SUMMARY
The entire functional even-skipped locus of Drosophila melanogaster is contained within a 16 kilobase region. As a transgene, this region is capable of rescuing even-skipped mutant flies to fertile adulthood. Detailed analysis of the 7.7 kb of regulatory DNA 3′ of the transcription unit revealed ten novel, independently regulated patterns. Most of these patterns are driven by non-overlapping regulatory elements, including ones for syncytial blastoderm stage stripes 1 and 5, while a single element specifies both stripes 4 and 6. Expression analysis in gap gene mutants showed that stripe 5 is restricted anteriorly by Krüppel and posteriorly by giant, the same repressors that regulate stripe 2. Consistent with the coregulation of stripes 4 and 6 by a single cis-element, both the anterior border of stripe 4 and the posterior border of stripe 6 are set by zygotic hunchback, and the region between the two stripes is ‘carved out’ by knirps. Thus the boundaries of stripes 4 and 6 are set through negative regulation by the same gap gene domains that regulate stripes 3 and 7 (Small, S., Blair, A. and Levine, M. (1996) Dev. Biol.175, 314–24), but at different concentrations. The 3′ region also contains a single element for neurogenic expression in ganglion mother cells 4-2a and 1-1a, and neurons derived from them (RP2, a/pCC), suggesting common regulators in these lineages. In contrast, separable elements were found for expression in EL neurons, U/CQ neurons and the mesoderm. The even-skipped3′ untranslated region is required to maintain late stage protein expression in RP2 and a/pCC neurons, and appears to affect protein levels rather than mRNA levels. Additionally, a strong pairing-sensitive repression element was localized to the 3′ end of the locus, but was not found to contribute to efficient functional rescue.
Keywords: even-skipped, CNS, Pattern formation, Segmentation, Translational control, Chromatin
INTRODUCTION
A primary pair-rule gene, even-skipped (eve) is expressed in a striped pattern in precellular embryos, as well as in several tissues at later stages of embryogenesis (Frasch et al., 1987; Macdonald et al., 1986). eve function is required in the early segmentation network to establish odd-numbered parasegment primordia, as well as to establish all of the parasegment boundaries (Macdonald et al., 1986). eve first appears as a single broad band in early nuclear division cycle 12, which then begins to split into stripes. By nuclear cycle 14, eve is expressed as a regular pattern of seven stripes (the early stripes) in the primordia of the odd-numbered parasegments. This pattern of broad early stripes then gives way during cycle 14 to narrow ‘late’ stripes with sharply demarcated anterior borders, where the segment polarity gene engrailed will be expressed (Ingham et al., 1988; Lawrence et al., 1987). eve is also required for engrailed expression in the anterior-most cell row of the even-numbered parasegments, where weak eve expression is observed (the minor stripes) at the same time as the late stripes. In addition, stripe 1 is required for cephalic furrow formation (Vincent et al., 1997) and eve function is required for proper germband extension (Irvine and Wieschaus, 1994).
Previous studies focused on cis-regulatory elements upstream of the eve-coding region. Individual elements were defined for early stripes 2 and 3, and a single element for all seven late stripes. Early stripe 7 can be driven by a region that includes either the stripe 2 or the stripe 3 element (Goto et al., 1989; Harding et al., 1989; Small et al., 1996).
Detailed analysis of eve expression in gap and pair-rule mutants established that gap genes regulate the early stripes directly, while pair-rule genes are required for the proper expression of late stripes (Frasch and Levine, 1987). Reporter transgenes driven by elements for stripes 2, 3 and 7 give the same response in gap gene mutants as the endogenous gene (Goto et al., 1989). The stripe 2 regulatory element requires both the Bicoid protein and the hunchback (hb) gap gene product for its activation, while the anterior and posterior borders are formed by the repressive action of giant (gt) and Krüppel (Kr), respectively (Small et al., 1991,1992; Stanojevic et al., 1989,1991; Wu et al., 1998). Stripes 3 and 7 are activated by ubiquitously distributed factors including D-STAT (Hou et al., 1996; Yan et al., 1996), and their borders are set through negative regulation by knirps (kni) and hb (Small et al., 1996; Stanojevic et al., 1989).
Expression of the eve late stripes is driven by a single upstream element. This ‘late element’ is regulated by the pair-rule genes paired (Fujioka et al., 1995,1996) and runt (Goto et al., 1989) as well as by early eve expression (Fujioka et al., 1995; Goto et al., 1989; Harding et al., 1989). The early, broad stripes of Eve protein act in a concentration-dependent manner to repress both the activator paired as well as repressors of late element expression. The repressors are sensitive to lower Eve concentrations, generating a narrow zone at the edge of each early stripe where a late stripe is activated (Fujioka et al., 1995). Early runt stripes overlap the posterior portion of early eve stripes and provide ‘polarity’ by preventing late expression there (Fujioka et al., 1995).
As germband extension proceeds, the seven late eve stripes begin to fade, while a new, 8th stripe appears in the posterior region (Frasch et al., 1987; Macdonald et al., 1986). The anterior border of this stripe corresponds with that of engrailed stripe 15 (Lawrence et al., 1987). While the germband is shortening, eve is expressed as a ring surrounding the anal plate (Frasch et al., 1987) and continues to be expressed there after shortening is complete. Posterior embryonic eve expression is apparently conserved through evolution. In the grasshopper, the eve homolog is expressed at the germband stage in a ring of tissue at the anal plate, as well as in patterns similar to those in Drosophila in identified neurons and in the dorsal mesoderm (Patel et al., 1992,1994). Additionally, eve homologs in Caenorhabditis elegans (Ahringer, 1996) and in zebrafish (Joly et al., 1993) were shown to function in the specification of posterior cell fates while, in the mouse, posteriorly biased expression is seen in the primitive streak and the tail bud (Bastian and Gruss, 1990; Dush and Martin, 1992).
Patterned eve expression is observed in the developing nervous system (Frasch et al., 1987; Patel et al., 1989). Ganglion mother cells (GMCs) 1-1a and 7-1a express eve at stage 10, and continue to do so while dividing to produce the aCC/pCC sibling neurons and the U/CQ/fpCC neurons, respectively (Bossing et al., 1996; Broadus et al., 1995). At early stage 11, expression is seen in GMC 4-2a. This GMC divides to produce the RP2 neuron, which continues to express eve, and the RP2 sibling, which extinguishes eve expression (Broadus et al., 1995). At late stage 12, expression occurs in a lateral cluster of neurons (EL cells; Patel et al., 1989) derived from neuroblast 3–3 (Schmidt et al., 1997). These cells maintain eve expression at high levels throughout embryogenesis. The CNS function of eve was analyzed in a subset of eve-expressing neurons using a temperature-sensitive eve allele (Doe et al., 1988). Removal of eve function during CNS development was found to change the axonal projections of the aCC and RP2 neurons. This led to the suggestion that eve controls the fates of these neurons, since their axons showed a preferred alternate morphology. From stage 11 onwards, eve is also expressed in a small subset of cells in the dorsal mesoderm, including some pericardial cells (Frasch et al., 1987) and the dorsal-most somatic muscle (Bodmer, 1993), DA1 (nomenclature of Bate, 1993).
Here, we show that a 15.6 kb genomic region is sufficient to rescue the lethality of eve null mutants, and we describe the localization of regulatory elements required for eve expression in several tissues. We show that previously uncharacterized early stripe elements are negatively regulated by the gap genes, with a separable elements for stripes 1 and 5 but a composite element for stripes 4 and 6. Stripes 4 and 6 are regulated by the same gap genes as stripes 3 and 7, but apparently at quite distinct concentrations. Neuronal elements are also separable for some lineages, but composite for others, suggesting an underlying commonality of upstream regulators. The 3′ untranslated region (UTR) is required for efficient late stage protein expression, while a chromatin control region at the edge of the locus did not apparently facilitate function.
MATERIALS AND METHODS
Drosophila strains
Injection procedures were as described previously (Rubin and Spradling, 1982) with some modifications (Fujioka et al., 1999). The alleles used for mutant analysis were hb14F, Kr2, gtX11, kni1, tllG, Df(2R)eve, eve3 (eveR13), eve1 (eveID19), runtLB5, hairyI22, prd4, fish87, mrl (stat92E), ftzKMQ, Df(2L)edsZ1(for slp) and odd[7L]
Construction of transgenes
All lacZ reporter constructs (unless noted) were made using a modified C3D vector (Fujioka et al., 1996) in which KpnI and Xba I sites were introduced downstream of the α-tubulin polyadenylation signal, providing cloning sites for the 3′ deletion fragments. All 3′ deletions were cloned into a modified pSP72 vector flanked 5′ by a KpnI site (also by NotI) and 3′ by an XbaI site, then transferred to the C3D vector. This places each fragment in an orientation and position relative to the promoter like that in the endogenous gene. In order to test the eve 3′ untranslated region (UTR) with the RP2+aCC/pCC element, a modified C3D vector was used in which NotI and XbaI sites were introduced upstream of lacZ, carrying either the α-tubulin 3′ UTR, or the eve 3′ UTR from +1306 (BstU1 site) to +1542 (KpnI site). The 5′ endpoint of the eve 3′ UTR was chosen based on a previous report (Kosman and Small, 1997). The RP2 element, from +7843 (EcoRI site) to +9235 (EcoRI site), was placed in opposite orientation relative to its normal 5′ to 3′ direction. Five to ten independent transgenic lines were analyzed for each lacZ construct.
