Abstract
A tetramer model for HIV-1 IN with DNA representing the LTR termini was previously assembled to predict the IN residues that interact with the LTR termini; these predictions were experimentally verified for nine amino acid residues (Chen et al, J. Biol. Chem. 281, 4173-4182 (2006)). In a similar strategy the unique amino acids found in ASV IN rather than HIV-1 or MPMV IN were substituted into the structurally related positions of HIV-1 IN. Substitutions of six additional residues (Q44, L68, E69, D229, S230 and D253) showed changes in the 3′ processing specificity of the enzyme verifying their predicted interaction with the LTR DNA. The newly identified residues extend interactions along a sixteen base pair length of the LTR termini and are consistent with known LTR DNA: HIV-1 IN cross-links. The tetramer model for HIV-1 IN with LTR termini was modified to include two IN binding domains for LEDGF/p75. The target DNA was predicted to bind in a surface trench perpendicular to the plane of the LTR DNA binding sites of HIV-1 IN and extending alongside LEDGF. This hypothesis is supported by the in vitro activity phenotype of HIV-1 IN mutant with a K219S substitution showing loss in strand transfer activity while maintaining 3′ processing on a HIV-1 substrate. Mutations at seven other residues reported in the literature have the same phenotype and all eight residues align along the length of the putative target DNA binding trench.
Keywords: HIV-1, integrase, model structure, DNA
INTRODUCTION
Integrase (IN) of human immunodeficiency virus type 1 (HIV-1) is an attractive target for therapeutic development as it is essential for early steps in viral replication and there are no homologues in the eukaryotic system for which inhibitors would negatively affect host viability. This enzyme is both necessary and sufficient to catalyze the insertion of viral into host DNA1-3. In the first step or 3′ processing reaction, two deoxyribonucleotides are removed from the 3′ end of the LTR strands containing the highly conserved CA dinucleotides. In the second step or strand transfer reaction, the newly created 3′ ends undergo a staggered nucleophilic attack on the two strands of the target DNA. These structures are resolved and repaired by host cell enzymes resulting in an integrated copy of the viral DNA with the gene encoding sequence co-linear to the viral RNA and flanked by a four to six base pair duplication of the target DNA, depending upon the viral IN.
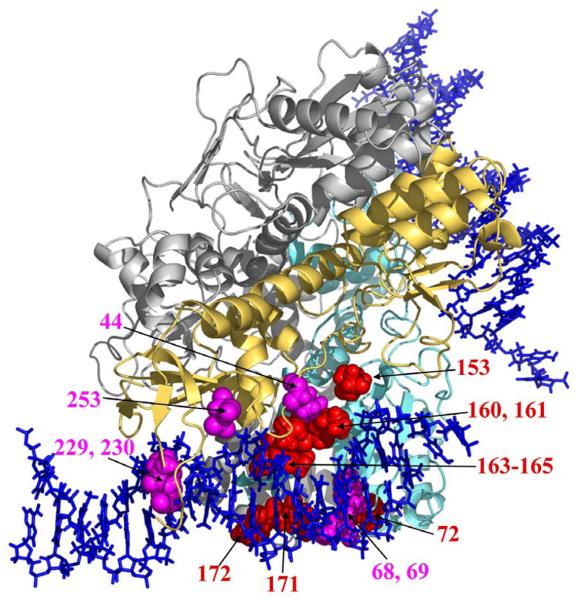
Design of inhibitors has been hampered by the lack of crystal structures available for full-length IN monomers or higher-order IN oligomers, let alone in complex with DNA. Partial structures with two of the three domains have been reported and these were used to assemble a model of a tetramer HIV-1 IN with bound LTR DNAs4. The model was then used to predict residues that were in close proximity to the viral DNA. To verify these predictions, a structural alignment of the primary sequences of HIV-1, SIV, and ASV INs5 was used to identify residues that were unique to each virus enzyme, because viral IN specifically recognizes its cognate LTR end substrates. The unique amino acids from ASV IN were substituted into the equivalent structural position of HIV-1 IN. Substitution of the ASV IN residues conferred on the HIV-1 IN mutants the partial ability to cleave an ASV substrate. Multiple residues of HIV-1 IN were demonstrated to alter specificity: V72, S153, K160, I161, G163, Q164, V165, H171, and L1724, shown in red in Figure 1 for only one of the two LTR ends. In this report we have identified six additional HIV-1 IN residues that influence the selection of the LTR end substrates for 3′ processing. These include Q44, L68, E69, D229, S230, and D253, which align along the two LTR binding grooves with the previous residues that alter recognition.
Figure 1.
HIV-1 IN homotetramer model with bound HIV-1 LTR substrates. IN tetramer model with subunits that interact with the two viral DNA ends colored cyan and yellow and two other subunits colored in gray. The LTR DNA ends are represented by blue helices. The amino acids that specifically recognize the viral DNA are shown in space fill model for only one viral DNA end. Those in red are from Chen et al.4; those newly identified are in magenta. Figures 1 and 5 were made using PyMol70.
In addition, the structural model was modified to include the host protein LEDGF/p75 integrase binding domains and predict a trench on the HIV-1 IN surface that may accommodate the target DNA. The putative binding site for target DNA is positioned roughly perpendicular to the LTR binding sites. Consistent with this interpretation, we have identified in the literature a series of amino acids along the length of one side of this trench where point amino acid substitutions result in enzymes that lose the ability to strand transfer with little or no effect on 3′ processing. In addition, we have identified a residue on the opposite wall of the trench, K219, where serine substitution displays the same phenotype. This residue is found in a peptide that was previously demonstrated to cross-link to the target DNA6.
RESULTS
Prediction of additional HIV-1 IN residues interacting with the LTR DNA ends
In the original selection of amino acids that could affect the recognition of LTR ends, we used the structural alignment of SIV, HIV-1, and ASV INs to identify those residues that were unique. We subsequently observed that HIV-1 IN was capable of 3′ processing a U5 SIV but not a MPMV LTR DNA substrate (data not shown). Therefore, we examined the structural alignment of HIV-1 and ASV INs with the MPMV IN sequence5. This analysis identified additional unique residues in the IN model near the LTR DNA ends: Ser39, Lys42, Gln44, Leu68, Glu69, Leu74, Lys156, Glu170, Tyr227, Asp229, Ser230, Asp253, Asn254, Lys258, and Arg262 (HIV-1 IN numbering). To test whether any of these residues were involved in recognition of the LTR ends, eleven HIV-1/ASV IN chimeras were constructed that substituted the amino acid from ASV IN into the structurally equivalent position of HIV-1 IN as described in the Materials and Methods. A list of chimeras is presented in Table 1.
