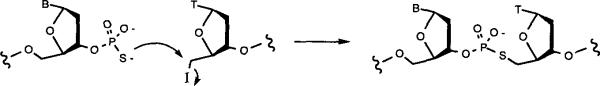Abstract
A new iodothymidine phosphoramidite enables the placement of a 5'-iodide into oligonucleotides; the iodide is stable to ammonia deprotection and allows nonenzymatic ligations of DNA.
Nonenzymatic approaches to the ligation of DNA strands have been the subject of study in several laboratories in recent years.1 Ligation is useful in convergent strategies for the synthesis of DNAs larger than can be assembled by synthesizer alone,1g as well as for incorporation of nonnatural residues into such DNAs. While enzymatic ligation of DNA has been important for the development of recombinant DNA methods,2 it can be expensive and relatively low-yielding as a preparative method.3 By comparison, nonenzymatic ligation strategies have the advantage of not requiring natural structure at the ligation site and, potentially, of proceeding in higher yields at lower cost.
Letsinger and coworkers have recently described an elegant and potentially very useful approach to the templated ligation of oligodeoxynucleotides.4 In this method, an oligonucleotide terminus substituted with a 5'-tosylate leaving group is reacted with a 3'-phosphorothioate group, giving SN2 displacement and resulting in natural DNA structure but with a sulfur replacing one of the bridging phosphodiester oxygen atoms. This method was used to ligate self-templated ends to yield dumbbell-type structures in good yields.4 However, due to the reactivity of the 5'-tosylate to ammonia, it was necessary to use labile protecting groups and rapid deprotection, and significant degradation was still observed for oligonucleotides carrying the reactive leaving group.5
In an ongoing program to construct larger linear and circular biologically active nucleic acids,1g we felt that the Letsinger approach was quite attractive, but we wished to avoid the lowered yields resulting from degradation as well as the requirement for specialized protecting groups and deprotection conditions. We now report that replacement of the tosyl leaving group with a 5'-iodo group on oligonucleotides gives good ligation yields (Scheme 1) and, because of the greater stability of the iodide, allows the use of standard deprotection methods.
Scheme 1.
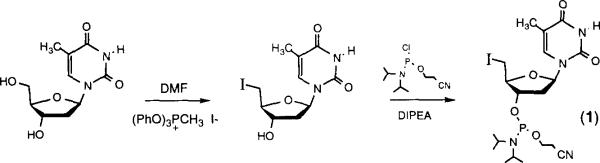
The iodophosphoramidite of thymidine (1) was synthesized in only two steps and in straightforward fashion (Scheme 2). Iodination of thymidine was performed as described,6 and subsequent phosphitylation proceeded normally to yield the iodide (1),7 suitable for automated DNA synthesis. Incorporation into oligonucleotides was carried out using the standard coupling cycle. Intact incorporation into DNA proceeded in ~85–95% yield, and was confirmed by HPLC analysis of oligonucleotide products, which have slower mobility on a reverse-phase column when iodinated, Analysis showed one major product (monitoring at 260nm) and only minor amounts of non-coupled (n-1)mer product, as confirmed by coinjection with authentic samples.8
Scheme 2.
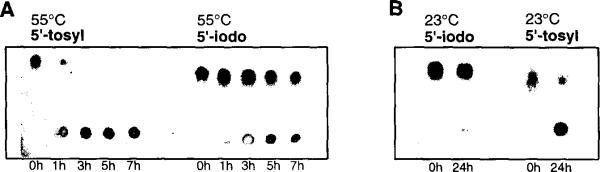
The stability of the 5'-iodothymidine in comparison to the 5'-tosylthyrnidine was analyzed by thin layer chromatography under varied conditions (Fig. 1). Results showed that the tosylnucleoside in concentrated ammonia (55°C) has a half-life of less than 1 h, whereas the iodonucleoside has a half-life of ca. 7 h. When treated at room temperature for 24 h (conc. NH3) the tosylnucleoside is >90% degraded, while the iodonucleoside is <2% degraded. The stability of the iodide in oligonucleotides was also analyzed by reverse-phase HPLC. Chromatograms revealed that the iodide (in the sequence 5'-I-TTCACGAGCCTG) has a half-life of >4 days in conc. NH3 at 23°C, similar to that of the nucleoside alone. Based on the HPLC analysis we chose the following conditions for deprotection: concentrated ammonia, 55°C for 1 h, followed by incubation at room temperature for 23 h, or treatment at room temperature alone for 24 h.9 It is anticipated that the iodide would also be stable to rapid deprotection conditions, although this was not explicity tested.