For rescue experiments, the region +1.85 to either +8.4 or +9.2 kb was added downstream of the eve-coding region in the localized rescue construct E+L-eve (Fujioka et al., 1995), so that the contiguous region was restored exactly as in the endogenous gene, from −6.4 to either +8.4 or +9.2 kb. Many of the rescued lines initially showed a faint, difficult to detect eye color. To avoid this problem, the region upstream of the DraI site in the mini-white gene promoter was replaced with Glass activator (Ellis et al., 1993) binding sites (a kind gift of Bruce Hay), resulting in much stronger white expression in the eye, and no detected changes in eve expression in embryos. Details are available on request.
Embryo analysis
In situ hybridization to whole-mount embryos using digoxigenin (DGG)-labeled antisense mRNA probes was performed as described (Tautz and Pfeifle, 1989). This was followed by antibody staining with anti-Eve antibody (kindly provided by Manfred Frasch). Biotinylated secondary antibody was detected using streptavidin-conjugated horseradish peroxidase (Chemicon International Inc.), as described (Mullen and DiNardo, 1995). Hatching rates were determined 36 hours after collection by counting both hatched and unhatched egg casings. Unhatched embryos were collected and subjected to cuticle preparation without devitellinization (data in Table 2; to determine phenotype of rescued embryos, cuticles were prepared prior to hatching). Briefly, after dechorionation, embryos were washed three times with the PBS/0.1% Tween 20, transferred onto slides, dried by blotting, and a 1:1 mixture of Hoyer’s reagent and lactic acid was added. The embryos were cleared by incubation at 55°C. All other incubations were at room temperature.
Table 2.
Hatching rates and cuticle phenotypes of rescued eve embryos
| Mutant Background |
Hatching Rate (%) |
Cuticle phenotype (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | Severe def. | Mild def. | Undeveloped | wg | No. scored | ||||
| EVE84 | A | Df(eve) | 54.8(1229) | 1.3 | 0.1 | 0.1 | 23.2 | 20.5 | 644 |
| R13 | 62.5 (566) | 4.2 | 0.0 | 0.3 | 12.4 | 20.6 | 217 | ||
| Df(eve) P+ | 35.9(1305) | 9.8 | 5.6 | 5.8 | 42.9 | NA | 321 | ||
| C | Df(eve)/R13 | 59.4(2000) | 12.4 | 0.0 | 0.0 | 28.2 | NA | 128 | |
| R13 | 61.1(1413) | 12.2 | 2.6 | 0.0 | 24.1 | NA | 332 | ||
| Df(eve) P+ | 46.0(3639) | 1.5 | 0.0 | 22.8 | 29.7 | NA | 246 | ||
| D | Df(eve) | 58.0(1170) | 2.7 | 0.0 | 0.4 | 17.2 | 21.7 | 611 | |
| R13 | 61.0 (515) | 2.4 | 0.2 | 0.0 | 20.3 | 16.1 | 181 | ||
| Df(eve) P+ | 40.8 (719) | 10.1 | 16.8 | 6.2 | 26.1 | NA | 258 | ||
| EVEG84 | A | Df(eve) | 69.8 (927) | 0.0 | 0.0 | 0.0 | 10.2 | 20.0 | 454 |
| R13 | 64.6(1572) | 2.2 | 1.0 | 0.4 | 13.3 | 18.5 | 251 | ||
| Df(eve) P+ | 64.3(1119) | 1.5 | 5.3 | 2.2 | 26.7 | NA | 315 | ||
| B | Df(eve)* | 26.1 (712) | 22.3 | 7.3/1.3 | 4.2 | 38.8 | NA | 298 | |
| R13 | 64.2 (662) | 3.7 | 0.0 | 0.0 | 12.5 | 19.6 | 221 | ||
| R13 P+ | 47.8(1165) | 6.9 | 26.1 | 5.7 | 13.5 | NA | 174 | ||
| D | Df(eve) | 64.9(1012) | 1.6 | 0.9 | 0.0 | 8.5 | 24.1 | 190 | |
| R13 | 67.4(1238) | 1.9 | 0.0 | 0.0 | 7.8 | 22.9 | 337 | ||
| Df(eve) P+ | 59.1(1193) | 1.6 | 7.3 | 3.3 | 28.7 | NA | 376 | ||
| E | Df(eve) | 75.5 (961) | 0.2 | 0.1 | 0.1 | 6.3 | 17.8 | 289 | |
| R13 | 68.5 (954) | 0.6 | 0.3 | 0.1 | 3.4 | 27.1 | 111 | ||
| EVE92 | G | Df(eve) | 47.3(1540) | 17.4 | 4.1 | 20.0 | 11.2 | NA | 335 |
| R13 | 14.1 (895) | 4.9 | 0.0 | 2.7 | 78.4 | NA | 355 | ||
| H | Df(eve) | 45.6 (851) | 14.5 | 3.1 | 0.3 | 36.5 | NA | 192 | |
| R13 | 68.0 (872) | 12.0 | 0.3 | 0.8 | 18.9 | NA | 125 | ||
| Df(eve) P+ | 55.9(1168) | 16.4 | 7.1 | 0.0 | 20.6 | NA | 326 | ||
| R13 P+ | 48.0(2020) | 6.0 | 33.4 | 0.0 | 12.6 | NA | 190 | ||
Transgenic constructs and lines are as in Table 1, and are indicated in the first column. Either one or two copies of each rescue construct were crossed into the indicated eve mutant background. Crosses in which all of the progeny carried one copy of the rescue transgene are indicated by P+ following the eve genotype. All others carried two copies of the transgene, except EVEG84 B Df(eve)*, where both parents carried one copy of the transgene over a TM3 balancer (used because lines homozygous for the transgene had low fertility); therefore, some eve-deficient progeny did not receive a copy of the transgene, which may account for cuticles with a lawn phenotype (% shown after / in severe def. column). The numbers in parentheses under hatching rate indicate the total number of eggs assayed. The hatching rate was determined about 36 hours after the end of the collection, then cuticles were prepared from unhatched eggs. Phenotypes of rescued embryos were determined by preparing cuticles prior to hatching, and they were indistinguishable from wild type (not shown) for all homozygous lines except EVE92-G, which did not rescue efficiently. Severe def. embryos showed a denticle phenotype characteristic of strong to intermediate eve hypomorphs (pair-rule defects to somewhat less severe). Mild def. embryos resembled weak eve hypomorphs (missing 1 or 2 denticle bands or had partial deletions of denticle bands in a pair-rule pattern). The wg phenotype is due to the homozygous wg-lacZ marked balancer. NA under wg indicates the absence of this balancer in the cross. The total number of cuticles scored is shown in the far right column.
Sequencing of the eveR13-coding region
eveR13 stocks were obtained from two different laboratories. Embryo DNA (a mixture of wild-type and eveR13 chromosomes) were extracted as described (Jowett, 1986). PCR reactions were done using primers from the transcription initiation site (+1) and from +1540, downstream of the polyadenylation site. PCR-amplified DNA from at least four independent reactions with template DNA from each stock were sequenced. After discovery of a single mutation in the eveR13 reactions, relative to wild-type sequences (Ludwig and Kreitman, 1995), which eliminates a PvuII site at +488, these PCR products were also cloned into the pSP72 vector (which does not have a PvuII site), and PvuII digestion was used as a selection for eveR13 DNA. Two independent clones were sequenced in both directions, and the alteration at the PvuII site was present in both. Genomic DNAs from two eveR13 lines and a wild-type line were cut by XhoI and PvuII and subjected to Southern analysis, using a probe made from the same region (+7 to +1542). With wild-type DNA, fragments were detected of the expected sizes, 406, 75 and 1138 bp. With eveR13 DNA, which contains both wild-type and mutant chromosomes, an additional fragment of about 1213 bp was detected, consistent with the absence of the PvuII site and confirming the point mutation on the R13 chromosome.