Purification of HIV-1/ASV IN Chimeras
HIV-1 IN mutants were constructed and purified as described in Materials and Methods. The chimeras were assembled in a 3CSF185H background, an enzyme with four amino acid substitutions (C56S, C65S, C280S, and F185H) to improve solubility. This enabled purification of the chimeras from the soluble faction. Individually these amino acid substitutions have little or no effect on viral replication7-9. Enzymes purified by this protocol were free of detectable non-specific nuclease4. When a HIV-1 G197I substitution, which caused a significant decrease in 3′ processing, was combined into chimeras that alter LTR recognition, this resulted in purified enzymes that did not efficiently 3′ process HIV-1 or ASV substrates. As further evidence for the purity of INs prepared by this protocol, 5′ 33P-end labeled DNA substrates representing the HIV-1 U5, ASV U3, and MLV U5 LTR termini, were individually incubated with wild type or selected IN chimeras. The 3CSF185H HIV-1 and ASV INs cleave their respective homologous substrates, but not heterologous substrates including the MLV sequence (Figure 2). The MLV U5 LTR DNA sequence is cleaved by the wild type MLV IN10. As previously reported4, the G163R Q164V V165L chimera in the 3CSF185H background cleaved both HIV-1 U5 and ASV U3 LTR end substrates (Figure 2). However, the chimera did not cleave the MLV U5 substrate indicating the ability to cleave the ASV substrate was not due to a non-specific nuclease activity. A second HIV-1 IN that contains a K211S substitution, also in the 3CSF185H background, cleaved the HIV-1 U5 but not the ASV U3 or MLV U5 substrates. This amino acid is positioned in the structural model at a distance from the LTR binding sites so that it was expected to maintain HIV-1 substrate specificity for 3′ processing.
Figure 2.
Purified chimeras do not cleave MLV substrates. 33P-end labeled duplex oligos representing the HIV-1 U5, ASV U3, and MLV U5 LTR ends were incubated with HIV-1 (3CSF185H), ASV, or mutant HIV-1 INs (the triple substitution mutant G163R Q164V V165L and K211S) and the products separated by denaturing gel electrophoresis as described in Materials and Methods. The production of the −2 cleavage product demonstrates the specific enzymatic processing of the viral DNA substrate.
Effect of Specific Amino Acid Changes on 3′ Processing
We screened each HIV-1/ASV IN chimera listed in Table 1 for their ability to 3′ process HIV-1 and ASV LTR duplex DNA substrates. Of the chimeras tested, S39T K42H, Y227I, N254D, K258T R262S showed decreased or no activity with HIV-1 substrates and did not cleave the ASV substrates (data not shown). As such, these residues were not further considered. In contrast, Q44N, L68E E69P, D229I S230E, and D253N maintained the ability to cleave the HIV-1 substrate, and gained the ability to cleave the ASV substrates to different extents (Figure 3). D229I S230E was unique among these mutants in that it possessed more 3′ processing activity towards the HIV-1 substrate than 3CSF185H IN (Figure 3A). Thus, the residues Q44, L68, E69, D229, S230, and D253 have been verified to interact with the viral LTR and are highlighted on the structural model in Figure 1 (magenta residues) along with the previously identified residues that affect 3′ processing specificity (red residues)4. Taken together, these residues strikingly define two linear trenches on the HIV-1 IN molecular surface that accommodate the two LTR ends. The binding trenches are asymmetrically positioned along the two strands of a 16 base pair length of the LTR ends. Twelve of the fifteen amino acid exchanges that affect LTR recognition in the processing reaction were combined into a single construct (V72W S153R K160D I161R G163R Q164V V165L H171K L172Q D229I S230E D253N) and purified from the soluble fraction. As shown in Figure 3B, this enzyme designated S7C is active and has substantially more specific 3′ processing activity towards the ASV than the HIV-1 DNA substrate. We also assembled a S7C IN chimera in which the substitutions at positions 185 and 280 were restored to wild-type residues. Unfortunately this resulted in an enzyme that was insoluble; 3′ processing activity, however, could be recovered by renaturation from urea and the activity of this enzyme was qualitatively similar to that shown for S7C. This indicates that these two substitutions did not affect the specificity of the 3′ processing reaction. Taken together, these results suggest that the sum of IN interactions along a 16 base pair length of the viral DNA ends determines its specificity.
Figure 3.
The 3′ processing of HIV-1 U5 and ASV U3 LTR duplex oligo substrates by wild type and HIV-1/ASV IN chimeras. (A). HIV-1 and ASV INs and Q44N, D229I S230E, D253N, N254D chimeras were incubated with HIV-1 U5 or ASV U3 LTR substrates and analyzed as described in the legend to Figure 2. (B) HIV-1 and ASV IN, S39T K42H, L68E E69P, K258I R262S, and S7C were analyzed as in panel A. S7C is in a HIV-1 IN background and has 12 amino acid substitutions as described in Table 1.
While we have been able to alter the specificity for 3′ processing of LTR substrates, we have not been able to demonstrate specificity changes in strand transfer activity. Several amino acid changes introduced into the HIV-1 IN (L68E and E69P, V72W, and H171K and L172Q) that alter 3′ processing disrupt the strand transfer activity towards the HIV-1 substrates. The reason for this is not known. Substitutions at the other ten residues that affect 3′ processing support strand transfer activity with HIV-1 substrates. None of the chimeras had strand transfer activity using the ASV substrate. When we combined multiple substitutions into the soluble form of S7C, it too was unable to support a strand transfer reaction with either the HIV-1 U5 LTR or ASV U3 LTR preprocessed end substrates. One possible explanation for this behavior might be that because IN is a tetramer, single substitutions of one residue will necessarily change all four subunits, which might be responsible for the loss in strand transfer activity.
