Abstract
Uterine leiomyomata (UL), the most common neoplasm in reproductive-age women, have recurrent cytogenetic abnormalities including del(7)(q22q32). To develop a molecular signature, matched del(7q) and non-del(7q) tumors identified by FISH or karyotyping from 11 women were profiled with expression arrays. Our analysis using paired t-tests demonstrates this matched design is critical to eliminate confounding effects of genotype and environment that underlie patient variation. A gene list ordered by genome-wide significance showed enrichment for the 7q22 target region. Modification of the gene list by weighting each sample for percent of del(7q) cells to account for the mosaic nature of these tumors further enhanced the frequency of 7q22 genes. Pathway analysis revealed two of the 19 significant functional networks were associated with development and the most represented pathway was protein ubiquitination, which can influence tumor development by stabilizing oncoproteins and destabilizing tumor suppressor proteins. Array CGH (aCGH) studies determined the only consistent genomic imbalance was deletion of 9.5 megabases from 7q22-7q31.1. Combining the aCGH data with the del(7q) UL mosacism-weighted expression analysis resulted in a list of genes that are commonly deleted and whose copy number is correlated with significantly decreased expression. These genes include the proliferation inhibitor HPB1, the loss of expression of which has been associated with invasive breast cancer, as well as the mitosis integrity-maintenance tumor suppressor RINT1. This study provides a molecular signature of the del(7q) UL subgroup and will serve as a platform for future studies of tumor pathogenesis.
Keywords: uterine leiomyomata, fibroids, del(7)(q22q32), expression, microarray
INTRODUCTION
Uterine leiomyomata (UL) are tumors commonly referred to as fibroids that arise from the uterine smooth muscle wall. Despite their non-malignant nature, UL represent a major concern in women’s health through their induction of significant morbidity in many of the approximately 25% of reproductive-age women in whom they are clinically detected (Buttram and Reiter, 1981). The overall prevalence is even higher, as systematic histological examination of hysterectomy specimens identified UL in approximately 77% of women (Cramer and Patel, 1990). This frequency, and symptoms including bladder dysfunction, abdominal pain, excessive menstrual bleeding and impaired fertility (Rein and Nowak, 1992; Coronado et al., 2000), leads UL to be the primary indication for hysterectomy and account for approximately 1 in 5 visits to a gynecologist, thereby resulting in expenditures of greater than 2.1 billion health care dollars annually in the U.S. (Lepine et al., 1997; Flynn et al., 2006; Hartmann et al., 2006).
Approximately 40% of UL have cytogenetic alterations including simple and recurrent deletions, inversions and translocations (Nibert and Heim, 1990; Meloni et al., 1992). These abnormalities were used to classify UL into subgroups and provide landmarks for gene discovery. One of the largest UL subgroups is defined by the presence of chromosome 7 long arm abnormalities, most commonly the interstitial deletion del(7)(q22q32), which represents approximately 15% of all UL and 20–35% of karyotypically abnormal UL (Nibert and Heim, 1990; Ozisik et al., 1993; Sargent et al., 1994; Xing et al., 1997). Deletion of 7q can sometimes be found as the sole alteration in a non-mosaic state, suggesting that it may play a primary early role in UL pathobiology.
Defining the del(7q) pathogenetic region has proven challenging. Initial work with rare translocations identified the gene-rich band 7q22 as the minimal cytogenetic region of importance (Ozisik et al., 1993; Sargent et al., 1994). Further refinement was attempted by multiple groups through loss of heterozygosity (LOH) analysis using polymorphic microsatellite markers; however, conflicting minimally deleted regions and inconsistent LOH maps have resulted. The most consistent common region of overlap based on the March 2006 assembly of the UCSC genome browser as defined by five previous studies is located between markers D7S2453 and D7S501 in 7q22.2-q22.3 (Zeng et al., 1997; Sell et al., 1998; van der Heijden et al., 1998; Saito et al., 2005; Vanharanta et al., 2005). Additional regions were suggested as separate tumorigenic targets in del(7q) UL such as 7q31.1 and 7q34, but such results only reflect one sample in each study and lack independent confirmation (Ishwad et al., 1997; Sell et al., 2005; Vanharanta et al., 2007).
UL provide a unique model for tumor pathobiology investigation as on average six to seven neoplasms are present in an individual woman and each is clonal as demonstrated by analysis of repeat polymorphisms in the X-linked androgen receptor and phosphoglycerokinase genes (Cramer and Patel, 1990; Mashal et al., 1994; Hashimoto et al., 1995). In addition, UL are homogenous and often of a size to provide an abundant sample. We have taken advantage of this for expression profiling to compare directly UL with del(7q) and UL without del(7q) obtained concurrently from the same uterus.
As we will show, this matched (or paired) study design is critical in identifying genetic events associated with the del(7q) abnormality. This design has not been exploited by any previous study and will nullify the confounding effect of patient to patient variability due to divergent genotype, environment, or interaction of genotype and environment.
MATERIALS AND METHODS
Clinical Material
GTG-banded karyotyping according to established protocols (Rein et al., 1991) or FISH (see below) were used to ascertain four UL with del(7q) (cases 1 to 4) and one UL that was mosaic for both del(7q) and t(12;14)(q15;q23-q24) (case 5) obtained from surgical specimens at Brigham and Women’s Hospital (BWH) through a Partners HealthCare IRB-approved protocol. Using the same abnormality detection strategy, six UL with del(7q) (cases 6 to 11) were identified from an IRB-approved tissue bank of over 100 consented 25–50 year-old women who underwent myomectomy or hysterectomy at BWH. Participants consented for the tissue bank also completed detailed epidemiological surveys ascertaining clinical, reproductive, sexual, dietary, and family history. For each of these 11 cases, matched uterine myometrium and a non-del(7q) UL were obtained concurrently with the karyotypically abnormal UL. Each case was grossly confirmed to be a UL or myometrial specimen and when possible hematoxylin- and eosin-stained tissue sections underwent histologic evaluation.
Fluorescence In Situ Hybridization (FISH)
End-sequenced and FISH-verified bacterial artificial chromosomes (BACs) (Cheung et al., 2001) were selected using the University of California Santa Cruz Biotechnology Genome Browser and Database (http://genome.ucsc.edu) (Karolchik et al., 2003) and then obtained from the RP11 library (BACPAC Resource Center at the Children’s Hospital Oakland Research Institute, Oakland, CA). DNA was isolated from bacterial cultures following a standard protocol consisting of alkaline lysis, neutralization and ethanol precipitation.
UL with del(7q) were identified by loss of probe RP11-374E17 at 7q22.2 with retention of the control probe RP11-71F18 at 7p21.1 by interphase FISH on nuclei from fresh fixed cell pellets as previously described (Moore et al., 2004). A total of 100 interphase nuclei were scored for each specimen. The probe set was validated on normal peripheral blood metaphases and on interphase nuclei from karyotype-confirmed del(7q) UL tumors. Each of the tumors was similarly screened by interphase FISH for another common chromosome abnormality in UL, t(12;14)(q15;q23-24), by assessing for the presence of a fusion signal of probes RP11-185D13 located at 12q15 and CTD-3225F7 at 14q24.
DNA Isolation
For the eight cases (2–3 and 6–11) for which tissue was available, a portion of each of the non-del(7q) UL and del(7q) UL was minced with scalpels and immediately placed in Buffer ATL (QIAGEN). Genomic DNA was isolated using the DNeasy Tissue kit with provided standard protocol (QIAGEN) and assessed for purity and quantity on a Nanodrop spectrophotometer (Thermo Scientific).
Array Comparative Genomic Hybridization (aCGH) Analysis
High quality genomic DNA from each of six cases (3, 6–9, and 11) was run on Agilent Human 244K CGH microarrays (Santa Clara, CA) using a standard direct method as described in the Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis Protocol version 4.0 at www.chem.agilent.com. Briefly, DNA was restriction digested and each del(7q) UL (test sample) was labeled with Cy5 and each matched myometrium (control sample) was labeled with Cy3 using the Genomic DNA Labeling Kit PLUS (Agilent). The labeled DNA was then washed, yield quantified, and appropriate control and test samples combined in equal amounts. After incubation in CotI DNA, blocking agent and hybridization buffer, each sample was applied to a 244K array to hybridize overnight followed by washing and array scanning. Data quality control measures were reviewed and array images created using Feature Extraction software v9.5 (Agilent). Data were imported into DNA Analytics software (Agilent) and analyzed for genomic copy number variation using the ADM2 algorithm with a 5.5 threshold, a 5 probe and ≥0.125 absolute log ratio filter, and fuzzy zero correction. The ADM2 algorithm gives a score that is proportional to the absolute log ratio within an interval and the number of probes that have a significantly different log ratio from that of the neighboring interval. The filter required that at least 5 consecutive probes had an absolute log ratio of ≥0.125 for an aberration to be called. The fuzzy zero correction took into account local and global data noise to reduce false positive calls. After establishing the aCGH-based DNA copy number aberrations, chromosome 7 data were aligned using the DNA Analytics software to the significantly expressed gene data (P < 0.01) from the del(7q) UL gene list weighted for percent of cells with the 7q deletion (the generation using Affymetrix arrays of which is described below).
RNA Isolation
A portion of each of the myometrial, non-del(7q) UL and del(7q) UL tissues in the 11 cases was frozen in liquid nitrogen immediately after surgical removal or placed directly into RNAlater solution (QIAGEN, Valencia, CA). RNA was isolated using the RNeasy Fibrous Tissue kit with provided standard protocol (QIAGEN) and assessed for purity and quantity on a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE).
Quantitative Polymerase Chain Reaction (Q-PCR)
Total RNA from the del(7q) and non-del(7q) UL from each of four women (cases 4, 7, 10 and 11) was examined for MLL5 gene expression. PCR was performed on the ABI PRISM 7900HT Sequence Detection System in a 384-well format. TaqMan Universal PCR MasterMix and a pre-designed and optimized Taqman Gene Expression Assay for quantitation of human MLL5 RNA (Applied Biosystems, Foster City, CA) were used according to the manufacturer’s instructions. Each RNA was run in quadruplicate and the Ct (cycle threshold) values of these replicates were averaged and then normalized by subtracting the Ct value of the co-amplified internal control housekeeping gene GAPDH for a ΔCt value. Data analysis used the comparative Ct method where the ΔCt of a non-del(7q) UL was used as a calibrator reference and subtracted from the ΔCt of the corresponding del(7q) UL to yield a ΔΔCt value. This was then converted into a fold-change relative to one using the following formula: MLL5 expression = 2(−ΔΔCt). This number was then averaged across the four samples.
Transcriptional Profiling
Total RNA isolated from the myometrial, non-del(7q) UL and del(7q) UL tissues from each of 11 cases was assessed for quality by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100. Standard protocols as described in the Affymetrix GeneChip Expression Analysis Technical Manual revision 4 (http://jaxservices.jax.org/Affymetrix_Gene_expression_manual_430.pdf) were employed at the Harvard Medical School - Partners HealthCare Center for Genetics and Genomics (HPCGG). Briefly, 5 µg total RNA template from each sample was reverse-transcribed into cDNA using oligo-dT primer containing T7 RNA polymerase binding sites using the GeneChip Expression 3’-Amplification Reagents One-Cycle cDNA Synthesis kit with subsequent purification of the double-stranded product with Affymetrix GeneChip Cleanup Module (Affymetrix, Santa Clara, CA). In vitro transcription to produce complementary RNA (cRNA) using T7 Polymerase and biotinylated dUTP and dCTP was performed with the GeneChip Expression Amplification Reagents kit (Affymetrix) and the biotin labeled product quantitated on a Bio-Tek UV plate reader (Bio-Tek, Winooski, VT). Following purification and fragmentation to reduce secondary structure, hybridization in a Model 640 hybridization chamber to GeneChip Human Genome U133 Plus 2.0 oligonucleotide expression microarrays (Affymetrix), which contain over 54,000 oligonucleotide probe sets representing more than 47,000 transcripts and 38,500 well-characterized genes, occurred overnight at 45°C. Arrays were washed using a Model 450 Fluidics station with GeneChip Operating Software (Affymetrix). The GeneChip Model 3000 7G was employed to scan the arrays and the probe set expression values were calculated by GeneChip software using the MAS 5.0 algorithm. Array images were inspected visually for experimental artifacts and various quality measurements such as presence calls and RNA degradation were examined to verify the quality of the data.
