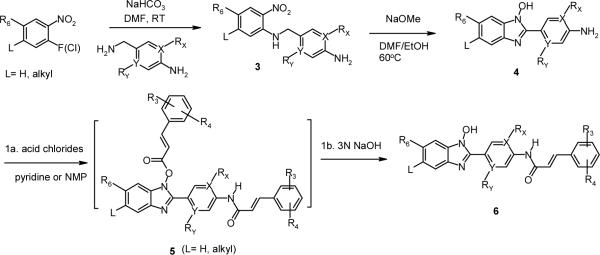
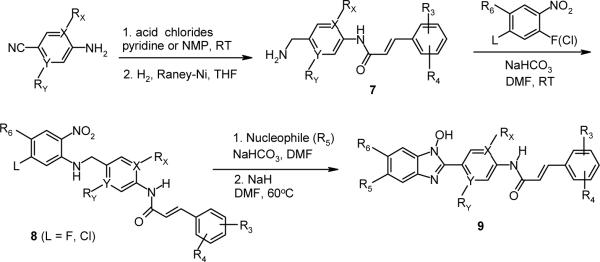
The N-hydroxybenzimidazoles in the lead series were synthesized using modified literature procedures (Scheme 1).33,34 A substituted o-nitro-fluorobenzene or the corresponding chloro analog was subjected to nucleophilic aromatic substitution (SNAr) reaction with a 4-aminobenzylamine to form 4-aminobenzyl-2-nitrophenylamine (3). Subsequent cyclization reaction in the presence of a base such as sodium hydride, potassium tert-butoxide, or sodium methoxide resulted in the formation of 2-(4-aminophenyl)-1-hydroxy-benzimidazole (4). When L is F or Cl, a non-nucleophilic base such as sodium hydride was used to avoid SNAr reaction at that position (see Scheme 2). Further reaction of the cyclized intermediate 4 with an appropriate acid chloride (ca. 2.5 equiv.) in either pyridine, or N-methylpyrrolidone generated bis-acylated product 5 (not isolated), which was then hydrolyzed in situ to afford the product 6. Alternatively, depending on commercial availability or functional group compatibility, N-hydroxybenzimidazoles were also prepared starting with a substituted 4-amino benzonitrile (Scheme 2). Amide formation with an acid chloride followed by catalytic hydrogenation provided 4-phenyl acrylic amido benzylamine derivative 7, which was then reacted with an activated nitrobenzene to give 8. Subsequently, with the intermediate 8 (for L = F or Cl), various functional groups R5 were introduced. For amino group substitutions at R5, compound 8 was reacted with the corresponding amines in the presence of sodium bicarbonate, and then following cyclization reaction provided 9. For alkoxy groups at R5, nucleophilic substitution reaction and cyclization of 8 occurred in one-pot in the presence of the corresponding alcohols (e.g. methanol, ethanol, etc.), and a base such as sodium hydride. This synthetic route proved to be highly efficient in generating R5 substituted N-hydroxybenzimidazole derivatives 9.
Scheme 1.
Method A: General synthesis of N-hydroxybenzimidazoles
Scheme 2.
Method B: Alternative synthesis of N-hydroxybenzimidazoles
Results and Discussion
The inhibitory activity of N-hydroxybenzimidazole derivatives was measured using an in vitro cell-free LcrF-DNA binding assay, and is reported as IC50 values (see Experimental section).
Among the previously identified inhibitors of SoxS-DNA binding, compound 10 (Table 1) displayed good inhibitory activity for LcrF (IC50 = 17.7 μM). As demonstrated in our earlier communication, the lead series of inhibitors in the SoxS-DNA binding assay consisted of a N-hyroxybenzimidazole core with a substituted phenyl amide.10 For the inhibition of LcrF-DNA binding, four different parts of the core structure were investigated to establish SAR: linker, terminal phenyl group (R3/R4), middle phenyl group (X, Y, RX and RY), and benzimidazole ring (R5, R6 and W) (general structure 2 in Figure 2).
Table 1.
Linker SAR of N-hydroxybenzimidazoles
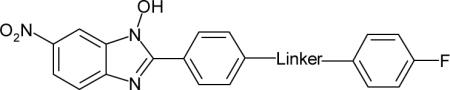
| Compound | Linker | LcrF IC50 (μM)a |
|---|---|---|
| 10 | NHCO | 17.7 |
| 11 | CH2NHCO | >58.4 |
| 12 | NHCOCH2 | >58.4 |
| 13 | NHCOCH2CH2 | >53.5 |
| 14 | NHCOCH=CH | 3.9 |
| 15 | CH2NHCOCH=CH | 14.0 |
IC50 was determined using a dose response analysis with a maximum concentration of 25 μmL. Data represent the average values from two independent experiments unless specified otherwise in the Experimental section.
To further improve the activity of 10, a variety of different linkers were introduced between the two phenyl groups (Table 1). According to our previous work, an amide linker in 10 provided better inhibitory activity than other linkers such as amino and ether groups.10 Therefore, in this linker SAR study, we focused on the evaluation of selected amide analogs (11–15). Extension of the amide linker in 10 with methylene groups in either direction (11, 12 and 13) resulted in derivatives with no measurable activity. Insertion of a vinyl group in compound 14 improved inhibition (IC50 = 3.9 μM), however, addition of another methylene group in 15 (IC50 = 14.0 μM) decreased activity. Within this linker series, the acrylic amide group in 14 was established as the most effective linker moiety, prompting further investigation of more substituted derivatives in acrylic amide series (Tables 2–5).
Table 2.
Substitution effect of R3 and R4
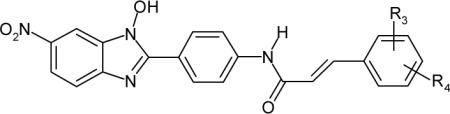
| Compound | R3/R4 | LcrF IC50 (μM)a |
|---|---|---|
| 14 | 4-F | 3.9 |
| 16 | H | 6.9 |
| 17 | 2,4-F/F | 5.0 |
| 18 | 3,4-F/F | 14.5 |
| 19 | 4-CN | 16.4 |
| 20 | 4-CH3 | 0.8 |
| 21 | 2-OCH3 | 20.9 |
| 22 | 4-OCH3 | 8.0 |
| 23 | 4-CF3 | 6.4 |
| 24 | 2,4-OCH3/OCH3 | 38.5 |
| 25 | 4-iso-propyl | >50.9 |
| 26 | 4-N(CH3)2 | 16.9 |
| 27 | 4-1H-[1,2,4]triazole | 35.7 |
| 28 | 4-1H-imidazole | >44.8 |
| 29 | 4-OH | >57.0 |
IC50 was determined using a dose response analysis with a maximum concentration of 25 μmL. Data represent the average values from two independent experiments unles specified otherwise in the Experimental section.
Table 5.
Substitution effect of R6
| Compound | W | R6 | LcrF IC50 (μM) |
|---|---|---|---|
| 14 | C | NO2 | 3.9a |
| 42 | C | H | >100b |
| 43 | N | - | >54.8 a |
| 44 | C | F | >100b |
| 45 | C | CN | 8.3 a |
| 46 | C | Cl | >100b |
| 47 | C | CF3 | 59.7b |
| 48 | C | COCH3 | 39.4b |
| 49 | C | SO2CH3 | 87.4b |
| 50 | C | 1H-pyrazole | 53.7 a |
| 51 | C | 1H-imidazole | 37.3 a |
| 52 | C | COOH | 38.5b |
IC50 was determined using a dose response analysis with a maximum concentration of 25 μg/mL
IC50 determined with a maximum concentration of 50 μg/mL. Data represent the average values from two independent experiments unless specified otherwise in the Experimental section.