Figure 1.
Thin layer chromatograms comparing stability of 5'-iodothymidine and 5'-tosylthymidine under concentrated ammonia deprotection conditions. A. Timecourse of incubation at 55°C. B. Incubation at room temperature for one day.
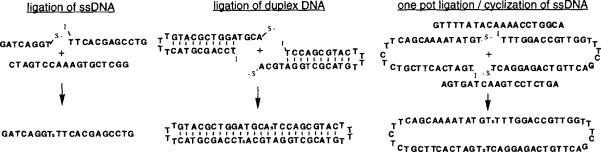
The ability of 5'-iodo-oligonucleotides to undergo template-directed ligations was also examined. Previous studies used the tosylnucleoside strategy to close self-templated dumbbell structures and to ligate a synthetic “cap” to close a hairpin structure.4,10 Our own goals involved the ligation of single-stranded oligonucleotides (using a complementary “splint”) to yield longer sequences, ligation of duplexes, and cyclization of oligonucleotides, also using a short splint sequence. We therefore examined three different ligation reactions on preparative scales: ligation of two short (8mer + 12mer) oligonucleotides using an 18mer splint, one-pot ligation of 30 + 33mers (20mer splint) followed by cyclization to yield a 63mer circular DNA, and dimerizationligation of self-complementary 28mer hairpin duplexes having 4-base overhanging “sticky” ends. The sequences tested, and expected products, are shown in Figure 2.
Figure 2.
Starting sequences and product sequences in the three types of ligations in this study.
Also required for ligations are 3'-phosphorothioate groups, which were incorporated as described.4 The standard conditions used for ligations were: 10mM MgC12, 10 mM Tris•acetate (pH 7.0), 23°C, and a DNA concentration of 50 μM for intermolecular reactions or 1.3 μM for intramolecular reactions. Splint concentrations were 1.1 times that of the DNAs being ligated. The results were analyzed by denaturing gel electrophoresis. A time course of the simple ligation showed that it proceeds over a period of 12–18 hr and reaches a plateau of ca. 90% ligation after ca. 18 hr (Fig. 3). The preparative reactions were carded out on 20–50 nmole scales over 18 hr using crude, unpurified starting materials and the products were isolated by preparative electrophoresis. Reaction conversions and isolated yields are shown in Table 1. In general, the ligation is found to proceed quite well, with apparent conversions (as judged by UV shadowing of the preparative gels) of ca. 45–95%, and isolated yields ranging from 20% for the combined two-step ligation and cyclization11,12 to 44% for the simple ligation. Although the same reactions were not compared, it appears that ligation rates may be somewhat slower for the less reactive iodiide than for a tosylate; however, both methods appear to give quite high ligation yields.
Figure 3.
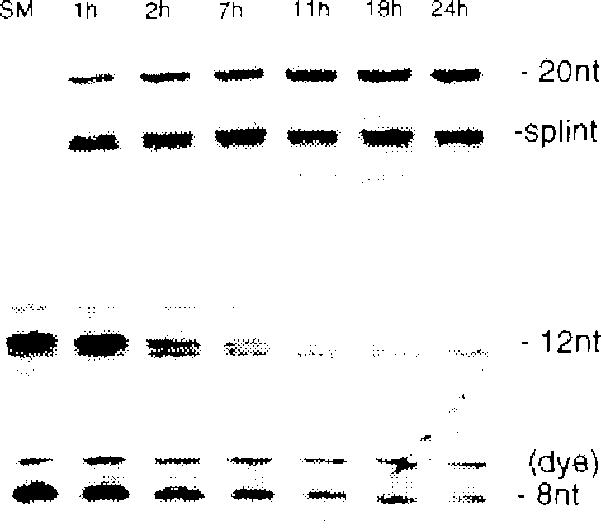
Time course of 8mer + 12mer ssDNA ligation followed by gel electrophoresis. Bands are visualized by staining.
Table I.