Data processing and analysis were carried out in the statistical language R (R Development Core Team, 2008), including the use of microarray analysis tools from the Bioconductor project (Gentleman et al., 2004). Normalization across arrays occurred by setting the trimmed (2% of each tail) mean of each array to 100. Probe sets with fewer than five present calls among the del(7q) UL, non-del(7q) UL, and myometrium arrays were excluded. Paired differential expression analysis (not accounting for percent mosaicism) between del(7q) UL and non-del(7q) UL was computed using paired t-tests in which tissue samples were analyzed as matched pairs based on patient status. Mosaicism-weighted paired differential expression analysis was implemented in the Bioconductor package limma (Smyth, 2005) by fitting a linear model (Gentleman et al., 2004) with weights equal to the percent mosaicism for each array. All differential expression analyses were corrected for multiple testing using the false discovery rate (Q-value).
Expression data were deposited at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/); the series entry number is GSE12814 and the specific accession identifiers are listed in Table 1.
Table 1.
Histopathology, Karyotype, and FISH Results of Uterine Leiomyomata for del(7q) Study
| Case Number | Accession Number | Sample Type | Histopathology | Karyotype | % del(7q) | Gene Expression Omnibus (GEO) Identifierb |
|---|---|---|---|---|---|---|
| ST91-026 | del(7q) UL | --a | 46,XX,del(7)(q22q32)[11]/46,XX[15] | 42d | GSM321965 | |
| 1 | ST91-027 | Non-del(7q) UL | -- | 46,XX[9] | 0d | GSM321966 |
| ST91-028 | Myometrium | -- | -- | -- | GSM321967 | |
| ST96-482 | del(7q) UL | Usual type, low mitotic index | 46,XX,del(7)(q22q32)[10]/46,XX[10] | 50d | GSM321969 | |
| 2 | ST96-481 | Non-del(7q) UL | Usual type, low mitotic index | 46,XX[20] | 0d | GSM321968 |
| ST96-483 | Myometrium | -- | -- | -- | GSM321970 | |
| ST99-045 | del(7q) UL | Usual type, low mitotic index | 46,XX,del(7)(q22q32)[8]/46,XX[2] | 80d | GSM321971 | |
| 3 | ST99-047 | Non-del(7q) UL | -- | 46,XX[8] | 0d | GSM321972 |
| ST99-050 | Myometrium | -- | -- | -- | GSM321973 | |
| ST99-216 | del(7q) UL | Cellular | 46,XX,del(7)(q22q32)[13] | 78d | GSM321974 | |
| 4 | ST99-219 | Non-del(7q) UL | Usual type, low mitotic index | 46,XX[14] | 0d | GSM321975 |
| ST99-220 | Myometrium | -- | -- | -- | GSM321976 | |
| ST04-065 | del(7q) UL | -- | -- | 25% del(7q)/20% t(12;14)e | GSM321979 | |
| 5c | ST04-066 | Non-del(7q) UL | -- | -- | 1% del(7q)/7% t(12;14)ef | GSM321978 |
| ST04-067 | Myometrium | -- | -- | -- | GSM321977 | |
| ST04-072F-2 | del(7q) UL | Usual type with extensive hyalinization, low mitotic index | -- | 62e | GSM321981 | |
| 6 | ST04-072F-1 | Non-del(7q) UL | Usual type, low mitotic index | -- | 0ef | GSM321982 |
| ST04-072M | Myometrium | Normal myometrium | -- | -- | GSM321980 | |
| ST04-120F-2 | del(7q) UL | Usual type, low mitotic index | -- | 52e | GSM321984 | |
| 7 | ST04-120F-3 | Non-del(7q) UL | -- | -- | 0ef | GSM321985 |
| ST04-120M | Myometrium | -- | -- | -- | GSM321983 | |
| ST05-004F-1 | del(7q) UL | Usual type, low mitotic index | -- | 34e | GSM321988 | |
| 8 | ST05-004F-2 | Non-del(7q) UL | Usual type, low mitotic index | -- | 6ef | GSM321987 |
| ST05-004M | Myometrium | -- | -- | -- | GSM321986 | |
| ST05-007F-4 | del(7q) UL | Usual type, low mitotic index | -- | 37e | GSM321990 | |
| 9 | ST05-007F-2 | Non-del(7q) UL | Usual type, low mitotic index | -- | 5ef | GSM321991 |
| ST05-007M | Myometrium | -- | -- | -- | GSM321989 | |
| ST05-024F-2 | del(7q) UL | Usual type with extensive hyalinization, low mitotic index | -- | 12e | GSM321993 | |
| 10 | ST05-024F-5 | Non-del(7q) UL | Usual type, low mitotic index | -- | 5ef | GSM321994 |
| ST05-024M | Myometrium | -- | -- | -- | GSM321992 | |
| ST05-025F-2 | del(7q) UL | Usual type, low mitotic index | -- | 71e | GSM321995 | |
| 11 | ST05-025F-4 | Non-del(7q) UL | -- | -- | 2ef | GSM321996 |
| ST05-025M | Myometrium | Normal myometrium | -- | -- | GSM321997 | |
Dash indicates unknown
Case 5 involves a mosaic del(7q)(q22q32)/t(12;14)(q15;q23-q24) tumor
Percentage of del(7q) cells determined by karyotype
Percentage of del(7q) cells determined by FISH
Value below FISH false positive cut-off for del(7q) of 9%
Ingenuity Pathways Analysis
Functional analyses of the top 300 probe sets from the del(7q) UL-specific gene list weighted for del(7q) cell mosaicism were performed using Ingenuity Pathways Analysis (IPA, Ingenuity Systems, Redwood City, CA, USA, www.Ingenuity.com) through uploading of the Affymetrix probe set identifiers and fold changes. Networks were generated by looking for interactions of the del(7q) UL-specific genes to others based on the published literature accumulated in the Ingenuity Pathways Knowledge Base. Fisher’s exact test was used to assign statistical significance, which is displayed as a score based on –log(P-value) and represents the probability of finding genes from the del(7q) UL-specific gene list in a network relative to genes being assembled into that network based on random chance. A score greater than two indicates less than a one in 100 likelihood (P –value < 0.01) that genes are assembled into a network by random chance. Networks including > 25 genes from the del(7q) UL-specific list are highly significant (giving a score of > 50).
RESULTS
Screening for del(7q) UL by Interphase FISH and Karyotyping
To identify UL with deletions in 7q22, interphase FISH or karyotype analysis was employed (Table 1). For del(7q) interphase FISH, a conservative false-positive cut-off of 9% monosomy was established by doubling the positive rate of 4.5% found in normal peripheral blood lymphocytes. Probe binding to the correct target region without cross-hybridization was validated on lymphocyte metaphases. The probe (RP11-374e17 at 7q22.2) was chosen based on its presence within the commonly deleted interval among five prior del(7q) UL LOH studies (Ishwad et al., 1997; Sell et al., 1998; van der Heijden et al., 1998; Saito et al., 2005; Vanharanta et al., 2005). Screening of 206 tumors identified 19 UL with at least partial deletion of 7q22 (9.2%). Of these 19 UL, those with parallel myometrium and non-del(7q) samples plus similar cases found through karyotyping were selected for further analysis resulting in a total of 11 cases. The level of mosaicism in these 11 cases of cells with del(7q) ranged from 12–80%. Case 5 also had a proportion of cells with another recurrent UL karyotypic abnormality, t(12;14)(q15;q23-q24).
Identification of del(7q)-Specific UL Genes
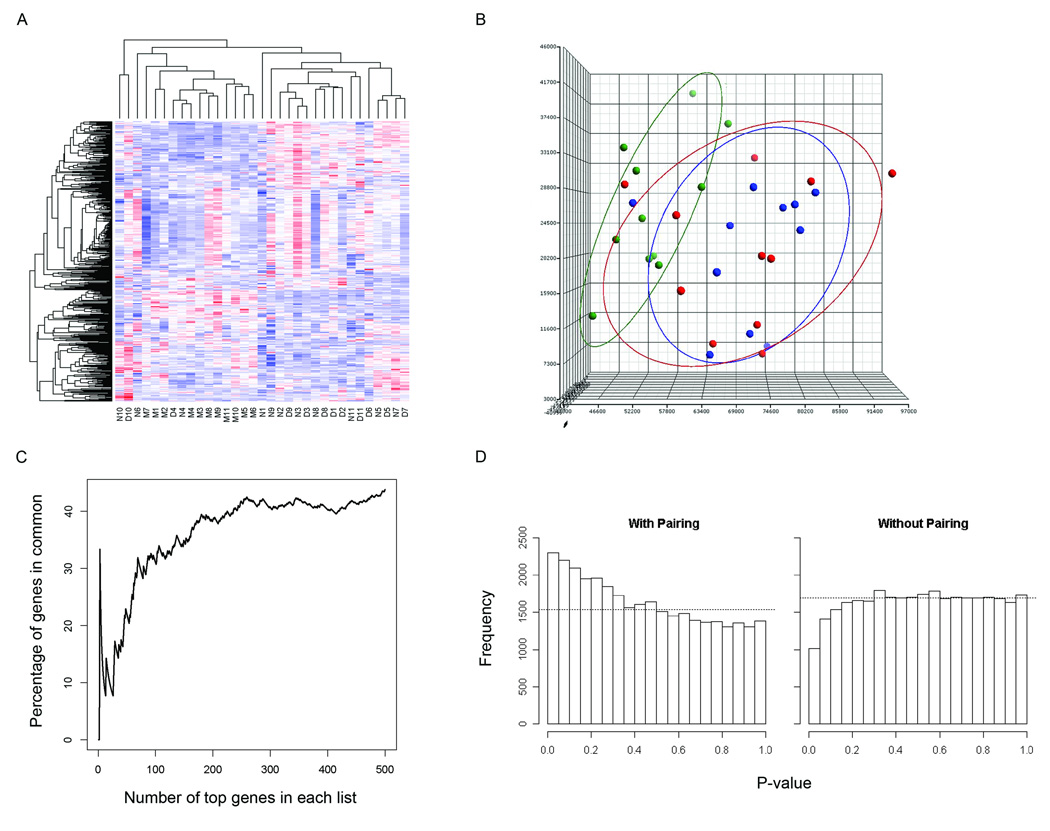
RNA from each del(7q) UL as well as from concurrently collected non-del(7q) UL and normal myometrial tissues from each of 11 cases was hybridized on Affymetrix GeneChip Human Genome U133 Plus 2.0 oligonucleotide arrays for expression analysis. Among these cases, multiple clinical features were variable such as UL size, race, and the patient age and stage of menstrual cycle at the time of surgical removal (Table 2). To control for such variables, a direct comparison of the array expression data was made between tissues obtained from each individual to identify differences in expression specifically resulting from the deletion. A heatmap from an unsupervised hierarchical cluster analysis of the 500 most variable genes demonstrates a tendency of the myometrial tissues to cluster separately from the UL samples and the del(7q) UL and non-del(7q) UL to cluster by patient rather than by presence or absence of the deletion (Fig. 1A). The separation of the myometrial samples from the overlapping UL groups can be visualized in three dimensions through principal component analysis (Fig. 1B). These results suggest incorporation of the myometrial array data is suited to determining genes that differentiate any UL from the normal myometrium tissue rather than identifying the del(7q)-specific UL genes. Therefore, the myometrial samples were not included in further analyses.
Table 2.
Clinical Features of Uterine Leiomyomata with del(7q)
| Case Number | Size of del(7q) Tumor (cm) | Size of Non-del(7q) Tumor (cm) |
Total Number of Tumors |
Race | Age of Onset (yrs) |
Age at Surgery (yrs) |
Menstrual Cycleb |
|---|---|---|---|---|---|---|---|
| 1 | --a | -- | >5 | White | 48 | 48 | Menstruation |
| 2 | 14 × 10 × 9 | 3.5 | 2 | Black | 22 | 33 | Secretory |
| 3 | 6 | 5.5 | 5 | Black | 36 | 49 | Menopausal |
| 4 | 3 | 3.5 | 4 | White | 41 | 42 | Proliferative |
| 5 | -- | -- | 3 | White | 40 | 43 | Menstruation |
| 6 | 4 × 4 × 3.5 | 5.5 × 4.5 × 4.5 | 2 | Hispanic | 41 | 41 | Secretory |
| 7 | 6.5 × 5.5 × 5.2 | 2.1 × 1.8 × 1.6 | 3 | White/Middle Eastern | 41 | 45 | Proliferative |
| 8 | 10 × 7.5 × 6.5 | 9 × 5.5 × 5.5 | 36+ | Black | 43 | 45 | Secretory |
| 9 | 3.5 × 2.5 × 2 | 6.5 × 5.6 × 3.5 | 14 | White | 36 | 39 | Menstruation |
| 10 | 7 × 7 × 6 | 5 × 4 × 4 | 10 | White | 51 | 59 | Menopausal |
| 11 | 3.6 × 3.5 × 2.9 | 2.2 × 1.7 × 1.6 | 4 | Black | 39 | 51 | Artificial Menopause (Megace-treated) |
Dash indicates unknown
Based on day one of last menstrual period relative to surgery date (days 1–5=Menstruation; 6–14=Proliferative; 14–28+=Secretory; >100 days=Menopausal)
Figure 1.