Activity complementation during drug selection
When HIV-1 is replicated in the presence of diketo-acid based compounds, a number of escape mutants are selected that are believed to act against the strand transfer reaction. We previously reported that V72 and S153 were among the residues that influence LTR selection and these residues were mutated in drug resistant INs4. Changes at position 230 in HIV-1 IN are also found in drug resistant INs11. When a substitution in integrase occurs that disrupt its catalytic activity (3′ processing or strand transfer), we hypothesize that second site mutations in IN are selected to compensate for the lost activity caused by the original mutation. In the case of the V72W IN mutant, which has reduced 3′ processing of HIV-1 substrates (79 % compared to wild-type, SD=8.6), second site substitutions at F121, T125, and V151 have been reported12. HIV-1 IN with a T125S substitution resulted in an enzyme that increased its 3′ processing reaction relative to 3CSF185H (126%, SD=11.7). When the V72W and T125S substitutions were combined into the same enzyme, the resultant chimera had a 3′ processing activity equivalent to 3CSF185H (97%, SD=2.0). This result demonstrates that at least one mutation at a second site associated with substitutions at position 72 can compensate for its decreased 3′ processing activity.
Model of IN tetramer with LTR ends and LEDGF/p75 integrase binding domains
The model of the IN tetramer with viral LTR ends was augmented with the host protein LEDGF. The LEDGF integrase binding domain from the crystal structure of LEDGF bound to the catalytic domain of IN (2B4J) was docked to the HIV-1 IN tetramer model. Two LEDGF subunits were accommodated, bound at opposite ends (Figure 4A). Structural data show the binding pocket for LEDGF integrase binding domain is formed by residues 102, 128, 129 and 132 in one IN subunit and residues 174 and 178 in a second subunit13. These residues, colored in red, are at the LEDGF/HIV-1 IN interface. Additional residues that may be involved in the interaction with LEDGF, colored in yellow, based on mutagenesis and structural data include 131, 161, 165, 166, 168, and 170-17313-15. These residues are also located at or near the interaction interface in our model. Therefore, the new model is consistent with information for the residues implicated in the IN interaction with LEDGF.
Figure 4.
Model of HIV-1 IN tetramer interacting with two LEDGF domains and two viral LTRs. (A) The LEDGF/p75 domains that interact with IN are shown in magenta, with the modeled tetramer of HIV-1 IN shown in gray. Residues reported to interact directly with LEDGF are visualized in red, while residues that affect the IN:LEDGF interaction when mutated are shown in yellow13-15. Only the residues closest to LEDGF are shown. The IN tetramer is formed by two dimers, so each pair of subunits have different conformations and interactions. LEDGF domains interact with opposite ends of the tetramer and the viral LTRs interact at the perpendicular ends of IN. The equivalent residues in other subunits of the IN tetramer may be closer to the LTR or other IN domains. The arrows indicate the proposed binding site for target DNA. (B) The four subunits of IN are shown in surface representation to reveal the proposed trench for binding target DNA in a similar orientation to Figure 4A. The IN subunits are colored as in Figure 1 with cyan and yellow indicating subunits interacting with the LTRs. The LTR DNA ends are shown in blue and LEDGF in magenta. Residues 94, 118-120, 123, 130, 132, 141, 159, 181, 185 and 203 that interact with target DNA are shown in green space fill models. Residue 219 (red) is in peptide 213-247 known to cross-link to target DNA in a disintegration model substrate6. The residues lie on either side of the trench that circles the IN tetramer running approximately perpendicular and between the modeled LTR ends.
Target DNA Binding Trench
A groove is observed on the IN tetramer model between the two LTR ends and approximately perpendicular to the long axis of the LTR DNA that could accommodate the target DNA. The positioning of LEDGF as an extension of the target DNA binding trench (indicated by arrows, Figure 4A) would be consistent with its role in interacting with chromatin to influence target site selection16-20. We predict from the structural model that mutations introduced into the target DNA binding site of IN, but distant from the catalytic and LTR binding sites, would have a phenotype where strand transfer would be inactivated without impairment of the 3′ processing reaction. Within the putative DNA binding trench, mutations of a series of residues including S11921, N12022, C13023,24, W13224, F181 and F18525 display this activity phenotype. These residues align along one wall of the trench (Figure 4B, green residues). If this trench is the binding site for the target DNA, we would predict that point mutations introduced at residues on the opposite wall would have the same phenotype. We therefore assembled HIV-1 IN mutants with K211S, K219S, and Q221S, substitutions, respectively. Each was tested for 3′ end processing and strand transfer against the homologous HIV-1 DNA substrates. The K211S mutant showed near wild-type 3′ processing while the K219S mutant still had 3′ processing activity towards a HIV-1 substrate though less than wild type (Figure 5). When tested in the strand transfer assay using pre-processed HIV-1 LTR DNA, we observed that the K211S mutant was as active as wild-type while the K219S mutant was inactive. The activity of the Q221S mutant in both 3′ end processing and strand transfer was similar to that of K211S (data not shown). Thus, the K219S enzyme lost the ability to strand transfer with a decrease in 3′ processing while the K211S and the Q221S mutants had no detectable affect on either activity. These results suggest K219 is involved in binding to the target DNA.
Figure 5.
The effect of amino acid substitutions on 3′ processing and strand transfer at residues in the putative DNA binding trench. HIV-1 IN, K211S, K219S IN mutants were incubated with HIV-1 U5 LTR end duplex substrate as in legend to Figure 2 (HIV 3′) or a preprocessed HIV-1 U5 LTR duplex substrate with the two bases removed adjacent to the CA dinucleotide (HIV ST) as described in Materials and Methods. In the ST lane, products migrating slower than the starting substrate represent integration of one oligo into another.
