Abstract
The Drosophila syncytial embryo uses multiple astral mitotic spindles that are specialized for rapid mitosis. The homotetrameric kinesin-5, KLP61F contributes to various aspects of mitosis in this system, all of which are consistent with it exerting outward forces on spindle poles. In principle, kinesin-5 could accomplish this by (i) sliding microtubules (MTs), minus end leading, relative to a static spindle matrix or (ii) crosslinking and sliding apart adjacent pairs of antiparallel interpolar (ip) MTs. Here, I critically review data on the biochemistry of purified KLP61F, its localization and dynamic properties within spindles, and quantitative modeling of KLP61F function. While a matrix-based mechanism may operate in some systems, the work tends to support the latter “sliding filament” mechanism for KLP61F action in Drosophila embryo spindles.
Introduction
It is a pleasure for me to write an essay on mitosis for this volume, not only because of Bill Brinkley's well-known pioneering contributions to the field [Heald 2007], but also personally, because he chaired the session in which I gave my first talk on mitotic motors. I was a new postdoc attending a meeting on “Mitosis, molecules and mechanisms” in the summer of 1983 at University College, London organized by Jerry Hyams. My P.I., Dick McIntosh had sent me over to discuss our progress in using microtubules (MTs) as affinity matrices for identifying mitotic MAPs and motors (aka ATPases) from echinoderm embryo extracts. In our session, I spoke immediately after our respected competitor, Richard Vallee, who was then pursuing similar studies [e.g. see Collins and Vallee, 1986; Scholey et al., 1984,1985; Vallee and Bloom, 1983]. After my talk, Richard and I entered into a vigorous debate over a MAP which migrated on SDS gels with Mr = 80kDa in our lab but at only 77kDa in his lab! To everyone's relief, chair Brinkley (who was then at Houston) eased the tension of the rapidly escalating disagreement, in characteristic fashion, with a comment about the large size of Texas Maps. Fortunately, nowadays we have more interesting issues to debate, since our understanding of the biochemistry of mitosis has advanced significantly during the subsequent quarter century.
For example, it is now generally accepted that the mitotic spindle uses dynamic MTs plus multiple kinesin and dynein motors as force generators to assemble itself and to accurately segregate chromosomes. It is thought that, in many systems, the action of these force generators relies on centrosomes and/or chromosomes, but the extent to which they also depend on a “spindle matrix” is less well understood [Brust-Mascher and Scholey, 2007; Johansen and Johansen, 2007; Karsenti and Vernos, 2001; Mitchison, 2001; Scholey et al., 2001; Wadsworth and Khodjakov, 2004; Walczak and Heald, 2008; Wells, 2001]. In this review, I present my lab's current perspective on whether the key mitotic motor, kinesin-5 in Drosophila embryo mitotic spindles, can perform its function solely by crosslinking and sliding adjacent MTs, or if its function must be augmented by a spindle matrix.
Kinesin-5 [Amos, 2008; Civelekoglu-Scholey and Scholey, 2007; Hildebrandt and Hoyt, 2000; Kashina et al., 1997; Valentine et al., 2006] was first identified using genetic screens in Aspergillus nidulans where, as in many organisms, its function is required for spindle bipolarity and loss of its function produces monoastral spindles that fail to complete mitosis [Enos and Morris, 1990]. Motility assays of a protein subfragment expressed in bacterial extracts from the cloned Xenopus gene revealed that kinesin-5 moves towards the plus ends of MTs at approximately 0.1 μm/s [Sawin et al., 1992]. The native kinesin-5 holoenzyme was independently purified from Drosophila embryo extracts in a pankinesin peptide antibody screen as a slow, plus-end directed, bipolar homotetrameric complex capable of crosslinking adjacent MTs [Cole et al., 1994; Kashina et al., 1996a]. Finally, pioneering motility assays showed that Xenopus kinesin-5 is capable of simultaneously moving along both MTs that it crosslinks [Kapitein et al., 2005]. Together, these results suggest that kinesin-5 motors could act via a “sliding filament mechanism” to crosslink adjacent MTs, organizing parallel MTs into bundles and sliding apart antiparallel MTs to push apart spindle poles [Sharp et al., 1999a]. For example, the sliding apart of antiparallel ipMTs by kinesin-5 motors could drive poleward flux and the elongation of astral anaphase B spindles [Brust-Mascher et al., 2004], as well as the poleward flux of chromosome-nucleated MTs in anastral spindles, where the pole-directed MT ends are focused at the poles [Burbank et al., 2007].