Paired analysis of the del(7q) UL and non-del(7q) UL microarray data to control for patient to patient variability without involvement of the myometrium is necessary to generate an accurate del(7q) UL-specific gene list. (A) Heatmap of an unsupervised hierarchical cluster analysis of the 500 most variable genes between myometrium (M), del(7q) UL (D) and non-del(7q) UL (N) from each of the 11 patients shows a trend of myometrial separation from all UL tissues and of UL clustering based on patient rather than del(7q) status. A similar result is obtained when more genes are included in the analysis. (B) Unsupervised principal component analysis illustrates in three dimensions the tendency of the myometrial samples (Liang et al.) to cluster and have only minimal overlap with the del(7q) (blue) and non-del(7q) tissues (red). (C) A comparison of the percent of genes in common between a paired and unpaired analysis of del(7q) UL versus non-del(7q) UL indicates the two modes of analysis produce different gene lists. (D) The distribution of p-values for two group comparison t-tests using a paired analysis (with pairing) includes a peak on the left side indicating more genes were found with significant p-values than expected in a random data set. In contrast, the unpaired analysis (without pairing) generates a nearly flat distribution suggesting the genes with significant p-values identified by such an assessment are not likely to be true positives.
The effect of controlling for patient to patient variability is further illustrated by a comparison of the percent of genes overlapping between gene lists generated by a paired and an unpaired analysis of the del(7q) UL and non-del(7q) UL expression data. Minimal overlap was found between the two analyses, particularly among the most significant genes (Fig. 1C). A comparison of the top 50 genes from each analysis showed only 20% in common, a percentage which did not increase above 40% when extended to include the top several hundred genes. Another demonstration of the need to take into account variability between patients is shown by examining the distribution of p-values from paired and unpaired t-tests for a two group comparison (Fig. 1D). In the paired case, the individual variation is accounted for by considering the difference between the samples of the same individual, resulting in a peak on the left side of the distribution indicating more genes with significant p-values were identified than expected from a random data set. In contrast, the unpaired analysis ignores the sample pairing and results in a nearly flat distribution, showing no clear evidence that genes appearing to have significant p-values from such a study analysis design would be true positives.
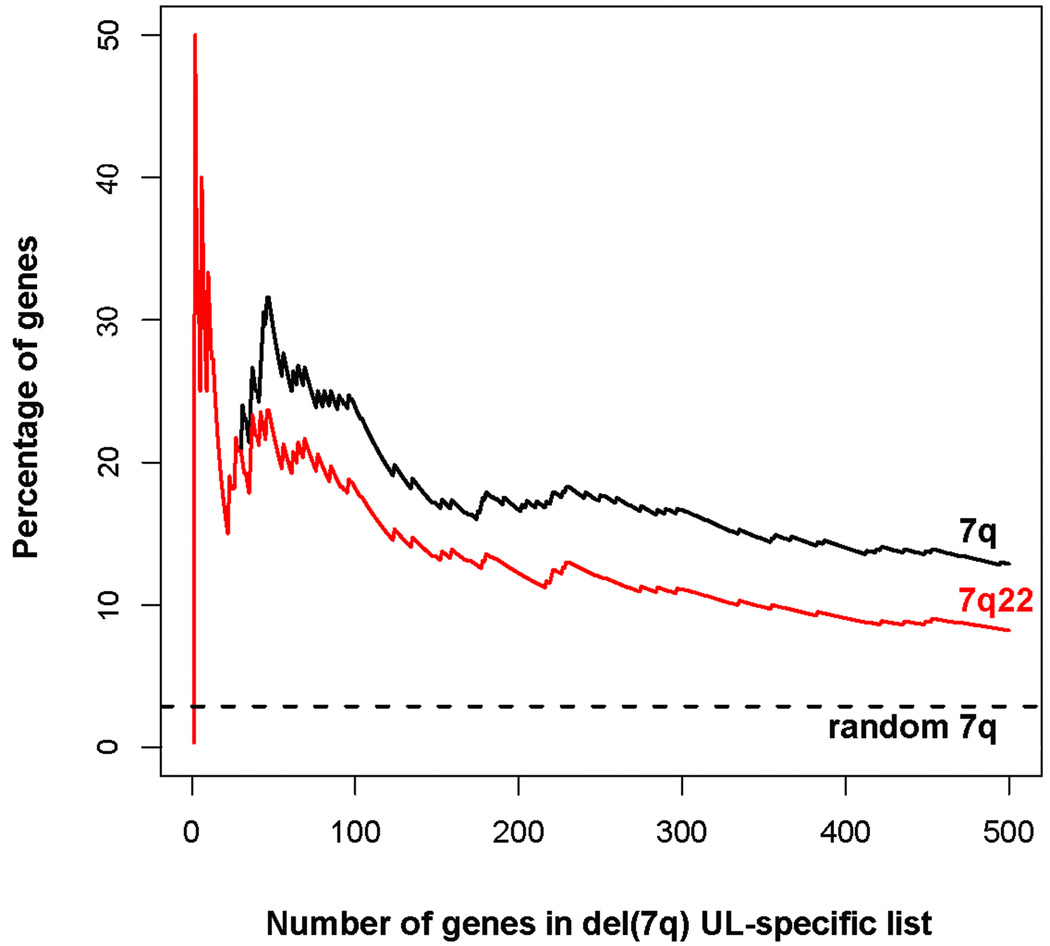
Based on these analyses, paired t-tests directly contrasting the del(7q) UL and the non-del(7q) UL from each individual were performed to determine del(7q) associated genes. This resulted in a list of genes ordered by their genome-wide significance levels corrected for multiple testing by the false discovery rate (Q-value) (Storey and Tibshirani, 2003). Of the 100 most significant del(7q) UL-specific genes, those with decreased expression are reported in Table 3 and those with increased expression in Table 4. A more extensive data set of 300 genes is provided as Supplementary Table S1. Importantly, the del(7q) UL-specific gene list is highly enriched for genes localized to 7q, most of which are in the proposed target region of 7q22 (Fig. 2). Further, all genes in 7q22 showed decreased expression. One of these genes, the vesicle transport mediator SYPL1 in 7q22.2, appears on the del(7q) UL-specific gene list at number 227 and is overlapped by the FISH probe employed to identify the del(7q) UL.
Table 3.
Genes Downregulated in del(7q) UL Compared to Non-del(7q) UL
| Numbera | Probe Set | Ref Seq | Gene Symbol | Gene Title | Fold Change | P-value | Q-value | Chromosome |
|---|---|---|---|---|---|---|---|---|
| 2 | 228138_at | NM_145115 | ZNF498 | zinc finger protein 498 | −1.42 | 4.56E-05 | 0.337 | 7q22.1 |
| 3 | 1555363_s_at | XR_041448 | MGC39821 | hypothetical protein MGC39821 | −1.67 | 4.90E-05 | 0.337 | 19p13.11 |
| 6 | 226100_at | NM_018682 | MLL5 | myeloid/lymphoid or mixed-lineage leukemia 5 | −1.37 | 7.84E-05 | 0.337 | 7q22.2 |
| 8 | 238494_at | NM_015650 | TRAF3IP1 | TNF receptor-associated factor 3 interacting protein 1 | −1.23 | 0.000149 | 0.492 | 2q37.3 |
| 9 | 200719_at | NM_006930 | SKP1 | S-phase kinase-associated protein 1 | −1.19 | 0.000235 | 0.541 | 5q31.1 |
| 10 | 223384_s_at | NM_033017 | TRIM4 | tripartite motif-containing 4 | −1.39 | 0.000304 | 0.541 | 7q22.1 |
| 12 | 233360_at | NM_003345 | UBE2I | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | −1.78 | 0.000377 | 0.541 | 16p13.3 |
| 13 | 226377_at | --- | --- | Transcribed locus | −1.3 | 0.000393 | 0.541 | 19p13.3 |
| 14 | 235634_at | NM_001015508 | PURG | purine-rich element binding protein G | −1.45 | 0.00041 | 0.541 | 8p12 |
| 16 | 227983_at | NM_145058 | RILPL2 | Rab interacting lysosomal protein-like 2 | −1.22 | 0.000437 | 0.541 | 12q24.31 |
| 23 | 226434_at | NM_145030 | C7orf47 | chromosome 7 open reading frame 47 | −1.42 | 0.000653 | 0.541 | 7q22.1 |
| 25 | 1557038_s_at | --- | --- | Clone IMAGE:110862, mRNA sequence | −1.35 | 0.000744 | 0.541 | 13q32.3 |
| 26 | 1561539_at | --- | --- | CDNA clone IMAGE:5303543 | −1.52 | 0.000801 | 0.541 | 11q14.2 |
| 27 | 218598_at | NM_021930 | RINT1 | RAD50 interactor 1 | −1.43 | 0.000828 | 0.541 | 7q22.2 |
| 30 | 226040_at | --- | --- | MRNA; cDNA DKFZp762N156 (from clone DKFZp762N156) | −1.34 | 0.000887 | 0.541 | 7q22.1 |
| 31 | 223982_s_at | NM_015723 | PNPLA8 | patatin-like phospholipase domain containing 8 | −1.27 | 0.000897 | 0.541 | 7q31.1 |
| 32 | 1553682_at | NM_152441 | FBXL14 | F-box and leucine-rich repeat protein 14 | −1.54 | 0.000904 | 0.541 | 12p13.33 |
| 33 | 229321_s_at | --- | --- | CDNA FLJ35002 fis, clone OCBBF2011914 | −1.27 | 0.000963 | 0.541 | 22q11.21 |
| 34 | 1554067_at | NM_152440 | FLJ32549 | hypothetical protein FLJ32549 | −1.25 | 0.000975 | 0.541 | 12q14.2 |
| 35 | 200899_s_at | NM_012215 | MGEA5 | meningioma expressed antigen 5 (hyaluronidase) | −1.21 | 0.000984 | 0.541 | 10q24.32 |
| 36 | 218785_s_at | NM_022777 | RABL5 | RAB, member RAS oncogene family-like 5 | −1.75 | 0.00102 | 0.541 | 7q22.1 |
| 37 | 238020_at | NM_002803 | PSMC2 | Proteasome (prosome, macropain) 26S subunit, ATPase, 2 | −1.44 | 0.00102 | 0.541 | 7q22.1 |
| 38 | 236917_at | NM_153353 | LRRC34 | leucine rich repeat containing 34 | −1.77 | 0.00103 | 0.541 | 3q26.2 |
| 40 | 225221_at | --- | --- | CDNA FLJ32068 fis, clone OCBBF1000114 | −1.41 | 0.00103 | 0.541 | 7q22.1 |
| 42 | 225945_at | NM_001009958 | ZNF655 | zinc finger protein 655 | −1.38 | 0.00104 | 0.541 | 7q22.1 |
| 43 | 204105_s_at | NM_001037132 | NRCAM | neuronal cell adhesion molecule | −1.54 | 0.00104 | 0.541 | 7q31.1 |
| 44 | 213018_at | NM_021167 | GATAD1 | GATA zinc finger domain containing 1 | −1.29 | 0.00108 | 0.541 | 7q21.2 |
| 45 | 233396_s_at | NM_020536 | CSRP2BP | CSRP2 binding protein | −1.4 | 0.00111 | 0.541 | 20p11.23 |
| 46 | 213097_s_at | NM_014377 | ZRF1 | zuotin related factor 1 | −1.43 | 0.00112 | 0.541 | 7q22.1 |
| 47 | 231436_at | --- | --- | Transcribed locus | −1.54 | 0.00122 | 0.541 | 15q13.3 |
| 52 | 38892_at | NM_015349 | KIAA0240 | KIAA0240 | −1.24 | 0.00126 | 0.541 | 6p21.1 |
| 55 | 213154_s_at | NM_001003800 | BICD2 | bicaudal D homolog 2 (Drosophila) | −1.2 | 0.00138 | 0.541 | 9q22.31 |
| 56 | 244534_at | --- | --- | Transcribed locus | −1.56 | 0.00138 | 0.541 | 7q22.1 |
| 57 | 201788_at | NM_007372 | DDX42 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | −1.19 | 0.00142 | 0.541 | 17q23.3 |
| 58 | 202054_s_at | NM_000382 | ALDH3A2 | aldehyde dehydrogenase 3 family, member A2 | −1.34 | 0.00145 | 0.541 | 17p11.2 |
| 59 | 232454_at | --- | --- | MRNA; cDNA DKFZp586N2224 (from clone DKFZp586N2224) | −1.54 | 0.00146 | 0.541 | 9q34.13 |
| 60 | 1556409_a_at | XM_001725148 | LOC100129932 | hypothetical protein LOC100129932 | −1.63 | 0.00148 | 0.541 | 11q21 |
| 61 | 231086_at | --- | --- | Transcribed locus | −1.2 | 0.00149 | 0.541 | 11q23.3 |