RESULTS
Characteristics of stripe elements
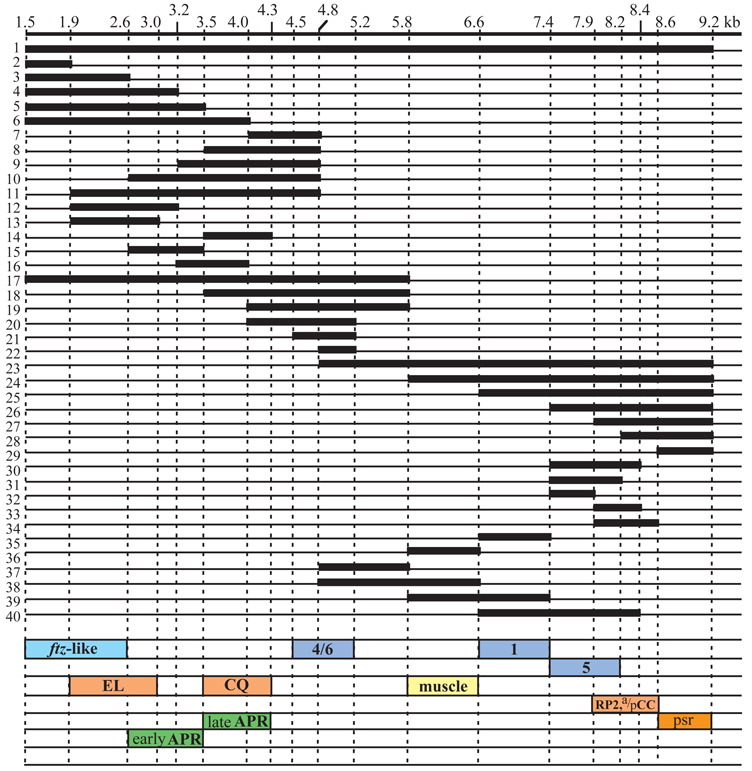
Previously, the regulatory elements for early stripes 1 and 5 were localized to between +6.6 and +8.4 kb, and for stripes 4 and 6 to between +4.8 and +6.6 kb (Sackerson et al., 1999). To test if each of the stripes could be driven individually by separable elements, deletions were made in each region in the context of a lacZ reporter (the deletions are summarized in Fig. 1). The results showed that there are separable elements for stripes 1 and 5. The stripe 1 element was localized to between +6.6 and +7.4 kb (Fig 1 #35, Fig 2A), and the stripe 5 element between +7.4 and +8.4 kb (Fig 1 #30, Fig 2B). The apparent extension of lacZ RNA expression into the posteriorly adjacent parasegment is due at least in part to the sensitivity of the assay. Endogenous protein expression can also be seen to extend into these regions, albeit at relatively low levels. Shortening the stripe 5 element to +7.9 kb (Fig. 1 #32) gave weaker but correctly localized expression, while shortening it to +8.2 kb (Fig. 1 #31) did not decrease the level of expression. Interestingly, both stripes 4 and 6 were driven by the region +4.5 to +5.2 kb (Fig. 1 #21), and also by the region +4.8 to +5.8 kb (Fig. 1 #37, this gave somewhat lower level expression than the former fragment), while the shared region of +4.8 to +5.2 kb (Fig. 1 #22) gave weak expression of both stripes, suggesting that these stripes share regulatory inputs.
Fig. 1.
A deletion analysis of the eve 3′ region. Numbers along the top show distance (in kb) to the transcription start site of the eve locus. Regions of the sequence included in each construct are indicated by thick lines, and deleted regions by thin lines. All fragments were placed downstream of an eve promoter-lacZ reporter in a pCaSpeR-based vector (see Materials and Methods). Five to ten independent transgenic lines were analyzed for each construct. Boxes at the bottom indicate minimal elements required to drive the following lacZ expression: ftz-like, 7 stripes in the even-numbered parasegments; EL, EL neurons in the central nervous system; CQ, GMC 7-1a and CQ neurons; early APR, early anal plate ring; late APR, late anal plate ring; 4/6, blastoderm stripes 4 and 6; muscle, mesodermal precursors; 1, blastoderm stripe 1; 5, blastoderm stripe 5; RP2,a/pCC, GMCs 4-2a and 1-1a, and neurons RP2, aCC and pCC. PSR indicates the region causing pairing-enhanced repression of the mini-white gene.
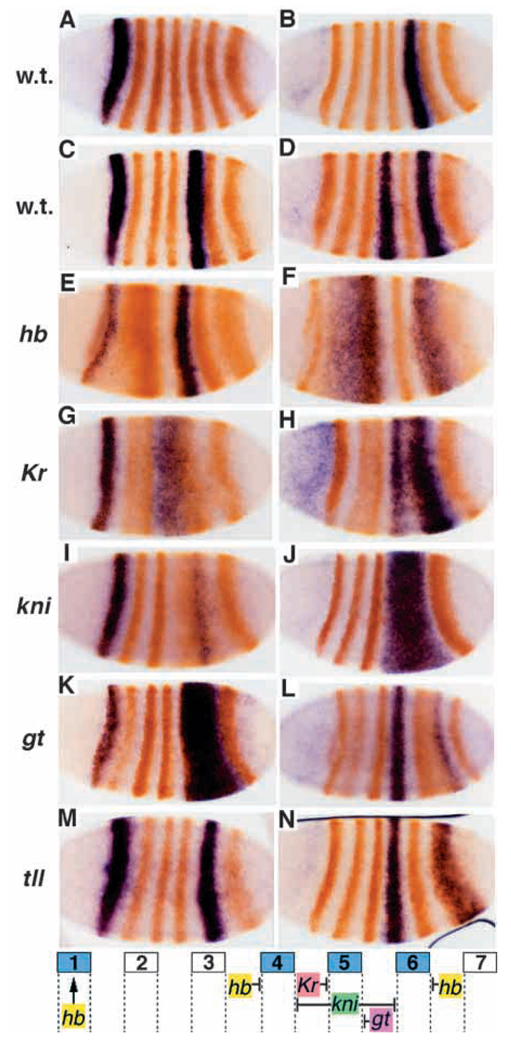
Fig. 2.
Elements for early stripes 1, 5 and 4+6 are negatively regulated by gap genes. In situ hybridization was performed for lacZ mRNA (blue), followed by staining with anti-Eve antibody (orange). (A) lacZ expression driven by the region +6.6 to +7.4 kb; (B) by the region +7.4 to +8.4 kb; (C) by the region +6.6 to +8.4 kb; (D) by the region +4.0 to 5.2 kb. The transgenic lines in C and D were crossed into several gap gene mutant backgrounds and stained as above. Staining patterns in these mutant embryos are shown below the corresponding wild-type pattern: hb (E,F), Kr (G,H), kni (I,J), gt (K,L) and tll (M,N). A summary of the apparent gap gene regulatory influences on stripes 1, 4, 5 and 6 is diagrammed at the bottom.
Genetic interactions with segmentation genes
In order to investigate the regulatory mechanisms responsible for establishment of these stripes, a lacZ reporter transgene carrying the elements for stripes 1 and 5 (+6.6 to +8.4 kb; Fig 1 #40, Fig 2C) and one carrying the stripe 4+6 element (+4.0 to +5.2 kb; Fig 1 #20, Fig 2D) were individually crossed into several gap and pair-rule mutant backgrounds.
In zygotic hb mutants, stripe 1 is weakened while stripe 5 is unaffected (or may be weakened slightly). Stripe 4 expands anteriorly and stripe 6 expands posteriorly, suggesting that hb sets the outside borders of expression of the stripe 4+6 element (Fig. 2E,F). The weakened expression may be explained by broadened Knirps expression (Hülskamp et al., 1990; Kraut and Levine, 1991b), since kni negatively regulates this element (see below).
In Kr mutant embryos, stripe 5 expands anteriorly and weakens (Fig. 2G), suggesting that its anterior border is defined by Kr. The weakening is consistent with an anterior shift of gt expression (Eldon and Pirrotta, 1991; Kraut and Levine, 1991a), which represses this element and appears to set the posterior border (see below). Alternatively, reduced expression of stripe 5 might be due to a change in the relative levels of activators (see below) and repressors within the stripe. Stripe 4 may also be weakened (Fig. 2H) and stripe 6 expands anteriorly. The latter effect is consistent with the reduction in kni expression that occurs in Kr mutants (Pankratz et al., 1989), since kni represses the 4+6 element (see below). The weakening of stripe 4 may be due to an anterior shift of the remaining kni expression. Alternatively, it may indicate that activators of the stripe 4+6 element are affected in Kr mutants (perhaps in common with those of stripe 5).
In kni mutant embryos, the 4+6 reporter is expressed in a single broad stripe covering the region between stripes 4 and 6, inclusive (Fig. 2J). Thus kni defines both the posterior border of stripe 4 and the anterior border of stripe 6. Stripe 5 is posteriorly shifted, overlapping with stripe 6, and weakened (Fig. 2I), indicating that kni also plays a role in regulating this stripe, possibly through its effects on the patterns of other genes, such as Kr (Gaul and Jäckle, 1987; see above and Discussion).
