Abstract
Background
Autophagy is a molecular process that breaks down damaged cellular organelles and yields amino acids for de novo protein synthesis or energy provision. Mechanical unloading with a left ventricular assist device (LVAD) decreases the energy demand of the failing human heart. We tested the hypothesis that LVAD support reverses activation of autophagy.
Methods and Results
Paired biopsies of left ventricular myocardium were obtained from 9 patients with idiopathic dilated cardiomyopathy (mean duration of LVAD support 214 days) at the time of implantation and explantation of the LVAD. Transcript and protein levels of markers and mediators of autophagy and apoptosis were measured by quantitative RT-PCR and Western blotting. TUNEL assays, C9 immunohistochemistry, and 20S proteasome activity assays were also performed. Mechanical unloading significantly decreased mRNA transcript levels of Beclin1, autophagy related gene 5 (Atg5), and microtubule-associated protein 1 light chain 3 (MAP1-LC3 or LC3) (p<0.02). Protein levels of Beclin 1, Atg5-Atg12 conjugate, and LC3-II were also significantly reduced after LVAD support (p<0.05). A significant increase in 20S proteasome activity was observed with unloading, in parallel to the decrease in autophagic markers. Although BNIP3 and the ratio of activated caspase 3 to procaspase 3 increased after LVAD support, Bcl-2 and TUNEL-positive nuclei were not significantly different between samples.
Conclusions
Mechanical unloading of the failing human heart decreases markers of autophagy. These findings suggest that autophagy may be an adaptive mechanism in the failing heart, and this phenomenon is attenuated by LVAD support.
Keywords: Heart Failure, Autophagy, Mechanical Unloading, LVAD
Introduction
In advanced stages of heart failure, ATP production from energy-providing substrates is impaired1. Amino acids are a potential source of energy for the stressed heart. For example, oxygen-deprived cardiomyocytes can metabolize aspartate and glutamate to succinate.2 Metabolism of succinate is coupled to substrate-level phosphorylation and formation of high-energy phosphates. One possible source of amino acids to sustain the failing heart is the degradation of intracellular proteins via autophagy.
Autophagy is a pathway for the degradation and recycling of long-lived proteins and cytoplasmic organelles through the lysosome and plays an important role in homeostasis and cell survival.3 The process of autophagy involves the sequestration of cytosolic material into autophagosomes. Beclin 1 (mammalian Atg6) is involved in the initial formation of the autophagosomal membrane, and two ubiquitin-like conjugation systems, Atg5-Atg12 and MAP1-LC3 (LC3; mammalian Atg8)-phosphatidylethanol-amine, are involved in elongation of the membrane. After processing and conjugation to phosphatidylethanolamine, LC3-I undergoes cleavage to LC3-II and localizes to the membrane of the autophagosome4. Autophagosomes subsequently fuse with lysosomes to form autophagolysomes and the contents are then degraded by lysosomal proteases. Stressors such as hypoxia5 and nutrient deprivation6 induce autophagy while activation of mammalian target of rapamycin (mTOR) inhibits autophagy. While autophagy has been observed in the failing myocardium of both murine models7 and human patients8,9, the effect of mechanical unloading on levels of autophagy in the failing heart is not known.
Mechanical unloading with a left ventricular assist device (LVAD) increases survival compared to medical treatment in patients with advanced heart failure.10 LVAD implantation leads to lowered cardiac pressure and reduction in volume overload in the myocardium. These changes are followed by decreased ventricular wall tension and reduced cardiomyocyte hypertrophy.11 At the cellular level, unloading decreases energy demand and simultaneously activates cardiomyocyte protein synthesis and degradation.12 By decreasing the heart’s energy requirement, the available supply of substrate is better matched to demand. We speculated that this may, in turn, lead to a downregulation of autophagy. We therefore examined whether LVAD support reverses activation of autophagy in the human failing heart.
Methods
Patient Population and Tissue Acquisition
We studied cardiac tissue samples from 9 patients diagnosed with idiopathic dilated cardiomyopathy (7 male, 2 female) who were referred to the Texas Heart Institute for heart transplantation and placed on LVAD support for a mean duration of 214 ±295 days. Each patient gave informed consent, and the protocol was approved by the Institutional Review Boards at St Luke’s Episcopal Hospital and at the University of Texas Health Science Center at Houston.
Left ventricular tissue was obtained from the apex during implantation of a left ventricular assist device and upon explantation of the device at the time of heart transplant (n=7) or at death (n=2). Tissue samples designated for RNA and protein analysis were immediately frozen in liquid nitrogen. Tissue samples used for histology and immunohistochemistry were placed in 10% neutral buffered formalin and embedded in paraffin.