| 62 | 223424_s_at | NM_145914 | ZSCAN21 | zinc finger and SCAN domain containing 21 | −1.31 | 0.00151 | 0.541 | 7q22.1 |
| 63 | 227899_at | NM_053276 | VIT | vitrin | −1.6 | 0.00152 | 0.541 | 2p16.3 |
| 65 | 201068_s_at | NM_002803 | PSMC2 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | −1.36 | 0.00156 | 0.541 | 7q22.1 |
| 69 | 222615_s_at | NM_024653 | PRKRIP1 | PRKR interacting protein 1 (IL11 inducible) | −1.38 | 0.00166 | 0.541 | 7q22.1 |
| 71 | 1559310_at | --- | --- | CDNA FLJ30875 fis, clone FEBRA2004331 | −1.57 | 0.00168 | 0.541 | 9p13.3 |
| 73 | 210305_at | NM_001002810 | PDE4DIP | phosphodiesterase 4D interacting protein (myomegalin) | −1.74 | 0.00169 | 0.541 | 1q21.1 |
| 75 | 233540_s_at | NM_001011649 | CDK5RAP2 | CDK5 regulatory subunit associated protein 2 | −1.29 | 0.00172 | 0.541 | 9q33.2 |
| 76 | 1563797_at | --- | --- | CDNA FLJ23730 fis, clone HEP14530 | −1.79 | 0.00176 | 0.541 | 6p12.1 |
| 77 | 209102_s_at | NM_012257 | HBP1 | HMG-box transcription factor 1 | −1.29 | 0.00176 | 0.541 | 7q22.3 |
| 78 | 224873_s_at | NM_022497 | MRPS25 | mitochondrial ribosomal protein S25 | −1.4 | 0.00177 | 0.541 | 3p24.3 |
| 80 | 227686_at | NM_138381 | OXNAD1 | oxidoreductase NAD-binding domain containing 1 | −1.18 | 0.00179 | 0.541 | 3p24.3 |
| 81 | 242981_at | --- | --- | Transcribed locus | −1.3 | 0.00182 | 0.541 | --- |
| 82 | 230619_at | NM_001668 | ARNT | aryl hydrocarbon receptor nuclear translocator | −1.21 | 0.00186 | 0.541 | 1q21.2 |
| 83 | 240201_at | --- | --- | Transcribed locus | −1.36 | 0.00188 | 0.541 | 15q25.2 |
| 85 | 1554480_a_at | NM_031905 | ARMC10 | armadillo repeat containing 10 | −1.58 | 0.00189 | 0.541 | 7q22.1 |
| 86 | 221192_x_at | NM_024311 | MFSD11 | major facilitator superfamily domain containing 11 | −1.32 | 0.0019 | 0.541 | 17q25.2 |
| 89 | 1568874_at | NM_014071 | NCOA6 | nuclear receptor coactivator 6 | −1.73 | 0.00192 | 0.541 | 20q11.22 |
| 90 | 202276_at | NM_006304 | SHFM1 | split hand/foot malformation (ectrodactyly) type 1 | −1.29 | 0.00193 | 0.541 | 7q21.3-q22.1 |
| 94 | 230837_at | XR_041430 | LOC647500 | phosphodiesterase 4D interacting protein-like | −1.39 | 0.00203 | 0.541 | 1q21.1 |
| 95 | 227572_at | NM_032663 | USP30 | ubiquitin specific peptidase 30 | −1.2 | 0.00209 | 0.541 | 12q24.11 |
| 96 | 218956_s_at | NM_015545 | PTCD1 | pentatricopeptide repeat domain 1 | −1.57 | 0.00211 | 0.541 | 7q22.1 |
| 98 | 238076_at | NM_020699 | GATAD2B | GATA zinc finger domain containing 2B | −1.45 | 0.00212 | 0.541 | 1q21.3 |
| 99 | 225136_at | NM_021623 | PLEKHA2 | pleckstrin homology domain containing, family A member 2 | −1.16 | 0.00214 | 0.541 | 8p11.23 |
Genes are those in the top 100 del(7q) UL-specific list (a more extensive list can be found as an online supplement)
Table 4.
Genes Upregulated in del(7q) UL Compared to Non-del(7q) UL
| Numbera | Probe Set | Ref Seq | Gene Symbol | Gene Title | Fold Change | P-value | Q-value | Chromosome |
|---|---|---|---|---|---|---|---|---|
| 1 | 200652_at | NM_003145 | SSR2 | signal sequence receptor, beta (translocon-associated protein beta) | 1.26 | 3.99E-05 | 0.337 | 1q22 |
| 4 | 225897_at | NM_002356 | MARCKS | myristoylated alanine-rich protein kinase C substrate | 1.32 | 5.29E-05 | 0.337 | 6q22.1 |
| 5 | 212552_at | NM_002149 | HPCAL1 | hippocalcin-like 1 | 1.47 | 7.43E-05 | 0.337 | 2p25.1 |
| 7 | 204360_s_at | NM_000263 | NAGLU | N-acetylglucosaminidase, alpha- (Sanfilippo disease IIIB) | 1.33 | 8.94E-05 | 0.337 | 17q21.31 |
| 11 | 225065_x_at | NM_152350 | C17orf45 | chromosome 17 open reading frame 45 | 1.89 | 0.000367 | 0.541 | 17p11.2 |
| 15 | 200052_s_at | NM_004515 | ILF2 | interleukin enhancer binding factor 2, 45kDa | 1.26 | 0.000412 | 0.541 | 1q21.3 |
| 17 | 211852_s_at | NM_139321 | ATRN | attractin | 1.66 | 0.000443 | 0.541 | 20p13 |
| 18 | 200700_s_at | NM_001100603 | KDELR2 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 | 1.36 | 0.000457 | 0.541 | 7p22.1 |
| 19 | 201317_s_at | NM_002787 | PSMA2 | proteasome (prosome, macropain) subunit, alpha type, 2 | 1.15 | 0.000459 | 0.541 | 7p14.1 |
| 20 | 214108_at | NM_002382 | MAX | MYC associated factor X | 1.49 | 0.000487 | 0.541 | 14q23.3 |
| 21 | 207998_s_at | NM_000720 | CACNA1D | calcium channel, voltage-dependent, L type, alpha 1D subunit | 1.82 | 0.000574 | 0.541 | 3p21.1 |
| 22 | 235290_at | NM_001001662 | ZNF782 | Zinc finger protein 782 | 1.28 | 0.000582 | 0.541 | 9q22.33 |
| 24 | 226390_at | NM_139164 | STARD4 | StAR-related lipid transfer (START) domain containing 4 | 1.64 | 0.000654 | 0.541 | 5q22.1 |
| 28 | 223241_at | NM_013321 | SNX8 | sorting nexin 8 | 1.69 | 0.000853 | 0.541 | 7p22.2 |
| 29 | 233419_at | --- | --- | CDNA FLJ11851 fis, clone HEMBA1006744 | 2.2 | 0.000856 | 0.541 | 9p24.2 |
| 39 | 242707_at | NM_004830 | MED23 | mediator complex subunit 23 | 1.93 | 0.00103 | 0.541 | 6q23.2 |
| 41 | 211864_s_at | NM_013451 | FER1L3 | fer-1-like 3, myoferlin (C. elegans) | 1.51 | 0.00103 | 0.541 | 10q23.33 |
| 48 | 204095_s_at | NM_006532 | ELL | elongation factor RNA polymerase II | 1.45 | 0.00122 | 0.541 | 19p13.11 |
| 49 | 223614_at | --- | C8orf57 | chromosome 8 open reading frame 57 | 1.74 | 0.00124 | 0.541 | 8q21.3 |
| 50 | 208708_x_at | NM_001969 | EIF5 | eukaryotic translation initiation factor 5 | 1.21 | 0.00124 | 0.541 | 14q32.32 |
| 51 | 217771_at | NM_016548 | GOLM1 | golgi membrane protein 1 | 1.42 | 0.00126 | 0.541 | 9q21.33 |
| 53 | 210868_s_at | NM_024090 | ELOVL6 | ELOVL family member 6, elongation of long chain fatty acids | 1.46 | 0.00135 | 0.541 | 4q25 |
| 54 | 212121_at | NM_015631 | TCTN3 | tectonic family member 3 | 1.18 | 0.00135 | 0.541 | 10q23.33 |
| 64 | 201267_s_at | NM_002804 | PSMC3 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 1.67 | 0.00155 | 0.541 | 11p11.2 |
| 66 | 204082_at | NM_006195 | PBX3 | pre-B-cell leukemia homeobox 3 | 1.66 | 0.00157 | 0.541 | 9q33.3 |
| 67 | 209188_x_at | NM_001938 | DR1 | down-regulator of transcription 1, TBP-binding (negative cofactor 2) | 1.32 | 0.0016 | 0.541 | 1p22.1 |
| 68 | 202687_s_at | NM_003810 | TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | 1.7 | 0.00163 | 0.541 | 3q26.31 |
| 70 | 219118_at | NM_016594 | FKBP11 | FK506 binding protein 11, 19 kDa | 1.33 | 0.00166 | 0.541 | 12q13.12 |
| 72 | 218384_at | NM_001042476 | CARHSP1 | calcium regulated heat stable protein 1, 24kDa | 1.76 | 0.00169 | 0.541 | 16p13.2 |
| 74 | 231948_s_at | NM_080678 | UBE2F | ubiquitin-conjugating enzyme E2F (putative) | 1.27 | 0.00171 | 0.541 | 2q37.3 |
| 79 | 200670_at | NM_001079539 | XBP1 | X-box binding protein 1 | 1.38 | 0.00178 | 0.541 | 22q12.1 |
| 84 | 200754_x_at | NM_003016 | SFRS2 | splicing factor, arginine/serine-rich 2 | 1.23 | 0.00189 | 0.541 | 17q25.2 |
| 87 | 243159_x_at | --- | --- | Transcribed locus | 15.9b | 0.0019 | 0.541 | 5p15.1 |
| 88 | 200753_x_at | NM_003016 | SFRS2 | splicing factor, arginine/serine-rich 2 | 1.31 | 0.00192 | 0.541 | 17q25.2 |
| 91 | 205499_at | NM_014467 | SRPX2 | sushi-repeat-containing protein, X-linked 2 | 2.02 | 0.00197 | 0.541 | Xq22.1 |
| 92 | 1555928_at | --- | --- | CDNA FLJ30680 fis, clone FCBBF2000123 | 4.83 | 0.00199 | 0.541 | --- |
| 93 | 228061_at | NM_138771 | CCDC126 | coiled-coil domain containing 126 | 1.65 | 0.002 | 0.541 | 7p15.3 |
| 97 | 216697_at | NM_007118 | TRIO | Triple functional domain (PTPRF interacting) | 2.47 | 0.00212 | 0.541 | 5p15.2 |
| 100 | 1558996_at | NM_001012505 | FOXP1 | forkhead box P1 | 1.5 | 0.00219 | 0.541 | 3p14.1 |
Genes are those in the top 100 del(7q) UL-specific list (a more extensive list can be found as an online supplement)
Removal of a single patient outlier for this probe set reduces the fold change to 3.9.
Figure 2.
The del(7q) UL-specific gene list is highly enriched for 7q genes. Examination of the top 500 genes based on false discovery rate shows a high percentage were localized to 7q (black line), the majority of which are in the proposed target region of 7q22 (red line). The more limited the gene list examined, the greater the percentage of genes mapped in 7q. This is in contrast to the expected percentage of genes in 7q if the list was generated randomly based on the fraction of genes in 7q in the total data (dotted line).