In Table 2, an additional fourteen compounds with varying substitutions R3/R4 were evaluated. Among the compounds examined, 4-H (16), 2,4-F/F (17), 4-CH3 (20), 4-OCH3 (22), and 4-CF3 (23) showed slightly improved or comparable inhibitory activity to that of 14, whereas relatively large substitutions in 24, 25, 27 and 28 resulted in loss of activity. No clear SAR emerged from the modification of R3/R4, however, the data in Table 2 suggest that a small lipophilic group at R3/R4 is a general requirement for a good inhibitor. Further analysis of SAR in this series would be difficult without having structural information on the binding site of LcrF protein. Compound 20 showed the best inhibitory activity (IC50 = 0.8 μM) in this series.
In the next set of N-hydroxybenzimidazole derivatives, selected modifications were made at the middle phenyl ring (Table 3). Substitution of nitrogen atoms at positions X (30) and Y (31) showed comparable to slightly decreased activity over their corresponding carbon analog 14. While substitution of a fluorine atom at RY (33) was tolerated, the fluorine at RX (32) was unfavorable for activity. Both the -CH3 group at RX (34) and the -OCH3 group at RY (35) displayed reduced activity compared to 14. As the SAR in Table 3 indicates, the middle phenyl ring is not likely to accommodate an extended range of substitutions.
Table 3.
Substitution effect at the middle phenyl ring
| Compound | X | Rx | Y | Ry | LcrF IC50 (μM)a |
|---|---|---|---|---|---|
| 14 | C | H | C | H | 3.9 |
| 30 | N | - | C | H | 7.4 |
| 31 | C | H | N | - | 10.8 |
| 32 | C | F | C | H | 20.7 |
| 33 | C | H | C | F | 6.5 |
| 34 | C | CH3 | C | H | 25.3 |
| 35 | C | H | C | OCH3 | 13.5 |
IC50 was determined using a dose response analysis with a maximum concentration of 25 μmL. Data represent the average values from two independent experiments unless specified otherwise in the Experimental section.
Table 4 summarizes the substitution effect of R5 groups. While the -CH3 group in 36 maintained the activity of 14, the -OCH2CH3 group in 37 resulted in four-fold decrease relative to 14. This may be linked to the increased steric effect of the -OCH2CH3 group in 37. The substitution of -F at R5 (38) showed much reduced activity. Considering the relatively small size of fluorine, its electron withdrawing effect may have contributed to the weak activity of 38. Aside from potentially unfavorable steric effect of the -N(CH3)2 group, factors contributing to the inactivity of 39 is not understood. The inhibitory activity of compound 40 is attributed to its non-specific binding to DNA, as determined using an agarose gel electrophoresis assay (see Experimental section), not to inhibition of LcrF-DNA binding. Given the similar basic nature of the amino group at R5, compound 41 is likely to bind to DNA as well.
Table 4.
Substitution effect of R5

| Compound | R5 | LcrF IC50 (μM) |
|---|---|---|
| 14 | H | 3.9a |
| 36 | CH3 | 7.6a |
| 37 | OCH2CH3 | 17.1a |
| 38 | F | 26.6b |
| 39 | N(CH3)2 | 66.0b |
| 40 | N(CH3)CH2CH2N(CH3)2 | 3.3c |
| 41 | OCH2CH2( 1 -methyl-4-piperazine) | 3.1c |
IC50 was determined using a dose response analysis with a maximum concentration of 25 μg/mL
IC50 determined with a maximum concentration of 50 μg/mL
Non-specific DNA binding. Data represent the average values from two independent experiments unless specified otherwise in the Experimental section.
The effects of various R6 groups are summarized in Table 5. Of the twelve R6 groups evaluated, the -NO2 group in 14 and the -CN group in 45 exhibited greater inhibitory activity than the others. In the literature -COCH3 and -SO2CH3 groups are described as isosteric replacements for a -NO2 group,35 whereas an imidazole group is indicated as a bioisostere of -NO2 and -CN groups.36 In our N-hydroxybenzimidazole series, however, none of these potential isosteres (in 48, 49 and 51) maintained the activity of 14. Of the other R6 groups, the -COOH group in 52 may have similar properties (e.g. lipophilicity, molar volume, and ionizability) to those of the -NO2 group in 14. Nonetheless, compound 52 lacks inhibitory activity, presumably because of H-bonding donor ability of the -COOH group.
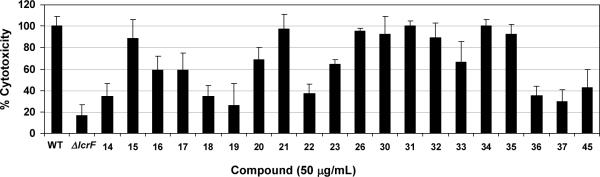
Select compounds in the acrylic amide series (IC50 ≤ 25 μM) were screened using a whole cell cytotoxicity assay (inhibition of T3SS-dependent Y. pseudotuberculosis killing of macrophages in vitro).37,38 As a control experiment, all compounds were tested for their intrinsic toxicity, and were non-cytotoxic in vitro to uninfected J774A.1 murine macrophages at a concentration of 50μg/mL. In this assay, an lcrF null mutant strain (ΔlcrF) typically induces only 15–25% of the cytotoxicity caused by a wild-type Y. pseudotuberculosis (WT) (Figure 3). Among 20 test compounds, 14, 18, 19, 22, 36, and 37 reduced Y. pseudotuberculosis cytotoxicity to a level approaching that of the ΔlcrF mutant (Figure 3). Activity (IC50 values) in the cell-free LcrF-DNA binding assay did not always correlate with activity in the whole cell assay. For example, compound 19 and 31 which exhibited similar inhibitory activity in the LcrF-DNA-binding assay (IC50 = 16.4 and 10.8 μM, respectively) had very different activities in the whole cell assay. While compound 19 strongly inhibited cytotoxicity, compound 31 was devoid of any measurable activity in the whole cell assay. Also, compound 20, the best inhibitor (IC50 =0.8 μM) identified from the LcrF-DNA-binding assay, was relatively inactive in the whole cell assay. This lack of whole cell activity may have resulted from poor membrane permeability; however, variables (e.g., physicochemical parameters) that affected the permeability of compounds are not clearly understood. There may also have been undetected differences in compound solubility in the whole cell assay media, which is distinct from the media used in the LcrF-DNA binding assay (see Experimental section). It is also possible that N-hydroxybenzimidazoles may exert their effects on targets other than LcrF in the whole cell assay. Overall, the results demonstrate that select compounds with good inhibitory activity against LcrF-DNA binding also effectively inhibited the virulence of Yersinia in the whole cell assay.
Figure 3.
Inhibition of cytotoxicity caused by Y. pseudotuberculosis in J774A.1 cells. WT, a wild-type strain (YPIIIpIB1) of Y. pseudotuberculosis; ΔlcrF, a lcrF null mutant strain (YPIIIpIB1ΔlcrF) of Y. pseudotuberculosis. Data are mean values ± standard deviation with n ≥ 4 (number of replicates).