In gt mutant embryos, stripe 5 expands posteriorly (Fig. 2K). While there is no apparent effect on stripe 4, stripe 6 is weakened (Fig. 2K,L), consistent with the posterior expansion of kni expression (Eldon and Pirrotta, 1991), which sets its anterior border (above). Stripe 1 may also be weakened.
In tll mutant embryos, all stripes are posteriorly shifted, especially stripe 6 (Fig. 2M,N), and stripe 7 is missing (Frasch and Levine, 1987), consistent with the shift of other gap gene expression patterns previously described (Bronner and Jäckle, 1991; Kraut and Levine, 1991a; Pankratz et al., 1989). tll may help to set the posterior border of stripe 6, since stripe 6 expands posteriorly in the mutant. However, this may be due to the absence of the posterior hb domain (Bronner and Jäckle, 1991), since hb appears to define this border (above).
There is no strong effect on the boundaries of stripe 1 expression in any of these gap gene mutants.
We also tested the effects of pair-rule mutations on the early stripe elements (data not shown). In runt embryos, endogenous Eve stripe 5 expression is delayed and weakened (Frasch and Levine, 1987). While the expression of lacZ driven by the stripe 5 element is slightly delayed, it recovers later. In eve null mutants, stripe 1 expression is somewhat weakened. In ftz, prd, h, odd and slp, there are no apparent effects on any of the stripes.
Consistent with previous results suggesting that fish-hook may be a direct positive regulator of early eve stripes 4, 5 and 6 (Ma et al., 1998; Nambu and Nambu, 1996; Russell et al., 1996), lacZ expression in these stripes showed some reduction in the mutant (not shown). When the marelle gene (encoding D-STAT) was removed both maternally and zygotically, stripe 5 element expression disappeared, while stripe 1 was unaffected (data not shown), consistent with previous observations (Hou et al., 1996; Yan et al., 1996).
Neuronal elements
eve is expressed in ganglion mother cells (GMCs) and neurons in the central nervous system. Deletion analysis of the 3′ region showed that distinct regulatory elements exist for some aspects of this pattern (the constructs used are summarized in Fig. 1). A regulatory element for GMC 7-1a and the CQ neurons was localized to between +3.5 and +4.3 kb (Fig 1 #14, Fig 3A,B), which overlaps the regulatory element for anal plate ring expression (see below). An element was identified for expression in GMCs 4-2a and 1-1a, and later in their derivatives the RP2, aCC and pCC neurons, between +7.9 and +9.2 kb (Fig 1 #27, Fig 3C,D). Truncating the latter element at +8.4 (Fig. 1 #33) reduced the overall level of expression, while truncating at +8.6 (Fig. 1 #34) did not cause an obvious reduction. Changing the 5′ endpoint to +8.2 (Fig. 1 #28) resulted in a complete loss of activity.
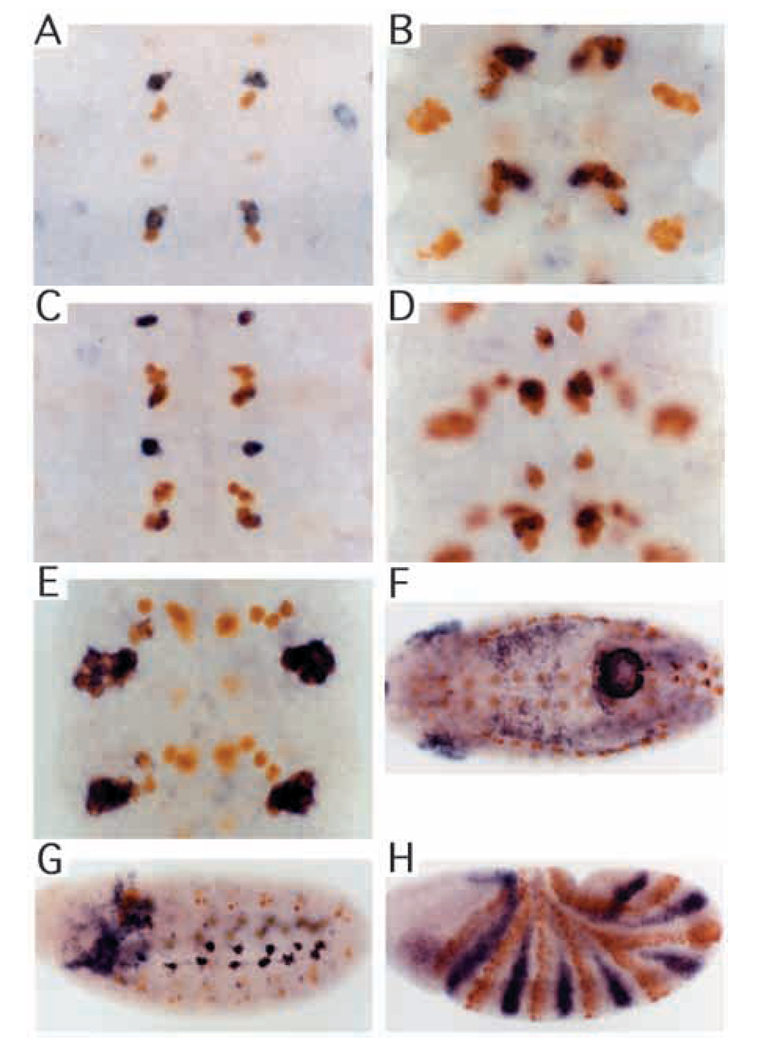
Fig. 3.
Isolated elements drive various components of tissue-specific expression. In situ hybridization was performed for lacZ mRNA (blue), followed by staining with anti-Eve antibody (orange). (A) lacZ expression in GMC 7-1a progeny at early stage 11 driven by the region + 3.5 to +4.3 kb. (B) lacZ expression in CQ neurons at stage 15 in the same transgenic line as in A. (C) lacZ expression in neurons RP2, aCC, and pCC at stage 11 driven by the region +7.9 to +9.2 kb. Note that at this stage lacZ expression is strong. (D) lacZ expression in RP2, aCC and pCC neurons at stage 15 in the same transgenic line as in C. Overall expression of lacZ at this stage is faint. (E) lacZ expression in EL neurons at stage 15 driven by the region +1.9 to +3.2 kb. (F) lacZ expression in the anal plate ring at stage 12 driven by the region +2.6 to +4.8 kb. (G) lacZ expression in muscle precursor cells at stage 11 driven by the fragment +5.8 to +6.6 kb. (H) lacZ expression in the anterior portion of even-numbered parasegments driven by the region +1.5 to +2.6 kb.
Either of the regions +1.5 to +3.2 kb (Fig 1 #4, Fig 3E) or +1.9 to +4.8 (Fig. 1, #11) could drive lacZ expression in EL cells, while neither the fragment +1.5 to +2.6 (Fig. 1 #3) nor +2.6 to +4.8 (Fig. 1 #10) could do so. This suggests a requirement for sequences both upstream and downstream of +2.6 kb. In fact, the region of overlap between the active regions, +1.9 to +3.2 (Fig. 1 #12) or to +3.0 (Fig. 1 #13), which still overlaps the early anal plate regulatory element (see below), was sufficient to drive lacZ expression in EL cells.
