Abstract
Microdialysis sampling is a widely used method to sample from complex biological matrices. Cytokines are important signaling proteins that are typically recovered with low relative recovery values during microdialysis sampling. Heparin was included in the microdialysis perfusion fluid as an affinity agent to increase in vitro recovery of different cytokines through polyethersulfone (PES) microdialysis membranes with 100 kDa molecular weight cutoff. No change in fluid volumes collected from the microdialysis probes occurred when heparin was included in the perfusion fluid up to concentrations of 10 μM. The loss of heparin (10 μM) across the dialysis membrane was minimal (2.7±0.9%, n=3). Additionally, heparin at these concentrations did not interfere with the cytokine immunoassays. The control and heparin-enhanced relative recoveries for five human cytokines using 0.1 μM heparin in the microdialysis perfusion fluid flowing at 0.5 μL min−1 were (n=3): interleukin-4 (IL-4), 4.2 ± 0.5% and 7.2±3.1%; interleukin-6 (IL-6), 1.4±0.8% and 3.6±1.3%; interleukin-7 (IL-7), 1.3±0.8% and 4.8±1.8%; monocyte chemoattractant protein-1 (MCP-1), 9.0±1.6% and 19.5±2.7%; and tumor necrosis factor-α (TNF-α), 7.4±1.3% and 16.9±1.6%, respectively. Heparin increased the microdialysis sampling relative recovery of several human cytokines in vitro.
Keywords: microdialysis, cytokines, heparin, in vitro
1. Introduction
Cytokines are vitally important signaling proteins that control various aspects of the immune response [1,2,3]. Knowing the imbalances of cytokines during various phases of disease is critical and for this reason cytokines are key biomarkers used in profiling many disease states [4-7]. The production of cytokines is transient and generally localized resulting in concentrations that can be in the picomolar to femtomolar range.
Microdialysis sampling is widely used in life science applications for collecting analytes, principally neurotransmitters, from the extracellular space of tissues [8]. Collected dialysates represent the fraction of the analyte concentration in the medium external to the dialysis probe. The performance of the microdialysis sampling collection process can be characterized via the relative recovery (RR) for an individual analyte shown in Equation 1, where Coutlet is the analyte concentration collected from the dialysis probe and Csample,∞ is the analyte concentration far from the microdialysis sampling probe.
| (1) |
Over the past few years, the application of microdialysis sampling to peptide and protein targets has increased [9]. Since microdialysis sampling relative recovery is dependent upon analyte diffusion, dialysis membranes with higher molecular weight cutoff (MWCO) and larger pore size are typically used for protein collection. For many protein collection applications, the RR obtained with commercially available microdialysis membranes is often low due to diffusive mass transport resistances [10]. Cytokines span a molecular weight range between approximately 8 to 80 kDa resulting in low microdialysis sampling RR [11,12]. The low endogenous concentrations of cytokines combined with low recovery make quantifying these important molecules challenging [13,14].
Cytokine collection into microdialysis sampling probes can be improved by addition of protein or dextran which balances the osmotic pressure and can also prevent non-specific adsorption of cytokines to the probe materials [15,16]. These agents are not used as affinity agents per se, but are necessary to prevent fluid loss across the large pore sized membranes that are required for recovery of the proteins. However, a concern with this approach in tissues such as the brain is that fluid recovery can sometimes be greater than 100%, i.e., fluid collected is greater than the fluid input.
An alternative approach to improving recovery is to add affinity agents into the dialysis perfusion fluid that serve to trap the targeted analytes [17]. The inclusion of antibody-immobilized microspheres in microdialysis perfusion fluids to capture cytokines allowed samples to be directly analyzed using flow cytometry-based immunoassay without further sample preparation [12]. However, during previous in vivo studies using this approach saturation of the antibodies (concentrations above 5,000 pg mL−1) was observed which led to semi-quantitative data, e.g. concentrations were over-range and samples could not be recovered from the waste and re-analyzed [12]. Therefore, an alternative approach was sought that decoupled the affinity-capture and detection steps.
In this work, heparin was chosen as an affinity agent to capture cytokines. Heparin, a highly sulfated, polydisperse, and negatively-charged glycosaminoglycan with an alternating copolymer structure consisting of uronic acid and amino sugars binds to many proteins and cytokines (Scheme A) [18-22]. The binding between heparin and cytokines can be attributed to the conformation of the individual cytokine, the structure of their heparin binding sites, and electrostatic interactions between positively-charged amino acids on the protein and the negatively-charged heparin molecule [23]. Compared with antibodies, heparin is stable, inexpensive, and readily available.
Scheme A.
Structures of heparin, dextran and β-cyclodextrin sulfate.
Five human cytokines (interleukin-4 [IL-4], interleukin-6 [IL-6], interleukin-7 [IL-7], monocyte chemoattractant protein-1 [MCP-1] and tumor necrosis factor-alpha [TNF-α]) that have been reported to bind to heparin were used for the in vitro study [24-28]. The choice of these cytokines reflects a range of molecular weight and structure. Negative controls (dextran and β-cyclodextrin sulfate) were included in the perfusion fluid to ensure that the binding of cytokines to heparin caused the increased relative recovery and not prevention of non-specific adsorption.