Preparative yields for the three reactions in Fig. 2.
| rxn. type | conversion | isolated yield |
|---|---|---|
| ssDNA ligation | >90% | 44% |
| duplex ligation | 75% | 36% |
| ligation/cyclization | >90% (1st step) 50% (2nd step) | 20% |
In summary, the results show that a 5'-iodide can be conveniently incorporated into DNA oligonucleotides in a thymidine derivative, and the reactive group undergoes little or no degradation under standard conditions of synthesis and deprotection. This makes possible several practically useful template-directed ligations, including the ligation of ssDNAs, cyclization of ssDNAs, and ligation of sticky-ended duplexes. These reactions proceed in good yield and without specialized protecting groups or deprotection conditions. The method further obviates the need for ligase enzyme, which is costly on a preparative scale.3
Acknowledgement
We thank the National Institutes of Health (GM46625) for support.
References and Notes
- 1.(a) Kanaya E, Yanagawa H. Biochemistry. 1986;25:7423. doi: 10.1021/bi00371a026. [DOI] [PubMed] [Google Scholar]; (b) Luebke KJ, Dervan PB. J. Am. Chem. Soc. 1989;111:8733. [Google Scholar]; (c) Dolinnaya NG, Sokolova NI, Asherbikova DT, Shabarova ZA. Nucleic Acids Res. 1991;19:3067. doi: 10.1093/nar/19.11.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kool ET. J. Am. Chem. Soc. 1991;113:6265. [Google Scholar]; (e) Ashley GW, Kushlan DM. Biochemistry. 1991;30:2927. doi: 10.1021/bi00225a028. [DOI] [PubMed] [Google Scholar]; (f) Gryaznov SM, Letsinger RL. Nucleic Acids Res. 1993;21:1403. doi: 10.1093/nar/21.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Rubin E, Rumney S, Kool ET. Nucleic Acids Res. 1995;23:3547. doi: 10.1093/nar/23.17.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Gryaznov SM. Nucleosides Nucleotides. 1995;14:1019. [Google Scholar]; (i) Li T, Weinstein D, Nicolaou K. Chem. Biol. 1997;4:209. doi: 10.1016/s1074-5521(97)90290-8. [DOI] [PubMed] [Google Scholar]
- 2.(a) Zoller MJ, Smith M. Nucleic Acids Res. 1982;10:6487. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hayashi K, Nakazawa M, Ishizaki Y, Hiraoka N, Obayashi A. Nucleic Acids Res. 1986;14:7617. doi: 10.1093/nar/14.19.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wu DY, Wallace RB. Gene. 1989;76:245. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- 3.Williamson R, Kool ET. manuscript in preparation.
- 4.Herrlein MK, Nelson JS, Letsinger RL. J. Am. Chem. Soc. 1995;117:10151. [Google Scholar]
- 5.It was found (ref. 4) that the 5'-tosyl-oligonucleotides reacted to a significant degree in ammonia, which lowers overall yields and necessitates the use of labile protecting groups, short deprotection times, and HPLC purification to remove degraded products.
- 6.Verheyden JPH, Moffatt JG. J. Org. Chem. 1970;35:2319. doi: 10.1021/jo00832a047. [DOI] [PubMed] [Google Scholar]
- 7.1H-NMR (δ,ppm, CDC13): 7.49(1H,d,J=8.0), 6.33(1H,t,J=7.0), 4.50-4.38(1H,m), 3.92-3.87(2H,m), 3.85-3.75(1H,m), 3.76-3.58(2H,m), 3.56-3.41(2H,m), 2.71-2.65(2H,m), 2.57-2.41(1H,m), 2.34-2.22(1H,m), 1.96(3H,s), 1.21(12H,d,J=7).
- 8.HPLC was carried out with an RP-C18 column (Alltech) using a gradient of 5->21% CH3CN in 100 mM aqueous triethylammonium acetate buffer (pH 7.0).
- 9.Herrlein MK, Letsinger RL. Angew. Chem. Int. Ed. Engl. 1997;36:599. [Google Scholar]
- 10.Deprotection for 24 h (rt) gives similar results and is sufficient for removal of standard protecting groups: Blackburn GM, Gait MJ. Nucleic Acids in Chemistry and Biology. IRL Press; Oxford: 1990. p. 119..
- 11.The one-pot two-step reaction was carried out using only the top splint (Fig. 2) as described in the text for the first step; after 18 hr (rt) the reaction was diluted with buffer to lower the strand concentration to 1.3μM, and the second splint was added. After another 18 hr the reaction was worked up and the products isolated.
- 12.The cyclic product was distinguished from undesired dimer by treatment with S 1 nuclease (Wang S, Kool ET. Nucleic Acids Res. 1995;23:1157. doi: 10.1093/nar/23.7.1157..)