In order to validate target specificity, select compounds (14, 22, and 36 with IC50 ≤ 10 μM) were further evaluated in both ExsA-DNA and SlyA-DNA binding assays (Table 6). ExsA is a MAR transcription factor found in Pseudomonas aeruginosa, and shares a high degree of homology with LcrF in the DNA binding domain (85% identity and 92% similarity).32 Therefore, if these inhibitors are indeed targeting the DNA binding domain of LcrF protein, they would be likely to inhibit ExsA-DNA binding with similar potency. As shown in Table 6, all three test compounds exhibited very close IC50 values for both LcrF and ExsA. The DNA binding motif of SlyA, a MarR family of transcription factor in Salmonella spp., is different from that of MAR proteins.39,40 Therefore, a SlyA-DNA binding assay was used to test the specificity of LcrF inhibitors. No measurable activity of the LcrF inhibitors was observed in the SlyA-DNA binding assay (Table 6). Most N-hydroxybenzimidazole derivatives tested in DNA binding-agarose gel electrophoresis assays displayed no detectable binding to DNA with the exception of compounds structurally related to 40 and 41. These results confirm the target specificity of N-hydroxybenzimidazole inhibitors for MAR transcription factors. In an in vitro antimicrobial susceptibility test, compounds 14, 22 and 36 showed no measurable antibacterial activity against Y. pseudotuberculosis as well as S. aureus and E. coli (Table 6), verifying that they do not target bacterial growth.
Table 6.
Target specificity and in vitro antibacterial activity
| Compound | IC50 (μM)a | MIC (μg/mL) | ||||
|---|---|---|---|---|---|---|
| LcrF | ExsAb | SlyAc | Y. pseudotuberculosisd | S. aureuse | E. colif | |
| 14 | 3.9 | 3.5 | >53.8 | >64 | >64 | >64 |
| 22 | 8.0 | 5.9 | >55.2 | >64 | >64 | >64 |
| 36 | 7.6 | 6.7 | >52 | >64 | >64 | >64 |
IC50 was determined using a dose response analysis with the maximum concentration of 25 μg/mL. Data represent the average values from two independent experiments unless specified otherwise in the Experimental section
MAR transcription factor in P. aeruginosa
MarR family transcription factor in Salmonella spp.
YPIIIpIB1 strain of Y. pseudotuberculosis
RN450 strain of S. aureus
ATCC25922 strain of E. coli.
Conclusions
A novel class of small molecules based on the N-hydroxybenzimidazole scaffold was developed as inhibitors of LcrF-DNA binding. They inhibit the virulence of Yersinia by targeting the LcrF protein, a MAR transcription factor. Select compounds in the acrylic amide series demonstrated inhibitory activity in a cell-free LcrF-DNA binding assay. Their anti-virulence activity was further confirmed by using a T3SS-dependent whole cell assay. This class of compounds, exemplified by 14, 22, and 36, is non-antibacterial, non-cytotoxic, non-DNA binding, and shows specificity for MAR proteins. This work suggests that a novel approach of targeting pathogen virulence could be achieved at the transcription level in Yersinia spp. The N-hydroxybenzimidazoles described in this work are effective anti-virulence agents in vitro, and have the potential to be developed into drug candidates against diseases caused by Yersinia spp.
Experimental Section
General
All chemical reactions were carried out under an atmosphere of either argon or nitrogen unless mentioned otherwise. All reagents and solvents were purchased from commercial vendors, and used without further purification. The reactions were monitored using an analytical HPLC and/or an LCMS. The analytical HPLC traces were recorded using a Luna C18-phenyl hexyl column (50 mm × 4.6 mm, 5 micron) with either acidic (0.1% v/v CF3COOH in water as solvent A, 0.1% v/v CF3COOH in acetonitrile as solvent B) or basic (20mM triethanolamine in water as solvent A and acetonitrile as solvent B) media with the λdet set at 280 nm. The LCMS (Shimadzu LCMS-2010 EV) spectra were recorded using 0.2% v/v HCOOH in water as solvent A and 0.2% v/v HCOOH in acetonitrile as solvent B (λdet = 280 nm) with Luna C18 Synergi column (50 mm × 4.66 mm, 5 micron) for LC, with the ESI-MS operating in positive mode, unless mentioned otherwise. The 1H NMR spectra were recorded in DMSO-d6 using a Bruker DPX300 NMR spectrometer operating at 300 MHz. The 1H NMR chemical shifts are reported in ppm (δ) relative to the residual protonated solvent peak. The purity of final compounds was assessed based on analytical HPLC, and the results were greater than 95% unless specified otherwise.
Method A: General synthesis of 4-aminobenzyl-(2,4-dinitro-phenyl)-amine derivatives (3)
To a solution of 4-aminobenzyl amine derivatives (25.5 mL, 225 mmol) and powdered NaHCO3 (94.5 g, 1125 mmol) in anhydrous DMF (300 mL) was added 2,4-dinitrofluoro benzene (18.8 mL, 150 mmol) dropwise at room temperature. After 2 h, the solution was slowly diluted with water (1000 mL) to precipitate the product, which was collected on a fritted funnel rinsing with water until the eluent was colorless. The solid was further dried under high vacuum to afford the product 3 as a bright orange solid (43 g, 99% yield). This crude material was used for the next step without further purification. 1H NMR (300 MHz, DMSO-d6): δ 9.18 (t, J = 5.7 Hz, 1H), 8.86 (d, J = 3.0 Hz, 1H), 8.22 (dd, J = 2.7, 9.6 Hz, 1H), 7.13 (d, J = 9.6 Hz, 1H), 7.05 (d, J = 8.4 Hz, 2H), 6.53 (d, J = 8.4 Hz, 2H), 5.05 (s, 2H), 4.54 (d, J = 6.0 Hz, 2H). MS (ESI, positive): calcd for [C13H12N4O4], 288.26; found, 330.10 [M+H+CH3CN]+.
General synthesis of 6-nitro-2-(4-aminophenyl)-1-hydroxybenzimidazole derivatives (4)
To a solution of N-(4-aminobenzyl)-2,4-dinitroaniline derivative 3 (21.6 g, 74.9 mmol) in anhydrous DMF (75 mL) was slowly added NaOMe (30% w/w in MeOH) (67.5 g, 375 mmol) at room temperature under argon atmosphere. After the addition, the solution was warmed to 60 °C for 2 h. After cooling to ambient temperature, the solution was transferred to an Erlenmeyer flask or tall beaker, diluted with water (700 mL), and then acidified with saturated citric acid. The resulting precipitate was collected on a sintered funnel rinsing with water. The crude product was purified by recrystallization in hot EtOH to afford 4 as a brown solid (18.1 g, 90% yield). 1H NMR (300 MHz, DMSO-d6): δ 12.36 (s, 1H), 8.22 (d, J = 2.4 Hz, 1H), 8.08–8.04 (m, 3H), 7.67 (d, J = 8.7 Hz, 1H), 6.68 (d, J = 8.7 Hz, 2H), 5.91 (br s, 2H). MS (ESI, positive): calcd for [C13H10N4O3], 270.25; found, 271.05 [M+H]+.
General synthesis of N-acyl-6-nitro-2-(4-aminophenyl)-1-hydroxybenzimidazole derivatives (6)
To a solution of 6-nitro-2-(4-aminophenyl)-1-hydroxybenzimidazole derivative 4 (1.0 mmol) in anhydrous pyridine (2.0 mL) was added an acid chloride (2.5 mmol) at room temperature. After stirring for 2–3 h, the solution was diluted with 3N NaOH (6.0 mL) and stirred for another hour. The deep amber solution was transferred to an Erlenmeyer flask or beaker through dilution with water (100 mL), and then acidified with saturated citric acid. The resulting precipitate was collected on a sintered funnel rinsing with water. The crude product 6 was further purified either by preparative HPLC or by recrystallization in hot EtOH.