2. Experimental
2.1 Materials
Human cytokine LINCOplex kits containing assays for human interleukin-4 [IL-4], interleukin-6 [IL-6], inerleukin-7 [IL-7], monocyte chemoattractant protein-1 [MCP-1] and tumor necrosis factor alpha [TNF-α] were purchased from LINCO Research (Charles, MO, USA).a The limits of detection for these kits are available from the manufacturer. In these studies, the suggested calibration range of 3.2 to 10,000 pg mL−1 was used. Table 1 lists the physicochemical properties for the cytokines used in this work. The kits include the capture antibody-immobilized beads, detection antibodies, assay buffer as well as standards and controls. The assay buffer contains 50 mM PBS with 25 mM EDTA, 0.08% sodium azide, 0.05% Tween 20, and 1% bovine serum albumin (BSA), pH 7.4. Human MCP-1 ELISA kits were purchased from BD Pharmingen (San Diego, CA, USA) and have a working range of 15.6 pg mL−1 to 1000 pg mL−1. The main components of the buffer from ELISA kits include 10 mM phosphate buffered saline (PBS) with 10% FBS (Fetal Bovine Serum), pH 7.0. Heparin sodium salt from porcine intestinal mucosa with an approximate 14 kDa average molecular weight was obtained from Sigma-Aldrich (St. Louis, MO, USA). Two negative controls, dextran with molecular weight 150,000 Da and β-cyclodextrin sulfate (18 mole sulfate per mole cyclodextrin) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Table 1.
Human cytokine physical properties.
| Predicted MW (kDa) of monomer | Conformation | Concentration (pM) for 2000 pg mL−1 standards | Reported Kd with Heparin | [Heparin] needed to achieve 90% bound based on reported Kd (μM) | |
|---|---|---|---|---|---|
| IL-4 | 15.0 | monomer | 133.3 | 80 nM 24 | 0.7 |
| IL-6 | 20.8 | monomer | 96.2 | 200 nM 25 | 1.8 |
| IL-7 | 17.4 | monomer | 114.9 | 25 nM 26 | 0.2 |
| MCP-1 | 8.7 | dimer | 114.9 | 1.5 μM 27 | 13.5 |
| TNF-α | 17.4 | trimer | 38.3 | 670 nM 28 | 6.0 |
2.2 Microdialysis and sample collection
CMA/20 (10 mm) polyethersulfone (PES) microdialysis probes with 100 kDa molecular weight cutoff (CMA Microdialysis, North Chelmsford, MA, USA) were used for all experiments. Probes were reused in these in vitro experiments. Different flow rates were controlled by a Bioanalytical Systems Bee microdialysis infusion pump (BASi, West Lafayette, IN, USA). Microdialysis experiments were performed using quiescent conditions at room temperature (22-23°C). Assay buffers from the cytokine detection kits were used as the perfusion fluids. When LINCOplex kits were used, the assay buffer contains 50 mM PBS with 25 mM EDTA, 0.08% sodium azide, 0.05% Tween-20 and 1% BSA, pH 7.4. The assay buffers for the ELISA kits contain 10 mM PBS and 10% FBS, pH 7.0.
During cytokine collection, standards from LINCOplex kits were dissolved in the assay buffer from the kit to create solutions containing 2000 pg mL−1 of each cytokine to ensure collection capability through the microdialysis sampling probes (molar concentrations are shown in Table 1). The probes were immersed into a 200 μL cytokine solution in a 0.25 mL tube. Heparin was dissolved in the LINCOplex assay buffer to concentrations of 0.1 μM, 1 μM or 10 μM and was perfused through the microdialysis probe. Control experiments were performed at the same time with only the assay buffer as the perfusion fluid. Samples (30 μL) were collected at each flow rate (0.5 μL min−1, 1.0 μL min−1 and 1.5 μL min−1) for controls and for each individual heparin concentration. From these samples, 25 μL were used for cytokine measurements with the Lincoplex kits. All the microdialysis experiments were performed in triplicate.
Human MCP-1 was used during single cytokine collection since it consistently gave improved relative recovery values with heparin within the flow rate range used. The assay diluent from the ELISA kit was used to prepare all the heparin and cytokine solutions to final MCP-1 concentrations of 2000 pg mL−1 to ensure collection capability through microdialysis sampling probes. Heparin was included in the perfusion fluid within the concentration range of 1 pM to 10 μM from lowest to highest concentration. Flow rates of 0.5 μL min−1, 1.0 μL min−1 and 2.0 μL min−1 were used in this order with samples collected for 4 hr (0.5 μL min−1), 2 hr (1.0 μL min−1) and 1 hr (2.0 μL min−1). Control experiments were performed with the assay diluent from the ELISA kits as the perfusion fluid and heparin did not interfere with this assay using these concentrations. All the experiments were performed in duplicate due to the 100 μL volume requirements of the ELISA kit.