While many aspects of kinesin-5 function can be explained by such a simple sliding filament mechanism, other plausible mechanisms have also been proposed. Many workers have hypothesized that the mitotic spindle contains a non-MT spindle matrix that could augment the activity of motors such as kinesin-5. Molecular candidates for this matrix include the complex of skeletor/megator/chromator, nuclear lamins, actin/titin and poly-(ADP ribose) [Chang et al., 2004; Fabian et al., 2007; Johansen and Johansen, 2007; Qi et al., 2004; Scholey et al., 2001; Tsai et al., 2006]. It is clearly plausible that such a matrix could act as a stable substrate, somewhat akin to a glass coverslip in a motility assay, against which kinesin-5 motors slide MTs with their minus-ends leading to exert outward forces on spindle poles [Kapoor and Mitchison, 2001; Sawin et al., 1992]. Furthermore, a new study proposes the intriguing hypothesis that kinesin-5 motors can control chromosome congression by mediating the length-dependent disassembly of kinetochore fibers [Gardner, 2008].
Here, I summarize our lab's studies on kinesin-5 in Drosophila embryos, where this motor has multiple mitotic functions, all of which are consistent with it exerting outward forces on spindle poles [Brust-Mascher et al., 2009; Sharp et al., 2000, 1999b]. Functional perturbation studies suggest that kinesin-5 contributes to prometaphase spindle maintenance, metaphase spindle length control, poleward flux, tight chromosome congression, the rate of anaphase chromatid-to-pole motility, and anaphase B spindle elongation,. As discussed below, in our opinion these functions are all consistent with the simple MT-MT sliding filament hypothesis, but there exist areas of uncertainty which leave open the possibility of alternative mechanisms, and which require further scrutiny.
Biochemistry of KLP61F
Kinesin-5 was identified in Drosophila using independent genetic (KLP61F; [Heck et al., 1993]) and biochemical (KRP130; [Cole et al., 1994]) approaches. cDNA sequence analysis predicted a tripartite, 1066 amino acid residue polypeptide chain, with N-terminal motor domains linked by a stalk to a C-terminal tail [Heck et al., 1993]. It was initially hypothesized that KLP61F and KRP130 are distinct kinesin-5 motors, possibly analogous to yeast Kip1p and Cin8p [Barton et al., 1995], but this idea was refuted by peptide mapping, mass spectroscopy and sequence analysis of purified KRP130, which showed that KRP130 and KLP61F are one and the same [Kashina et al., 1996b]. Subsequent examination of the Drosophila genome sequence supports the idea that this organism contains only a single member of the kinesin-5 family, now referred to as KLP61F [Goldstein and Gunawardena, 2000; Kashina et al., 1996b].
Like the Xenopus kinesin-5, Eg5, purified KLP61F is a slow (0.04 μm/s), plus-end directed motor that potentially could slide spindle MTs relative to a stationary matrix with their minus ends leading, thereby moving spindle poles attached to the MT minus ends outward at the rates similar to those observed during fruitfly embryo mitosis [Brust-Mascher et al., 2004; Cole et al., 1994; Sawin et al., 1992; Tao et al., 2006]. However, our thinking has been strongly influenced by observations that native KLP61F purified from embryonic cytosol [Cole et al., 1994; Kashina et al., 1996a, 1996b] and recombinant KLP61F purified from baculovirus infected sf9 cells [Tao et al., 2006; van den Wildenberg, et al. 2008] behave as homotetrameric complexes in solution, with 4 motor polypeptides plausibly organized as a bipolar “minifilament” with pairs of motor domains at opposite ends of a 4-strand coiled-coil rod [Kashina et al., 1996a]. Residues 631–920 within the C-terminal region of the stalk are thought to direct “tetramerization” [Tao et al., 2006].