4-Fluoro-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-benzyl]-benzamide (11)
1H NMR (300 MHz, DMSO-d6): δ 12.64 (s, 1H), 9.19 (t, J = 5.7 Hz, 1H), 8.39 (d, J = 2.1 Hz, 1H), 8.29 (d, J = 8.4 Hz, 2H), 8.14 (dd, J = 2.4, 9 Hz, 1H), 8.01 (dd, J = 5.7, 8.7 Hz, 2H), 7.85 (d, J = 9 Hz, 1H), 7.55 (d, J = 8.4 Hz, 2H), 7.34 (t, J = 9 Hz, 2H), 4.59 (d, J = 6 Hz, 2H). MS (ESI, positive): calcd for [C21H15F1N4O4], 406.38; found, 407.18 [M+H]+.
2-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acetamide (12)
1H NMR (300 MHz, DMSO-d6): δ 12.56 (s, 1H), 10.41 (s, 1H), 8.23 (d, J = 2.3 Hz, 1H), 8.20 (d, J = 8.8 Hz, 2H), 8.01 (dd, J = 2.3, 8.9 Hz, 1H), 7.71 (d, J = 8.8 Hz, 2H), 7.70 (d, J = 8.9 Hz, 1H), 7.28 (dd, J = 5.7, 8.6 Hz, 2H), 7.06 (t, J = 8.9 Hz, 2H), 3.60 (s, 2H). MS (ESI, positive): calcd for [C21H15F1N4O4], 406.38; found, 407.17 [M+H]+.
3-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-propionamide (13)
1H NMR (300 MHz, DMSO-d6): δ 12.63 (s, 1H), 10.39 (s, 1H), 8.34 (d, J = 2.2 Hz, 1H), 8.30 (d, J = 8.8 Hz, 2H), 8.12 (dd, J = 2.2, 8.9 Hz, 1H), 7.81 (d, J = 8.9 Hz, 2H, and s, 1H), 7.31 (dd, J = 8.5, 5.8 Hz, 2H), 7.12 (t, J = 8.8 Hz, 2H), 2.94 (t, J = 7.4 Hz, 2H), 2.69 (t, J = 7.8 Hz, 2H). MS (ESI, positive): calcd for [C22H17F1N4O4], 420.40; found, 421.09 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (14)
1H NMR (300 MHz, DMSO-d6): δ 12.61 (s, 1H), 10.54 (s, 1H), 8.37 (d, J = 2.0 Hz, 1H), 8.35 (d, J = 8.7 Hz, 2H), 8.14 (dd, J = 2.2, 8.9 Hz, 1H), 7.94 (d, J = 8.8 Hz, 2H), 7.83 (d, J = 8.9 Hz, 1H), 7.73 (dd, J = 5.7, 8.6 Hz, 2H), 7.66 (d, J = 15.7 Hz, 1H), 7.31 (t, J = 8.8 Hz, 2H), 6.83 (d, J = 15.7 Hz, 1H). MS (ESI, positive): calcd for [C22H15F1N4O4], 418.39; found, 419.08 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-benzyl]-acrylamide (15)
1H NMR (300 MHz, DMSO-d6): δ 12.65 (s, 1H), 8.75 (t, J = 5.9 Hz, 1H), 8.39 (d, J = 2.4 Hz, 1H), 8.30 (d, J = 8.4 Hz, 2H), 8.14 (dd, J = 2.4, 9 Hz, 1H), 7.86 (d, J = 9 Hz, 1H), 7.66 (dd, J = 5.7, 8.4 Hz, 2H), 7.53 (d, J = 8.4 Hz, 2H), 7.51 (d, J = 16.2 Hz, 1H), 7.27 (t, J = 9 Hz, 2H), 6.68 (d, J = 15.9 Hz, 1H), 4.52 (d, J = 6 Hz, 2H). MS (ESI, positive): calcd for [C23H17F1N4O4], 432.41; found, 433.23 [M+H]+.
(E)-N-[4-(1-Hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-phenyl-acrylamide (16)
1H NMR (300 MHz, DMSO-d6): δ 12.65 (s, 1H), 10.51 (s, 1H), 8.32 (d, J = 2.2 Hz, 1H), 8.30 (d, J = 8.8 Hz, 2H), 8.09 (dd, J = 2.3, 8.9 Hz, 1H), 7.90 (d, J = 8.8 Hz, 2H), 7.78 (d, J = 8.9 Hz, 1H), 7.62 (d, J = 8.5 Hz, 2H), 7.61 (d, J = 15.2 Hz, 1H), 7.47–7.37 (m, 3H), 6.85 (d, J = 15.8 Hz, 1H). MS (ESI, positive): calcd for [C22H16N4O4], 400.40; found, 401.11 [M+H]+.
(E)-3-(2,4-Difluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (17)
1H NMR (300 MHz, DMSO-d6): δ 12.62 (brs, 1H), 10.62 (s, 1H), 8.34 (d, J = 2.2 Hz, 1H), 8.33 (d, J = 8.7 Hz, 2H), 8.11 (dd, J = 2.4, 9 Hz, 1H), 7.93 (d, J = 9 Hz, 2H), 7.81 (d, J = 9 Hz, 1H), 7.78 (t, J = 8.7 Hz, 1H), 7.63 (d, J = 15.9 Hz, 1H), 7.39 (dt, J = 2.7, 9.6 Hz, 1H), 7.21 (dt, J = 2.1, 8.4 Hz, 1H), 6.94 (d, J = 15.9 Hz, 1H). MS (ESI, positive): calcd for [C22H14F2N4O4], 436.38; found, 437.08 [M+H]+.
(E)-3-(3,4-Difluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (18)
1H NMR (300MHz, DMSO-d6): δ 12.64 (br, 1H), 10.62 (s, 1H), 8.37 (d, J = 2.4 Hz, 1H), 8.35 (d, J = 8.7 Hz, 2H), 8.14 (dd, J = 2.4, 9 Hz, 1H), 7.94 (d, J = 9.0 Hz, 2H), 7.84 (d, J = 9.0 Hz, 1H), 7.77 (m, 1H), 7.63 (d, J = 15.9 Hz, 1H), 7.57–7.53 (m, 2H), 6.87 (d, J = 15.6 Hz, 1H). MS (ESI, positive): calcd for [C22H14F2N4O4], 436.38; found, 437.15 [M+H]+.
(E)-N-[4-(1-Hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-p-tolyl-acrylamide (20)
1H NMR (300 MHz, DMSO-d6): δ 12.63 (s, 1H), 10.52 (s, 1H), 8.37 (d, J = 2.4 Hz, 1H), 8.35 (d, J = 9 Hz, 2H), 8.13 (dd, J = 2.1, 8.7 Hz, 1H), 7.94 (d, J = 9 Hz, 2H), 7.83 (d, J = 9 Hz, 1H), 7.61 (d, J = 15.9 Hz, 1H), 7.55 (d, J = 8.1 Hz, 2H), 7.28 (d, J = 7.8 Hz, 2H), 6.83 (d, J = 15.6 Hz, 1H), 2.35 (s, 3H). MS (ESI, positive): calcd for [C23H18N4O4], 414.42; found, 415.15 [M+H]+.
(E)-N-[4-(1-Hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-(2-methoxy-phenyl)-acrylamide (21)
1H NMR (300 MHz, DMSO-d6): δ 12.58 (s, 1H), 10.46 (s, 1H), 8.30 (d, J = 2.0 Hz, 1H), 8.29 (d, J = 8.6 Hz, 2H), 8.08 (dd, J = 2.2, 8.9 Hz, 1H), 7.89 (d, J = 8.8 Hz, 2H), 7.81 (d, J = 14.4 Hz, 1H), 7.77 (d, J = 8.8 Hz, 1H), 7.56 (d, J = 7.6 Hz, 1H), 7.37 (t, J = 8.3 Hz, 1H), 7.07 (d, J = 8.3 Hz, 1H), 6.99 (t, J = 7.5 Hz, 1H), 6.88 (d, J = 15.8 Hz, 1H), 3.86 (s, 3H). MS (ESI, positive): calcd for [C23H18N4O5], 430.42; found, 431.14 [M+H]+.