2.3 Cytokine analysis
The five different human cytokines were measured using a LUMINEX 100 IS™ system (Luminex Corp., Austin, TX, USA). The principles for these types of assays have been previously reviewed [29]. Data analysis and regression was performed using BeadView Multiplex Data Analysis Software, Version 1.0 (Upstate Scientific, Lake Placid, NY, USA). In some experiments, human MCP-1 was quantified using an ELISA set with a Tecan SPECTRA Fluor plate reader (Tecan Group Ltd., Männedorf, Switzerland) at 450 nm. Cytokine concentrations were determined using normal standard curves for control probes. Dialysates containing heparin were compared against standard curves that were prepared with identical concentrations of heparin spiked into the standards.
2.4 Heparin loss during microdialysis sampling
To determine the extent of heparin loss from the microdialysis sampling probe, heparin (0.1, 1, and 10 μM) was included in the perfusion fluid and delivered at a flow rate of 0.5 μL min−1 for 7 hours at room temperature. The microdialysis probes containing separate heparin concentrations were immersed into individual 1 mL solution containing LINCOplex assay buffer in a 1.5 mL volume microcentrifuge tube. The solutions were stirred and 200 μL sample (both dialysate and solution external to the probe) was collected at each concentration. The collected heparin was hydrolyzed to glucosamine using 4.8 M HCl. The amount of glucosamine was used as an indirect measure of heparin concentration [30]. The calibration range for glucosamine is 0.05-0.7 μg mL−1 which gives an approximate heparin concentration of 14 to 194 nM).
Glucosamine was quantified using high performance liquid chromatography (SHIMADZU Scientific Instruments. INC. Columbia, MD, USA) combined with pulsed amperometric detection (ANTEC Leyden B.V., Leiden, The Netherlands) (HPLC-PAD). The separations of hydrolysates were performed on a Dionex CarboPac PA1 column, 2×250 mm, and CarboPac PA1-Guard column, 2×50 mm (Dionex, Sunnyvale, CA). The mobile phase contained 95 mM sodium hydroxide and 10 mM sodium acetate. For pulsed amperometric detection the potentials were E1= +0.05 V vs. Ag/AgCl, t1= 400 ms; E2= +0.75 V vs. Ag/AgCl, t2= 200 ms; E3= −0.75 V vs. Ag/AgCl, t3= 400 ms.
2.5 Ultrafiltration effects
The ultrafiltration across the microdialysis membrane was measured directly by weighing the dialysate after perfusing the heparin through the microdialysis probe on a tared Ohaus Analytical Plus Electronic Balance sensitive to 0.00001 g (Ohaus Corporation, Florham Park, NY, USA). Heparin was included into the perfusion fluid (assay buffer from the LINCOplex kit) with concentrations of 0.1 μM, 1 μM and 10 μM. The flow rate was 1.0 μL min−1 and samples were collected over a 30 minute time period.
2.6 Negative controls
Negative control experiments with MCP-1 as the target cytokine were performed using dextran and β-cyclodextrin sulfate. Microdialysis probes were immersed in individual quiescent solutions containing 2000 pg mL−1 of MCP-1 and flow rates of 0.5, 1.0 and 2.0 μL min−1 were used. In one set of probes, dextran (1 μM) in the ELISA assay buffer was used. Dextran was chosen because of its linear oligosaccharide structure. In another set β-cyclodextrin of probes sulfate (50 μM) in the ELISA assay buffer was infused with the choice of 50 μM β-cyclodextrin sulfate exceeding the total sulfate content in 10 μM heparin. The control the last set of probes were perfused with the the ELISA assay buffer.
2.7 Statistical analysis
All statistical tests were performed using Microsoft Excel (Office 2003 edition) or Microcal Origin 8.
3. Results and Discussion
3.1 Ultrafiltration
Fluid loss through 100 kDa MWCO membranes during microdialysis sampling is well documented. To overcome this potential problem, osmotic agents such as Dextran-70 or bovine serum albumin (BSA) are typically added [15]. The fluid weight collected during 30 minutes for the control probe was 31.61 ± 0.14 mg. For probes containing 0.1, 1.0 and 10 μM heparin, the weights were 31.82 ± 0.42, 31.76 ± 0.39 and 31.43 ± 0.07 mg, respectively. Among these sets, the collected weights and thus volumes are not different.
3.2 Heparin loss
A significant loss of heparin across the 100 kDa MWCO PES membrane may interfere with cytokine collection or with data interpretation. The percent heparin lost across the microdialysis sampling membrane using a 0.5 μL min−1 flow rate was determined to be not detectable (0.1 μM), 1.4 ± 1.9% (1 μM), and 2.7 ± 0.9% (10 μM), n=3. The loss values that could be measured are not statistically different and the amount of heparin detected in the external solution is at or near the quantitation limit (14 nM) for this assay. The rigid linear polymer structure of heparin that gives it a large hydrodynamic radius is the likely reason for the low loss of heparin across the dialysis membrane. In other words, it simply cannot diffuse easily through the pores. The measured radius of gyration for heparin is 3.2 to 5.5 nm and its characteristic length values are 15-33 nm [31]. These values are larger than those for the cytokines, whose radius of gyration values are compiled in reference 12. With such large radius of gyration and length values, it is unlikely that heparin linear polymers will diffuse through the dialysis pores easily. Therefore, it is vitally important to realize that in addition to molecular weight that shape and size determine the relative recovery of molecules across microdialysis membranes. This is especially true for the 100 kDa PES membranes that have an inner membrane pore size of 9 nm reported by the manufacturer.