The hypothesis that this bipolar homotetrameric ultrastructure allows kinesin-5 motors to crosslink and slide adjacent MTs in relation to one another was supported using elegant motility assays of Eg5 [Kapitein et al., 2005] and subsequently confirmed using KLP61F itself [van den Wildenberg et al., 2008] (Fig. 1A, B). In these assays, KLP61F moves non-aligned MTs along immobilized MTs at half the rate of aligned MTs, as expected [van den Wildenberg et al., 2008]. However, the aligned MTs moved more slowly than expected based on gliding assays (i.e. at 0.03 rather than 2 × 0.04 = 0.08 μm/s) which requires explanation. While the different buffers used for MT gliding assays (20mM Tris, 75mM KCl, 2mM MgCl2, 2mM DTT, pH8.0) and MT-MT sliding (80mM PIPES, 1mM EGTA, 2mM MgCl2, pH 6.8) could contribute to the difference, this possibility has not yet been directly tested.
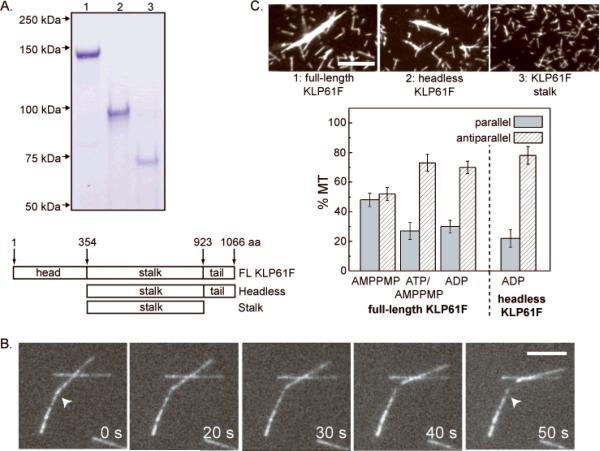
Fig. 1. Biochemistry of KLP61F.
(A) SDS gels and corresponding maps of full length (FL) KLP61F, motorless (headless) KLP61F and the KLP61F stalk purified using the baculovirus/sf9 cell system. (B) MT-MT sliding driven by purified FL-KLP61F (Fig. 1A, lane 1) in motility assays. (C) MT bundling assays (upper panels) and MT-MT orientation preference displayed by purified KLP61F and its subfragments (collaboration between Li Tao from my laboratory and Lukas Kapitein and Siet Van den Wildenberg from the Peterman/Schmidt laboratories, as detailed in van den Wildenberg et al. [2008]).
Further work revealed that KLP61F's MT crosslinking and sliding activities may contribute to the organization of spindle MTs into specific polarity patterns [Mastronarde et al., 1993; McIntosh et al., 1979; Sharp et al., 1999a]. Purified KLP61F has robust MT bundling activity in physiological buffers containing MgATP [Tao et al., 2006]. Using polarity marked MTs, it was observed that KLP61F crosslinks pairs of MTs into both parallel and antiparallel orientations, but in the presence of nucleotides that support movement or diffusion of the motor along the MT polymer lattice, KLP61F displays a 3-fold higher preference for the antiparallel orientation [van den Wildenberg et al., 2008] (Fig. 1C). These results are consistent with the hypothesis that KLP61F can crosslink spindle MTs of both orientations throughout the spindle, but will accumulate at the spindle equator where antiparallel ipMTs overlap [Sharp et al., 1999a]. Thus we propose that KLP61F crosslinks parallel MTs into bundles within each half spindle and slides apart the antiparallel ipMTs with which it associates at the spindle midzone, thereby exerting outward forces on spindle poles [Sharp et al., 1999a; van den Wildenberg et al., 2008] .
Interestingly, the C-terminal tail domains of kinesin-5 appear to play key roles in its MT crosslinking and sliding activities (Fig. 1A, C) since, remarkably, a homotetrameric “motorless” subfragment of KLP61F crosslinks MTs into bundles and also displays the same preference for the antiparallel orientation as the full length motor [Tao et al., 2006; van den Wildenberg et al., 2008]. This tail domain contains the “bimC box” [Heck et al., 1993] whose phosphorylation by cyclin-dependent kinase (cdk) enhances MT binding by the kinesin-5, Eg5 [Cahu et al., 2008]. Such enhanced spindle MT binding by phospho-KLP61F could provide a simple explanation for its cdk-dependent targeting to metaphase spindles, which would be consistent with it binding solely to spindle MTs and functioning via a MT crosslinking and sliding mechanism [Cheerambathur et al., 2008; Sawin and Mitchison, 1995; Sharp et al., 1999a].