DISCUSSION
The homotetramer form of IN catalyzes all of its known enzymatic activities. While dimers of IN are capable of catalyzing 3′ processing and strand transfer reactions, they do not support a concerted DNA integration reaction26. For this reason, a homotetramer model was assembled. In the model, two of the four subunits are depicted with major contacts with the LTR and target DNAs. The remaining two subunits are available for contacts with proteins that interact with integrase27,28 including the host protein LEDGF as shown in our new augmented IN tetramer model. There are at least fifteen residues in the LTR binding groove on the IN surface that are associated with LTR specificity. They are predicted to interact asymmetrically along a 15-16 base length of DNA duplex (see Figure 1). When LTR DNA is bound to IN and the complex treated with DNase, a 16 base pair duplex length of the LTR is protected from digestion29,30. Thus there is agreement between the size of the protected DNA and residues in close proximity to the LTR DNA that change specificity for substrate (Figure 1). Moreover, when mutations are introduced into HIV-1 LTR duplex DNA substrates at 16 base pairs from the ends, changes in concerted DNA integration in vitro are detected61. Interactions between viral DNA and HIV-1 IN have also been demonstrated in cross-linking studies for residues 143, 148, 156, 159, 160, 230, 246, 262, 263, and 26431-35. These residues are highlighted in Figure 6. A group cluster in and around the catalytic site in close proximity to the first 6 bases/base-pairs of the processed LTR ends. A second group which includes residues 246, 262, 263, and 264 are in close proximity to base-pairs 15-16 of the LTR DNA that interact with residues 229, 230 and 253 identified in this study. Finally, Agapkina, et al. used substrate analogs to probe contacts between HIV-1 IN and LTR DNA substrates36. In this study they identified eleven contacts with the sugar phosphate backbones from residues 5-9 and interactions with four bases asymmetrically distributed between the two strands of the LTR ends. The fifteen residues that influence specificity for 3′ processing reactions are spatially in close proximity to all of these sugar phosphate backbone contacts and most of the base contacts.
Figure 6.
Residues reported in the literature to cross-link to viral LTR DNA. Interactions between viral DNA and HIV-1 IN have been demonstrated for residues Tyr143, Gln148, Lys156, Lys159, Lys160, Ser230, Glu246, Arg262, Arg263, and Lys264 31-34,71. These residues are colored red in space-fill representation and shown only for one LTR end.
There are a series of naphthyridine carboxamide and diketo acid related drugs that act in the nanomolar range to inhibit HIV-1 IN11,12,37-44. Drug resistant enzymes with changes at more than ten different sites were identified. Five of these residues were unique in the structural alignment of different INs and were located near the LTR ends in the structural model. While these drugs were thought to act at strand transfer and not 3′ processing, we found that HIV-1 IN residues S153 and V724 as well as S230 were among those positions involved in LTR end recognition and 3′ processing. Because drug resistant sites affect specific recognition of the viral DNA ends and change the rate of processing of HIV-1 substrates, we predict that amino acid changes at some of these sites will lead to partially defective INs in cells. This should subsequently result in selection of second site substitutions that compensate for the loss in 3′ processing activity caused by the initial drug resistant amino acid substitutions. If correct, we predict that depending upon the extent of change in 3′ processing observed towards HIV-1 duplex substrates4, our data will be correlated to the appearance of individual or multiple residue substitutions detected in drug resistant enzymes. For example, in the case of a position 153 chimera, it gains the ability to 3′ process the ASV LTR end duplex with only a small decrease in its ability to 3′ process the HIV-1 duplex substrate4. As such, we would predict that this mutation would be found by itself and should have only a small affect on replication of HIV-1 in cells, as observed44,45. In contrast, the substitution at position 72 (V72W) caused a larger decrease in the ability to process the HIV-1 U5 duplex substrate4. On this basis, we would predict that the V72I drug resistant mutation would appear in the presence of other substitutions that compensate for the loss in its 3′ processing towards HIV-1 substrates. Second site mutations of F121Y and T125K subsequently appear in HIV-1 IN containing the V72I mutation12. A T125S substitution increases the 3′ processing of U5 HIV-1 duplex4 as well as joining of a HIV-1 preprocessed substrate. When combined with the V72W mutation, this produces an enzyme with near wild type levels of 3′ processing, suggesting that a second site mutation compensates for the decrease in 3′ processing caused by the initial drug resistant mutation. Another illustrative example involves position 230 where substitutions at this residue also affect recognition of the viral DNA ends. With the caveat that the exchange of S230E was analyzed as a double mutant in combination with D229I, it gained the ability to cleave the ASV substrate, but in contrast to chimeras with changes at positions 72 or 153, it displayed an increase in activity towards HIV-1 substrates. Changes at position 230 are reported to appear in conjunction with T66I and M74L substitutions in cells11. We have not analyzed substitutions at position 66 in vitro, but Lee and Robinson reported that the T66I substitution caused a small decrease in 3′ processing44. We tested a M74A substitution that resulted in a significant loss of 3′ processing of HIV-1 substrates. Taken together, this suggests that substitution at position 230 might compensate for the loss in 3′ processing caused by mutations at positions 74 and 66.
In examining the structural model, we identified a trench on the HIV-1 IN surface with its long axis almost perpendicular to those accommodating the viral DNA ends4. We speculate that the target DNA fits into this groove. Moreover, interactions with LEDGF will further stabilize this IN: target DNA complex and would be consistent with the role for LEDGF in promoting the interaction of the integration complex with host chromosomal DNA14,17,28,46-48. The target DNA is positioned between the viral DNA ends and this location will facilitate the nucleophilic attack of the 3′ hydroxyl ends of the respective CA strands into each strand of the target DNA. There are several lines of evidence that support this hypothesis. First, amino acid residues S119, N120, C130, W132, and K159 are reported to interact with target DNA based upon activity and drug sensitivity data21,22,24,45,49. These residues strikingly align along one surface wall of the proposed target DNA binding trench. A N120S mutant is reported to increase 3′ processing and strand transfer activities while N120Q and N120K mutants show little effect on processing but some decrease in strand transfer22,45. The C130S and W132A/G/R substitutions are reported to have normal 3′ processing but little or no joining activity24. A C130S substitution in combination with three other mutations shows loss in strand transfer but with some decrease in 3′ processing9. More recently, four HIV-1 IN mutants with W132Y, M178C, F181G, and F185G substitutions, respectively, were constructed25. The enzymes with mutations at positions W132, F181, and F185 did not support a strand transfer reaction but had wild type or near wild type levels of 3′ processing. In contrast, the mutation at M178 showed decreases in both activities25. This latter residue lies below the surface of the target DNA binding trench (data not shown). An ASV IN mutant, structurally equivalent to HIV-1 IN S119, has normal 3′ processing but barely detectable strand transfer activity21. Similarly, Asp substitutions in ASV IN equivalent to HIV-1 IN G94 and S123 show the same activity phenotype (Michael Katzman, Penn State College of Medicine, personal communication). Finally, HIV-1 IN substitutions of I141K, I203P or I203K (Corinne Ronfort, Universite de Lyon, personal communication) also show a loss in strand transfer with little affect on 3′ processing. As shown in Figure 4B, G94, S119, S123, W132, I141, F181, F185 and I203 lie on the IN surface in the putative target DNA binding trench (green residues) and are aligned with other residues that cause similar activity defects. As reported here, the K219S mutation also loses strand transfer but maintains 3′ processing activity. In contrast to the above residues, K219 is found on the opposite wall of the trench (see Figure 4B, red residues). A K219A mutation has been analyzed for its effect on HIV-1 replication and was reported to have a limited affect50. It is not known why it did not show a stronger phenotype. This may be related to alanine rather than serine being substituted in this study or reflects differences in sensitivity between in vitro and in cell assays.