3.3 Heparin interference
To determine if heparin would interfere with the assays, standard curves containing heparin were prepared for both the standard ELISA kits and the LINCOplex kits (data not shown). At heparin concentrations up to 10 μM, no alterations in the standard curve for the LINCOplex kits were observed. This suggests that the binding site between heparin, these cytokines, and their associated antibodies are different. Additionally, no change in the standard curve was observed with up to 10 μM heparin added to standards measured using the MCP-1 ELISA kit. However, it is important not to extrapolate the lack of interference results obtained with this subset of cytokines and these particular assay kits to all cytokines and/or growth factors and their assays since these should be properly validated on a case-by-case basis.
3.4 Enhanced relative recovery of cytokines
Table 2 shows the cytokine RR values for controls vs. different concentrations of heparin. In general, the increase in recovery using heparin in the perfusion fluid was approximately two-fold compared to controls. When comparing the four treatments (control, 0.1 μM, 1 μM, and 10 μM heparin) at the 0.5 μL min−1 flow rate, a one-way ANOVA shows that among the five cytokines tested the RR values for controls are statistically different from the RR values obtained using heparin for all the cytokines with the exception of IL-4. For IL-4, higher variance values within each treatment group as compared to between groups were obtained thus negating its significance. A second one-way ANOVA (0.5 μL min−1 flow rate) that compares the three heparin concentrations shows that for all five cytokines there is no statistical difference in the mean values obtained among the heparin treatments. With two exceptions at 0.5 μL min−1 (1 μM heparin for IL-6 and IL-7), the recovery for heparin-containing probes compared to the controls are statistically different when compared using a Tukey post hoc test between the control and heparin-containing probes. Similar ANOVA and Tukey post hoc analyses were performed with the 1.0 and 1.5 μL min−1 data. For these flow rates, the variability in the data resulted in RR values that were not statistically different values with the exception of IL-7 and MCP-1. Control recovery values differ between the cytokines and this is likely caused by variations due to different molecular weights, tertiary structures, and diffusive mass transport.
Table 2.
Enhanced vs. control relative recovery (RR) for human cytokines
| Cytokines | Control RR (Mean±SD) | Enhanced RR (0.1 μM heparin) (Mean±SD) | Enhanced RR (1 μM heparin) (Mean±SD) | Enhanced RR (10 μM heparin) (Mean±SD) |
|---|---|---|---|---|
| 0.5 μL min−1 | ||||
| IL-4 | 4.2 ± 0.5% | 7.2 ± 3.1% | 5.3 ± 2.9% | 7.2 ± 3.0% |
| IL-6 | 1.4 ± 0.8% | 3.6 ± 1.3% | 2.6 ± 0.9% | 4.9 ± 0.1%* |
| IL-7 | 1.3 ± 0.8% | 4.8 ± 1.8%* | 3.0 ± 0.9% | 4.6 ± 0.6%* |
| MCP-1 | 9.0 ± 1.6% | 19.5 ± 2.7%* | 15.7 ± 1.8% | 21.1 ± 5.7%* |
| TNF-α | 7.4 ± 1.3% | 16.9 ± 1.6%* | 15.0 ± 4.7%* | 15.8 ± 1.3%* |
| 1.0 μL min−1 | ||||
| IL-4 | 2.5 ± 0.4% | 7.3 ± 3.3% | 5.4 ± 0.2% | 3.4 ± 2.0% |
| IL-6 | 0.7 ± 0.1% | 2.6 ± 1.4% | 1.3 ± 0.8% | 2.3 ± 0.6% |
| IL-7 | 0.8 ± 0.2% | 3.1 ± 1.5%* | 2.5 ± 0.6% | 3.0 ± 0.8% |
| MCP-1 | 6.3 ± 0.6% | 13.1 ± 3.0%* | 10.1 ± 1.6% | 13.1 ± 3.0%* |
| TNF-α | 4.8 ± 0.7% | 10.8 ± 3.0% | 9.4 ± 2.1% | 9.4 ± 3.0% |
| 1.5 μL min−1 | ||||
| IL-4 | 2.1 ± 0.7% | 5.7 ± 3.2% | 4.9 ± 0.5% | 3.4 ± 2.1% |
| IL-6 | 0.8 ± 0.2% | 3.3 ± 1.8% | 1.8 ± 0.3% | 3.3 ± 1.9% |
| IL-7 | 0.9 ± 0.2% | 3.2 ± 1.7% | 2.5 ± 0.7% | 2.8 ± 0.5% |
| MCP-1 | 5.1 ± 1.4% | 12.4 ± 2.6%* | 10.3 ± 1.2% | 14.1 ± 3.9%* |
| TNF-α | 3.5 ± 1.5% | 6.1 ± 1.6% | 8.3 ± 2.4% | 8.3 ± 3.5% |
denotes a statistically significant difference between the control and the heparin-perfused probe. Significance determined via a one-way ANOVA followed by a Tukey post hoc test at the 95% confidence level. The data represent means ± S.D., n=3.