Stage-specific post-transcriptional regulation
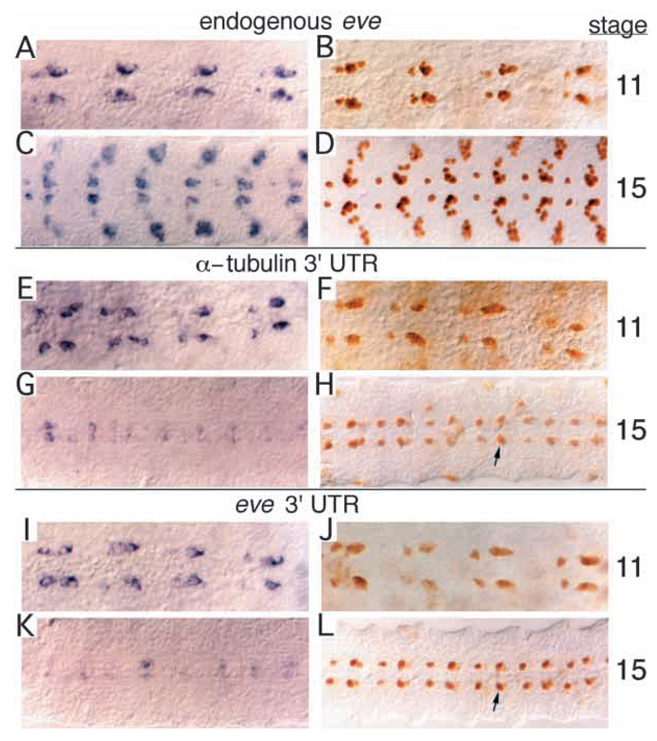
Endogenous Eve expression normally persists strongly in the developing CNS through stage 15 (Fig. 4A–D). The same was true for rescue transgenes (see below) containing the region −6.4 to +8.4 kb. In contrast, when the RP2+a/pCC element was used to drive lacZ, although all transgenic lines showed quite high level expression at stage 11, the level of staining faded by stage 15 (Fig. 3C,D). Since the eve 3′ untranslated region (UTR) was not contained in the lacZ transgenes, we reasoned that it might be required for high level expression in these neurons, particularly after stage 11. To test this possibility, we constructed a vector containing the eve 3′ UTR (see Materials and Methods) in place of the α-tubulin 3′ UTR of the original vector. In this construct, and in controls with the original 3′ UTR, the element was placed upstream of lacZ, in order to reduce ectopic expression due to position effects. The relative levels of mRNA and protein were compared at late stage 11 and at stage 15 in multiple lines for each construct. Endogenous eve mRNA is expressed strongly at stage 11 as a 6-cell cluster (consisting of aCC, pCC, the two CQs and two of unknown origin; Broadus et al., 1995) and slightly more weakly in the RP2 neuron (Fig. 4A), while the apparent level drops by stage 15 in all cells, especially in RP2 (Fig. 4C). Eve protein is expressed strongly through stage 15 (Fig. 4B,D), with no apparent reduction at the later stage. Staining for lacZ mRNA in transgenic lines without the eve 3′ UTR appeared similar to that of the endogenous mRNA at stage 11, marking the aCC/pCC and RP2 neurons strongly (Fig. 4E). Like eve mRNA, the level dropped by stage 15 (Fig. 4G), perhaps more severely than did the eve mRNA, particularly in the aCC/pCC cluster. Likewise, lacZ protein at stage 11 showed strong expression in all of the cells (Fig. 4F). However, unlike endogenous Eve, expression at stage 15 was quite severely reduced relative to the earlier stage (Fig. 4H).
Fig. 4.
The eve 3′ UTR is required for high level protein expression in RP2 and aCC/pCC neurons at later stages. (A-D) Wild type; (E-H) transgenic line carrying lacZ construct driven by the +7.9 to + 9.2 kb fragment, with the α-tubulin 3′ UTR (See Materials and Methods); diffuse ectopic expression due to a position effect is visible. (I-L) Transgenic line carrying lacZ construct driven by the same +7.9 to + 9.2 kb fragment, but with the eve 3′ UTR; again, there is diffuse ectopic expression, particularly at stage 11. (A,C,E,G,I,K) mRNA expression visualized by in situ hybridization with appropriate probe; (B,D,F,H,J,L) protein expression visualized by staining with appropriate antibodies. (A,C) eve mRNA; (B,D) Eve protein; (E,G,I,K) lacZ mRNA; (F,H,J,L) β-galactosidase protein. Expression in RP2, aCC and pCC neurons, at stage 11 (E,F,I,J); at stage 15 (G,H,K,L). Arrows, a representative neuron in a representative line, showing increased protein staining at stage 15 with the eve 3′ UTR (in L) relative to that with the α-tubulin 3′ UTR (in H).
With the eve 3′ UTR, the transgene showed fairly strong mRNA expression at stage 11 (albeit perhaps weaker than with the α-tubulin 3′ UTR), and again the level had dropped by stage 15 (Fig. 4I,K). The protein level at stage 11, like that of the mRNA, appeared to be slightly lower than without the eve 3′ UTR (Fig. 4J, compared to F). However, in contrast to transgenes with the α-tubulin 3′ UTR, with the eve 3′ UTR the level of lacZ protein staining at stage 15 increased relative to stage 11 (Fig. 4L, compared to J), resulting in protein expression that was clearly higher than without the eve 3′ UTR (Fig. 4L, compared to H). This was true for multiple lines with each construct, with the staining performed in parallel with complete sets of embryos, as shown in Fig. 4. Although there is some variation from one neuromere to another, the strong trend in all neuromeres, as well as in all lines examined, is that protein levels are consistently higher at stage 15 with the eve 3′ UTR than without it. This maintenance of high protein levels at stage 15 despite an apparent decrease in mRNA levels (relative to stage 11) is similar to the situation with endogenous Eve, indicating that there is a mechanism to maintain the level of protein that requires the eve 3′ UTR. Since the eve 3′ UTR causes either no change or a slight decrease in mRNA levels at stage 15, relative to the α-tubulin 3′ UTR, while protein levels are quite strongly increased, the mechanism would seem to involve translational control.
Localization of other elements
Expression in the anal plate ring is driven by sequences between +2.6 and +4.8 kb (Fig 1 #10, Fig 3F). Detailed analysis revealed complicated regulation in this region. Posterior expression starts as the germband becomes fully elongated as an ‘8th stripe’ driven by the eve late element (Goto et al., 1989). This expression fades at stage 10 (when eve-positive GMCs appear) but, at early stage 11, expression from the +2.6 to +4.8 element begins. This expression may overlap the posterior extent of the earlier 8th stripe expression, or may lie just posterior to it (data not shown). This element can be partially separated into two overlapping elements, one for early anal plate ring (early APR) expression through germband shortening, and another for expression after dorsal closure is completed (late APR). Late without early APR expression can be driven by the region from +3.5 to +4.3 (Fig. 1 #14), a region also sufficient to drive CQ neuronal expression (described above). Early APR expression can be driven by the region +2.6 to +3.5 (Fig. 1 #15), but this also drives weak late APR expression, indicating some redundancy in its specification. The region +3.2 to +4.0 (Fig. 1 #16) drives, in addition to late APR expression, weak early APR expression. The weakness of the latter indicates that something upstream of +3.2 is required for the proper level of early APR expression.
Mesodermal cells that include pericardial and other muscle precursors express eve. The element for this expression was found to lie between +5.8 and +6.6 kb (Fig 1 #36, Fig 2G), which does not overlap other elements. However, at the time of dorsal closure, expression from this element becomes weaker, unlike that of endogenous eve (see Discussion).
The fragment from +1.5 to +3.2 kb (Fig. 1 #4) drove strong lacZ expression in the ftz domain in stage 7–9 embryos, where eve is not normally expressed strongly. In the endogenous eve gene, the activity of this element may be suppressed by the upstream late element (Sackerson et al., 1999). The ftz stripe element can be shortened to between +1.5 and +2.6 (Fig 1 #3, Fig 2I), while the region +1.5 to +1.9 (Fig. 1 #2) drives very weak ftz domain expression. This shorter fragment also gives short-lived and faint lacZ expression in GMC 1-1a at stage 10.
The region +7.9 to +9.2 kb contains a strong pairing-sensitive repression (PSR) element. When this fragment was present upstream of the promoter, 50–70% of homozygous viable lines showed weaker mini-white expression in homozygotes than in heterozygotes, as reflected in the eye color (data not shown; normally, homozygotes show stronger expression due to the increased copy number). A similar effect was seen even when the element was placed 3′ of the lacZ-coding region, 3.8 kb downstream of the promoter, or when it was present in its normal position downstream of the promoter, in the EVE92 rescue construct. The fragment was dissected into two parts, +7.9 to +8.6 (Fig. 1 #34) and +8.6 to +9.2 kb (Fig. 1 #29). The former fragment, when placed upstream of the mini-white promoter, showed PSR in one out of 12 lines (8%), while the latter (Fig. 1 #29) caused PSR in 5 out of 7 lines (71%). Most, if not all, of the PSR activity is therefore separable from the minimal element for RP2+aCC/pCC expression.
Complete rescue of eve function
Since all eve regulatory elements seem to be localized between −6.4 and +9.2 kb, we tested whether this region, which is flanked by DNaseI hypersensitive sites (Sackerson et al., 1999), can rescue the lethality of eve mutants. We tested both the region −6.4 to +9.2 (EVE92) and the region −6.4 to +8.4 (EVE84, Table 1). We used the latter endpoint because it also gave a complete pattern of lacZ reporter expression (above), albeit with possibly reduced levels in the nervous system (in aCC/pCC and RP2 neurons). Transgenic lines with insertions on the 3rd chromosome were crossed into either Df(2R)eve [Df(eve)] or eveR13 (R13) mutant backgrounds, generating flies of the genotypes Df(eve)/CyO, P[wg-lacZ (or hb-lacZ)]; P[EVE]/P[EVE] and b, R13/CyO, P[wg-lacZ (or hb-lacZ)]; P[EVE]/P[EVE]. R13 is an apparent null mutation that truncates the protein within the homeodomain (see below). One transgenic insertion on the 2nd chromosome was recombined onto these eve mutant chromosomes. Adults from the R13 lines were scored for both a wild-type wing phenotype (non-CyO) and the black (b) phenotype (indicating R13 homozygotes). Df(eve) lines were stained with anti-Eve antibody to determine the pattern and level of Eve expression. In addition, cuticles from both lines were analyzed to determine the overall degree of rescue (see Materials and Methods).