Cardiomyocyte Size
Average cardiomyocyte size at the time of LVAD implantation was compared to average cardiomyocyte size at the time of device explantation in two paired samples. Cardiomyocyte size was assessed by measuring cardiomyocyte cross-sectional area in paraffin-embedded left ventricular sections stained with hematoxylin and eosin. Five to eight random fields (x400) were selected from each slide and imaged with Olympus BX61 microscope, equipped with a color digital camera (Olympus DP70, 12.5 mega-pixel resolution). Image analysis was performed with Olympus MicrosuiteTM Biological Suite software (version 5.0) on a Dell Optiplex GX270 computer. Criteria for suitable cross-sections were defined as having nearly circular nuclei and capillaries.
Quantification of TUNEL- and C9-Positive Myocytes
Formalin-fixed, paraffin-embedded tissue was cut into 5 um-thick sections and used for the detection of apoptosis via TUNEL assay and necrosis via immunohistochemistry for complement component C9 (in both assays: n=4 for implant samples, n=6 for explant samples). The number of myocyte nuclei per mm2 was calculated from the mean of 5 randomly chosen fields of vision (x200) (185±34 myocytes/mm2 for implant samples and 192±41myocytes/mm2 for explant samples). Apoptotic myocytes were identified by the TUNEL method using the ApoTag Plus Peroxidase in Situ Apoptosis Detection kit (Millipore, Billerica, MA). TUNEL-positive nuclei were counted for the entire section and expressed as percentage of the total number of cardiomyocyte nuclei. The same counting procedure was performed for cells staining positive for C9 (Novocastra, Newcastle-upon-Tyne, UK). In both experiments, data from the implant samples and explant samples were pooled within their respective groups for analysis.
Gene Expression
Quantitative RT-PCR was performed as previously described.13 Sequences for probes and primers are shown in Table 1. Transcripts were normalized to total RNA.
Table 1.
Sequences of Quantitative RT-PCR Primers and Probes
| Gene | Primer/Probe | Sequence |
|---|---|---|
| Atg5 | Forward | 5’-TCTGGATGGGATTGCAAAATG -3’ |
| Reverse | 5’-TTTCTTCTGCAGGATATTCCATGA-3’ | |
| Probe | 5’-ATTTGACCAGTTTTGGGCC-TAMRA-3’ | |
| Beclin 1 | Forward | 5’-GCCGCACCTTCGAACAAA-3’ |
| Reverse | 5’-TCGTTCTATTATCACCGGGATTTT-3’ | |
| Probe | 5’AAGATGTCCGACTTATTCGA-TAMRA-3’ | |
| MAP1LC3A | Forward | 5’-CCGACCGCTGTAAGGAGGTA-3’ |
| Reverse | 5’-ACCCTTGTAGCGCTCGATGAT-3’ | |
| Probe | 5’-AGCAGATCCGCGACCAGC-TAMRA-3’ | |
| MAP1LC3B | Forward | 5’-GCCGCACCTTCGAACAAA-3’ |
| Reverse | 5’-TCGTTCTATTATCACCGGGATTTT-3’ | |
| Probe | 5’AAGATGTCCGACTTATTCGA-TAMRA-3’ |
Atg5 indicates autophagy related gene 5; MAP1LC3A, microtubule associated protein 1 light chain 3 A, MAP1LC3B, microtubule associated protein light chain 3 B.
Protein Expression
Protein was extracted from frozen, powdered heart tissue as described previously.13 The protein concentration of each sample was determined using the Bradford assay (Sigma, St. Louis, MO). Western blots were performed using the antibodies for Beclin1, Atg5, Bcl-2, BNIP3, and ubiquitin (Santa Cruz, Santa Cruz, CA); mammalian target of rapamycin (mTOR) and phospho-mTOR (Cell Signaling, Beverly, MA); caspase 3 (R&D Biosystems, Minneapolis, MN); LC3 (Novus, Littleton, CO); and gamma-amino butyric acid receptor-associated protein-like 1 (Gabarapl1) (ProteinTech, Inc, Tucson, AZ). To normalize for protein loading, blots were stripped and reprobed with mouse monoclonal anti-GAPDH (Fitzgerald, Concord, MA). Quantification was performed by scanning blots and using ImageJ software (NIH, Bethesda, MD).
Assay of 20S Proteasome Chymotrypsin-like Activity
20S proteasome activity was measured using a commercially available kit (20S Proteasome Activity Assay kit from Chemicon, Temecula, CA). This assay is based on the detection of the fluorophore 7-Amino-4-methylcoumarin (AMC, absorption=351nm, emission=430nm), which is released from the substrate Suc-LLVY-AMC upon cleavage by the 20S proteasome. At the end of the reaction, the free AMC fluorescence in each sample was measured using a 380/460 nm filter set in a fluorometer. Results were expressed as chymotrypsin activity in units of fluorescence/min/ug of protein.