Although the reported binding constants between heparin and each individual cytokine are quite different (Table 1), the trends show that cytokines exhibiting the highest control recovery also gave the highest enhanced recovery. This suggests that binding affinities to heparin only account for a part of the enhancement. Other properties of cytokines including their membrane-protein interactions and kinetic interactions with the affinity agent may also be important factors influencing the extent of RR enhancement.
Table 1 shows the minimum heparin concentration that would be needed to achieve a 90% bound state for the cytokines if the system were able to reach equilibrium within the perfusion fluid and assuming a minimum of 1:1 binding stoichiometry. It should be noted that some chemokines do form complex oligomers upon binding with heparin/heparan sulfate; however, these interactions have not been studied for every single chemokine with heparin. Thus, the 1:1 binding stoichiometry is being used as an estimate for the approximate heparin concentration needed to achieve a 90% bound state. Except for MCP-1, the minimum heparin concentrations needed to reach a 90% bound state lie between 0.1 and 10 μM suggesting a maximum in recovery could be reached using these heparin concentrations. It is important to note that in the short time span (~25 s at 1.0 μL min−1 based on the known internal tubing dimensions provided by the manufacturer) that analyte diffuses into the probe lumen and is carried away by the perfusion fluid that achieving an equilibrium state between the external solution and the solution within the probe lumen is unlikely. This is well known for microdialysis sampling since equilibrium between the perfusion fluid and the external solution would be achieved for controls if the relative recovery values were nearly 100%, which is not the case in these studies. To determine if these heparin concentrations are sufficient to maximize the relative recovery, human MCP-1 was chosen as a model cytokine to further explore the effect of heparin concentration on recovery increases. This cytokine has been chosen because among the five cytokines it consistently had significant improvements in recovery with heparin at all three perfusion fluid flow rates.
3.5 Quantification of heparin/human MCP-1 binding
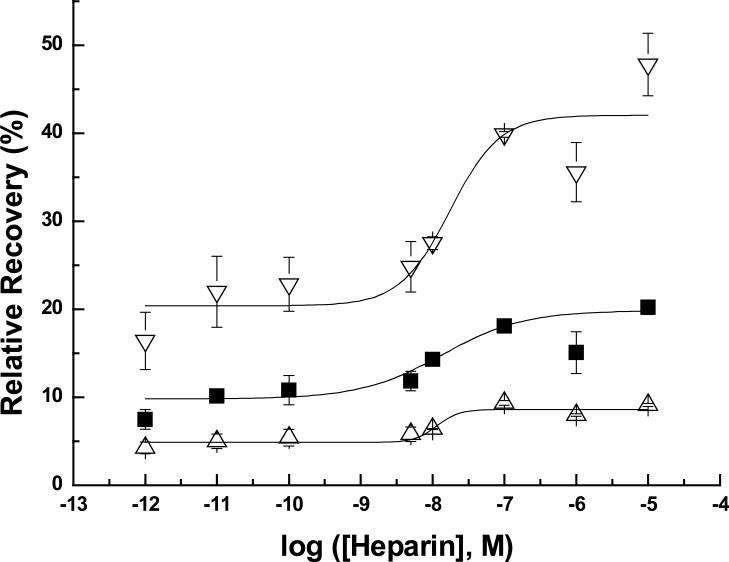
The RR values of human MCP-1 obtained using a large concentration range (1 pM to 10 μM) of heparin in the microdialysis sampling perfusion fluid are shown in Figure 1. Among the different flow rates, the enhanced RRs showed the same trend as with the multiple cytokines collection in that lower flow rates give higher recovery values. By including 10 μM, 1 μM and 0.1 μM heparin in the perfusion fluid, the heparin-perfused probe RR values are almost twice the control RR values which were 17.7±2.0%, 10.5±2.0%, 4.5±1.2%, at 0.5, 1.0, and 2.0 μL min−1, respectively (n=8). Heparin concentrations ranging from 0.1 μM to 100 pM exhibit reduced RR values compared to the higher concentrations. When the heparin concentration was in the range of 10 pM or 1 pM, there is no obvious enhancement in the RR values compared to the controls. The enhanced RRs are proportional to the heparin concentration, which suggests binding between MCP-1 and heparin. This data shows that the maximum recovery possible for MCP-1 is achieved using heparin concentrations in the micromolar range. The fitted sigmoidal curves to the data in Figure 1 show similar inflection points among the different flow rates. The inflection points on these curves are roughly 1.5 to 2 orders below the literature reported Kd value between human MCP-1 and heparin. Relative recovery for any analyte involves numerous factors in addition to the concentration of the affinity agent. Whether or not other cytokines also exhibit the same relative recovery trend between heparin concentrations, inflection points on the relative recovery curve, and reported Kd values requires further investigation.