In our view, therefore, studies of purified KLP61F are consistent with this motor functioning mainly by crosslinking and sliding adjacent MTs, thereby driving an antiparallel ipMT “sliding filament” mechanism that could push apart spindle poles. However, further work on purified KLP61F, including the determination of an atomic structure for the homotetrameric complex, is needed before this sliding filament mechanism can be considered proven. If correct, it is easy to see how such a sliding filament mechanism could operate without the need for a spindle matrix, but even so, perhaps a matrix could be used to augment the sliding mechanism by somehow controlling the spatial organization of KLP61F motors in a manner somewhat analogous to the way M-line proteins may hold thick filaments in register within the muscle sarcomere.
Localization and Dynamics of KLP61F
Valuable information on the 3-dimensional arrangement of MTs throughout several mitotic spindles has been obtained using serial section EM and MT tracking methods [e.g. Mastronarde et al., 1993]. Although this has not been accomplished in the case of the Drosophila embryo spindle, we have determined that these spindles have a fairly well-defined organization, consisting of either 9 or 10, highly dynamic, interpolar MT bundles based on deconvolved light microscopic images (Fig. 2, top left panel) [Brust-Mascher et al., 2004; Cheerambathur et al., 2007], each containing approximately 30 ipMTs per half spindle based on 3D reconstructions from EM sections (Fig. 2 center) [Sharp et al., 1999a]. As expected, these ipMTs are parallel towards the pole and overlap in an antiparallel orientation at the equator (Fig. 2 lower left panel) and as discussed below, KLP61F localizes along the length of these ipMT bundles (Fig. 2 right) [Cheerambathur et al., 2008; Sharp et al., 1999a; van den Wildenberg et al., 2008].
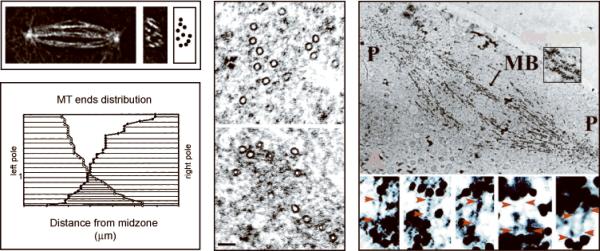
Fig. 2. Localization of KLP61F Crosslinkers.
Top left panel: Deconvolved fluorescence microscopy of an anaphase B spindle in longitudinal (left micrograph) and transverse (right micrograph) section. There are ~10 ipMT bundles per spindle (cartoon). Lower left and center panels: Organization of MTs within a single ipMT bundle from an anaphase B spindle (lower left) based on MT tracking from EMs (center). There are approximately 30 MTs emanating from each pole that overlap antiparallel. Right panel: ImmunoEM using gold anti-KLP61F labeling showing KLP61F all along an ipMT bundle (MB, upper micrograph) running from pole (P) to pole, where it may form electron dense MT-MT crosslinks (lower micrograph). See Brust-Mascher et al. [2004], Sharp et al. [1999a].
KLP61F co-localizes with tubulin within spindles of the syncytial embryo, based on immunofluorescence microscopy of fixed embryos [Barton et al.. 1995; Sharp et al.. 1999a] and time-lapse fluorescence microscopy of living transgenic embryos expressing functional KLP61F-GFP (Fig 3A) [Cheerambathur et al., 2008]. This spindle localization appears to depend on the presence of the cdk-phosphorylatable T933 within the bimC box, based on studies with phospho-specific peptide antibodies [Sharp et al., 1999a] and mutant KLP61F-GFP constructs [Cheerambathur et al., 2008]. Careful observations revealed that KLP61F-GFP localizes to half spindles, asters and along ipMT bundles throughout mitosis, except for a curious “gap”, possibly the Flemming body, that appears at the central midzone during telophase [Cheerambathur et al., 2008]. During metaphase, the motor appears concentrated around spindle poles, as previously reported for Eg5 in Xenopus extract metaphase spindles (though its localization in anaphase extracts is not known) [Sawin et al., 1992]. ImmunoEM and immunofluorescence of Drosophila embryo spindles reveal a striking localization of phospho-KLP61F all along ipMT bundles, with a relatively high concentration at the overlap zone. In these studies, gold-conjugated anti-phospho BimC box antibodies were observed to decorate sites spaced 60nm apart on opposite ends of fine filaments that plausibly represent bipolar KLP61F tetramers crosslinking adjacent ipMTs (Fig. 2 right) [Sharp et al., 1999a]. While this interpretation is consistent with the biochemical properties of KLP61F, these studies are performed at the limits of EM technology and confirmation would require the development of more reliable techniques for visualizing KLP61F molecules within spindles.