Table 1.
The region −6.4 to +8.4 kb can rescue the lethality of eve mutants
| % rescued (no. scored) |
||||
|---|---|---|---|---|
| R13/R13 | R13/Df(eve) | ID19/Df(eve) | ||
| EVE92 | G | 0.7 (136) | 2.0 (525) | ND |
| H | 21.4 (341) | 23.1 (981) | ND | |
| EVE84 | A | 29.0 (376) | 32.2 (421) | 28.1 (466) |
| C | 15.8 (183) | 21.3 (1058) | 22.9 (280) | |
| D | 17.2 (174) | 34.2 (372) | 33.1 (320) | |
| EVEG84 | A | 28.1 (473) | 35.6 (988) | 18.5 (417) |
| B | 28.8 (163) | 31.1 (283) | ND | |
| D | 32.3 (572) | 34.9 (1224) | 26.9 (579) | |
| E | 19.3 (351) | 25.8 (1161) | ND | |
Two copies of each rescue construct were crossed into the eve mutant background indicated at the top. EVE92 carries the −6.4 to +9.2 kb fragment, EVE84 has a 3′ endpoint of +8.4 kb instead and EVEG84 is the same as EVE84, except that it has Glass activator binding sites upstream of the mini-white gene, which strongly enhance the eye color of transgenic flies. Each letter designation in the second column indicates an independent insertion. All were homozygous viable, and all were on chromosome III except EVEG84-E, which was on chromosome II and was recombined onto each eve mutant chromosome prior to the experimental cross. Adult flies either R13/R13, R13/Df(eve) or ID19/Df(eve) were identified by their wild-type (non-Curly) wing phenotype. The percentages of adult flies showing this phenotype are shown. The total number of flies counted is shown in parentheses.
Df(eve) is a deficiency that includes at least three lethal complementation groups (O’Brien et al., 1994) and, therefore, as expected, was not rescued by eve transgenes. However, both the EVE92 and EVE84 transgenes rescued the lethality of R13 (Table 1). For EVE92, two lines were analyzed. One (EVE92-H) produced adults with both the wild-type wing and black phenotypes (always together) at 21.4% of the total adult population, while the other line (EVE92-G) gave these phenotypes at 0.7%. For EVE84, seven lines were analyzed, and all showed the rescued phenotype at frequencies of between 15% and 32%. The Eve expression pattern and the cuticle phenotype of each of the efficiently rescued lines were indistinguishable from wild type (data not shown, see Table 2 for details). Thus the rescued phenotype did not differ significantly between EVE84 and EVE92 based on cuticle and expression patterns. The line EVE92-G, which did not rescue efficiently, showed a weak hypomorphic eve cuticle phenotype (Nusslein-Volhard et al., 1985). Each of these transgenes (except EVE92-G) also rescued the heterozygous mutant combination Df(eve)/R13 to adulthood at a frequency of between 21% and 36%. When rescued R13/R13 flies were self-crossed, they showed poor fertility, while rescued Df(eve)/R13 showed normal fertility, indicating the presence of recessive mutations on the R13 chromosome that affect fertility. In many cases, rescue to adulthood required two copies of the transgene, for both R13/R13 and Df(eve)/R13. Cuticles of the single copy rescued lines showed a high frequency of defects characteristic of eve hypomorphic mutants (Table 2, P+).
Even though the transgenes rescued R13 lethality, they failed to rescue the eveID19 mutant (ID19) to adult viability. ID19 is a hypomorphic allele that has a single amino acid substitution in the homeodomain (Frasch et al., 1988). However, the heteroallelic combination ID19/Df(eve) was efficiently rescued (Table 1), indicating the existence of at least one additional recessive lethal mutation on the ID19 chromosome.
Many of the lines carrying rescue constructs showed very faint eye color from the mini-white gene (some lines required aging to identify transgenic flies). To avoid this problem, Glass activator binding sites were introduced upstream of the mini-white gene (see Methods). Lines with this construct (EVEG84) showed strong eye color without apparent effects on the efficiency of either transformation or rescue, in combination with either R13/R13, R13/Df(eve) or ID19/Df(eve).
eveR13 protein is truncated within the homeodomain
R13 shows a null cuticle phenotype and was found to lack detectable Eve protein expression (Frasch et al., 1988). eve RNA is present and we do detect very weak protein staining under some circumstances (data not shown). Since this mutant was used extensively in our analysis of eve regulatory function, we sequenced the R13-coding region (see Materials and Methods). A single alteration from the wild-type sequence was found: a C-to-T transition, which would create a termination codon in place of Gln 106. Fortuitously, this change also eliminates a PvuII restriction site, and Southern analysis showed that R13 DNA (from stocks of two different laboratories) was missing the PvuII site (not shown, see Materials and Methods). This mutation is consistent with the R13 null phenotype, as the resulting truncated protein would be missing the ‘recognition helix’ (helix 3) of the homeodomain required for DNA binding, as well as a transcriptional repression domain (Han and Manley, 1993). The very low level of antibody staining in mutant embryos suggests that this truncated protein is relatively unstable.
DISCUSSION
Concentration-dependent regulation of eve by gap genes
Recent studies showed that regulatory elements for all known aspects of eve expression are located within a 15.6 kb region, spanning −6.4 to +9.2 kb from the transcription start site (Sackerson et al., 1999). Based on the regions of overlap of larger transgenes, these studies suggested that early stripes 1 and 5 might be driven by the region from +4.8 to +8.4 kb, and stripes 4 and 6 by the +4.8 to +6.6 kb region. In order to determine whether these stripes were regulated independently, and to define minimal elements required for each of them, we further dissected these regions. Elements for stripes 1 and 5 proved to be separable, while a single element drives stripes 4 and 6 (see Fig. 1 for details). Extensive analysis has been done of how gap genes regulate early eve stripes 2, 3 and 7 (Goto et al., 1989; Harding et al., 1989; Small et al., 1991, 1992, 1996; Stanojevic et al., 1989, 1991). We now have an opportunity to compare their regulation with that of the remaining stripes. The growing knowledge of segmentation genes in other insects (Brown et al., 1997; Patel et al., 1992, 1994), when combined with these data, may provide an understanding of how pattern formation has evolved. As a first step in this direction, we examined how the stripe elements are regulated by gap genes. Consistent with a composite element driving stripes 4 and 6, both the anterior border of stripe 4 and the posterior border of stripe 6 are determined by zygotic hb expression. In addition, in a kni mutant, the isolated stripe 4+6 element drives expression throughout the interstripe region. The spatial and temporal expression patterns of zygotic hb and kni (Kraut and Levine, 1991b) are consistent with the products of these loci exerting direct repression on the element. Similarly, the anterior and posterior borders of expression of the stripe 5 element are set by Kr and gt, respectively. Again, these regulators are expressed in an appropriate pattern to direct repression of this stripe element. More subtle effects of these mutations and of tll on these elements are consistent with the above interpretation, when previously observed crossregulation among the gap genes is taken into account (see Results). Thus, as for stripes 2, 3 and 7, much of the spatial regulation of stripes 4, 5 and 6 appears to be due to repression by gap gene products. The sequences of these regulatory elements contain potential binding sites for the gap gene products that we suggest may directly regulate them (our unpublished observations). However, further analysis will be required to determine if these regulatory interactions are indeed direct.
The above observations concerning regulation of stripes 4 and 6 include a striking parallel with the regulation of stripes 3 and 7. The stripe 7 element is not separable from that of stripe 3, although full activation of stripe 7 requires sequences outside of the minimal stripe 3 element (Small et al., 1996). Like the 4+6 element, a combined stripe 3+7 element directs expression throughout the interstripe region in a kni mutant, and both the anterior and posterior borders (of stripes 3 and 7, respectively) are set by hb-dependent repression (Small et al., 1996). Thus, an intriguing situation exists in which the stripe 4+6 element is repressed by a higher concentration of Knirps protein than is the stripe 3+7 element and, at the same time, by a lower concentration of Hunchback protein. The differential sensitivity of these elements to repressor concentrations might be due to simple mechanisms, such as differential affinities of binding sites, or to more complex mechanisms, such as combinatorial interactions with different cofactors. Whatever the mechanism, this differential sensitivity is precise enough to allow three gap protein domains (those of Knirps and the anterior and posterior Hunchback domains), acting as repressor gradients, to regulate the positioning of eight distinct expression boundaries, thus helping to define four of the early stripes of eve expression.