Fig 1.
Increased RRs of human MCP-1 by using different concentrations of heparin (n=2; 2000 pg mL−1 for human MCP-1; ELISA DATA; Symbols and error bars denote the mean and range.) Symbols denote flow rates of : ▽ 0.5 μL min−1 , ■ 1.0 μL min−1, △ 2.0 μL min−1. The sigmoidal fit is to show the inflection points and not a particular model or fit.
3.6 Negative controls
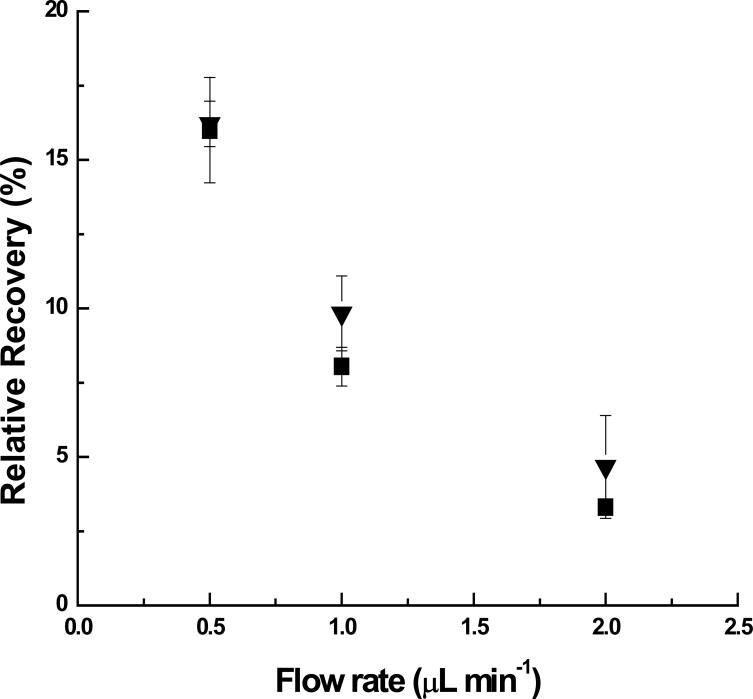
Negative controls were performed to ensure that prevention of non-specific binding was not leading to the improved recoveries obtained with heparin in the perfusion fluid. Dextran (Scheme 1.B.) at a concentration of 1 μM was chosen as one of the negative controls, because it has a similar polysaccharide structure when compared to heparin, but possesses no sulfate and carboxyl group substitution, (i.e., it is not negatively charged). By perfusing dextran through the dialysis probe at the same concentration as heparin, there was no alteration in recovery between dextran and controls as shown in Figure 2.
Fig 2.
Control RRs (▼) and dextran-perfused RR (■) by perfusing dextran as an affinity agent. (Human MCP-1: 2000 pg/mL; Dextran: 1 μM; n=2; ELISA DATA; Symbols and error bars denote Mean ±SD.)
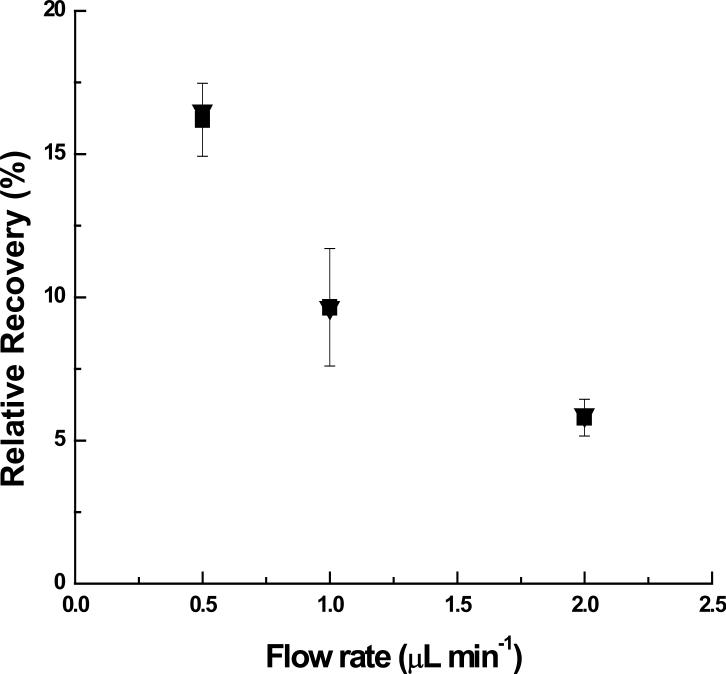
β-cyclodextrin sulfate (Scheme 1.C) was another perfusion-fluid additive used as a negative control. Compared with the heparin used, which has approximately 48 mole sulfate groups per 1 mole heparin, β-cyclodextrin sulfate has 18 mole sulfate groups per mole of cyclodextrin, but does not possess a linear polysaccharide structure. If interactions between the protein and the sulfate groups are mostly ionic rather than structural, then β-cyclodextrin sulfate should improve the recovery of the protein. Concentrations of up to 50 μM β-cyclodextrin sulfate included in the perfusion fluid did not improve the recovery of human MCP-1 as shown in Figure 3. The potential for interference of either dextran or β-cyclodextrin to the ELISA standard curve was also tested and no changes in the calibration curves were observed using these concentrations. The results from both of these negative controls point to the binding between heparin and MCP-1 as the source for improved microdialysis recovery.