Identification of del(7q)-Specific UL Genes Weighted for Percent Mosaicism of del(7q) Cells
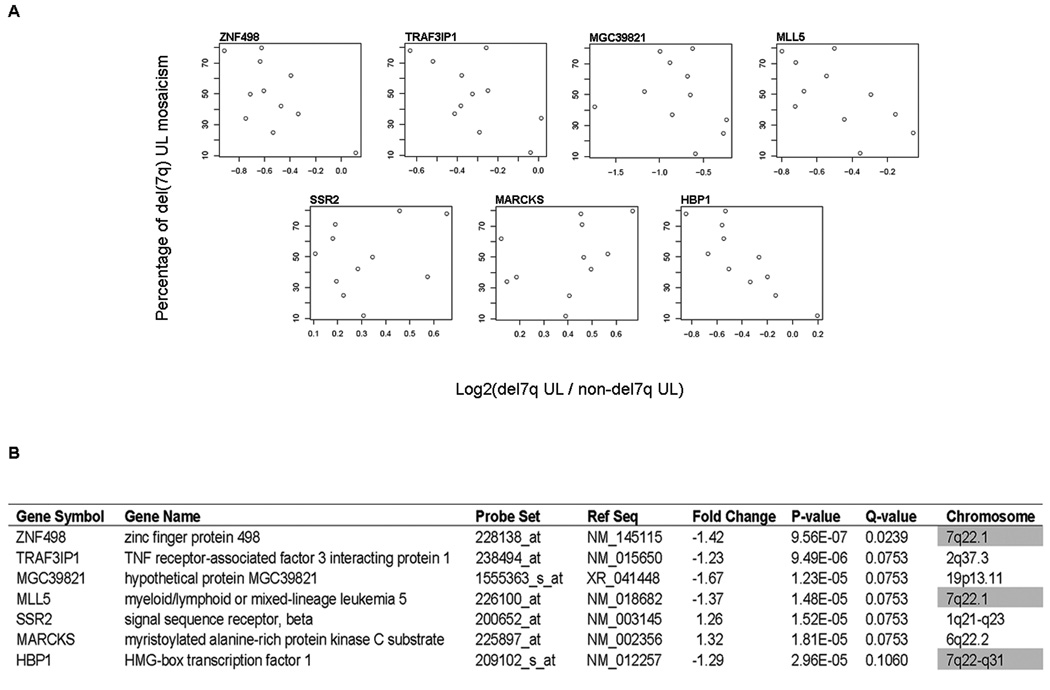
Cells with 7q abnormalities in UL are usually present in a mosaic form with karyotypically normal cells (Xing et al., 1997). In contrast to previous studies, the impact of this biology was integrated by weighting each sample pair [del(7q) UL and non-del(7q) UL from the same woman] for percent del(7q) mosaicism of the tumor in a paired differential expression analysis: the higher the percentage of del(7q) cells present, the more heavily weighted was that sample. The 50 most significant genes based on Q-value in this modified del(7q) UL-specific gene list are given in Table 5. A more expansive list of 300 genes is also presented (Supplementary Table S2). The purpose of weighting the samples for mosaicism level is to compensate for background noise caused by the karyotypically normal cells in order to identify those genes specific to the del(7q) abnormality. The validity of this approach is supported by the two-fold increase in the proportion of genes in 7q22 within the top 50 of the mosaicism-weighted list relative to the non-weighted list (from 10 up to 20 genes). A comparison of the expression log ratios of genes and percent mosaicism identified seven genes with a Q-value ≤ 0.1 which are illustrated as scatterplots (Fig. 3A). Three of these seven significant genes are located in 7q22 (Fig. 3B).
Table 5.
Genes Upregulated or Downregulated in del(7q) UL Compared to Non-del(7q) UL Weighted for Percent Mosaicism
| Numbera | Non-weightedb | Probe Set | Ref Seq | Gene Symbol | Gene Title | Fold Change | P-value | Q-value | Chromosome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 228138_at | NM_145115 | ZNF498 | zinc finger protein 498 | −1.42 | 9.56E-07 | 0.0239 | 7q22.1 |
| 2 | 8 | 238494_at | NM_015650 | TRAF3IP1 | TNF receptor-associated factor 3 interacting protein 1 | −1.23 | 9.49E-06 | 0.0753 | 2q37.3 |
| 3 | 3 | 1555363_s_at | XR_041448 | MGC39821 | hypothetical protein MGC39821 | −1.67 | 1.23E-05 | 0.0753 | 19p13.11 |
| 4 | 6 | 226100_at | NM_018682 | MLL5 | myeloid/lymphoid or mixed-lineage leukemia 5 | −1.37 | 1.48E-05 | 0.0753 | 7q22.2 |
| 5 | 1 | 200652_at | NM_003145 | SSR2 | signal sequence receptor, beta (translocon-associated protein beta) | 1.26 | 1.52E-05 | 0.0753 | 1q22 |
| 6 | 4 | 225897_at | NM_002356 | MARCKS | myristoylated alanine-rich protein kinase C substrate | 1.32 | 1.81E-05 | 0.0753 | 6q22.1 |
| 7 | 77 | 209102_s_at | NM_012257 | HBP1 | HMG-box transcription factor 1 | −1.29 | 2.96E-05 | 0.106 | 7q22.3 |
| 8 | 17 | 211852_s_at | NM_139321 | ATRN | attractin | 1.66 | 8.79E-05 | 0.207 | 20p13 |
| 9 | 175 | 214670_at | NM_003439 | ZKSCAN1 | zinc finger with KRAB and SCAN domains 1 | −1.46 | 8.90E-05 | 0.207 | 7q22.1 |
| 10 | 9 | 200719_at | NM_006930 | SKP1 | S-phase kinase-associated protein 1 | −1.19 | 9.40E-05 | 0.207 | 5q31.1 |
| 11 | 10 | 223384_s_at | NM_033017 | TRIM4 | tripartite motif-containing 4 | −1.39 | 9.50E-05 | 0.207 | 7q22.1 |
| 12 | 62 | 223424_s_at | NM_145914 | ZSCAN21 | zinc finger and SCAN domain containing 21 | −1.31 | 9.94E-05 | 0.207 | 7q22.1 |
| 13 | 7 | 204360_s_at | NM_000263 | NAGLU | N-acetylglucosaminidase, alpha- (Sanfilippo disease IIIB) | 1.33 | 0.000128 | 0.225 | 17q21.31 |
| 14 | 37 | 238020_at | NM_002803 | PSMC2 | Proteasome (prosome, macropain) 26S subunit, ATPase, 2 | −1.44 | 0.000132 | 0.225 | 7q22.1 |
| 15 | 42 | 225945_at | NM_001009958 | ZNF655 | zinc finger protein 655 | −1.38 | 0.000151 | 0.225 | 7q22.1 |
| 16 | 23 | 226434_at | NM_145030 | C7orf47 | chromosome 7 open reading frame 47 | −1.42 | 0.000164 | 0.225 | 7q22.1 |
| 17 | 11 | 225065_x_at | NM_152350 | C17orf45 | chromosome 17 open reading frame 45 | 1.89 | 0.000168 | 0.225 | 17p11.2 |
| 18 | 5 | 212552_at | NM_002149 | HPCAL1 | hippocalcin-like 1 | 1.47 | 0.000176 | 0.225 | 2p25.1 |
| 19 | 46 | 213097_s_at | NM_014377 | ZRF1 | zuotin related factor 1 | −1.43 | 0.000176 | 0.225 | 7q22.1 |
| 20 | 95 | 227572_at | NM_032663 | USP30 | ubiquitin specific peptidase 30 | −1.2 | 0.00018 | 0.225 | 12q24.11 |
| 21 | 16 | 227983_at | NM_145058 | RILPL2 | Rab interacting lysosomal protein-like 2 | −1.22 | 0.000191 | 0.228 | 12q24.31 |
| 22 | 19 | 201317_s_at | NM_002787 | PSMA2 | proteasome (prosome, macropain) subunit, alpha type, 2 | 1.15 | 0.00021 | 0.235 | 7p14.1 |
| 23 | 40 | 225221_at | --- | --- | CDNA FLJ32068 fis, clone OCBBF1000114 | −1.41 | 0.000241 | 0.235 | 7q22.1 |
| 24 | 180 | 201405_s_at | NM_006833 | COPS6 | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | −1.34 | 0.000243 | 0.235 | 7q22.1 |
| 25 | 553 | 219155_at | NM_012417 | PITPNC1 | phosphatidylinositol transfer protein, cytoplasmic 1 | 1.52 | 0.000249 | 0.235 | 17q24.2 |
| 26 | 179 | 221998_s_at | NM_001025778 | VRK3 | vaccinia related kinase 3 | −1.24 | 0.000253 | 0.235 | 19q13.33 |
| 27 | 14 | 235634_at | NM_001015508 | PURG | purine-rich element binding protein G | −1.45 | 0.000254 | 0.235 | 8p12 |
| 28 | 34 | 1554067_at | NM_152440 | FLJ32549 | hypothetical protein FLJ32549 | −1.25 | 0.000294 | 0.249 | 12q14.2 |
| 29 | 153 | 201682_at | NM_004279 | PMPCB | peptidase (mitochondrial processing) beta | −1.38 | 3.00E-04 | 0.249 | 7q22.1 |
| 30 | 81 | 242981_at | --- | --- | Transcribed locus | −1.3 | 0.000303 | 0.249 | --- |
| 31 | 56 | 244534_at | --- | --- | Transcribed locus | −1.56 | 0.00031 | 0.249 | 7q22.1 |
| 32 | 107 | 208808_s_at | NM_002129 | HMGB2 | high-mobility group box 2 | −1.26 | 0.000318 | 0.249 | 4q34.1 |
| 33 | 13 | 226377_at | --- | --- | Transcribed locus | −1.3 | 0.000341 | 0.258 | 19p13.3 |
| 34 | 43 | 204105_s_at | NM_001037132 | NRCAM | neuronal cell adhesion molecule | −1.54 | 0.000366 | 0.258 | 7q31.1 |
| 35 | 31 | 223982_s_at | NM_015723 | PNPLA8 | patatin-like phospholipase domain containing 8 | −1.27 | 0.000377 | 0.258 | 7q31.1 |
| 36 | 44 | 213018_at | NM_021167 | GATAD1 | GATA zinc finger domain containing 1 | −1.29 | 0.000388 | 0.258 | 7q21.2-q22 |
| 37 | 130 | 228346_at | --- | --- | Transcribed locus | −1.22 | 0.000395 | 0.258 | 19p13.2 |
| 38 | 55 | 213154_s_at | NM_001003800 | BICD2 | bicaudal D homolog 2 (Drosophila) | −1.2 | 0.000399 | 0.258 | 9q22.31 |
| 39 | 96 | 218956_s_at | NM_015545 | PTCD1 | pentatricopeptide repeat domain 1 | −1.57 | 0.000403 | 0.258 | 7q22.1 |
| 40 | 27 | 218598_at | NM_021930 | RINT1 | RAD50 interactor 1 | −1.43 | 0.000437 | 0.266 | 7q22.2 |
| 41 | 18 | 200700_s_at | NM_001100603 | KDELR2 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 | 1.36 | 0.000445 | 0.266 | 7p22.1 |
| 42 | 57 | 201788_at | NM_007372 | DDX42 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | −1.19 | 0.000457 | 0.266 | 17q23.3 |
| 43 | 168 | 227187_at | --- | --- | Transcribed locus | −1.34 | 0.000457 | 0.266 | 7q31.1 |
| 44 | 158 | 1569077_x_at | NM_001102657 | FLJ16287 | FLJ16287 protein | −1.34 | 0.000481 | 0.266 | 19q13.33 |
| 45 | 91 | 205499_at | NM_014467 | SRPX2 | sushi-repeat-containing protein, X-linked 2 | 2.02 | 0.000494 | 0.266 | Xq22.1 |
| 46 | 133 | 203484_at | NM_001012456 | SEC61G | Sec61 gamma subunit | 1.31 | 0.000515 | 0.266 | 7p11.2 |
| 47 | 114 | 206659_at | XR_041868 | FLJ14082 | hypothetical protein FLJ14082 | −1.39 | 0.000524 | 0.266 | 2q11.1 |
| 48 | 85 | 1554480_a_at | NM_031905 | ARMC10 | armadillo repeat containing 10 | −1.58 | 0.000531 | 0.266 | 7q22.1 |
| 49 | 159 | 242621_at | NM_145115 | ZNF498 | zinc finger protein 498 | −1.34 | 0.00054 | 0.266 | 7q22.1 |
| 50 | 90 | 202276_at | NM_006304 | SHFM1 | split hand/foot malformation (ectrodactyly) type 1 | −1.29 | 0.000541 | 0.266 | 7q21.3-q22.1 |
Genes are those in the top 50 del(7q) UL-specific list that have been weighted for the percent mosaicism (a more extensive list can be found as an online supplement)
Number on non-weighted del(7q) UL-specific gene list
Figure 3.
Seven most significant genes identified by weighting the microarray data for the level of del(7q) cell mosaicism in each UL. (A) Scatter plots show the relationship between the percent of del(7q) cells in each UL and the log ratio of gene expression in del(7q) UL relative to the non-del(7q) UL in each of the 11 women. (B) The significance of these seven genes is reflected by a Q-value ≤ 0.10, and three of the genes are in the region of interest at 7q22.