Statistical Analysis
Normalized expression values were used for the two-tailed paired Student’s t-test to compare means between the two groups of patients before LVAD insertion and after removal. The Wilcoxon test was used for comparison of parameters without normal distribution (Atg5-Atg12 conjugate and the ratio of phospho-mTOR to total mTOR). The Pearson correlation coefficient was used to assess the relation among the parameters. A p value <0.05 was considered statistically significant.
Results
Clinical Data of Heart Failure Patients
These studies were performed on heart tissue samples from 9 patients diagnosed with idiopathic dilated cardiomyopathy (Table 2). Prior to and during LVAD support, all patients were treated with diuretics, angiotensin-converting enzyme inhibitors, and a low dose of β blockers.
Table 2.
Clinical Data for 9 Patients Diagnosed with Idiopathic Dilated Cardiomyopathy
| Gender | 7 male, 2 female |
| Ethnicity | 5 Caucasian, 4 African-American |
| Mean Age | 46±8 years (range 31–56 years) |
| Mean Time on LVAD | 214±295 days (range 4–797 days) |
| Mean LVEDD prior to LVAD | 7.10±0.62 cm |
| Mean LVEDD post LVAD | 6.12±1.32cm |
| Mean EF prior to LVAD | <20% for all patients |
LVAD indicates left ventricular assist device; LVEDD, left ventricular end-diastolic diameter; EF, ejection fraction.
Cardiomyocyte Size
Average cardiomyocyte volume appeared to decrease with unloading, consistent with our previous observations13 (data not presented).
Autophagy
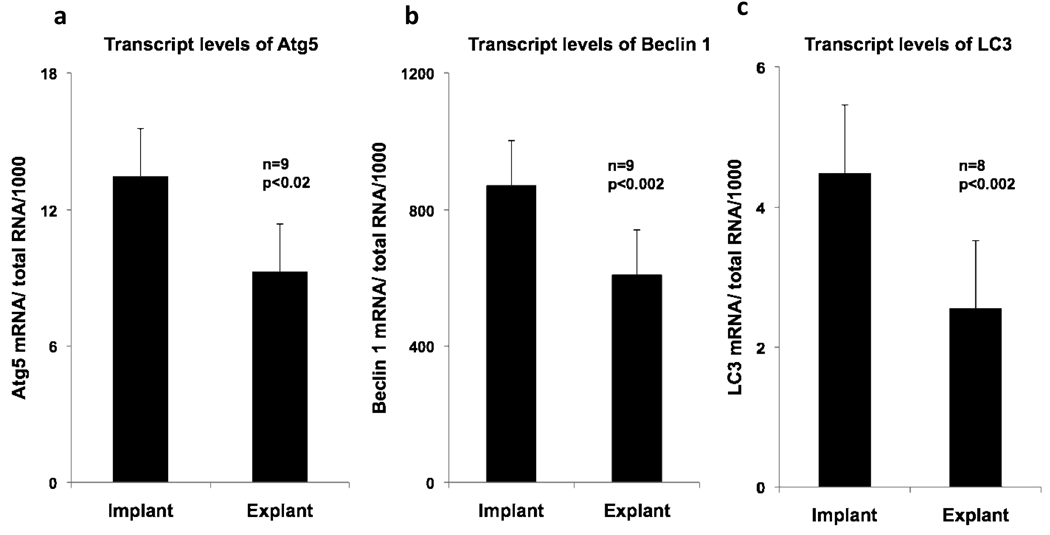
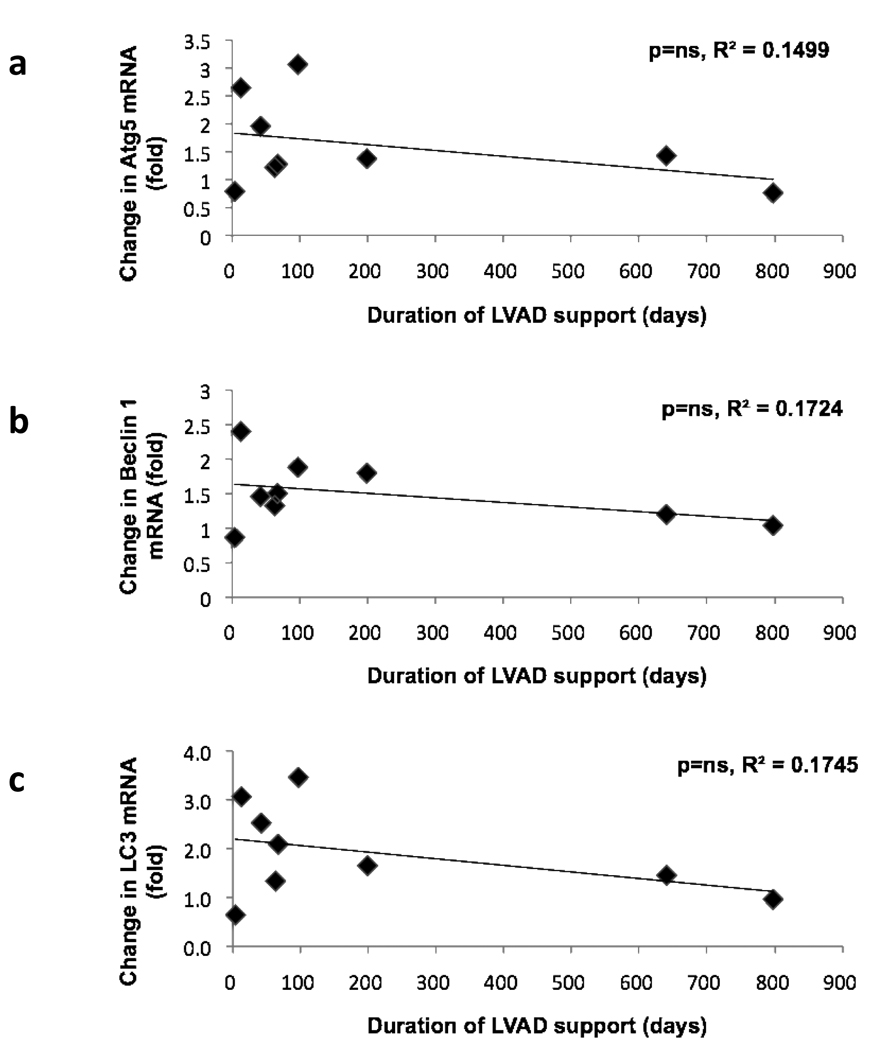