(E)-N-[4-(1-Hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-(4-methoxy-phenyl)-acrylamide (22)
1H NMR (300 MHz, DMSO-d6): δ 12.57 (s, 1H), 10.40 (s, 1H), 8.31 (d, J = 2.3 Hz, 1H), 8.28 (d, J = 9.0 Hz, 2H), 8.08 (dd, J = 2.3, 8.9 Hz, 1H), 7.88 (d, J = 8.9 Hz, 2H), 7.77 (d, J = 8.9 Hz, 1H), 7.56 (d, J = 9.0 Hz, 2H), 7.55 (d, J = 14.6 Hz, 1H), 6.97 (d, J = 8.7 Hz, 2H), 6.68 (d, J = 15.7 Hz, 1H), 3.76 (s, 3H). MS (ESI, positive): calcd for [C23H18N4O5], 430.42; found, 431.14 [M+H]+.
(E)-3-(2,4-Dimethoxy-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (24)
1H NMR (300 MHz, DMSO-d6): δ 12.62 (brs, 1H), 10.43 (s, 1H), 8.36 (d, J = 2.4 Hz, 1H), 8.33 (d, J = 9 Hz, 2H), 8.13 (dd, J = 2.1, 9 Hz, 1H), 7.94 (d, J = 8.7 Hz, 2H), 7.83 (d, J = 9 Hz, 1H), 7.78 (d, J = 16.8 Hz, 1H), 7.54 (d, J = 8.4 Hz, 1H), 6.81 (d, J = 15.6 Hz, 1H), 6.65–6.62 (m, 2H), 3.91 (s, 3H), 3.83 (s, 3H). MS (ESI, positive): calcd for [C24H20N4O6], 460.45; found, 461.25 [M+H]+.
(E)-N-[4-(1-Hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-(4-isopropyl-phenyl)-acrylamide (25)
1H NMR (300 MHz, DMSO-d6): δ 12.64 (brs, 1H), 10.54 (s, 1H), 8.36 (br s, 1H), 8.35 (d, J = 8.7 Hz, 2H), 8.12 (dd, J = 2.3, 8.9 Hz, 1H), 7.93 (d, J = 8.8 Hz, 2H), 7.81 (d, J = 8.9 Hz, 1H), 7.62 (d, J = 15.5 Hz, 1H), 7.58 (d, J = 8.0 Hz, 2H), 7.34 (d, J = 8.1 Hz, 2H), 6.84 (d, J = 15.7 Hz, 1H), 2.93 (m, J = 6.9 Hz, 1H), 1.23 (dd, J = 6.9 Hz, 6H). MS (ESI, positive): calcd for [C25H22N4O4], 442.48; found, 443.26 [M+H]+.
(E)-3-(4-Dimethylamino-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (26)
1H NMR (300 MHz, DMSO-d6): δ 12.61 (s, 1H), 10.28 (s, 1H), 8.29 (d, J = 2.0 Hz, 1H), 8.27 (d, J = 8.7 Hz, 2H), 8.07 (dd, J = 2.3, 8.9 Hz, 1H), 7.86 (d, J = 8.9 Hz, 2H), 7.76 (d, J = 8.9 Hz, 1H), 7.48 (d, J = 15.5 Hz, 1H), 7.42 (d, J = 8.9, 2H), 6.71 (d, J = 8.9, 2H), 6.55 (d, J = 15.5 Hz, 1H), 2.93 (s, 6H). MS (ESI, positive): calcd for [C24H21N5O4], 443.47; found, 444.14 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[5-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-pyridin-2-yl]-acrylamide (30)
1H NMR (300 MHz, DMSO-d6): δ 12.81 (s, 1H), 11.10 (s, 1H), 9.27 (d, J = 1.8 Hz, 1H), 8.71 (dd, J = 2.4, 8.7 Hz, 1H), 8.47 (d, J = 9.0 Hz, 1H), 8.41 (d, J = 1.8 Hz, 1H), 8.15 (dd, J = 2.4, 8.7 Hz, 1H), 7.87 (d, J = 9.0 Hz, 1H), 7.73–7.68 (m, 3H), 7.32 (t, J = 8.7 Hz, 2H), 7.03 (d, J = 15.6 Hz, 1H). MS (ESI, positive): calcd for [C21H14F1N5O4], 419.38; found, 420.10 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[6-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-pyridin-3-yl]-acrylamide (31)
1H NMR (300 MHz, DMSO-d6): δ 12.83 (brs, 1H), 10.79 (s, 1H), 9.09 (br s, 1H), 8.42–8.33 (m, 3H), 8.16 (dd, J = 1.8, 8.7 Hz, 1H), 7.88 (d, J = 8.7 Hz, 1H), 7.75 (dd, J = 5.4, 8.4 Hz, 2H), 7.70 (d, J = 15.6 Hz, 1H), 7.32 (t, J = 8.7 Hz, 2H), 6.82 (d, J = 15.6 Hz, 1H). MS (ESI, positive): calcd for [C21H14F1N5O4], 419.38; found, 420.15 [M+H]+.
(E)-N-[3-Fluoro-4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-(4-fluoro-phenyl)-acrylamide (33)
1H NMR (300 MHz, DMSO-d6): δ 12.47 (s, 1H), 10.72 (s, 1H), 8.39 (d, J = 2.1 Hz, 1H), 8.17 (dd, J = 2.2, 8.9 Hz, 1H), 7.99–7.84 (m, 1H), 7.88 (d, J = 8.8 Hz, 2H), 7.76–7.56 (m, 4H), 7.31 (t, J = 8.8 Hz, 2H), 6.80 (d, J = 15.7 Hz, 1H). MS (ESI, positive): calcd for [C22H14F2N4O4], 436.38; found, 437.15 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-2-methyl-phenyl]-acrylamide (34)
1H NMR (300 MHz, DMSO-d6): δ 12.68 (s, 1H), 9.58 (s, 1H), 8.36 (d, J = 2.2 Hz, 1H), 8.22–8.18 (m, 2H), 8.13 (dd, J = 2.3, 8.9 Hz, 1H), 8.03 (dd, J = 8.4 Hz, 1H), 7.83 (d, J = 8.9 Hz, 1H), 7.73 (dd, J = 5.6, 8.7 Hz, 2H), 7.64 (d, J = 15.7 Hz, 1H), 7.31 (t, J = 8.8 Hz, 2H), 7.05 (d, J = 15.7 Hz, 1H), 2.41 (s, 3H). MS (ESI, positive): calcd for [C23H17F1N4O4], 432.41; found, 433.10 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-5-methyl-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (36)
1H NMR (300 MHz, DMSO-d6): δ 12.59 (s, 1H), 10.52 (s, 1H), 8.31 (d, J = 8.7 Hz, 2H), 8.17 (s, 1H), 7.91 (d, J = 8.8 Hz, 2H), 7.75–7.62 (m, 4H), 7.31 (t, J = 8.8 Hz, 2H), 6.82 (d, J = 15.7 Hz, 1H), 2.63 (s, 3H). MS (ESI, positive): calcd for [C23H17F1N4O4], 432.41; found, 433.10 [M+H]+.