Fig. 3. Dynamics of KLP61F Crosslinkers.
(A) Comparison of localization of KLP61F-GFP and rhodamine-tubulin throughout mitosis in living Drosophila embryo spindles. (B) FRAP recovery curves of fluorescent KLP61F and tubulin in transgenic embryo spindles. (C) KLP61F-GFP speckle behavior in three distinct regions of an embryo mitotic spindle. (the boundaries of the 3 regions, left, center and right are marked on the micrograph in the inset of the left panel) . The speckle dynamics at the equator (mean velocity = zero, i.e. motors stay put while moving on antiparallel MTs) is consistent with ensembles of dynamic KLP61F crosslinkers attaching transiently to antiparallel ipMT overlaps, moving approximately 100nm on the MTs they crosslink and thereby sliding them apart, and then detaching. See Cheerambathur et al. [2008].
The availability of transgenic lines expressing functional KLP61F-GFP allowed an examination of this motor's dynamic properties using FRAP (fluorescence recovery after photobleaching) and FSM (fluorescence speckle microscopy) (Fig. 3) [Cheerambathur et al., 2008]. In general, KLP61F is highly dynamic, displaying a turnover halftime in FRAP experiments of only about 5s, which is basically indistinguishable from that of tubulin (Fig. 3B). Moreover, using FSM we observe stationary KLP61F speckles that we interpret as representing KLP61F-GFP motors crosslinking adjacent MTs and moving along them for about 100nm at an equal and opposite rate to the MTs themselves (Fig 3C). For example, it is easy to imagine that the stationary speckles at the spindle equator in Fig 3C could be dynamic KLP61F MT crosslinkers caught in the act of sliding apart adjacent antiparallel ipMTs while remaining in place relative to the laboratory frame of reference [Cheerambathur et al., 2008]. Stationary fluorescent Eg5 speckles in extract spindles were initially interpreted as representing motors bound to a static spindle matrix [Kapoor and Mitchison. 2001], but there is a growing consensus that they also could simply represent kinesin-5 motors crosslinking and sliding adjacent MTs in the absence of any matrix [Cheerambathur et al., 2008; Uteng et al., 2008]. Further, the dynamic properties of KLP61F-GFP during Drosophila embryo mitosis conform to a simple reaction-diffusion model in which dynamic KLP61F crosslinkers partition only between MT bound and freely diffusing states (see below), but again, further work is required to rigorously exclude the possibility of a fraction binding to a (possibly dynamic) spindle matrix [Cheerambathur et al., 2008].
Modeling the Mitotic Functions of KLP61F
One of the functions that we propose for KLP61F in Drosophila embryo mitosis is to drive anaphase B spindle elongation (Fig. 4) [Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Cheerambathur et al., 2007]. Functional data in support of this hypothesis was obtained using microinjected antibodies that dissociate KLP61F to a variable, measurable extent from spindles within distinct regions of the syncytial embryo [Brust-Mascher et al., 2009]. In null mutants lacking the antagonistic kinesin-14 motor, Ncd, we encountered spindles partially depleted of KLP61F that retained sufficient KLP61F function to maintain the prometaphase spindle in a bipolar state, yet insufficient to drive anaphase B, and consequently these spindles failed to elongate (e.g. Fig. 2D of Brust-Mascher et al. [2009]). This is consistent with previous time lapse fluorescence microscopy analysis of budding yeast mutants depleted of kinesin-5 function, which also display defects in anaphase B spindle elongation [Straight et al., 1998].