To determine how levels of autophagy are influenced by mechanical unloading, transcripts levels of autophagy markers and mediators prior to and following mechanical unloading were measured (Figure 1). Transcript levels of Atg5 decreased by 31% (p<0.02) in the explant samples, and Beclin 1 transcripts declined by 30% (p<0.002) after unloading. Similarly, the expression of LC3 transcripts was reduced by 43% (p<0.002). The results were similar for both MAP1LC3A and MAP1LC3B mRNA, two human isoforms of MAP-LC3.14 Negative correlations which failed to reach statistical significance were noted between the duration of LVAD support and the changes in transcript levels of Atg5 mRNA, Beclin 1 mRNA and LC3 mRNA (Figure 2). Patients who received LVAD support for less than 200 days had notable variability in transcript levels in the explant tissue; in the two patients supported for a longer duration, the fold change in mRNA transcripts in explant samples was more uniform. Changes in autophagy-related transcript levels following LVAD support likely proceed on a time course that is unique to each individual patient.
Figure 1.
Transcripts of autophagy markers and mediators before (implant) and after mechanical unloading (explant). a) Atg5 transcripts, b) Beclin 1 transcripts, c) LC3 transcripts. Atg5 indicates autophagy related gene 5 and LC3 indicates microtubule associated protein 1 light chain 3. Error bars represent standard error of the mean.
Figure 2.
Fold change of mRNA transcripts of autophagy markers and mediators following mechanical unloading plotted as a function of the duration of LVAD support. a) Atg5 transcript (fold change), b) Beclin 1 transcript (fold change), c) LC3 transcript (fold change). Abbreviations are the same as those used in Figure 1.