Fig 3.
Control RRs (▼) and RRs by perfusing β-cyclodextrin sulfate (■)as an affinity agent (Human MCP-1: 2000 pg/mL; β-cyclodextrin sulfate: 50 μM; n=2; ELISA DATA; Symbols and error bars denote Mean ±SD.)
3.7. RR Variability between Luminex and ELISA assay
An interesting observation during the course of these studies was that different RR values were obtained with different proteins used in the perfusion fluid. Both cytokine detection methods gave MCP-1 RR values that were twice that for controls with heparin in the perfusion fluid. However, the absolute values of the control and enhanced RR between the Luminex assay and the ELISA are quite different. Table 3 shows the RR comparison between the Luminex and ELISA assay buffers that were passed through the dialysis probes. All the MCP-1 RR values obtained from samples measured with the ELISA were higher than those measured using the Luminex.
Table 3.
Control and Enhanced RRs of human MCP-1 in different measurement systems (Luminex and ELISA) by using 0.1 μM heparin.
| Luminex DATA(n=3) | ELISA DATA(n=3) | |
|---|---|---|
| Control RR 0.5 μL/min | 9.0±1.6% | 19.9±0.8%* |
| Enhanced RR 0.5 μL/min | 19.5±2.7% | 39.9±0.3%* |
| Control RR 1.0 μL/min | 6.0±2.5% | 10.5±0.6% |
| Enhanced RR 1.0 μL/min | 13.1±3.0% | 18.0±0.4% |
The Luminex and ELISA data are statistically different at the p < 0.05 level using a Student's t-test.
The Luminex system assay buffer contains 50 mM PBS with 25 mM EDTA, 0.08% sodium azide, 0.05% Tween-20 and 1% bovine serum albumin (BSA), pH 7.4. The ELISA kits contain assay diluent with 10 mM PBS and 10% fetal bovine serum (FBS), pH 7.0. When the Luminex assay buffer was used as the perfusion fluid for samples that were then analyzed using the ELISA assay, the control MCP-1 RR was 9.4±0.6% and with 0.1 μM the RR was 20.1±2.5%, n=3. These results are different at the p>0.05 level using a Student's t-test. These results suggest that for protein collection via microdialysis sampling, the perfusion buffer affects relative recovery through either analyte binding with different serum proteins or alterations in non-specific adsorption to device materials.
The level at which heparin would improve in vivo relative recovery of cytokines is difficult to predict a priori. Since cytokines are transient in nature and exact concentrations of cytokines within tissues are unknown, determining the microdialysis relative recovery of cytokines in vivo would not be trivial. Additionally, the level of recovery would be different from tissue to tissue depending on both how the cytokine is supplied to the tissue space as well as its mass transport properties including diffusion and potential binding to the extracellular matrix. Predicting approximate in vivo relative recovery values from in vitro relative recovery is not possible at this time for cytokines or many other analytes since all these unknown tissue properties affect the relative recovery [32].
For human in vivo studies, typically much longer microdialysis sampling membranes (20-30 mm) are used for human studies with lower perfusion fluid flow rates. Our past experience with cylcodextrins as affinity agents to improve hydrophobic compounds as well as this work has shown that lower flow rates will provide the greatest level of relative recovery improvement [17]. This occurs due to the flow dynamics and residence times that occur within the dialysis tubing during the sampling process.
4. Conclusions
The use of heparin in the perfusion fluid during in vitro microdialysis sampling increased the relative recovery of IL-6, IL-7, MCP-1 and TNF-α. For these cytokines and their ELISA detection kits, heparin did not interfere with the analysis.
5.0 Acknowledgements
Funding from NIH (EB001441) is gratefully acknowledged. Discussions with Professor Robert Linhardt are gratefully acknowledged. Dr. Jia Duo is acknowledged for helpful comments on the manuscript.
Abbreviations
- BSA
bovine serum albumin
- EDTA
ethylenedinitrilotetraacetic acid
- FBS
fetal bovine serum
- IL-4
interleukin-4
- IL-6
interleukin-6
- IL-7
interleukin-7
- MCP-1
monocyte chemoattractant protein-1
- MWCO
molecular weight cutoff
- PBS
phosphate buffered saline
- RR
relative recovery
- TNF-α
tumor necrosis factor-α
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LINCO has been purchased by Millipore.