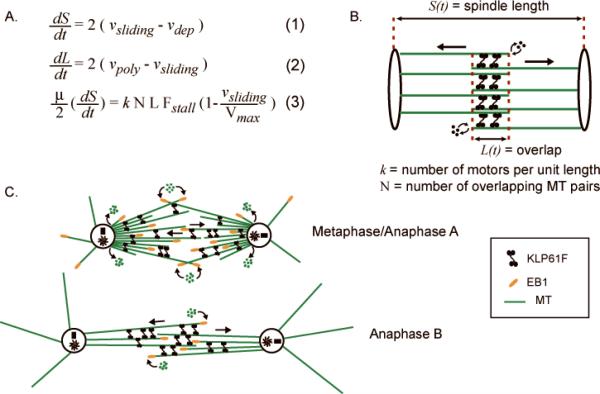
Fig. 4. Model for Anaphase B.
(A) Model equations describing the dynamics of an idealized spindle (B) in terms of : (1) the kinematics of spindle pole separation (S(t)) in which the rate of pole-pole separation (dS/dt) is related to the difference between the outward ipMT sliding rate and ipMT depolymerization at poles; (2) the kinematics of the overlap zone (L(t)) in which dL/dt is related to the net polymerization of ipMTs at the equator (a term that incorporates MT dynamic instability parameters) and the rate of sliding apart of ipMTs; and (3) a force-balance equation in which a mean number, k, of KLP61F motors with force-velocity relations as indicated, act on a number, N, of ipMT overlaps of length L(t) to push apart the spindle poles at a rate dS/dt against viscous drag, μ. A complex system of such equations was solved for realistic spindle geometry and showed that KLP61F can slide apart highly dynamic ipMTs to drive steady, linear anaphase spindle elongation. (C) Working hypothesis for the functions of KLP61F during metaphase and anaphase in Drosophila embryo mitosis. See Brust-Mascher et al. [2004]; Cheerambathur et al. [2008]; Cheerambathur et al. [2007].
Based on FSM studies, we hypothesized that KLP61F drives the persistent sliding apart of antiparallel ipMTs throughout metaphase and anaphase. During metaphase and anaphase A, this sliding is balanced by the depolymerization of ipMTs at spindle poles so that the spindle is maintained at a steady state length of approximately10μm. Then, at the onset of anaphase B, ipMT depolymerization at spindle poles is turned off, so that the KLP61F-driven ipMT sliding can now exert force on the poles to elongate the spindle at a steady, linear rate to 14μm [Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002] (Fig 4). In Drosophila embryos, the sequential destruction of cyclins A, B and B3 is required for progression through mitosis [Parry and O'Farrell, 2001] and using non-degradable constructs, we obtained evidence for a role for a cyclin B/cdk signaling cascade in turning off this ipMT depolymerization to initiate anaphase B spindle elongation [Cheerambathur et al., 2007]. In a subsequent study, a simple reaction-diffusion model resulted in an excellent fit to the dynamic turnover data of this motor obtained from transgenic flies expressing GFP-labeled KLP61F, and suggested that the motors partition into a MT-bound and freely diffusing fraction, with the MT-bound fraction forming the major portion[Cheerambathur et al., 2008]. Perhaps the rapid dynamics of the spindle MTs contributes to the high turnover rate of these motors in the spindle, which will be tested in future studies (looking at KLP61F turnover, by FRAP and by FSM, in taxol stabilized spindles). It will therefore be interesting to test by modeling if the rapid MT-dynamics has a role in maintaining a robust, rapid and linear rate of anaphase B by relieving possible road-blocks that could be created by these bipolar motors, if they were to remain on more stable tracks for prolonged periods of time.
The proposal that KLP61F drives anaphase spindle elongation via an antiparallel ipMT sliding filament mechanism was challenged by our finding that, during anaphase B, overlapping MTs at the spindle equator are highly dynamic, plausibly because they are stochastically switching between phases of growth and shrinkage in accordance with the principles of MT dynamic instability [Mitchison and Kirschner, 1984]. How could KLP61F drive the steady, linear elongation of the anaphase B spindle by a sliding filament mechanism if the tracks it slides apart are constantly growing and shrinking under its feet (aka motor domains)? Surely it is far easier to imagine that a motor held in place by attachment to a static spindle matrix could act on such dynamic MT tracks to drive the steady movement of spindle poles that is observed? While the idea of a sliding filament mechanism utilizing such highly dynamic MTs seems somewhat counter-intuitive, modeling suggests that it is highly plausible (Fig 4) [Brust-Mascher et al., 2004]. Briefly, we solved a system of force-balance and kinematic equations that describes the dynamics of the spindle poles in terms of underlying spindle geometry and biochemistry (Fig. 4A, B). The model results confirmed the plausibility of the idea that KLP61F can indeed act on highly dynamic, growing and shrinking antiparallel ipMT tracks, in the absence of a stabilizing matrix, to elongate the spindle at the steady linear rate observed in vivo [Brust-Mascher et al., 2004]. This model is very robust, with the rate of spindle pole separation being insensitive to changes of most model parameters. Significantly, the model predicts that the rate of spindle elongation is controlled only by the free sliding rate of KLP61F and the extent of suppression of ipMT depolymerization at the poles.