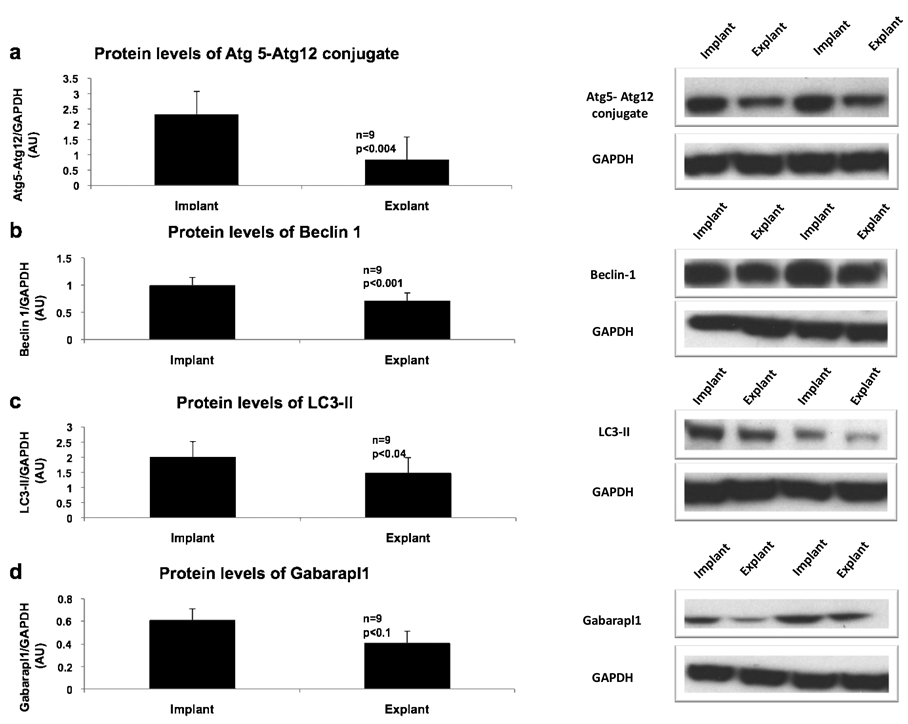
To obtain a more complete picture of autophagy in the unloaded, failing heart, protein levels of autophagy markers and mediators were evaluated (Figure 3). Levels of the Atg5-Atg12 conjugate were decreased with mechanical unloading (p<0.004), and beclin-1 was also significantly reduced by 28% (p<0.001). Similarly, LC3-II, a specific marker of autophagosomes, was reduced after unloading by 26% (p<0.04) while there was no change in LC3-I. The ratio of phospho-mTOR to total mTOR (an approximation of mTOR activity) was not significantly different between implant and explant samples (data not presented). Gabarapl1, one of the mammalian homologs of yeast Atg7, showed a non-significant trend to decrease in the explant samples (p<0.1). Taken together, the protein and transcript data strongly suggest that autophagy is decreased after unloading in the failing heart.
Figure 3.
Protein expression of autophagy markers and mediators in the heart before and after mechanical unloading. Representative western blots from two patients are depicted on the right. a) Atg5-Atg12 conjugate, b) Beclin 1, c) LC3-II, d) Gabarapl1. Gabarapl1 indicates gamma-amino butyric acid receptor-associated protein-like 1. GAPDH was used as a loading control. Other abbreviations are as in Figure 1. Error bars represent standard error of the mean.
Apoptosis and Necrosis
To determine whether any correlation could be established between autophagy and apoptosis or necrosis, we also examined markers of these modes of cell death. Tissue obtained at implantation and upon explantation of the LVAD was assayed for the presence of apoptotic nuclei by TUNEL labeling. The percentage of TUNEL (+) nuclei was very small, in agreement with data from other investigators8, and there was no significant difference between implant and explant specimens (data not presented). Immunohistochemistry for marker of necrosis complement component C9 was also performed. Analysis revealed no significant difference between groups (data not presented).
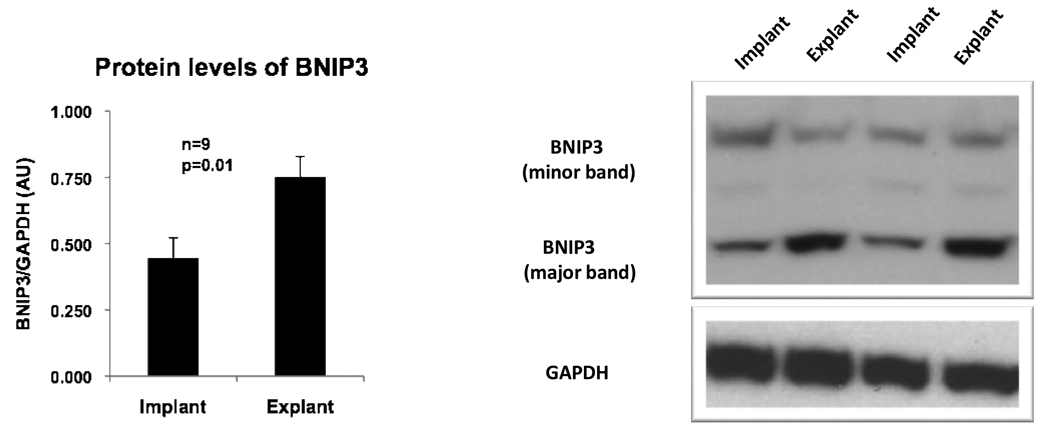
Indicators and influencers of apoptosis were analyzed using western blotting. The ratio of activated caspase 3 to procaspase 3 increased in the explant samples although it was not significant (p=0.12) (data not presented). BNIP3, a pro-apoptotic protein that may also induce autophagy in certain settings15, was significantly increased after mechanical unloading (p=0.01) (Figure 4). However, there was no significant difference in the level of the anti-apoptotic protein Bcl2 between the implant and explant samples (data not presented).