Second, the target DNA binding site contains peptides previously shown to be cross-linked to the target DNA portion of a disintegration substrate modified with an azidophenacyl group6. After UV photo activation, cross-links were established between the DNA substrate and six endoproteinases Glu-C digested peptides. In terms of our model, the peptide 139-152 represents the active site between the two LTR ends and contains Q148 as well as Q137, Q146, and N144. A second peptide, 213-247 is found at a distance from the catalytic site in the putative target DNA binding site and contains K219 but not K211. The K211S mutation has no affect on activity of IN in vitro. The other peptides identified in that report were implicated in binding both viral and target DNA substrates or only to the viral DNA substrate in agreement with the model's predictions.
Third, we find a series of residues (Arg, Lys, Gln, and Asn) that appear along the length of the putative DNA binding pocket, which are found in known DNA binding sites of other enzymes51-53 and could therefore be involved in binding to the target DNA. In contrast to the residues interacting with the LTR ends, these residues are conserved among INs to different extents. This would be consistent with IN inserting the viral DNA into many sites in the target DNA. Five of these residues (Q62, N117, Q148, N155, K159) have been mutated and cause defects to 3′ processing, strand transfer, and disintegration8,22,33-35. These residues are predicted to lie in the catalytic site between the ends of the two LTRs so they could interact with both the viral and the target DNAs. This conclusion is supported by a recent study that showed that Q148 cross-linked to the ends of the LTRs54.
Fourth, when we examined the positions in the structural model of 50 amino acid residues described in the literature8,22,35,55-58 where mutations result in either little or no effect on or decreases in both 3′ processing and joining activities, none lay in the proposed target DNA binding trench. The only exception is when the targeted amino acids were positioned between the two LTR ends where they could interact with both viral and target DNAs. Additionally, Puglia et al.59 reported an analysis of HIV-1 IN where in-frame insertions of small peptides were placed at 56 sites. The mutants were analyzed for changes in the joining reaction (but not 3′ processing because a preprocessed substrate was used). We examined the positions of these mutations in our structural model and can interpret their reported activity changes. For example, when the bulky insertions are near the enzyme surface but not near the viral DNA ends or the proposed target DNA binding site, the model predicts and Puglia et al.59 report that there is no effect on activity. In contrast, when the peptide insertions are at the surface near either the viral DNA or target DNA binding sites, we predict that there should be a disruption to the joining reaction as observed. When insertions are buried within the structure we predict distortions that disrupt all activities and this too is seen.
Finally, we mapped the naphthyridine carboxamide and diketo acid related drug resistant sites on the structural model. The drug resistant sites associated with diketo acid compounds (residues 66, 74, 92, 143, 148, and 151-155) map in the active site region of the target DNA binding trench near or between the two LTRs. The napthyridine carboxamide related drug resistant sites (residues 121 and 125) map in the target DNA binding trench near to the LEDGF binding sites shown in Fig. 4. This observation suggests that the napthyridine carboxamide related drugs might interfere with formation of the LEDGF-HIV-1 IN complex. Others (residues 72, and 150) map in the target DNA binding site near the LTRs. Taken together, these results are consistent with the model and the hypothesis for the binding of target DNA.
MATERIALS AND METHODS
Reagents
[γ-33P]-ATP (2500 Ci/mmole) was purchased from Perkin Elmer Life Sciences. HiTrapTM Chelating HP resin and HiTrapTM Heparin HP resin were purchased from GE Healthcare Life Sciences (Piscataway, NJ). T4 polynucleotide kinase was from USB (Cleveland, Ohio). IPTG was from Roche (Indianapolis, IN). The Slide-A-Lyzer Dialysis cassette (10kD MWCO) was obtained from Pierce (Rockford, IL). CentriPrep centrifugal filter devices with YM-10 MW membranes were from Millipore (Bedford, MA). Acrylamide and bisacrylamide solutions were from Bio-Rad (Hercules, CA). SimplyBlue Safe Stain was from Invitrogen (Carlsbad, CA). DE81 filters were purchased from Whatman International Ltd (Kent, UK). Unless specified, all restriction enzymes were purchased from New England Biolabs (Beverly, MA). ASV IN was provided by Dr. Ann Skalka (Fox Chase Cancer Center, Phil, PA). An expression construct for HIV-1 IN 1-288 residues (p28bIN-3CS-F185H) was also obtained from the laboratory of Dr. Ann Skalka and contains the wild type NY5 HIV-1 sequence (Parke Davis clone) from the NdeI to the HindIII site in the pET28b plasmid vector. The IN sequence encodes 4 substitutions (C56S, C65S, C280S, and F185H) to increase solubility and a six amino acid His-tag separated from the N-terminus of IN by a thrombin cleavage site. Two translation stop codons were added after residue D288. MLV oligo substrates were a gift from Monica Roth (UMDMJ).