Conclusions
Our studies of the kinesin-5, KLP61F, in Drosophila embryos tend to support the hypothesis that this mitotic motor is a bipolar homotetramer, capable of crosslinking MTs throughout the spindle, thereby organizing parallel MTs into bundles and sliding apart antiparallel MTs (Fig. 4C). However, uncertainties remain. Definitive testing of this model would require the elucidation of the atomic structure of the KLP61F complex, and the time-resolved determination of nucleotide-dependent conformational changes in the motor as it functions within spindles. Such studies are being pioneered for myosin-2 function during striated muscle contraction [Huxley, 2004], but for mitotic spindles, this is obviously not feasible at present, even though structural studies are progressing. There also exist some inconsistencies in our current data that need resolving, including the basis for the aforementioned discrepancy in the rates of MT gliding versus aligned MT-MT sliding in KLP61F motility assays, and the functional significance of KLP61F's accumulation around the metaphase spindle poles, along with its depletion from the telophase spindle equator in localization studies. The precise function of the C-terminal tail domains in controlling KLP61F activity also merits further study.
Whether the activity of KLP61F and other kinesin-5 motors is augmented by a spindle matrix remains difficult to rule out, even though we find little evidence for this in Drosophila embryos. It has become clear that natural selection has designed a diversity of spindle types to accomplish chromosome segregation in different systems. For example, within a single organism, Drosophila, the centrosome-controlled syncytial embryo spindle appears streamlined for fast mitosis, whereas the cultured S2 cell spindle is much slower, less streamlined and relies to a lesser extent on centrosomes. Among the various types of spindle that exist throughout nature, it is easy to imagine that in some cases, but not others, the core machinery consisting of MTs and motors is supplemented with accessory cytoskeletal elements and polymers that serve as “matrices”. Thus, it remains unclear to what extent motors like kinesin-5 and MTs act alone and to what extent they recruit other subcellular components to augment their function in different spindles.
Acknowledgements
I thank the many members of my lab who contributed to our work on KLP61F over the years, especially Ingrid Brust-Mascher, Dave Carlson, Dhanya Cheerambathur, Gul Civelekoglu-Scholey, Doug Cole, Anna Kashina, Dave Sharp, and Li Tao. I also thank our numerous great collaborators from the labs of Bill(s) Saxton and Sullivan (Drosophila embryo biology), Erwin Peterman and Christoph Schmidt (KLP61F sliding/polarity preference assays), Henning Stahlberg and Kent McDonald (EM) and Alex Mogilner (modeling). Finally, I thank Tim Mitchison and the colleagues who participated in the ASCB “spindle matrix” subgroup session, December 2008. Our work on mitosis is generously supported by NIH grant GM55507.