Figure 4.
Protein expression of BNIP3 in the heart before and after mechanical unloading. Representative western blots from two patients are depicted. GAPDH was used as a loading control. Error bars represent standard error of the mean.
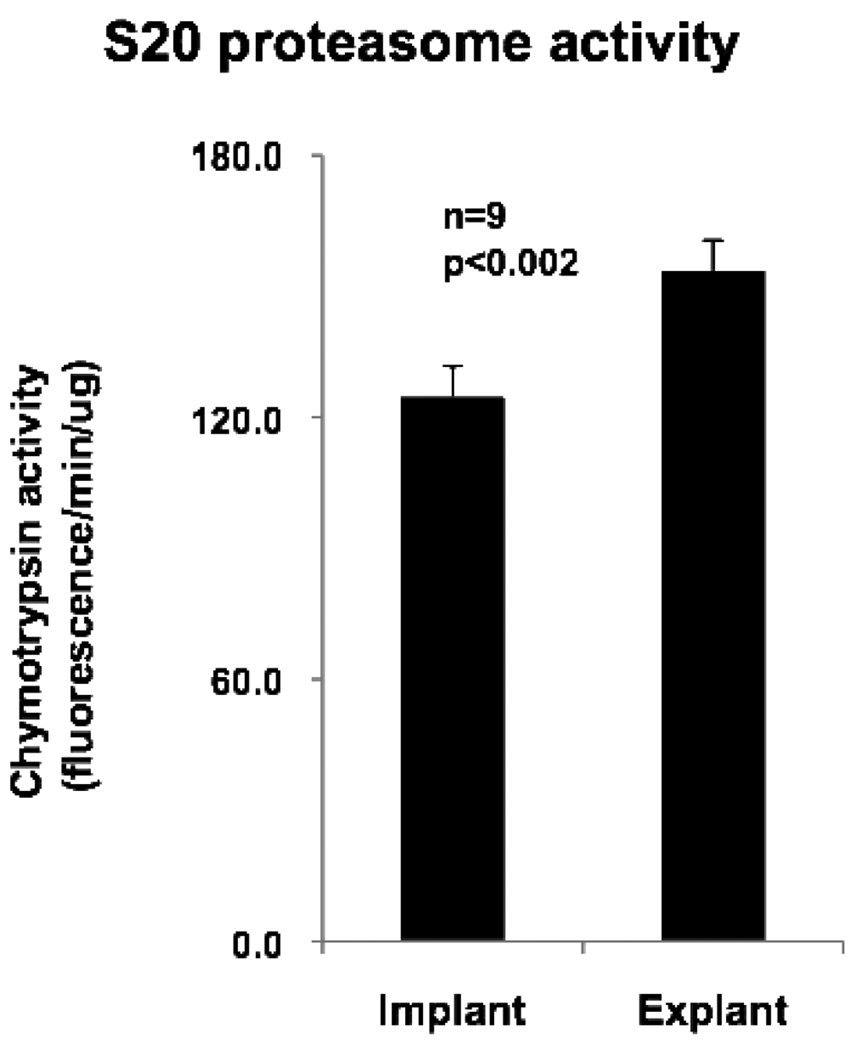
Proteasome Activity and Polyubiquitinated Proteins
Proteasome activity was assessed directly by 20S proteasome activity assays and indirectly by western blotting for polyubiquitinated proteins. A significant increase in 20S proteasomal activity was noted in the heart with mechanical unloading (Figure 5). A non-significant increase in polyubiquitinated proteins was also observed following LVAD support (p=0.2) (data not presented). These results suggest that proteasomal activity is increased with mechanical unloading of the failing heart.
Figure 5.
20S proteasome activity in myocardial tissue before and after mechanical unloading. Error bars represent standard error of the mean.
Discussion
The key finding of this study is that both transcript and proteins levels of autophagy markers and mediators are decreased in the failing human heart with mechanical unloading. Autophagy has previously been observed in the failing human heart8,9, and other studies have shown upregulation of this process in animal models of pressure overload-induced heart failure.16–18 We now describe the effect of mechanical unloading on markers and mediators of autophagy in the failing human heart.