Determination of Deletion Size Using aCGH and Alignment to Gene Expression Data
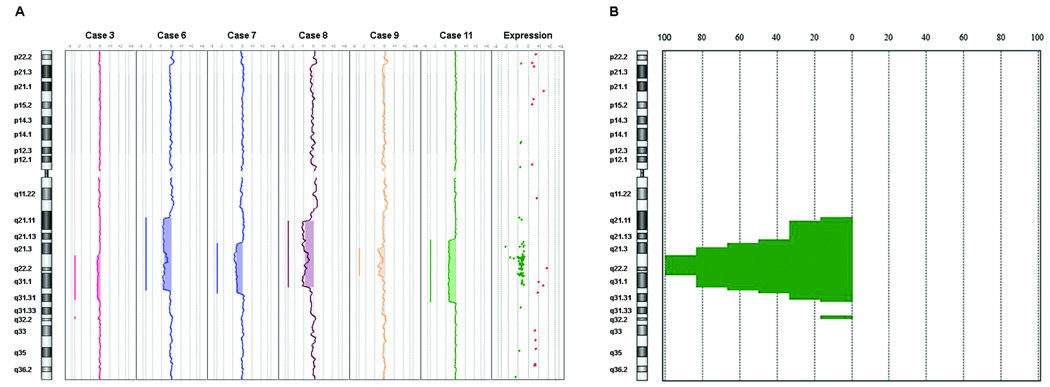
Genomic copy number changes in the del(7q) UL of six cases for which DNA was available was assessed using array comparative genomic hybridization (aCGH). The matched myometrium from each woman was used as the control against which each del(7q) UL DNA was compared to remove any copy number variants inherent in the patient but not related to tumorigenesis. All samples tested showed genomic deletion of 7q (Fig. 4A). The smallest commonly deleted region on chromosome 7 in all six samples spanned approximately 9.5 megabases from 7q22.1-q31.1 (Fig. 4B). No other deletion or amplification was universal to all samples (Supplementary Table S3). Alignment of this aCGH-defined commonly deleted region on 7q to the significant genes (P < 0.01) from the percent mosaicism-weighted del(7q) UL-specific list demonstrated a high correlation between genes located in the deletion interval and significantly downregulated expression (Fig. 4A). This correlation supports the accuracy of both the aCGH and expression microarray data. The significant genes in the commonly deleted interval are listed in Table 6.
Figure 4.
Detection of a commonly deleted region using aCGH analysis of six del(7q) UL cases and alignment with microarray expression data showed a strong correlation between the 7q deletion and decreased gene expression. (A) aCGH analysis demonstrated variable 7q deletion sizes in each of the six cases. Alignment of the common deletion interval to genes with significant expression from the mosaicism-weighted del(7q) UL-specific gene list revealed a cluster of downregulated genes in the deleted interval, supporting the accuracy of the microarray expression analysis. Green dots represent downregulated genes and red dots upregulated genes. (B) Genomic penetrance summary of chromosome 7 showing the affected regions and in what percentage of the six cases they were found to be abnormal. Skewing of data to the left of zero indicates deletion. The common region of genomic loss for the six cases, as indicated by the 100% line, spans approximately 9.5 megabases from 7q22.1-q31.1 (basepairs 98,598,014–108,112,352 based on the UCSC genome browser March 2006 assembly; aCGH probe A_16_P18041391 to A_16_P18063689).
Table 6.
Genes in the aCGH-defined Smallest Commonly Deleted Region on 7q with Significant (p < 0.01) Expression in del(7q) UL Relative to Non-del(7q) UL
| Probe Set | Ref Seq | Gene Symbol | Gene Title | Fold Changea | Chromosome |
|---|---|---|---|---|---|
| 205690_s_at | NM_003910 | BUD31 | BUD31 homolog (S. cerevisiae) | −0.567 | 7q22.1 |
| 218956_s_at | NM_015545 | PTCD1 | pentatricopeptide repeat domain 1 | −1.05 | 7q22.1 |
| 206688_s_at | NM_001081559 | CPSF4 | cleavage and polyadenylation specific factor 4, 30kDa | −0.529 | 7q22.1 |
| 202961_s_at | NM_001003713 | ATP5J2 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | −0.379 | 7q22.1 |
| 214714_at | NM_032164 | ZNF394 | zinc finger protein 394 | −0.541 | 7q22.1 |
| 203730_s_at | NM_014569 | ZKSCAN5 | zinc finger with KRAB and SCAN domains 5 | −0.544 | 7q22.1 |
| 225945_at | NM_001009958 | ZNF655 | zinc finger protein 655 | −0.624 | 7q22.1 |
| 242621_at; 228138_at | NM_145115 | ZNF498 | zinc finger protein 498 | −0.584; −0.602 | 7q22.1 |
| 205765_at; 214235_at | NM_000777 | CYP3A5 | cytochrome P450, family 3, subfamily A, polypeptide 5 | −1.79; −0.956 | 7q22.1 |
| 223384_s_at; 1554287_at | NM_033017 | TRIM4 | tripartite motif-containing 4 | −0.558; −0.941 | 7q22.1 |
| 214670_at; 1557953_at | NM_003439 | ZKSCAN1 | zinc finger with KRAB and SCAN domains 1 | −0.83; −0.869 | 7q22.1 |
| 225221_at | --- | GATS | opposite strand transcription unit to STAG3 | −0.593 | 7q22.1 |
| 223424_s_at | NM_145914 | ZSCAN21 | zinc finger and SCAN domain containing 21 | −0.503 | 7q22.1 |
| 232497_at | NM_017715 | ZNF3 | zinc finger protein 3 | −1.4 | 7q22.1 |
| 201405_s_at; 213504_at | NM_006833 | COPS6 | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | −0.555; −0.479 | 7q22.1 |
| 210983_s_at | NM_005916 | MCM7 | minichromosome maintenance complex component 7 | −0.495 | 7q22.1 |
| 227313_at | NM_152755 | CNPY4 | canopy 4 homolog (zebrafish) | −0.669 | 7q22.1 |
| 224890_s_at | NM_001008395 | LOC389541 | similar to CG14977-PA | −0.457 | 7q22.1 |
| 225321_s_at; 220954_s_at | NM_013440 | PILRB | paired immunoglobin-like type 2 receptor beta | −0.694; −0.573 | 7q22.1 |
| 220618_s_at | NM_017984 | ZCWPW1 | zinc finger, CW type with PWWP domain 1 | −0.679 | 7q22.1 |
| 219798_s_at | NM_019606 | MEPCE | methylphosphate capping enzyme | −0.529 | 7q22.1 |
| 226434_at | NM_145030 | C7orf47 | chromosome 7 open reading frame 47 | −0.577 | 7q22.1 |
| 209482_at | NM_005837 | POP7 | processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) | −0.482 | 7q22.1 |
| 209129_at | NM_003302 | TRIP6 | thyroid hormone receptor interactor 6 | −0.569 | 7q22.1 |
| 214808_at | --- | --- | --- | −0.56 | 7q22.1 |
| 226040_at | --- | --- | --- | −0.489 | 7q22.1 |
| 222742_s_at; 218785_s_at | NM_022777 | RABL5 | RAB, member RAS oncogene family-like 5 | −0.645; −0.929 | 7q22.1 |
| 218378_s_at; 222615_s_at | NM_024653 | PRKRIP1 | PRKR interacting protein 1 (IL11 inducible) | −0.816; −0.553 | 7q22.1 |
| 212706_at; 208534_s_at | NM_001079877 | RASA4 | RAS p21 protein activator 4 | −0.661; −0.805 | 7q22.1 |
| 1554480_a_at; 223328_at | NM_031905 | ARMC10 | armadillo repeat containing 10 | −0.829; −0.589 | 7q22.1 |
| 226041_at; 242229_at | NM_001122838 | NAPEPLD | N-acyl phosphatidylethanolamine phospholipase D | −0.617; −0.477 | 7q22.1 |
| 201682_at | NM_004279 | PMPCB | peptidase (mitochondrial processing) beta | −0.654 | 7q22.1 |
| 213097_s_at | NM_014377 | ZRF1 | zuotin related factor 1 | −0.623 | 7q22.1 |
| 244534_at | --- | RELN | reelin | −0.685 | 7q22.1 |
| 201067_at; 238020_at | NM_002803 | PSMC2 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | −0.537; −0.593 | 7q22.1 |
| 204957_at | NM_002553 | ORC5L | origin recognition complex, subunit 5-like (yeast) | −0.652 | 7q22.2 |
| 236761_at | NM_199000 | LHFPL3 | lipoma HMGIC fusion partner-like 3 | 1.82 | 7q22.2-q22.3 |
| 226100_at | NM_018682 | MLL5 | myeloid/lymphoid or mixed-lineage leukemia 5 | −0.54 | 7q22.3 |
| 230091_at; 203181_x_at; 203182_s_at | --- | SRPK2 | SFRS protein kinase 2 | −0.848; −0.743; −0.789 | 7q22.3 |
| 218984_at | NM_019042 | PUS7 | pseudouridylate synthase 7 homolog (S. cerevisiae) | −0.642 | 7q22.3 |
| 218598_at | NM_021930 | RINT1 | RAD50 interactor 1 | −0.594 | 7q22.3 |
| 209102_s_at | NM_012257 | HBP1 | HMG-box transcription factor 1 | −0.499 | 7q22.3 |
| 203629_s_at; 203630_s_at | NM_006348 | COG5 | component of oligomeric golgi complex 5 | −0.721; −0.629 | 7q22.3 |
| 205761_s_at; 205762_s_at | NM_181581 | DUS4L | dihydrouridine synthase 4-like (S. cerevisiae) | −0.718; −0.989 | 7q22.3 |
| 225674_at; 225677_at; 217657_at | NM_001008405 | BCAP29 | B-cell receptor-associated protein 29 | −0.604; −0.618; −0.716 | 7q22.3 |
| 227187_at | --- | LAMB1 | laminin, beta 1 | −0.578 | 7q31.1 |
| 209095_at | NM_000108 | DLD | dihydrolipoamide dehydrogenase | −0.655 | 7q31.1 |
| 236437_at | --- | LAMB4 | laminin beta 4 | −0.856 | 7q31.1 |
| 204105_s_at | NM_001037132 | NRCAM | neuronal cell adhesion molecule | −0.84 | 7q31.1 |
| 223310_x_at; 223982_s_at | NM_015723 | PNPLA8 | patatin-like phospholipase domain containing 8 | −0.43; −0.399 | 7q31.1 |
| 227636_at | NM_182529 | THAP5 | THAP domain containing 5 | −0.57 | 7q31.1 |
Expression is the log2(fold change) between del(7q) UL relative to non-del(7q) UL weighted for percent of del(7q) cell mosaicism
Quantitative PCR (Q-PCR) Confirmation of MLL5 Expression
Cases 4, 7, 10 and 11 for which additional RNA was available were evaluated by Q-PCR for expression of MLL5 (Mixed-Lineage Leukemia-5), a gene in 7q22.3 which had decreased expression in del(7q) UL by microarray analysis (sixth gene in the del(7q)-specific gene list). In addition, MLL5 was significant with a Q-value of 0.0753 and a P-value of 0.0000148 in the fourth position in the gene list produced by weighting for percent mosaicism of del(7q) cells (Table 5). Q-PCR for MLL5 confirmed the microarray data of a 1.4-fold reduction by showing a 1.5-fold decrease in RNA expression in the del(7q) UL compared to non-del(7q) UL after normalization to GAPDH (data not shown).