References
- Amos L. Spindle Assembly: Kinesin-5 Is in Control. Curr Biol. 2008;18(24):R1146–R1149. doi: 10.1016/j.cub.2008.10.045. [DOI] [PubMed] [Google Scholar]
- Barton NR, Pereira AJ, Goldstein LS. Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol Biol Cell. 1995;6(11):1563–74. doi: 10.1091/mbc.6.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci U S A. 2004;101(45):15938–43. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Scholey JM. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol Biol Cell. 2002;13(11):3967–75. doi: 10.1091/mbc.02-05-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Scholey JM. Mitotic spindle dynamics in Drosophila. Int Rev Cytol. 2007;259:139–72. doi: 10.1016/S0074-7696(06)59004-7. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent Poleward Flux and Spindle Length Control in Drosophila Embryo Mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17(16):1373–83. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS ONE. 2008;3(12):e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432(7017):645–9. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Cheerambathur DK, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM. Dynamic partitioning of mitotic kinesin-5 crosslinkers between microtubule-bound and freely diffusing states. J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol. 2007;177(6):995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Scholey JM. Mitotic motors: kinesin-5 takes a brake. Curr Biol. 2007;17(14):R544–7. doi: 10.1016/j.cub.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269(37):22913–6. [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Vallee RB. A microtubule-activated ATPase from sea urchin eggs, distinct from cytoplasmic dynein and kinesin. Proc Natl Acad Sci U S A. 1986;83(13):4799–803. doi: 10.1073/pnas.83.13.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60(6):1019–27. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Fabian L, Xia X, Venkitaramani DV, Johansen KM, Johansen J, Andrew DJ, Forer A. Titin in insect spermatocyte spindle fibers associates with microtubules, actin, myosin and the matrix proteins skeletor, megator and chromator. J Cell Sci. 2007;120(Pt 13):2190–204. doi: 10.1242/jcs.03465. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Bouck D, Paliulis L, Meehl JB, O'Toole E, Haase J, Soubry A, Joglekar A, Winey M, Salmon ED, Bloom K, Odde DJ. Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS, Gunawardena S. Flying through the drosophila cytoskeletal genome. J Cell Biol. 2000;150(2):F63–8. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R. Brinkley-fest of mitosis. Dev Cell. 2007;13(2):168–76. doi: 10.1016/j.devcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123(3):665–79. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1496(1):99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Huxley HE. Fifty years of muscle and the sliding filament hypothesis. Eur J Biochem. 2004;271(8):1403–15. doi: 10.1111/j.1432-1033.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Cell and molecular biology of the spindle matrix. Int Rev Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–8. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mitchison TJ. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J Cell Biol. 2001;154(6):1125–33. doi: 10.1083/jcb.200106011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294(5542):543–7. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996a;379(6562):270–2. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Rogers GC, Scholey JM. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta. 1997;1357(3):257–71. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Scholey JM, Leszyk JD, Saxton WM. An essential bipolar mitotic motor. Nature. 1996b;384(6606):225. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123(6 Pt 1):1475–89. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, McDonald KL, Edwards MK, Ross BM. Three-dimensional structure of the central mitotic spindle of Diatoma vulgare. J Cell Biol. 1979;83(2 Pt 1):428–42. doi: 10.1083/jcb.83.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mitchison TaES. Mitosis: a history of division. Nat Cell Biol. 2001;3:E17–21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Parry DH, O'Farrell PH. The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr Biol. 2001;11(9):671–83. doi: 10.1016/s0960-9822(01)00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15(11):4854–65. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359(6395):540–3. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92(10):4289–93. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Neighbors B, McIntosh JR, Salmon ED. Isolation of microtubules and a dynein-like MgATPase from unfertilized sea urchin eggs. J Biol Chem. 1984;259(10):6516–25. [PubMed] [Google Scholar]
- Scholey JM, Porter ME, Grissom PM, McIntosh JR. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature. 1985;318(6045):483–6. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- Scholey JM, Rogers GC, Sharp DJ. Mitosis, microtubules, and the matrix. J Cell Biol. 2001;154(2):261–6. doi: 10.1083/jcb.200101097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11(1):241–53. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999a;144(1):125–38. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999b;1(1):51–4. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143(3):687–94. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16(23):2293–302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311(5769):1887–93. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Uteng M, Hentrich C, Miura K, Bieling P, Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J Cell Biol. 2008;182(4):715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Block SM. Eg5 steps it up! Cell Div. 2006;1:31–38. doi: 10.1186/1747-1028-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Bloom GS. Isolation of sea urchin egg microtubules with taxol and identification of mitotic spindle microtubule-associated proteins with monoclonal antibodies. Proc Natl Acad Sci U S A. 1983;80(20):6259–63. doi: 10.1073/pnas.80.20.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The Homotetrameric Kinesin-5 KLP61F Preferentially Crosslinks Microtubules into Antiparallel Orientations. Curr Biol. 2008;18(23):1860–4. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P, Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14(8):413–9. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–58. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Wells WA. Searching for a spindle matrix. J Cell Biol. 2001;154(6):1102–4. doi: 10.1083/jcb.200108139. [DOI] [PMC free article] [PubMed] [Google Scholar]