In a similar vein, stripe 5 is negatively regulated by the same gap genes that regulate stripe 2. The Kr domain represses both the posterior border of stripe 2 and the anterior border of stripe 5, while the anterior and posterior domains of gt expression are involved in setting the anterior and posterior borders of stripes 2 and 5, respectively.
Previous computer modeling of the regulatory circuitry upstream of eve predicted specific negative regulatory interactions of gap genes on stripes 4 and 5 (Reinitz and Sharp, 1995). This model predicted that kni would be the primary regulator of both the posterior border of stripe 4 and the anterior border of stripe 5, and that Kr and gt would repress the anterior border of stripe 4 and the posterior border of stripe 5, respectively. Two of these proposals are consistent with our data, and two are not. Rather than Kr setting the anterior boundary of stripe 4 (at a high concentration), it sets that of stripe 5 (at lower concentration). Similarly, rather than setting the stripe 5 anterior boundary at high concentration, a lower concentration of kni sets the stripe 6 anterior boundary. Apparently, the independent behavior of the stripe elements allows one enhancer to essentially ‘ignore’ a high repressor concentration, while a different stripe enhancer responds to the same repressor at a lower concentration. Further analysis of specific binding sites within identified enhancers may support more detailed models of eve regulation.
Recently, several genes were reported to show stripe-specific effects on eve activation. lacZ expression driven in stripes 4, 5 and 6 by the eve 3′ region were weakened in a fish-hook mutant (fish) (Ma et al., 1998). It was also shown that the product of this gene can bind within this large regulatory region, as determined by gel mobility shift assays. While we observed that expression from the minimal elements also showed some reduction in fish embryos, expression from these elements is clearly activated by other proteins as well. In a marelle mutant (encoding D-STAT), lacZ expression from a stripe 3 element was seen to be weakened (Hou et al., 1996; Yan et al., 1996). We observed that D-STAT is a primary activator of stripe 5, since expression from the stripe 5 element was absent in this mutant. Consistent with a direct effect on this element, we find several consensus sequences for D-STAT binding within the +7.4 to +8.2 kb region (M. F. and J. B. J., unpublished observations).
None of the gap and pair-rule mutants that we tested had a strong effect on stripe 1 element expression (see Results). hb mutants weakened reporter gene expression, but not severely. In a buttonhead mutant (btd), endogenous eve expression in the stripe 1 region was seen to be reduced (Vincent et al., 1997). In beetles, as in Drosophila, eve forms stripes with anterior borders that coincide with parasegment boundaries but, rather than forming multiple stripes at once, stripe 1 is formed first, followed by sequential progression toward the posterior (Patel et al., 1994). Further analysis of the regulation of stripe 1 may reveal regulatory relationships that predated the divergence of Diptera. Recent analyses of eve stripe elements among Drosophila species (Fujioka et al., 1996; Ludwig and Kreitman, 1995; Ludwig et al., 1998; Sackerson, 1995) suggested that many of the regulatory mechanisms are evolutionarily conserved. The growing body of information from various species may soon support detailed hypotheses for how the regulatory mechanisms of segmentation evolved.
Nervous system regulation and eve 3′ UTR function
eve is expressed in the nervous system, initially in GMCs 1-1a, 4-2a and 7-1a, and later in the aCC/pCC, RP2, CQ and EL neurons. We were unable to separate elements for GMC 1-1a, its cellular progeny the aCC/pCC neurons, GMC 4-2a and its progeny neuron RP2. This was surprising, since these cells originate from different neuroblasts. A single element drives lacZ expression strongly in these neurons at least through stage 11. However, by stage 15, transgene expression is reduced, particularly at the protein level, when both endogenous Eve expression and expression from rescue constructs (in an eve− background) remain strong. The eve 3′ UTR, which the initial lacZ transgenes did not contain, appears to affect the efficiency of translation in these cells. Transgenic lines in which the standard 3′ UTR (from the α-tubulin gene) is replaced by that of eve, while they show reduced mRNA levels at stage 11 and similar levels at later stages, give lacZ protein levels that remain high through stage 15 in the RP2 and aCC/pCC neurons. The eve 3′ UTR was previously reported to confer a rapid turnover rate in early cycle 14 (Kosman and Small, 1997) of the blastoderm stage. Thus it appears that the eve 3′ UTR has functions in controlling protein levels in several tissues, at various stages, and probably through multiple mechanisms.
Elements for EL cells and for GMC 7-1a and its progeny CQ neurons were also localized. However, the CQ and EL elements overlap those for posterior region expression and for even-numbered parasegment expression, respectively (see Fig. 1 and below), suggesting that common activators may be utilized in these different tissues.
Expression in posterior regions and the mesoderm
An element for muscle precursor cell expression is separable from those of other tissues. However, its expression becomes weaker than that of endogenous Eve at stage 15, as observed for the RP2+aCC/pCC element. The eve 3′ UTR may provide for a high level of protein expression in this tissue, at a similar time to that in the nervous system.
Expression in the posterior region of the embryo is apparently a highly conserved feature of eve function, since it is shared by eve homologs in C. elegans, zebrafish and mice (Ahringer, 1996; Bastian and Gruss, 1990; Dush and Martin, 1992; Joly et al., 1993). While it was reported that posterior structures are not affected in eveID19 mutants at the non-permissive temperature (Sato and Denell, 1986), it remains a possibility that eve has some function in this region. eve homologs have been shown to have important functions in specifying posterior cell fates in C. elegans and zebrafish (Ahringer, 1996; Joly et al., 1993). The regulation of eve expression in the posterior region is complex. Initially, the late stripe element is responsible for expression in this region, which appears as an 8th stripe corresponding to parasegment 15 (Lawrence et al., 1987; Frasch et al., 1987; Macdonald et al., 1986). Later, expression is driven in a ring near the posterior end of the embryo by two separable elements, one active through germband retraction and the other after dorsal closure. The latter expression corresponds to the anal plate ring.
Just downstream of the eve-coding region (+1.5 to +2.6 kb) lies an element that, when assayed by itself, drives lacZ expression strongly in the even-numbered parasegments, where only very weak eve expression is normally observed. As suggested previously, the upstream late element may be responsible for long-range repression of these ‘ftz-like stripes’ (Sackerson et al., 1999) in the endogenous eve gene. The biological function of this element, if any, is unclear, although eve expression does extend into this region, where it is required to clear odd-skipped expression from the anterior ftz domain, allowing activation of engrailed (Fujioka et al., 1995). This element may serve a function in this context.
Rescue of the eve mutant phenotype
The regulatory DNA that we have characterized downstream of the transcription unit, in combination with upstream regions previously analyzed, is sufficient to functionally rescue eve null mutants. In most cases, a single copy of the rescue transgene was not sufficient for full rescue. Many mutant embryos exhibited a weak eve hypomorphic phenotype when they carried only one copy of the transgene, suggesting that transgene expression is below that of the endogenous gene when inserted at most chromosomal locations. This might indicate that the transgene is missing a general enhancer of early eve expression. Alternatively, sequences within the Pelement vector may repress eve expression at early stages. It is also possible that a chromosomal environment exists around the eve locus that is required for full activity which most insertion sites do not provide. The PSR element described below might participate in providing such an environment.
The genomic region downstream of the RP2+aCC/pCC element causes strong pairing-sensitive repression (PSR) of the mini-white gene. Similar PSR is observed when Polycomb-group gene responsive elements are introduced into the genome with mini-white. Recently, it was reported that a region from the engrailed gene that exhibits PSR is bound directly by the Drosophila YY1 homolog, Pho, encoded by the pleiohomeotic gene (Brown et al., 1998). Consensus sites for YY1/Pho binding, as well as for GAGA factor, which are also seen in the engrailed element, exist within this region (M. F., J. Kassis, and J. B. J., unpublished observation). Consistent with chromatin-based regulation of eve, a Polycomb-group protein, Polyhomeotic, was found to bind to polytene chromosomes in the region of the eve locus (DeCamillis et al., 1992), and eve expression in the NB4-2 lineage was seen to be affected by Polycomb-group activity (Weigmann and Lehner, 1995). Nonetheless, the function of the eve PSR element is unclear, since rescue transgenes that lack it do not show abnormal eve expression, and since including it did not appear to enhance either expression or rescue (see Results). It is possible that the PSR element is only required in the context of the eve locus, perhaps to prevent inappropriate activation of eve by enhancers from a neighboring gene, or of a neighboring gene by eve enhancers (see Sackerson et al., 1999). Other regions of the eve locus are also capable of repressing mini-white expression, since the −6.4 to +8.4 kb transgenes consistently gave transformants with very weak eye color. The utilization of Glass activator binding sites to enhance mini-white expression facilitated identification of transformants, but these also showed weak eye color relative to other Glass-mini-white transgenes. Although this repression was not consistently pairing sensitive, it may represent a function that is redundant with that of the PSR region in some aspect of eve regulation.