Bacterial Strains and Growth Conditions
The protein expression host strain BL21(DE3) were purchased from Novagen (Madison, WI). Selection of mutagenesis for chimera construction was performed in Supercompetent XL1-Blue cells from Stratagene (LaJolla, CA). Storage of confirmed clone DNA was carried out in DH5α at −80°C using competent cells from Invitrogen (Carlsbad, CA). Unless otherwise noted, bacteria were selected for using LB+Kan50 media at 37°C.
Preparation of Duplex Oligo Substrates
The following oligos were used in the integrase 3′ processing activity assay:
- HIV-1 U5
- 5′ TGTGGAAAATCTCTAGCAGT 3′ (+)
- 3′ ACACCTTTTAGAGATCGTCA 5′ (−)
- ASV U3
- 5′ GTATTGCATAAGACTACATT 3′ (+)
- 3′ CATAACGTATTCTGATGTAA 5′ (−)
- MLV U5
- 5′ CCGTCAGCGGGGGTCTTTCATT 3′ (+)
- 3′ GGCAGTCGCCCCCAGAAAGTAA 5′ (−)
The following oligos were used in the integrase strand transfer activity assay to simulate pre-processed LTR ends:
- HIV-1 pre-processed U5
- 5′ TGTGGAAAATCTCTAGCA 3′ (+)
- 3′ ACACCTTTTAGAGATCGTCA 5′ (−)
- ASV pre-processed U3
- 5′ GAGTATTGCATAAGACTACA 3′ (+)
- 3′ CTCATAACGTATTCTGATGTAA 5′ (−)
The plus-strand substrates (100pmoles, containing the conserved ‘CA’ dinucleotides) were 5’ end labeled using T4 polynucleotide kinase (30U) and [γ-33P]-ATP as previously described4. The specific activity of the radiolabeled substrates was diluted to 105 cpm/pmol using unlabeled plus-strand oligo and the mixture was purified and recovered from a 20% denaturing polyacrylamide gel. Duplex oligos were formed by annealing to a molar excess of unlabeled complementary strand as described4.
Construction of HIV-1 IN Mutants
Mutagenesis oligos were obtained from Integrated DNA Technologies Inc (Coralville, Iowa) and are listed in Supplementary Data Table 1. The mutations were constructed using QuikChange® Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA) according to manufacturer's directions. Codon preferences for E. coli were used in the oligo design. The presence of all mutations was confirmed by sequencing the complete individual DNA clones. The Wizard® Plus SV Miniprep DNA Purification System (Promega, Madison, WI) was used to prepare DNA for cloning.
Purification of HIV-1 IN chimeras
His-tagged HIV-1 IN chimeras were purified from the soluble fraction as previously described60 with some modification. Briefly, proteins were induced in BL21 (DE3) cells at 20°C by adding IPTG to 0.5mM after the bacteria had grown to optical density at 600nm of 0.8. The bacteria were lysed in 25mM Bis Tris, pH 6.1, 1M KCl, 1M urea, 1% thiodiglycol, 5mM imidazole and then filtered through 0.22μm membrane from Millipore (Billerica, MA). The lysate fraction was applied to a HiTrap™ Chelating HP Ni-affinity column (5ml) and IN was eluted with a 5mM to 1.0M linear imidazole gradient. Fractions containing IN, as detected by absorbance at 280nm and confirmed by SDS-PAGE with staining using the SimplyBlue Safe Stain, were applied to a HiTrap™ Heparin HP column (5ml) and eluted with a 0.25 - 1.0M linear KCl gradient. Selected fractions were concentrated using a Centriprep filter with YM-10 MW membrane and then dialyzed against 25mM Bis Tris, pH 6.1, 0.5M KCl, 1% Thiodiglycol, 1mM DTT, 0.1mM EDTA, and 40% glycerol. The purified protein was aliquoted and stored at −80°C. The protein concentration was determined using a Bio-Rad protein assay as described by the manufacturer.
Integrase 3′ end processing or strand transfer assay using duplex oligodeoxyribonucleotide substrates
The processing reactions for the HIV-1 U5 or ASV U3 LTR substrates were carried out as previously described 61. These two substrates were used because they are the “dominant” LTR ends where substitutions into these sequences caused large decreases in the rate of concerted integration in vitro62-64. Substitutions introduced in the HIV-1 U3 or ASV U5 LTR substrates change the mechanism of concerted DNA integration in vitro from two to one-ended insertion events. Reactions were in a volume of 12μl with 25mM MOPs, pH 7.2, 10mM DTT, 15mM potassium glutamate, 5% PEG8000, 5% DMSO, 500ng of HIV-1 or HIV-1 chimeras and 1pmol of labeled duplex substrate as indicated. Reaction mixtures were assembled from individual components and preincubated overnight at 4°C. To start the processing reaction, MgCl2 was added to final concentration at 10mM and reaction mixtures incubated at 37°C for 90 minutes. The reactions were stopped by the addition of 3μl o f Stop buffer (95% formamide, 20 mM EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue), heated to 95°C for 5 min, and then placed on ice. Products of the reaction were separated through a 20% polyacrylamide denaturing sequencing gel. Labeled reaction products were visualized using KODAK MR film by exposure overnight. For reactions containing ASV IN the final reaction mixture contained 20mM MOPs, pH 7.2, 3mM DTT, 100μg/ml BSA, 500ng ASV IN, and 1pmol labeled duplex substrates as indicated. For the strand transfer assay, the reaction conditions are identical to those used for 3′ end processing, however the substrates used mimic the preprocessed LTR end (designed with 5′-CA dinucleotide overhang) in order to promote strand transfer. The reaction products were analyzed by denaturing gel electrophoresis4,65.