(E)-N-[4-(6-Cyano-1-hydroxy-1H-benzoimidazol-2-yl)-phenyl]-3-(4-fluoro-phenyl)-acrylamide (45)
1H NMR (300 MHz, DMSO-d6): δ 12.44 (brs, 1H), 10.53 (s, 1H), 8.31 (d, J = 8.8 Hz, 2H), 8.08 (br s, 1H), 7.92 (d, J = 8.8 Hz, 2H), 7.81 (d, J = 8.4 Hz, 1H), 7.75–7.60 (m, 4H), 7.31 (t, J = 8.8 Hz, 2H), 6.83 (d, J = 15.7 Hz, 1H). MS (ESI, positive): calcd for [C23H15F1N4O2], 398.40; found, 399.17 [M+H]+.
(E)-3-(4-Fluoro-phenyl)-N-[4-(1-hydroxy-6-pyrazol-1-yl-1H-benzoimidazol-2-yl)-phenyl]-acrylamide (50)
To a solution of 4-aminobenzyl amine (35.4 mL, 313 mmol) and powdered NaHCO3 (158 g, 1875 mmol) in anhydrous DMF (500 mL) at room temperature was added a solution of 4-bromo-1-fluoro-2-nitrobenzene (31.4 mL, 250 mmol) in anhydrous DMF (50 mL) dropwise via addition funnel over 1 h period. After another 4 h, the solution was diluted with anhydrous absolute EtOH (1000 mL) and powdered KOtBu (140 g, 1250 mmol) was added in portions. This solution was subsequently heated to 60 °C for 6 h. After cooling to room temperature, the solution was poured into a stirring solution of water (4 L), and then adjusted to pH 6 with 1M HCl. The slowly stirring suspension was cooled with an ice bath to facilitate solidification. The suspended product was collected on a fine fritted funnel, and rinsed with water until the eluent was colorless. The orange solid was further dried under high vacuum. A 20 mL Biotage microwave vial was charged with 6-bromo-2-(4 aminophenyl)-1-hydroxybenzimidazole (1.5 g, 5.0 mmol), N,N'-dimethylethylenediamine (1.1 mL, 10.0 mmol), CuI (0.9 g, 5.0 mmol), pyrazole (1.4 g, 20.0 mmol) and KOtBu (2.3 g, 20.0 mmol) and anhydrous DMSO (20 mL). The secured vial was placed into a Biotage microwave reactor with a temperature setting of 195 °C for 45 min. After cooling, the vial was opened and poured into a rapidly stirring water solution. The resulting suspension was filtered through a plug of Celite® rinsing with 0.5 M NaOH. The water solution was loaded onto a prepared DVB column. After loading, the product was eluted with acetonitrile. The acetonitrile was removed under reduced pressure. The resulting water solution was cooled to 0 °C with an ice bath, and then the solution pH was adjusted to 6 with 1 M HCl to precipitate the product. The resulting solid was collected, and rinsed with cold water. After further drying under vacuum, the product 50 was obtained as a light brown solid. 1H NMR (300 MHz, DMSO-d6): δ 12.22 (br, 1H), 10.50 (s, 1H), 8.59 (s, 1H), 8.26 (d, J = 8.1 Hz, 2H), 7.93 (s, 1H), 7.90 (d, J = 8.4 Hz, 2H), 7.75–7.66 (m, 5H), 7.63 (d, J = 15.9 Hz, 1H), 7.29 (t, J = 8.4 Hz, 2H), 6.82 (d, J = 15.6 Hz, 1H), 6.55 (s, 1H). MS (ESI, positive): calcd for [C25H18F1N5O2], 439.45; found, 440.20 [M+H]+.
Method B: General synthesis of 4-phenylamidobenzylamine derivatives (7)
To a solution of 4-cyanoaniline derivative (225 mmol) in pyridine or N-methylpyrrolidone (180 mL), was added a cinnamoyl chloride (225 mmol) over a period of 3–5 min with vigorous stirring. After stirring the reaction mixture for 5 h (until HPLC monitoring of the reaction indicated a complete consumption of the starting materials), it was poured into 1400 mL of water at room temperature, and the resulting suspension was stirred for 1 h. The precipitate was filtered, washed with 4 × 500 mL portions of water, and dried. A second crop of solid was obtained from the filtrate and washings. The solids were combined, and used for the next step without further purification. In a pressure reactor, 4-phenylamido benzonitrile intermediate (98 mmol) was dissolved in anhydrous THF (940 mL), and the solution was purged with argon for 2–3 min, followed by the addition of 11 mL of the uniformly suspended catalyst (Raney® nickel 2400, suspension in water). After addition of a small amount of MeOH to the suspension, the reactor was pressurized at 55 psi of H2 while stirring vigorously. LC-MS monitoring of the reaction indicated a complete conversion of the starting material to the corresponding amine within 2.5 h. The reaction mixture was filtered over a bed of diatomaceous earth (e.g., Celite®), and washed with 3 × 100 mL portions of anhydrous THF. The combined filtrates were evaporated to dryness, and further dried under high vacuum to afford 7 as a white colored solid. The crude product was used for the next step without further characterization.
(E)-N-{4-[(5-Fluoro-2,4-dinitro-phenylamino)-methyl]-phenyl}-3-phenyl-acrylamide (8)
To a solution of 1,5-difluoro-2,4-dinitrobenzene (0.8 g, 3.9 mmol) in 30 mL of DMF, was added sodium bicarbonate (3.3 g, 39 mmol) and the intermediate 7 (1.5 g, 3.9 mmol). The reaction mixture was stirred at room temperature for 3 h, and then poured into ice-water to give a precipitate. The precipitate was filtered, washed with water, and dried under vacuum to give the desired product 8 (1.6 g, 90% yield). This material was used for the next step without further purification. 1H NMR (300 MHz, DMSO-d6): δ 10.21 (s, 1H), 9.40 (t, J = 6 Hz, 1H), 8.89 (d, J = 8.1 Hz, 1H), 7.71–7.66 (m, 4H), 7.58 (d, J = 15.6 Hz, 1H), 7.37 (d, J = 8.4 Hz, 2H), 7.28 (t, J = 8.7 Hz, 2H), 7.00 (d, J = 15 Hz, 1H), 6.76 (d, J = 15.6 Hz, 1H), 4.70 (d, J = 6 Hz, 2H). MS (ESI, positive): calcd for [C22H16F2N4O5], 454.39; found, 455.15 [M+H]+.
(E)-N-[4-(5-Ethoxy-1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-(4-fluoro-phenyl)-acrylamide (37)
To a solution of 8 (0.2 g, 0.4 mmol) in EtOH (10 mL) and DMF (10 mL), was added NaH (60% w/w in mineral oil) (88 mg, 2.2 mmol). The reaction mixture was heated at 60 °C for 3 h. After cooling to room temperature, it was poured into ice-water, and acidified with aqueous citric acid. The resulting precipitates were collected, washed with water, and purified by preparative HPLC. Fractions showing >95% purity by analytical HPLC were combined, and the volatiles were evaporated under vacuum. The resulting solid was collected, and washed with water. After drying under vacuum, the product 37 was obtained as a yellow solid. 1H NMR (300 MHz, DMSO-d6): δ 12.41 (s, 1H), 10.51 (s, 1H), 8.29 (d, J = 8.7 Hz, 2H), 8.05 (s, 1H), 7.91 (d, J = 8.8 Hz, 2H), 7.72 (dd, J = 5.7, 8.5 Hz, 2H), 7.65 (d, J = 15.7 Hz, 1H), 7.50 (s, 1H), 7.30 (t, J = 8.8 Hz, 2H), 6.82 (d, J = 15.7 Hz, 1H), 4.22 (qr, J = 6.9 Hz, 2H), 1.37 (t, J = 6.9 Hz, 3H). MS (ESI, positive): calcd for [C24H19F1N4O5], 462.44; found, 463.15 [M+H]+.