The reduction in autophagic markers observed with unloading may be related to a favorable effect of the LVAD on energy requirements of the myocardium. Previously, we have shown that mechanical unloading only partially reverses the depressed metabolic gene expression in the failing human heart.19 If cardiac energy demand is lessened, the need to generate substrates for energy provision through autophagy may be decreased, and the lack of dramatic changes in metabolic gene expression with mechanical unloading would be explained. In mammalian skeletal muscle, atrophy induced by fasting results in significant upregulation of autophagic markers LC3-II and Gabarapl1 in vivo.20 Increased protein levels of Beclin1, LC3-II, and Atg5-Atg12 conjugate along with Atg5 transcripts have been observed in the heterotopically transplanted rat heart (KW, unpublished observations). Because we have shown here that mechanical unloading decreases autophagy, we suggest that the process of autophagy is already maximally stimulated in end-stage heart failure. The lack of a control group consisting of non-failing hearts is, however, a limitation of the present study.
Changes in protein turnover could also contribute to the difference in autophagic markers and mediators between implant and explant samples. We have previously shown that unloading activates both pathways of protein synthesis and the ubiquitin proteasome system in the heterotopically transplanted rat heart.12 This was associated with an increase in polyubiquitinated content and ubiquitin conjugating enzymes. Recently, Tannous et al reported that intracellular protein aggregation resulting from pressure overload can trigger cardiomyocyte autophagy.21 The same group also demonstrated that inhibition of proteasome activity can lead to increased autophagy and vice-versa. In our study, a significant increase in proteasome activity was noted in parallel with the reduction of markers of autophagy with mechanical unloading. We speculate that the observed increase in proteasome activity in the explant samples could, indirectly or directly, lead to a reduced need for autophagy.
The interplay of signals regulating pathways of autophagy and apoptosis in the heart has been the subject of a number of recent reviews.22, 23 We therefore examined markers of apoptosis in samples obtained pre- and post-mechanical unloading. There was no significant difference in Bcl-2 levels or TUNEL-positive nuclei between groups. However, the level of BNIP3 and the ratio of activated caspase 3 to procaspase 3 were both increased in our explant samples. Based on these results, we cannot conclusively say how mechanical unloading affects apoptosis in the failing heart. The lack of definitive differences in the levels of apoptosis between implant and explant samples is common in LVAD studies. Previously, Bartling et al24 found a significant increase in Bcl-XL transcript levels and a non-significant decrease in DNA fragmentation after LVAD implantation but the investigators found no significant differences in transcript levels of Bak, Bax, Bcl-2 or Fas. The observed discrepancies between studies may be due to differences in each patient’s underlying cause of heart failure, length of time on LVAD support, and endpoint (transplantation or death) precipitating collection of explant tissue.
Increased expression of BNIP3 in the failing heart after LVAD support is a particularly interesting finding, given the ability of BNIP3 to induce both apoptosis and autophagy. BNIP3 mediates apoptosis during hypoxia and ischemia/reperfusion25 but also modulates FoxO3-induced autophagy in atrophied skeletal muscle.21 Context likely dictates whether BNIP3 induces apoptosis or autophagy 25. Our finding that BNIP3 is increased in the unloaded, failing human heart is almost paradoxical: one would expect it to be accompanied by clear-cut evidence showing either that it corresponded to upregulation of either apoptosis or autophagy. When BNIP3’s ability to regulate apoptosis is considered along with the ratio of caspase 3 to procaspase 3, it is tempting to speculate that apoptosis may actually be increased in our explant samples. This would be extremely hard to reconcile with the well-established role of LVAD support in preserving or improving the cardiac function of end-stage heart failure patients. Further study is needed to elucidate BNIP3’s role in the regulation of autophagy and/or apoptosis in the failing and unloaded human heart.
Study Limitations
There were a number of limitations to our study. First and foremost, the lack of samples from normal hearts in our study prevented us from determining whether unloading brings levels of autophagy closer to those found in normal heart tissue. Shimomura et al found increased histological evidence of autophagy in the myocardium of patients with dilated cardiomyopathy (DCM) compared to normal controls 26; while his group interpreted the presence of autophagy as a precipitant of cell death, it is possible that autophagy in DCM represents a last stand by cardiomyocytes to meet energy demand. If that were the case, increased autophagy in DCM would be an adaptive response to heart failure. Additionally, we were unable to include samples from patients whose heart function improved while on LVAD support. This would be a very interesting group to add to our study but this clinical outcome is very rare.11 Our study was also hampered by the small number of patients. Lastly, we included in our analysis only patients with the diagnosis of idiopathic dilated cardiomyopathy. Therefore the results cannot be extrapolated to any other type of heart failure.
Conclusions
Mechanical unloading of the failing human heart decreases the expression of autophagy markers and mediators with a concomitant increase in proteasomal activity. These findings suggest that autophagy may be an adaptive mechanism in the failing heart, and this phenomenon is attenuated by LVAD support.
Acknowledgements
We thank Sylvia Carranza for expert help in tissue bank management, and Dr. A. J. Marian for his helpful comments.