Functional Significance of del(7q)-Specific UL Genes
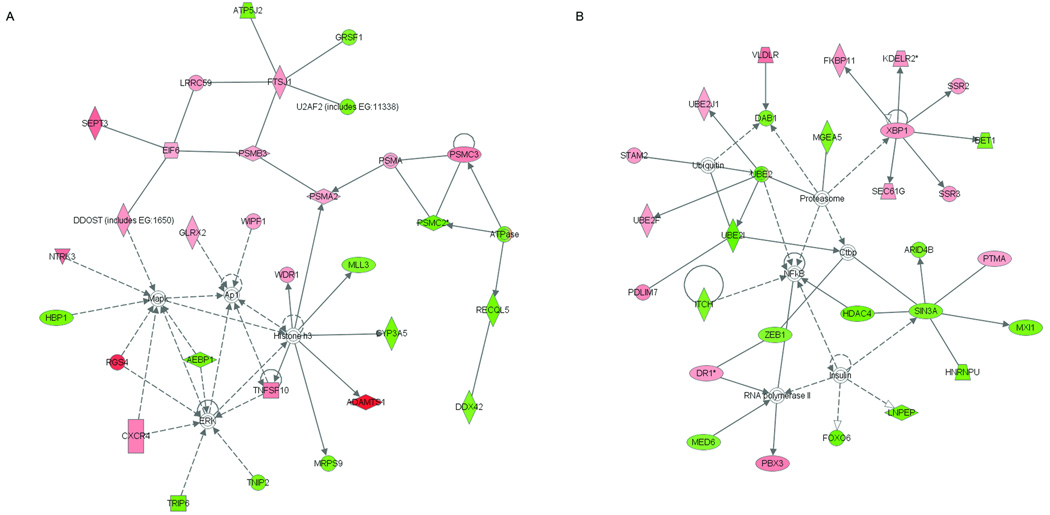
To extract biological insight from the transcriptional profile of del(7q) UL, the top 300 probe sets from the del(7q) mosaicism-weighted gene list, which represent the majority of genes with altered expression in the target region of 7q22, were investigated with the Ingenuity Pathways Analysis (IPA) System. IPA is a web-based entry tool developed by systematic encoding of manually curated functional relationships between genes presented in hundreds of thousands of scientific publications. Of the 300 probe sets, 197 were assigned to networks. There were 19 networks of functional dependencies generated of which two were highly significant (score > 50, genes from deletion 7q list > 25); nodes in the network correspond to a gene and each arc to a published article reporting a functional relationship between those two linked genes. The most significant networks (Fig. 5A and 5B) are principally associated with development. The other networks involving 11 to 18 del(7q) UL-specific genes (scores 15 to 28) are associated with multiple functions including but not limited to cell cycle, cell growth, cancer, cell morphology, DNA replication and repair, reproductive system disease, gene expression, and additional development pathways.
Figure 5.
Identification of highly significant networks of functional dependencies of genes from the top 300 del(7q) UL-specific gene list weighted for percent of del(7q) cells using Ingenuity Pathways Analysis. The functions of the two networks of highest significance are involved in (A) drug metabolism, organismal development and carbohydrate metabolism (score = 55, genes from deletion 7q list = 29) and (B) embryonic, nervous system and tissue development (score = 52, genes from deletion 7q list = 28). Pink shading indicates upregulated gene expression, green downregulated gene expression, solid lines a direct relationship between connected genes, and dotted lines an indirect relationship between linked genes.
In addition to functional networks, IPA was also used to identify which well-characterized canonical pathways are most relevant across the entire del(7q) UL dataset. The significance is based on a p-value calculated using the right-tailed Fisher’s Exact Test by comparing the number of user-supplied genes that participate in a given function or pathway relative to the total number of occurrences of these genes in all function/pathway annotations stored in the Ingenuity pathways knowledge base. The protein ubiquitination pathway was the most significantly associated (P-value = 2.76×10−5), involving 11 genes from the del(7q) UL-specific list. The genes from network 1 involved in protein ubiquitination include PSMA2, PSMB3, PSMC2, PSMC3 while those in network 2 include UBE2I and UBE2J1.
DISCUSSION
Multiple recurrent cytogenetic abnormalities have been described in UL, suggesting these tumors develop from several distinct genetic pathways. This necessitates examination of each major cytogenetic subgroup for its role in UL tumorigenesis. One of the most common abnormalities, deletion or rearrangement of 7q22, remains largely undefined as determination of the causative gene(s) has been complicated by the uncertainty of the smallest commonly deleted region and the gene-dense nature of the target region 7q22.
Rearrangements of 7q22 are found more consistently in UL but have been observed in other solid tumors such as lipomas and endometrial polyps as well as some hematological malignancies (Dal Cin et al., 1995; Dal Cin et al., 1997; Liang et al., 1998). Such frequent deletion or rearrangement of a specific chromosomal region is generally thought to indicate involvement of a tumor suppressor gene where tumorigenesis results from the structural loss of one copy and subsequent mutation of the other allele. However, it remains unclear if UL with chromosome 7 abnormalities follow a loss of function pattern either due to deletion or disruption at the translocation breakpoint. Alternatively, haploinsufficiency may be the underlying molecular mechanism. Another possibility is that del(7q) UL arise through a gain of function, resulting from either production of a fusion gene or a positional effect due to rearranging sequences within chromosome 7 such as by an interstitial deletion. A gain of function is not as likely because the del(7q) breakpoints are variable. It also has yet to be determined if the predisposing gene(s) at 7q22 is the same or divergent between UL, other mesenchymal solid tumors, and myeloid cells (AML and MDS).
Multiple studies have targeted del(7q) UL and shown LOH of microsatellite markers or altered expression of genes within 7q22 such as CUTL1 (repressor of c-MYC expression), ORC5L (DNA replication initiation factor), LAMB1 (extracellular matrix component), LHFPL3 (transmembrane protein of unknown function), and PAI1 (hemostasis and smooth muscle cell expression) (Sourla et al., 1996; Zeng et al., 1997; Quintana et al., 1998; Saito et al., 2005; Ptacek et al., 2007). No alteration in expression was found by RT-PCR for NRCAM, DLD, PIK3G, PBEF or SRPK2 (Saito et al., 2005). However, despite these varied efforts, no consistent gene expression changes have been identified and the causative gene(s) in the pathogenesis of the del(7q) subgroup of UL remain to be established.
It is notable that UL with 7q abnormalities are often mosaic with karyotypically normal 46,XX cells and when cultured grow poorly and frequently lose the chromosomally aberrant cell line (Xing et al., 1997). These observations indicate a gene(s) deleted from the 7q region likely plays a role in regulating cellular growth (Rein et al., 1998). Such mosaic UL were demonstrated to be clonal tumors, which may suggest the del(7q) abnormality is not likely to be the primary pathogenetic event. Deletion of 7q has however been observed as the sole abnormality in a non-mosaic state, and it is possible there are submicroscopic pathogenetic events such as small deletions or point mutations in the regulatory or coding sequences occurring in the same genes as those indicated by chromosomal rearrangement (Xing et al., 1997). In fact, submicroscopic deletions have been identified by the finding of LOH in 7q in a subset of karyotypically normal UL through microsatellite allelotyping (Ishwad et al., 1997).
Further inconsistency has been noticed when attempting to correlate LOH and karyotype data. LOH was not detected in a proportion of UL with cytogenetically visible 7q deletions (Ishwad et al., 1997). This discrepant result may be due to the mosaic nature of these UL where the karyotypically normal cells dilute the ability of aCGH to detect the change in DNA copy number of the involved genes. Another explanation is that del(7q) tumors identified solely by interphase FISH may have rare complex chromosomal rearrangements which result in some 7q genes being integrated elsewhere in the genome.
In addition to an inability to detect complex rearrangements, FISH is not able to quantify precisely the deletion interval size. Defining the deletion boundaries was therefore addressed in the current study by employing aCGH analysis on six of the del(7q) UL. In contrast to earlier work that employed aCGH using a general female normal DNA as the control (Vanharanta et al., 2007), we used the normal myometrial tissue from the same patient as the control in each case to eliminate any confounding effect of germline copy number variation. The only consistent copy number change among the six cases analyzed was deletion on chromosome 7, the smallest common region of which spanned approximately 9.5 megabases from 7q22.1-q31.1. Alignment of genes significantly expressed in del(7q) UL relative to non-del(7q) UL after correction for mosaicism demonstrated a high correlation between gene presence in the common deletion interval and decreased expression. This provides a further validation of the accuracy of the microarray expression data. Our results also confirm the finding that small homozygous deletions are not observed in del(7q) UL, arguing against the target gene(s) in 7q being a tumor suppressor. In fact, microarray expression analysis and coding region sequencing of many genes across the region have previously failed to identify a gene of interest (Vanharanta et al., 2005; Vanharanta et al., 2007).
Other microarray expression analyses of UL relied on a comparison of the tumors to the normal myometrial tissue, not taking into account the known cytogenetic variation among UL. To identify genes specific to the del(7q) abnormality rather than those that distinguish myometrium from any UL, the expression profile of del(7q) UL needs to be compared directly to that of non-del(7q) UL. The current study takes this approach, and in contrast to recent work (Vanharanta et al., 2005), the del(7q) and non-del(7q) tumors were from the same rather than different patients to eliminate the genotype, environment, and genotype x environment confounding effects that underlie patient to patient variation. In addition, the aforementioned study employed the HG-U133A array which has less extensive genome coverage than the U133 plus 2.0 microarray used in the present work, and even after reducing the stringency to identify any contrasting genes of significance, none were discovered in their 7q commonly deleted region (Vanharanta et al., 2005).
The validity and necessity of our approach to compare UL with and without the abnormality from the same patient is supported by the finding of a del(7q) UL-specific gene list that is highly enriched for genes in 7q22, all of which showed significantly decreased expression. It is interesting that the chromatin-modulating gene MLL5 (Mixed Lineage Leukemia-5), which is ranked sixth in the del(7q) UL-specific gene list and was examined by Q-PCR to confirm the decreased expression observed by microarray data analysis, has been found to inhibit cancer cell cycle progression when ectopically overexpressed or when knocked down by small interfering RNAs (Deng et al., 2004; Cheng et al., 2008). This suggests cells are very sensitive to MLL5 dosage, and haploinsufficiency due to deletion at 7q22.3 in UL may explain at least in part the poor growth and frequent loss of the abnormal cell line in culture as well as a relatively smaller size of del(7q) tumors compared to another UL subgroup, those with a t(12;14)(q15;q23-q24) (Rein et al., 1998).
MLL5 was found to reach statistical significance and rank fourth on the del(7q) UL-specific gene list when the data were weighted for the percent of cells containing the 7q deletion. Previous studies had not accounted for the mosaic nature of most del(7q) UL, and the doubling of genes at 7q22 in the top 50 of the mosaicism-weighted gene list relative to the unweighted list (from 10 up to 20 genes) suggests the importance of implementing this data correction to reduce the noise introduced by karyotypically normal cells. Six other genes (ZNF498, TRAF3IP1, MGC39821, SSR2, MARCKS, and HBP1) were also identified as being significant. HBP1 at 7q22.3 is of interest as loss of expression of this proliferation repressor has been associated with invasive breast cancer, suggesting the decreased expression found in del(7q) UL may contribute to the proliferative capacity of these tumors (Paulson et al., 2007). A similarly functioning gene, SIN3A at 15q24.2, also has decreased expression in del(7q) UL and is present at number 155 on the mosaicism weighted gene list (also in network 2 as described below). SIN3A is a core component of a complex with histone deacetylase enzyme activity which is employed by multiple factors such as p53 to repress their target genes such that loss of SIN3A activity is linked to proliferation and cell survival (Dannenberg et al., 2005). Downregulation of SIN3A has been demonstrated in human cancer, specifically non-small cell lung type (Suzuki et al., 2008). Another gene of significance in the del(7q) UL mosaicism-weighted list is RINT1, which has been shown through RNA interference studies and the development of multiple tumors in haploinsufficient mice to serve a novel tumor suppressor function by maintaining integrity of the Golgi apparatus and centrosome necessary for proper cell division (Lin et al., 2007).
Biological insight into del(7q) UL was pursued further through Ingenuity Pathways Analysis (IPA) of the top 300 probe sets based on the Q-value from the del(7q) mosaicism-weighted gene list. The two networks of highest significance were associated with development, which may reflect the need of the tumor cells to revert towards a more embryonic phenotype to proliferate. This could be related to the hypothesis that UL arise from an inappropriate activation of myometrial cell proliferation deriving from the inherent abilities of the uterine tissue during pregnancy (Andersen et al., 1995). Interestingly, the most represented canonical pathway was protein ubiquitination, with 11 of the top 300 del(7q) UL-specific genes. Genes from network 1 involved in protein ubiquitination include PSMA2, PSMB3, PSMC2, and PSMC3 while those in network 2 include UBE2I and UBE2J1. Ubiquitination is of interest because >80% of cellular proteins are tagged with ubiquitin for proteasome degradation and cancer can develop through disruption of this system either by stabilizing oncoproteins or destabilizing tumor suppressor genes (Burger and Seth, 2004). The hierarchical nature of the ubiquitination enzyme cascade with transfer from E1 to any of a multitude of E2s and then E3s as well as a number of different subunits present on the proteasome allows for specialization targeting of proteins for degradation. It is therefore likely that disruption of multiple E2 enzymes (EBE2 genes) and proteasome subunits (PSM genes) reflects an altered protein homeostasis in the del(7q) UL which potentially contributes to tumorigenesis.
In conclusion, this study provides a genome-wide expression profile of the 7q deletion cytogenetic subgroup of UL. The unique design employed to target del(7q) UL-specific genes included a paired comparison to non-del(7q) UL from the same women and weighting of the data for percent of del(7q) cells to account for the mosaicism usually present in this UL subgroup. Although the full implications and biological significance of the differentially expressed genes and networks remain to be fully elucidated, the resultant gene list, which is dense with genes from the target region of 7q22, may serve as a platform to explore further relevant mechanisms of tumor pathogenesis and understanding of the molecular basis of UL.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank David Zahrieh at Dana-Farber Cancer Institute for helpful discussions and exploration of the data by ANOVA and Karen T.Cuenco at the University of Pittsburgh for a thoughtful review of the manuscript.