(E)-N-[4-(5-Dimethylamino-1-hydroxy-6-nitro-1H-benzoimidazol-2-yl)-phenyl]-3-(4-fluoro-phenyl)-acrylamide (39)
To a solution of 8 (0.9 g, 2 mmol) in DMF (50 mL), was added NaHCO3 (1.7 g, 20 mmol) followed by 4 mL of 2.0 M dimethylamine solution in THF (8 mmol). After stirring at RT for 3 h, the reaction mixture was poured into water (500 mL). After filtration, the collected solid was washed with water, and dried under vacuum, resulting in (E)-N-{4-[(5-Dimethylamino-2,4-dinitro-phenylamino)-methyl]-phenyl}-3-(4-fluoro-phenyl)-acrylamide intermediate as a yellow solid (0.8g, 84% yield). This crude material was used for the next step. To a solution of the aforementioned intermediate (0.8 g, 1.7 mmol) in DMF (30 mL) was added NaOMe (30% w/w in MeOH) (1.5 g, 8.4 mmol). The reaction mixture was heated at 60 °C for 3 h. After cooling to room temperature, it was poured into ice-water, and acidified with aqueous citric acid. The resulting precipitation was collected, washed with water, and purified by preparative HPLC. Fractions showing greater than 95% purity by analytical HPLC were combined, and concentrated. The resulting solid was collected, and washed with water. After drying under vacuum, the product 39 was obtained as a yellow solid. 1H NMR (300 MHz, DMSO-d6): δ 12.32 (s, 1H), 10.52 (s, 1H), 8.28 (d, J = 8.8 Hz, 2H), 7.98 (s, 1H), 7.91 (d, J = 8.9 Hz, 2H), 7.74–7.70 (m, 2H), 7.65 (d, J = 15.7 Hz, 1H), 7.49 (s, 1H), 7.31 (t, J = 8.8 Hz, 2H), 6.82 (d, J = 15.7 Hz, 1H), 2.75 (s, 6H). MS (ESI, positive): calcd for [C24H20F1N5O4], 461.46; found, 462.15 [M+H]+.
Cell-free LcrF-DNA binding assay
A biotinylated double-stranded DNA molecule (2 nM), containing the LcrF binding sequences, was incubated with 6His-LcrF (20 nM) in a streptavidin coated 96-well microtiter plate (Thermo Lab Systems, Waltham, MA) in the presence of varying concentrations of an inhibitor, and in assay buffer (20 mM Hepes, 10 mM ammonium sulfate, 30 mM potassium chloride, 1 mM EDTA, 0.2% polysorbate 20, and 0.5% non-fat dry milk). Unbound DNA and the protein were removed by washing, and a primary monoclonal anti-6His antibody was then added. Bound DNA-protein complexes were detected by a secondary horseradish peroxidase (HRP)-conjugated antibody acting on a chemiluminescence substrate (Cell Signaling Technology, Beverly, MA). Luminescence was measured on a Victor V plate reader (PerkinElmer Life Sciences, Wellesley, MA). Luminescence data were converted to % LcrF binding inhibition by comparison to control wells. The % LcrF binding inhibition data for each test compound titration were plotted in Microsoft Excel, and XLfit (IDBS) was used to fit the dose response curve and reported as IC50 (test compound concentration that inhibited LcrF binding by 50%). ExsA-DNA binding and SlyA-DNA binding assays were carried out by following a similar procedure. Data represent the average values from two independent experiments for most compounds. IC50 data for compounds 10, 14, 17, 24, 26, 45, and 52 are the median values from at least three independent experiments (see supporting information).
Whole cell assay: Inhibition of Yersinia cytotoxicity in J774A.1 macrophages
The cytotoxicity assay was based on the protocol by Monack et al. described previously.24 J774A.1 macrophages were plated at 2 × 104 cells per well in 96-well plates on the day prior to infection. Overnight cultures of wild-type (YPIIIpIB1)23 or the lcrF deletion mutant (YPIIIpIB1ΔlcrF)25 strains of Y. pseudotuberculosis were diluted into 2-YT broth supplemented with 20 mM magnesium chloride and 20 mM sodium oxalate. Cultures were grown for 90 min at 26 °C and then shifted to 37 °C for 90 min to induce LcrF expression. Bacteria were added to cells in DMEM (Invitrogen, Carlsbad, CA), and incubated at 37 °C with 5% CO2 for 2 h. The multiplicity of infection was 5-10 bacteria per J774A.1 cell. Gentamicin was added at 50 μg/mL to kill extracellular bacteria, and incubations continued overnight (~18 h). Supernatants containing released LDH (lactate dehydrogenase), an indication of cell death, were assayed colorimetrically using the Cytox 96® kit from Promega (Madison, WI). Test compounds at 50 μg/mL were added to the Yersinia culture media and to the J774A.1 cell wells during infections. Control cultures and wells received an equal volume of vehicle (DMSO with 0.4% ethanolamine). Results are expressed as % wild-type cytotoxicity (the amount of LDH released in wells infected with wild-type Y. pseudotuberculosis in the presence of vehicle alone).
In vitro antimicrobial susceptibility test
The minimum inhibitory concentration (MIC) of test compounds were determined using Clinical Laboratory Standards Institute (CLSI) methodology.41 On each day of testing, serial dilutions of compounds were prepared in microdilution plates using a Tecan robotic workstation. Mueller Hinton broth cultures of strains were grown or adjusted to match the turbidity of a 0.5 McFarland standard. 1:100 dilutions were made in an appropriate broth to achieve a final inoculum of 5 × 105 cells/mL. Plates were incubated at 35 °C in ambient air for 18–24 h, read spectrophotometrically, and checked manually for evidence of bacterial growth. The lowest dilution of a compound that inhibited bacterial growth was recorded as the MIC.
In vitro DNA binding assay
To assess non-specific DNA binding, the effects of compounds on the migration of a supercoiled plasmid DNA during agarose gel electrophoresis were tested. Compounds were diluted in DMSO with 0.4% ethanolamine to a concentration of 1 mg/mL. In a clear 96 well plate, 2 μL of 1 mg/mL compound solution was added to 18 μL assay buffer (20 mM Hepes, pH 7.6, 10 mM ammonium sulfate, 30 mM potassium chloride, 1mM EDTA, 0.2% polysorbate-20) containing 100 ng of pET-15b supercoiled plasmid DNA. The final concentration of test compounds was therefore 100 μg/mL. The plate was incubated at room temperature for 30 min. Ten microliters of compound and DNA solutions were loaded onto a 0.8% agarose gel and subjected to electrophoresis. The gel was then stained with ethidium bromide and photographed. Control tests with drug vehicle (DMSO with 0.4% ethanolamine) did not affect DNA migration through the gel. In contrast, tests with 100 μg/mL of positive control Hoechst 33342 dye, a known DNA intercalator, resulted in a faint smear on the gel rather than the discrete bands seen when unperturbed DNA is electrophoresed.
Supplementary Material
Acknowledgment
This work was supported in part by a grant from the National Institutes of Allergy and Infectious Disease (NIAID) (Grant R43 AI058627-01 A1). We thank Joan C. Mecsas and Warangkhana Songsungthong at Tufts University School of Medicine for providing Y. pseudotuberculosis strains. We also appreciate the helpful suggestions of Michael P. Draper and Haregewein Assefa in preparing the manuscript.