6.0 References
- 1.Dinarello CA. Eur. J. Immunol. 2007;37:S34. doi: 10.1002/eji.200737772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meager A. The Molecular Biology of Cytokines. John Wiley & Sons; New York: 1998. [Google Scholar]
- 3.Dinarello CA. Chest. 2000;118:503. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- 4.Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Eur. J. Cancer. 2005;41:2213. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 5.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. Mol.Cell. Proteomics. 2005;4:1569. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Gouwy M, Struyf S, Proost P, Van Damme J. Cytokine Growth Factor Rev. 2005;16:561. doi: 10.1016/j.cytogfr.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Tripepi G, Mallamaci F, Zoccali C. J. Am. Soc. Nephrology. 2005;16:S83. doi: 10.1681/asn.2004110972. [DOI] [PubMed] [Google Scholar]
- 8.Plock N, Kloft C. Eur. J. Pharm. Sci. 2005;25:1. doi: 10.1016/j.ejps.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Clough GF. AAPS Journal. 2005;7:E686. doi: 10.1208/aapsj070369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schutte RJ, Oshodi SA, Reichert WM. Anal. Chem. 2004;76:6058. doi: 10.1021/ac0493626. [DOI] [PubMed] [Google Scholar]
- 11.Rosenbloom AJ, Ferris R, Sipe DM, Riddler SA, Connolly NC, Abe K, Whiteside TL. J. Immunol. Methods. 2006;309:55. doi: 10.1016/j.jim.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Ao X, Stenken JA. Methods. 2006;38:331. doi: 10.1016/j.ymeth.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Hutchinson PJ, O'Connell MT, Nortje J, Smith P, Al-Rawi PG, Gupta AK, Menon DK, Pickard JD. Physiol. Meas. 2005;26:423. doi: 10.1088/0967-3334/26/4/008. [DOI] [PubMed] [Google Scholar]
- 14.Clough GF, Jackson CL, Lee JJP, Jamal SC, Church MK. J. Invest Dermatol. 2007;127:2799. doi: 10.1038/sj.jid.5700930. [DOI] [PubMed] [Google Scholar]
- 15.Trickler WJ, Miller DW. J. Pharm. Sci. 2003;92:1419. doi: 10.1002/jps.10410. [DOI] [PubMed] [Google Scholar]
- 16.Helmy A, Carpenter KLH, Skepper JN, Kirkpatrick PJ, Pickard JD, Hutchinson PJ. J. Neurotrauma. 2009;26:549. doi: 10.1089/neu.2008.0719. [DOI] [PubMed] [Google Scholar]
- 17.Duo J, Fletcher H, Stenken JA. Biosens. Bioelectron. 2006;22:449. doi: 10.1016/j.bios.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Capila I, Linhardt RJ. Angew. Chem. Int. Ed. 2002;41:390. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 19.Gibbs RV. Adv. Exp. Med. Biol. 2003;535:125. doi: 10.1007/978-1-4615-0065-0_9. [DOI] [PubMed] [Google Scholar]
- 20.Handel TM, Johnson Z, Crown SE, Lau EK, Sweeney M, Proudfoot AE. Ann. Rev. Biochem. 2005;74:385. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 21.Rider CC. Biochem. Soc. Trans. 2006;34:458. doi: 10.1042/BST0340458. [DOI] [PubMed] [Google Scholar]
- 22.Nika K, Mulloy B, Carpenter B, Gibbs R. Eur. J. Phycology. 2003;38:257. [Google Scholar]
- 23.Powell AK, Yates EA, Fernig DG, Turnbull JE. Glycobiology. 2004;14:17R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 24.Lortat-Jacob H, Garrone P, Banchereau J, Grimaud JA. Cytokine. 1997;9:101. doi: 10.1006/cyto.1996.0142. [DOI] [PubMed] [Google Scholar]
- 25.Mummery RS, Rider CC. J. Immunol. 2000;165:5671. doi: 10.4049/jimmunol.165.10.5671. [DOI] [PubMed] [Google Scholar]
- 26.Clarke D, Katoh O, Gibbs RV, Griffiths SD, Gordon MY. Cytokine. 1995;7:325. doi: 10.1006/cyto.1995.0041. [DOI] [PubMed] [Google Scholar]
- 27.Chakravarty L, Rogers L, Quach T, Breckenridge S, Kolattukudy PE. J. Biol. Chem. 1998;273:29641. doi: 10.1074/jbc.273.45.29641. [DOI] [PubMed] [Google Scholar]
- 28.Yu H. Doctoral dissertation. University of Iowa; 2005. Studies of the interaction of heparin with proteins using improved sensor chips. [Google Scholar]
- 29.Vignali DAA. J. Immunol. Methods. 2000;243:243. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 30.Ander B, Karlsson A, Ohrlund A. J. Chromatogr.A. 2001;917:105. doi: 10.1016/s0021-9673(01)00661-6. [DOI] [PubMed] [Google Scholar]
- 31.Pavlov G, Finet S, Tatarenko K, Korneeva E, Ebel C. Eur. Biophys. J. 2003;32:437. doi: 10.1007/s00249-003-0316-9. [DOI] [PubMed] [Google Scholar]
- 32.Bungay PM, Morrison PF, Dedrick RL. Life Sciences. 1990;46:105. doi: 10.1016/0024-3205(90)90043-q. [DOI] [PubMed] [Google Scholar]