Acknowledgments
We thank Manfred Frasch for anti-Eve antibody, Bruce Hay for Glass activator binding sites, Douglas Coulter, J. Peter Gergen, Douglas A. Harrison, John Nambu, Yuri Sedkov, Stephen Small, and the Bloomington Stock Center for fly stocks. We thank James B. Skeath, Stephen Small, and John Reinitz for helpful comments on the manuscript. We also thank Hannes Alder in the Kimmel Cancer Institute for advice on sequencing and design of oligonucleotides, and Deepali Deka for technical assistance. This work was supported by NSF grant IBN9507406 to T. G. and J. B. J.
This work is dedicated to the memory of Tadaatsu Goto.
Footnotes
In memory of Tadaatsu Goto
REFERENCES
- Ahringer J. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev. 1996;10:1120–1130. doi: 10.1101/gad.10.9.1120. [DOI] [PubMed] [Google Scholar]
- Bastian H, Gruss P. A murine even-skipped homologue, Evx-1, is expressed during early embryogenesis and neurogenesis in a biphasic manner. EMBO J. 1990;9:1839–1852. doi: 10.1002/j.1460-2075.1990.tb08309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate M. The mesoderm and its derivatives. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 1013–1090. [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech. Dev. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Bronner G, Jäckle H. Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech. Dev. 1991;35:205–211. doi: 10.1016/0925-4773(91)90019-3. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb Group Gene pleiohomeotic Encodes a DNA Binding Protein With Homology to the Transcription Factor YY1. Molecular Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Parrish JK, Beeman RW, Denell RE. Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mech. Dev. 1997;61:165–173. doi: 10.1016/s0925-4773(96)00642-9. [DOI] [PubMed] [Google Scholar]
- DeCamillis M, Cheng NS, Pierre D, Brock HW. The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Smouse D, Goodman CS. Control of neuronal fate by the Drosophila segmentation gene even-skipped. Nature. 1988;333:376–378. doi: 10.1038/333376a0. [DOI] [PubMed] [Google Scholar]
- Dush MK, Martin GR. Analysis of mouse Evx genes: Evx-1 displays graded expression in the primitive streak. Dev. Biol. 1992;151:273–287. doi: 10.1016/0012-1606(92)90232-6. [DOI] [PubMed] [Google Scholar]
- Eldon ED, Pirrotta V. Interactions of the Drosophila gap gene giant with maternal and zygotic pattern-forming genes. Development. 1991;111:367–378. doi: 10.1242/dev.111.2.367. [DOI] [PubMed] [Google Scholar]
- Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987;1:981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Frasch M, Warrior R, Tugwood J, Levine M. Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev. 1988;2:1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Bejsovec A, Weir M. Production of Transgenic Drosophila. In: Tuan RS, Lo CW, editors. Developmental Biology Protocols. Totowa, New Jersey: Humana Press Inc.; 1999. [Google Scholar]
- Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Miskiewicz P, Raj L, Gulledge AA, Weir M, Goto T. Drosophila Paired regulates late even-skipped expression through a composite binding site for the paired domain and the homeodomain. Development. 1996;122:2697–2707. doi: 10.1242/dev.122.9.2697. [DOI] [PubMed] [Google Scholar]
- Gaul U, Jäckle H. How to fill a gap in the Drosophila embryo. Trends Genet. 1987;3:127–131. [Google Scholar]
- Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even-skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. The EMBO J. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Pfeifle C, Tautz D. A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Krüppel and knirps in the early Drosophila embryo. Nature. 1990;346:577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Baker NE, Martinez AA. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even-skipped. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. Cell intercalation during Drosophila germ band extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- Joly JS, Joly C, Schulte-Merker S, Boulekbache H, Condamine H. The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development. 1993;119:1261–1275. doi: 10.1242/dev.119.4.1261. [DOI] [PubMed] [Google Scholar]
- Jowett T. Preparation of nucleic acids. In: Roberts DB, editor. Drosophila A Practical Approach. Oxford, UK: IRL Press, Limited; 1986. pp. 275–286. [Google Scholar]
- Kosman D, Small S. Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development. 1997;124:1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- Kraut R, Levine M. Spatial regulation of the gap gene giant during Drosophila development. Development. 1991a;111:601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- Kraut R, Levine M. Mutually repressive interactions between the gap genes giant and Krüppel define middle body regions of the Drosophila embryo. Development. 1991b;111:611–621. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Macdonald P, Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skipped genes. Nature. 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Kreitman M. Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Molecular Biology & Evolution. 1995;12:1002–1011. doi: 10.1093/oxfordjournals.molbev.a040277. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- Ma Y, Niemitz EL, Nambu PA, Shan X, Sackerson C, Fujioka M, Goto T, Nambu JR. Gene regulatory functions of Drosophila Fish-hook, a high mobility group domain Sox protein. Mech. Dev. 1998;73:169–182. doi: 10.1016/s0925-4773(98)00050-1. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Mullen JR, DiNardo S. Establishing parasegments in Drosophila embryos: roles of the odd-skipped and naked genes. Dev. Biol. 1995;169:295–308. doi: 10.1006/dbio.1995.1145. [DOI] [PubMed] [Google Scholar]
- Nambu PA, Nambu JR. The Drosophila fish-hook gene encodes a HMG domain protein essential for segmentation and CNS development. Development. 1996;122:3467–3475. doi: 10.1242/dev.122.11.3467. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Kluding H, Jurgens G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harbor Symposia On Quantitative Biology. 1985;50:145–154. doi: 10.1101/sqb.1985.050.01.020. [DOI] [PubMed] [Google Scholar]
- O’Brien MA, Roberts MS, Taghert PH. A genetic and molecular analysis of the 46C chromosomal region surrounding the FMRFamide neuropeptide gene in Drosophila melanogaster. Genetics. 1994;137:121–137. doi: 10.1093/genetics/137.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz MJ, Hoch M, Seifert E, Jäckle H. Krüppel requirement for knirps enhancement reflects overlapping gap gene activities in the Drosophila embryo. Nature. 1989;341:337–340. doi: 10.1038/341337a0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Ball EE, Goodman CS. Changing role of even-skipped during the evolution of insect pattern formation. Nature. 1992;357:339–342. doi: 10.1038/357339a0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Condron BG, Zinn K. Pair-rule expression patterns of even-skipped are found in both short-and long-germ beetles. Nature. 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Schafer B, Goodman CS, Holmgren R. The role of segment polarity genes during Drosophila neurogenesis. Genes Dev. 1989;3:890–904. doi: 10.1101/gad.3.6.890. [DOI] [PubMed] [Google Scholar]
- Reinitz J, Sharp DH. Mechanism of eve stripe formation. Mech. Dev. 1995;49:133–158. doi: 10.1016/0925-4773(94)00310-j. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Russell SR, Sanchez-Soriano N, Wright CR, Ashburner M. The Dichaete gene of Drosophila melanogaster encodes a SOX-domain protein required for embryonic segmentation. Development. 1996;122:3669–3676. doi: 10.1242/dev.122.11.3669. [DOI] [PubMed] [Google Scholar]
- Sackerson C. Patterns of conservation and divergence at the even-skipped locus of Drosophila. Mech. Dev. 1995;51:199–215. doi: 10.1016/0925-4773(95)00365-7. [DOI] [PubMed] [Google Scholar]
- Sackerson CM, Fujioka M, Goto T. The even-skipped locus is contained in a 16 kb chromatin domain. Dev. Biol. 1999 doi: 10.1006/dbio.1999.9301. in press. [DOI] [PubMed] [Google Scholar]
- Sato T, Denell RE. Segmental identity of caudal cuticular features of Drosophila melanogaster larvae and its control by the bithorax complex. Dev. Biol. 1986;116:78–91. [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. Dev. Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. 1989;341:331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Vincent A, Blankenship JT, Wieschaus E. Integration of the head and trunk segmentation systems controls cephalic furrow formation in Drosophila. Development. 1997;124:3747–3754. doi: 10.1242/dev.124.19.3747. [DOI] [PubMed] [Google Scholar]
- Weigmann K, Lehner CF. Cell fate specification by even-skipped expression in the Drosophila nervous system is coupled to cell cycle progression. Development. 1995;121:3713–3721. doi: 10.1242/dev.121.11.3713. [DOI] [PubMed] [Google Scholar]
- Wu X, Vakani R, Small S. Two distinct mechanisms for differential positioning of gene expression borders involving the Drosophila gap protein giant. Development. 1998;125:3765–3774. doi: 10.1242/dev.125.19.3765. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]