Supporting Information Available: Compound yields, purities, HPLC conditions for purity determination, additional spectral data for final compounds, individual IC50 values. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations
- MAR
Multiple Adaptational Response
- SAR
structure-activity relationship
- MDR
multi-drug resistance
- HTH
helix-turn-helix
- T3SS
type III secretion system
- Yops
Yersinia outer proteins
- LD50
50% lethal dose
- CFU
colony forming unit
- IC50
50% inhibitory concentration
- MIC
minimum inhibitory concentration
- ATCC
American type culture collection
- His
histidine
- HRP
horseradish peroxidase
- DMEM
Dulbecco's minimal essential medium
- LDH
lactate dehydrogenase
- CLSI
Clinical and Laboratory Standards Institute
References and Notes
- (1).Perry RD, Fetherston JD. Yersinia pestis--etiologic agent of plague. Clin. Microbiol. Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hawley RJ, Eitzen EM. Biological weapons-a primer for microbiologists. Annu. Rev. Microbiol. 2001;55:235–253. doi: 10.1146/annurev.micro.55.1.235. [DOI] [PubMed] [Google Scholar]
- (3).Welch TJ, Fricke WF, McDermott PF, White DG, Rosso M-L, Rasko DA, Mammal MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, LeClerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE. 2007;2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Levy SB. Antibiotic resistance-the problem intensifies. Adv. Drug. Delivery Rev. 2005;57:1446–1450. doi: 10.1016/j.addr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- (5).Talbot GH, Bradley J, Edwards JE, Jr., Gilbert D, Scheld M, Bartlett JG. Bad bugs need drugs: An update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Disease Society of America. Clin. Infect. Dis. 2006;42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- (6).Projan SJ, Bradford PA. Late stage antibacterial drugs in the clinical pipeline. Curr. Opin. Microbiol. 2007;10:441–446. doi: 10.1016/j.mib.2007.08.007. [DOI] [PubMed] [Google Scholar]
- (7).Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- (8).Alekshun MN, Levy SB. Targeting virulence to prevent infection: to kill or not to kill? Drug Discov. Today Ther. Strategies. 2004;1:483–489. [Google Scholar]
- (9).Alksne LE, Projan SJ. Bacterial virulence as a target for antimicrobial chemotherapy. Curr. Opin. Biotechnol. 2000;11:625–636. doi: 10.1016/s0958-1669(00)00155-5. [DOI] [PubMed] [Google Scholar]
- (10).Bowser TE, Bartlett VJ, Grier MC, Verma AK, Warchol T, Levy SB, Alekshun MN. Novel anti-infection agents: Small-molecule inhibitors of bacterial transcription factors. Bioorg. Med. Chem. Lett. 2007;17:5652–5655. doi: 10.1016/j.bmcl.2007.07.072. [DOI] [PubMed] [Google Scholar]
- (11).Multiple Adaptational Response (MAR) proteins are defined as a subgroup of the AraC family transcription factors that control the virulence of bacteria. A subset of MAR proteins may also control antibiotic resistance (see reference 12).
- (12).Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Casaz P, Garrity-Ryan LK, McKenney D, Jackson C, Levy SB, Tanaka SK, Alekshun MN. MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology. 2006;152:3643–3650. doi: 10.1099/mic.0.2006/000604-0. [DOI] [PubMed] [Google Scholar]
- (14).Egan SM. Growing repertoire of AraC/XylS activators. J. Bacteriol. 2002;184:5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gallegos M, Schleif R, Bairoch A, Hofmann K, Ramos J. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Flashner Y, Mamroud E, Tidhar A, Ber R, Aftalion M, Gur D, Lazar S, Zvi A, Bino T, Ariel N, Velan B, Shafferman A, Cohen S. Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect. Immun. 2004;72:908–915. doi: 10.1128/IAI.72.2.908-915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Champion GA, Neely MN, Brennan MA, DiRita VJ. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol. Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- (18).Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- (19).Hoe NP, Minion FC, Goguen JD. Temperature sensing in Yersinia pestis: Regulation of yopE transcription by lcrF. J. Bacteriol. 1992;174:4275–4286. doi: 10.1128/jb.174.13.4275-4286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Mota LJ, Cornelis GR. The bacterial injection kit: type III secretion systems. Ann. Med. 2005;37:234–249. doi: 10.1080/07853890510037329. [DOI] [PubMed] [Google Scholar]
- (21).Cornelis GR, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- (22).Logsdon LK, Mecsas J. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect. Immun. 2003;71:4595–4607. doi: 10.1128/IAI.71.8.4595-4607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Mecsas J, Bilis I, Falkow S. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 2001;69:2779–2787. doi: 10.1128/IAI.67.5.2779-2787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Monack DM, Mecsas J, Bouley D, Falkow S. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J. Exp. Med. 1998;188:2127–2137. doi: 10.1084/jem.188.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Unpublished results: In our own experiment, deletion of the lcrF gene exhibited a similar effect of attenuating virulence of Y. pseudotuberculosis: Garrity-Ryan LK, Kim OK, Balada-Llasat J-M, Bartlett VJ, Verma AK, Fisher ML, Castillo C, Songsungthong W, Tanaka SK, Levy SB, Mecsas J, Alekshun MN.
- (26).Nordfelth R, Kauppi AM, Norberg HA, Wolf-Watz H, Elofsson M. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 2005;73:3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. Targeting bacterial virulence: Inhibitors of type III secretion in Yersinia. 2003;10:241–249. doi: 10.1016/s1074-5521(03)00046-2. [DOI] [PubMed] [Google Scholar]
- (28).Pan NJ, Brady MJ, Leong JM, Goguen JD. Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 2009;53:385–392. doi: 10.1128/AAC.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Of all peer-reviewed articles searched through SciFinder and Google Scholar, Elofsson et al. suggested that select small molecule inhibitors of T3SS in Yersinia might directly interact with LcrF. However, based on the experimental data, they either ruled out the possibility, or concluded it unknown (see references 26 and 27).
- (30).Kwon HJ, Bennik MHJ, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nature Struct. Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- (31).Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: The first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32). Proteins were aligned using the BLASTp program on the National Center for Biotechnology Information (NCBI) website: http://www.ncbi.nlm.nih.gov.
- (33).Gardiner JM, Procter J. Synthesis of N-alkoxybenzimidazoles with differentiated C2 and O-substituents. Tet. Lett. 2001;42:5109–5111. [Google Scholar]
- (34).DeStevens G, Brown AB, Rose D, Chernov HI, Plummer AJ. Derivatives of 1-hydroxybenzimidazoles and 1-hydroxyindoles and their central depressant effects. J. Med. Chem. 1967;10:211–214. doi: 10.1021/jm00314a018. [DOI] [PubMed] [Google Scholar]
- (35).Wermuth CG. Molecular variations based on isosteric replacements. In: Wermuth CG, editor. The Practice of Medicinal Chemistry. Academic Press Limited; 1996. pp. 203–247. [Google Scholar]
- (36).Ohmori J, Sakamoto S, Kubota H, Shimizu-Sasamata M, Okada M, Kawasaki S, Hidaka K, Togami J, Furuya T, Murase K. 6-(l H-imidazol-l-y1)-7-nitro-2,3(1H,4H)-quinoxalinedione hydrochloride (YM90K) and related compounds: Structure-activity relationships for the AMPA-type non-NMDA receptor. J. Med. Chem. 1994;37:467–475. doi: 10.1021/jm00030a006. [DOI] [PubMed] [Google Scholar]
- (37).In the whole cell cytotoxicity assay, Y. pseudotuberculosis was used instead of Y. pestis in order to address biosafety concerns associated with Y. pestis (Category A pathogen). The chromosomes of Y. pestis and Y. pseudotuberculosis are highly similar, and these two pathogens possess a common set of virulence factors including LcrF (see reference 38).
- (38).Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- (40).Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- (41).Clinical and Laboratory Standards Institute . Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. approved standard-7th edition 2006. pp. M7–A7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.