Supported by: This work was supported by NIH grants RO1HD046226 and RO1CA78895 (to CCM) and JCH was supported by T32GM007748.
REFERENCES
- Andersen J, DyReyes VM, Barbieri RL, Coachman DM, Miksicek RJ. Leiomyoma primary cultures have elevated transcriptional response to estrogen compared with autologous myometrial cultures. J Soc Gynecol Investig. 1995;2:542–551. doi: 10.1016/1071-5576(94)00053-4. [DOI] [PubMed] [Google Scholar]
- Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur J Cancer. 2004;40:2217–2229. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Buttram VC, Jr, Reiter RC. Uterine leiomyomata: etiology, symptomatology, and management. Fertil Steril. 1981;36:433–445. doi: 10.1016/s0015-0282(16)45789-4. [DOI] [PubMed] [Google Scholar]
- Cheng F, Liu J, Zhou SH, Wang XN, Chew JF, Deng LW. RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int J Biochem Cell Biol. 2008;40:2472–2481. doi: 10.1016/j.biocel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, Kim UJ, Kuo WL, Olivier M, Conroy J, Kasprzyk A, Massa H, Yonescu R, Sait S, Thoreen C, Snijders A, Lemyre E, Bailey JA, Bruzel A, Burrill WD, Clegg SM, Collins S, Dhami P, Friedman C, Han CS, Herrick S, Lee J, Ligon AH, Lowry S, Morley M, Narasimhan S, Osoegawa K, Peng Z, Plajzer-Frick I, Quade BJ, Scott D, Sirotkin K, Thorpe AA, Gray JW, Hudson J, Pinkel D, Ried T, Rowen L, Shen-Ong GL, Strausberg RL, Birney E, Callen DF, Cheng JF, Cox DR, Doggett NA, Carter NP, Eichler EE, Haussler D, Korenberg JR, Morton CC, Albertson D, Schuler G, de Jong PJ, Trask BJ. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado GD, Marshall LM, Schwartz SM. Complications in pregnancy, labor, and delivery with uterine leiomyomas: a population-based study. Obstet Gynecol. 2000;95:764–769. doi: 10.1016/s0029-7844(99)00605-5. [DOI] [PubMed] [Google Scholar]
- Cramer SF, Patel A. The frequency of uterine leiomyomas. Am J Clin Pathol. 1990;94:435–438. doi: 10.1093/ajcp/94.4.435. [DOI] [PubMed] [Google Scholar]
- Dal Cin P, Van den Berghe H, Sciot R, de Wever I. Deletion of the long arm of chromosome 7 in lipoma. Cancer Genet Cytogenet. 1997;96:85–86. doi: 10.1016/s0165-4608(96)00261-0. [DOI] [PubMed] [Google Scholar]
- Dal Cin P, Vanni R, Marras S, Moerman P, Kools P, Andria M, Valdes E, Deprest J, Van de Ven W, Van den Berghe H. Four cytogenetic subgroups can be identified in endometrial polyps. Cancer Res. 1995;55:1565–1568. [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Develop. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng LW, Chiu I, Strominger JL. MLL 5 protein forms intranuclear foci, and overexpression inhibits cell cycle progression. Proc Natl Acad Sci U S A. 2004;101:757–762. doi: 10.1073/pnas.2036345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn M, Jamison M, Datta S, Myers E. Health care resource use for uterine fibroid tumors in the United States. Am J Obstet Gynecol. 2006;195:955–964. doi: 10.1016/j.ajog.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann KE, Birnbaum H, Ben-Hamadi R, Wu EQ, Farrell MH, Spalding J, Stang P. Annual costs associated with diagnosis of uterine leiomyomata. Obstet Gynecol. 2006;108:930–937. doi: 10.1097/01.AOG.0000234651.41000.58. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Azuma C, Kamiura S, Kimura T, Nobunaga T, Kanai T, Sawada M, Noguchi S, Saji F. Clonal determination of uterine leiomyomas by analyzing differential inactivation of the X-chromosome-linked phosphoglycerokinase gene. Gynecol Obstet Invest. 1995;40:204–208. doi: 10.1159/000292336. [DOI] [PubMed] [Google Scholar]
- Ishwad CS, Ferrell RE, Hanley K, Davare J, Meloni AM, Sandberg AA, Surti U. Two discrete regions of deletion at 7q in uterine leiomyomas. Genes Chromosomes Cancer. 1997;19:156–160. doi: 10.1002/(sici)1098-2264(199707)19:3<156::aid-gcc4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepine LA, Hillis SD, Marchbanks PA, Koonin LM, Morrow B, Kieke BA, Wilcox LS. Hysterectomy surveillance--United States, 1980–1993. MMWR CDC Surveill Summ. 1997;46:1–15. [PubMed] [Google Scholar]
- Liang H, Fairman J, Claxton DF, Nowell PC, Green ED, Nagarajan L. Molecular anatomy of chromosome 7q deletions in myeloid neoplasms: evidence for multiple critical loci. Proc Natl Acad Sci U S A. 1998;95:3781–3785. doi: 10.1073/pnas.95.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liu CC, Gao Q, Zhang X, Wu G, Lee WH. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol Cell Biol. 2007;27:4905–4916. doi: 10.1128/MCB.02396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal RD, Fejzo ML, Friedman AJ, Mitchner N, Nowak RA, Rein MS, Morton CC, Sklar J. Analysis of androgen receptor DNA reveals the independent clonal origins of uterine leiomyomata and the secondary nature of cytogenetic aberrations in the development of leiomyomata. Genes Chromosomes Cancer. 1994;11:1–6. doi: 10.1002/gcc.2870110102. [DOI] [PubMed] [Google Scholar]
- Meloni AM, Surti U, Contento AM, Davare J, Sandberg AA. Uterine leiomyomas: cytogenetic and histologic profile. Obstet Gynecol. 1992;80:209–217. [PubMed] [Google Scholar]
- Moore SD, Herrick SR, Ince TA, Kleinman MS, Cin PD, Morton CC, Quade BJ. Uterine leiomyomata with t(10;17) disrupt the histone acetyltransferase MORF. Cancer Res. 2004;64:5570–5577. doi: 10.1158/0008-5472.CAN-04-0050. [DOI] [PubMed] [Google Scholar]
- Nibert M, Heim S. Uterine leiomyoma cytogenetics. Genes Chromosomes Cancer. 1990;2:3–13. doi: 10.1002/gcc.2870020103. [DOI] [PubMed] [Google Scholar]
- Ozisik YY, Meloni AM, Surti U, Sandberg AA. Deletion 7q22 in uterine leiomyoma. A cytogenetic review. Cancer Genet Cytogenet. 1993;71:1–6. doi: 10.1016/0165-4608(93)90195-r. [DOI] [PubMed] [Google Scholar]
- Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, Huang CY, Giri D, Kaufman S, Dugan JM, Blum J, Netto G, Wazer DE, Summerhayes IC, Yee AS. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67:6136–6145. doi: 10.1158/0008-5472.CAN-07-0567. [DOI] [PubMed] [Google Scholar]
- Ptacek T, Song C, Walker CL, Sell SM. Physical mapping of distinct 7q22 deletions in uterine leiomyoma and analysis of a recently annotated 7q22 candidate gene. Cancer Genet Cytogenet. 2007;174:116–120. doi: 10.1016/j.cancergencyto.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Quintana DG, Thome KC, Hou ZH, Ligon AH, Morton CC, Dutta A. ORC5L, a new member of the human origin recognition complex, is deleted in uterine leiomyomas and malignant myeloid diseases. J Biol Chem. 1998;273:27137–27145. doi: 10.1074/jbc.273.42.27137. [DOI] [PubMed] [Google Scholar]
- Rein MS, Friedman AJ, Barbieri RL, Pavelka K, Fletcher JA, Morton CC. Cytogenetic abnormalities in uterine leiomyomata. Obstet Gynecol. 1991;77:923–926. [PubMed] [Google Scholar]
- Rein MS, Nowak RA. Biology of uterine myomas and myometrium in vitro. Sem Repro Endocrin. 1992;10:310–319. [Google Scholar]
- Rein MS, Powell WL, Walters FC, Weremowicz S, Cantor RM, Barbieri RL, Morton CC. Cytogenetic abnormalities in uterine myomas are associated with myoma size. Mol Hum Reprod. 1998;4:83–86. doi: 10.1093/molehr/4.1.83. [DOI] [PubMed] [Google Scholar]
- Saito E, Okamoto A, Saito M, Shinozaki H, Takakura S, Yanaihara N, Ochiai K, Tanaka T. Genes associated with the genesis of leiomyoma of the uterus in a commonly deleted chromosomal region at 7q22. Oncol Rep. 2005;13:469–472. [PubMed] [Google Scholar]
- Sargent MS, Weremowicz S, Rein MS, Morton CC. Translocations in 7q22 define a critical region in uterine leiomyomata. Cancer Genet Cytogenet. 1994;77:65–68. doi: 10.1016/0165-4608(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Sell SM, Altungoz O, Prowse AA, Meloni AM, Surti U, Sandberg AA. Molecular analysis of chromosome 7q21.3 in uterine leiomyoma: analysis using markers with linkage to insulin resistance. Cancer Genet Cytogenet. 1998;100:165–168. doi: 10.1016/s0165-4608(97)00032-0. [DOI] [PubMed] [Google Scholar]
- Sell SM, Tullis C, Stracner D, Song CY, Gewin J. Minimal interval defined on 7q in uterine leiomyoma. Cancer Genet Cytogenet. 2005;157:67–69. doi: 10.1016/j.cancergencyto.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Smyth G, editor. Limma: linear models for microarray data. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Sourla A, Polychronakos C, Zeng WR, Nepveu A, Kukuvitis A, Naud F, Koutsilieris M. Plasminogen activator inhibitor 1 messenger RNA expression and molecular evidence for del(7)(q22) in uterine leiomyomas. Cancer Res. 1996;56:3123–3128. [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ouchida M, Yamamoto H, Yano M, Toyooka S, Aoe M, Shimizu N, Date H, Shimizu K. Decreased expression of the SIN3A gene, a candidate tumor suppressor located at the prevalent allelic loss region 15q23 in non-small cell lung cancer. Lung Cancer. 2008;59:24–31. doi: 10.1016/j.lungcan.2007.08.002. [DOI] [PubMed] [Google Scholar]
- van der Heijden O, Chiu HC, Park TC, Takahashi H, LiVolsi VA, Risinger JI, Barrett JC, Berchuck A, Evans AC, Behbakht K, Menzin AW, Liu PC, Benjamin I, Morgan MA, King SA, Rubin SC, Boyd J. Allelotype analysis of uterine leiomyoma: localization of a potential tumor suppressor gene to a 4-cM region of chromosome 7q. Mol Carcinogenesis. 1998;23:243–247. doi: 10.1002/(sici)1098-2744(199812)23:4<243::aid-mc7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Vanharanta S, Wortham NC, Laiho P, Sjoberg J, Aittomaki K, Arola J, Tomlinson IP, Karhu A, Arango D, Aaltonen LA. 7q deletion mapping and expression profiling in uterine fibroids. Oncogene. 2005;24:6545–6554. doi: 10.1038/sj.onc.1208784. [DOI] [PubMed] [Google Scholar]
- Vanharanta S, Wortham NC, Langford C, El-Bahrawy M, van der Spuy Z, Sjoberg J, Lehtonen R, Karhu A, Tomlinson IP, Aaltonen LA. Definition of a minimal region of deletion of chromosome 7 in uterine leiomyomas by tiling-path microarray CGH and mutation analysis of known genes in this region. Genes Chromosomes Cancer. 2007;46:451–458. doi: 10.1002/gcc.20427. [DOI] [PubMed] [Google Scholar]
- Xing YP, Powell WL, Morton CC. The del(7q) subgroup in uterine leiomyomata: genetic and biologic characteristics. Further evidence for the secondary nature of cytogenetic abnormalities in the pathobiology of uterine leiomyomata. Cancer Genet Cytogenet. 1997;98:69–74. doi: 10.1016/s0165-4608(96)00406-2. [DOI] [PubMed] [Google Scholar]
- Zeng WR, Scherer SW, Koutsilieris M, Huizenga JJ, Filteau F, Tsui LC, Nepveu A. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene. 1997;14:2355–2365. doi: 10.1038/sj.onc.1201076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.