Funding Sources
Funding for this study was provided in part by a grant from the National Heart, Lung and Blood Institute (RO1 HL/AG-61483) to H.T. K.W. is a Howard Hughes Medical Institute Medical Research Training Fellow.
Footnotes
Conflict of Interest Disclosures
O.H.F. has received an honorarium from Thoratec. The other authors have nothing to disclose.
Presented at the American Heart Association Scientific Sessions, November 8–12, 2008, New Orleans, LA.
References
- 1.Taegtmeyer H, Razeghi H, Young ME. Alterations in cardiac metabolism in heart failure. In: Mann D, editor. Heart Failure: a companion to Braunwald’s Heart Disease. Philadelphia, PA: Saunders; 2008. pp. 315–337. [Google Scholar]
- 2.Taegtmeyer H. Metabolic responses to cardiac hypoxia: Increased production of succinate by rabbit papillary muscles. Circ Res. 1978;43:808–815. doi: 10.1161/01.res.43.5.808. [DOI] [PubMed] [Google Scholar]
- 3.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death and Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 4.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 6.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 8.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 9.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, Bauer EP, Klovekorn WP, Schaper J. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–991. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 10.Rose EA, Gelijns AC, Moskowitz AJ, Heitjan DF, Stevenson LW, Dembitsky W, Long JW, Ascheim DD, Tierney AR, Levitan RG, Watson JT, Meier P, Ronan NS, Shapiro PA, Lazar RM, Miller LW, Gupta L, Frazier OH, Desvigne-Nickens P, Oz MC, Poirier VL. Long-term mechanical left ventricular assistance for end-stage heart failure. N Engl J Med. 2001;345:1435–1443. doi: 10.1056/NEJMoa012175. [DOI] [PubMed] [Google Scholar]
- 11.Razeghi P, Myers TJ, Frazier OH, Taegtmeyer H. Reverse remodeling of the failing human heart with mechanical unloading. Emerging concepts and unanswered questions. Cardiology. 2002;98:167–174. doi: 10.1159/000067313. [DOI] [PubMed] [Google Scholar]
- 12.Razeghi P, Sharma S, Ying J, Li YP, Stepkowski S, Reid MB, Taegtmeyer H. Atrophic remodeling of the heart in vivo simultaneously activates pathways of protein synthesis and degradation. Circulation. 2003;108:2536–2541. doi: 10.1161/01.CIR.0000096481.45105.13. [DOI] [PubMed] [Google Scholar]
- 13.Depre C, Shipley GL, Chen W, Han Q, Doenst T, Moore ML, Stepkowski S, Davies PJ, Taegtmeyer H. Unloaded heart in vivo replicates fetal gene expression of cardiac hypertrophy. Nat Med. 1998;4:1269–1275. doi: 10.1038/3253. [DOI] [PubMed] [Google Scholar]
- 14.He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JO, Yu L. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 15.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 16.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razeghi P, Young ME, Ying J, Depre C, Uray IP, Kolesar J, Shipley GL, Moravec CS, Davies PJ, Frazier OH, Taegtmeyer H. Downregulation of metabolic gene expression in failing human heart before and after mechanical unloading. Cardiology. 2002;97:203–209. doi: 10.1159/000063122. [DOI] [PubMed] [Google Scholar]
- 20.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr., Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorn GW., 2nd Apoptotic and non-apoptotic programmed cardiomyocyte death in ventricular remodeling. Cardiovasc Res. 2009;81:465–473. doi: 10.1093/cvr/cvn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishida K, Yamaguchi O, Otsu K. Crosstalk between autophagy and apoptosis in heart disease. Circ Res. 2008;103:343–351. doi: 10.1161/CIRCRESAHA.108.175448. [DOI] [PubMed] [Google Scholar]
- 24.Bartling B, Milting H, Schumann H, Darmer D, Arusoglu L, Koerner MM, El-Banayosy A, Koerfer R, Holtz J, Zerkowski HR. Myocardial gene expression of regulators of myocyte apoptosis and myocyte calcium homeostasis during hemodynamic unloading by ventricular assist devices in patients with end stage heart failure. Circulation. 1999;100:II216–II223. doi: 10.1161/01.cir.100.suppl_2.ii-216. [DOI] [PubMed] [Google Scholar]
- 25.Shaw J, Kirshenbaum LA. Molecular regulation of autophagy and apoptosis during ischemic and non-ischemic cardiomyopathy. Autophagy. 2009;4:427–434. doi: 10.4161/auto.5901. [DOI] [PubMed] [Google Scholar]
- 26.Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65:965–968. doi: 10.1253/jcj.65.965. [DOI] [PubMed] [Google Scholar]