Modeling of LEDGF Complex with IN Tetramer and LTR DNAs
The crystal structure of LEDGF (residues 345-426) bound to the IN catalytic domain dimer (2B4J) was superimposed on the IN catalytic domains in the tetramer model. Then, the program AMMP66 was used to produce and minimize hydrogen atom positions for the two LEDGF monomers and the monomers were minimized using conjugate gradients with all non-bonded and geometric terms. The all-atom sp4 potential set67,68 was used with the charge generation parameters from Bagossi et al.69 and a dielectric of 1.0. The LEDGF monomers were combined with the model consisting of the tetramer of full-length IN with Zn and Mg atoms, two 20-mer LTRs, as described in Chen et al.4. The new model with LEDGF was optimized by 500 cycles of conjugate gradients minimization in AMMP to ensure good non-bonded interactions. The standard AMMP conjugate gradients algorithm with the Polak-Ribere beta and inexact line search was used for optimization. The total non-bonded energy was minimized from 11,754,460.0 kcal/mol to −20,446.6 kcal/mol, and the maximum magnitude (l1 norm) of the derivative of the energy (gradient) was reduced from 1,734,389,397.9 kcal/(mol A) to 50.03 kcal/(mol A) in the final model comprising 22,162 atoms. Figures of the model were made using PyMol70.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yuan-Fang Wang for assistance with the modeling and preparing structural Figures. This work was supported in part by United States Public Health Service Grants AI054143 (to J.L.), GM6290 and GM065762 (to I.T.W. and R.W. H.). J.D. is supported in part by the Training Program in Viral Replication T32 AI060523.
Abbreviations used
- HIV-1
human immunodeficiency virus, type 1
- IN
integrase
- LTR
long terminal repeat
- ASV
avian sarcoma virus
- SIV
simian immunodeficiency virus
- MPMV
Mason Pfizer monkey virus
- MLV
murine leukemia virus
- oligo
oligodeoxyribonucleotide
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 2.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 2005;280:29334–9. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J Virol. 2005;79:8208–16. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen A, Weber IT, Harrison RW, Leis J. Identification of amino acids in HIV-1 and avian sarcoma virus integrase subsites required for specific recognition of the long terminal repeat Ends. J Biol Chem. 2006;281:4173–82. doi: 10.1074/jbc.M510628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snasel J, Krejcik Z, Jencova V, Rosenberg I, Ruml T, Alexandratos J, Gustchina A, Pichova I. Integrase of Mason-Pfizer monkey virus. Febs J. 2005;272:203–16. doi: 10.1111/j.1432-1033.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 6.Heuer TS, Brown PO. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–65. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 7.Bischerour J, Tauc P, Leh H, de Rocquigny H, Roques B, Mouscadet JF. The (52-96) C-terminal domain of Vpr stimulates HIV-1 IN-mediated homologous strand transfer of mini-viral DNA. Nucleic Acids Res. 2003;31:2694–702. doi: 10.1093/nar/gkg364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman A, Liu Y, Chen H, Farzan M, Dyda F. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J Virol. 1997;71:3507–14. doi: 10.1128/jvi.71.5.3507-3514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu K, Dobard C, Chow SA. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol. 2004;78:5045–55. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villanueva RA, Jonsson CB, Jones J, Georgiadis MM, Roth MJ. Differential multimerization of Moloney murine leukemia virus integrase purified under nondenaturing conditions. Virology. 2003;316:146–60. doi: 10.1016/s0042-6822(03)00559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fikkert V, Van Maele B, Vercammen J, Hantson A, Van Remoortel B, Michiels M, Gurnari C, Pannecouque C, De Maeyer M, Engelborghs Y, De Clercq E, Debyser Z, Witvrouw M. Development of resistance against diketo derivatives of human immunodeficiency virus type 1 by progressive accumulation of integrase mutations. J Virol. 2003;77:11459–70. doi: 10.1128/JVI.77.21.11459-11470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, Fisher TE, Embrey M, Guare JP, Jr., Egbertson MS, Vacca JP, Huff JR, Felock PJ, Witmer MV, Stillmock KA, Danovich R, Grobler J, Miller MD, Espeseth AS, Jin L, Chen IW, Lin JH, Kassahun K, Ellis JD, Wong BK, Xu W, Pearson PG, Schleif WA, Cortese R, Emini E, Summa V, Holloway MK, Young SD. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc Natl Acad Sci U S A. 2004;101:11233–8. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A. 2005;102:17308–13. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280:17841–7. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 15.Rahman S, Lu R, Vandegraaff N, Cherepanov P, Engelman A. Structure-based mutagenesis of the integrase-LEDGF/p75 interface uncouples a strict correlation between in vitro protein binding and HIV-1 fitness. Virology. 2007;357:79–90. doi: 10.1016/j.virol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Raghavendra NK, Engelman A. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: Implications for the mechanism of viral cDNA integration. Virology. 2007;360:1–5. doi: 10.1016/j.virol.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–4. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 18.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–73. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 19.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–95. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J Cell Sci. 2005;118:1733–43. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 21.Konsavage WM, Jr., Burkholder S, Sudol M, Harper AL, Katzman M. A substitution in rous sarcoma virus integrase that separates its two biologically relevant enzymatic activities. J Virol. 2005;79:4691–9. doi: 10.1128/JVI.79.8.4691-4699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerton JL, Ohgi S, Olsen M, DeRisi J, Brown PO. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J Virol. 1998;72:5046–55. doi: 10.1128/jvi.72.6.5046-5055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Mawsawi LQ, Sechi M, Neamati N. Single amino acid substitution in HIV-1 integrase catalytic core causes a dramatic shift in inhibitor selectivity. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 24.Al-Mawsawi LQ, Fikkert V, Dayam R, Witvrouw M, Burke TR, Jr., Borchers CH, Neamati N. Discovery of a small-molecule HIV-1 integrase inhibitor-binding site. Proc Natl Acad Sci U S A. 2006;103:10080–5. doi: 10.1073/pnas.0511254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Mawsawi LQ, Hombrouck A, Dayam R, Debyser Z, Neamati N. Four-tiered pi interaction at the dimeric interface of HIV-1 integrase critical for DNA integration and viral infectivity. Virology. 2008;377:355–63. doi: 10.1016/j.virol.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 26.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–86. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Cellular cofactors of HIV-1 integration. Trends Biochem Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–81. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. Embo J. 2006;25:1295–304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vora A, Grandgenett DP. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. J Virol. 2001;75:3556–67. doi: 10.1128/JVI.75.8.3556-3567.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drake RR, Neamati N, Hong H, Pilon AA, Sunthankar P, Hume SD, Milne GW, Pommier Y. Identification of a nucleotide binding site in HIV-1 integrase. Proc Natl Acad Sci U S A. 1998;95:4170–5. doi: 10.1073/pnas.95.8.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao K, Butler SL, Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. Embo J. 2001;20:3565–76. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins TM, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. Embo J. 1997;16:6849–59. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. Embo J. 1998;17:5832–43. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutzke RA, Vink C, Plasterk RH. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–31. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agapkina J, Smolov M, Barbe S, Zubin E, Zatsepin T, Deprez E, Le Bret M, Mouscadet JF, Gottikh M. Probing of HIV-1 integrase/DNA interactions using novel analogs of viral DNA. J Biol Chem. 2006;281:11530–40. doi: 10.1074/jbc.M512271200. [DOI] [PubMed] [Google Scholar]
- 37.Guare JP, Wai JS, Gomez RP, Anthony NJ, Jolly SM, Cortes AR, Vacca JP, Felock PJ, Stillmock KA, Schleif WA, Moyer G, Gabryelski LJ, Jin L, Chen IW, Hazuda DJ, Young SD. A series of 5-aminosubstituted 4-fluorobenzyl-8-hydroxy[1,6]naphthyridine-7-carboxamide HIV-1 integrase inhibitors. Bioorg Med Chem Lett. 2006;16:2900–4. doi: 10.1016/j.bmcl.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Embrey MW, Wai JS, Funk TW, Homnick CF, Perlow DS, Young SD, Vacca JP, Hazuda DJ, Felock PJ, Stillmock KA, Witmer MV, Moyer G, Schleif WA, Gabryelski LJ, Jin L, Chen IW, Ellis JD, Wong BK, Lin JH, Leonard YM, Tsou NN, Zhuang L. A series of 5-(5,6)-dihydrouracil substituted 8-hydroxy[1,6]naphthyridine-7-carboxylic acid 4-fluorobenzylamide inhibitors of HIV-1 integrase and viral replication in cells. Bioorg Med Chem Lett. 2005;15:4550–4. doi: 10.1016/j.bmcl.2005.06.105. [DOI] [PubMed] [Google Scholar]
- 39.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–50. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 40.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci U S A. 2002;99:6661–6. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchand C, Zhang X, Pais GC, Cowansage K, Neamati N, Burke TR, Jr, Pommier Y. Structural determinants for HIV-1 integrase inhibition by beta-diketo acids. J Biol Chem. 2002;277:12596–603. doi: 10.1074/jbc.M110758200. [DOI] [PubMed] [Google Scholar]
- 42.Pluymers W, Pais G, Van Maele B, Pannecouque C, Fikkert V, Burke TR, Jr., De Clercq E, Witvrouw M, Neamati N, Debyser Z. Inhibition of human immunodeficiency virus type 1 integration by diketo derivatives. Antimicrob Agents Chemother. 2002;46:3292–7. doi: 10.1128/AAC.46.10.3292-3297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fikkert V, Hombrouck A, Van Remoortel B, De Maeyer M, Pannecouque C, De Clercq E, Debyser Z, Witvrouw M. Multiple mutations in human immunodeficiency virus-1 integrase confer resistance to the clinical trial drug S-1360. Aids. 2004;18:2019–28. doi: 10.1097/00002030-200410210-00006. [DOI] [PubMed] [Google Scholar]
- 44.Lee DJ, Robinson WE., Jr Human immunodeficiency virus type 1 (HIV-1) integrase: resistance to diketo acid integrase inhibitors impairs HIV-1 replication and integration and confers cross-resistance to L-chicoric acid. J Virol. 2004;78:5835–47. doi: 10.1128/JVI.78.11.5835-5847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu R, Limon A, Ghory HZ, Engelman A. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. J Virol. 2005;79:2493–505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–39. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 47.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–9. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 48.Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, Christ F, Rain JC, Dargemont C, Debyser Z, Benarous R. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem. 2005;280:25517–23. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 49.Harper AL, Skinner LM, Sudol M, Katzman M. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J Virol. 2001;75:7756–62. doi: 10.1128/JVI.75.16.7756-7762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu R, Limon A, Devroe E, Silver PA, Cherepanov P, Engelman A. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J Virol. 2004;78:12735–46. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeltsch A, Wenz C, Wende W, Selent U, Pingoud A. Engineering novel restriction endonucleases: principles and applications. Trends Biotechnol. 1996;14:235–8. doi: 10.1016/0167-7799(96)10030-5. [DOI] [PubMed] [Google Scholar]
- 52.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. Embo J. 1993;12:1781–95. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z, Horton JR, Maunus R, Wilson GG, Roberts RJ, Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson AA, Santos W, Pais GC, Marchand C, Amin R, Burke TR, Jr., Verdine G, Pommier Y. Integration requires a specific interaction of the donor DNA terminal 5′-cytosine with glutamine 148 of the HIV-1 integrase flexible loop. J Biol Chem. 2006;281:461–7. doi: 10.1074/jbc.M511348200. [DOI] [PubMed] [Google Scholar]
- 55.Bischerour J, Leh H, Deprez E, Brochon JC, Mouscadet JF. Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J Virol. 2003;77:135–41. doi: 10.1128/JVI.77.1.135-141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Priet S, Navarro JM, Querat G, Sire J. Reversion of the lethal phenotype of an HIV-1 integrase mutant virus by overexpression of the same integrase mutant protein. J Biol Chem. 2003;278:20724–30. doi: 10.1074/jbc.M301768200. [DOI] [PubMed] [Google Scholar]
- 57.Sayasith K, Sauve G, Yelle J. Characterization of mutant HIV-1 integrase carrying amino acid changes in the catalytic domain. Mol Cells. 2000;10:525–32. doi: 10.1007/s10059-000-0525-3. [DOI] [PubMed] [Google Scholar]
- 58.Vincent KA, Ellison V, Chow SA, Brown PO. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–37. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puglia J, Wang T, Smith-Snyder C, Cote M, Scher M, Pelletier JN, John S, Jonsson CB, Roth MJ. Revealing domain structure through linker-scanning analysis of the murine leukemia virus (MuLV) RNase H and MuLV and human immunodeficiency virus type 1 integrase proteins. J Virol. 2006;80:9497–510. doi: 10.1128/JVI.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yi J, Asante-Appiah E, Skalka AM. Divalent cations stimulate preferential recognition of a viral DNA end by HIV-1 integrase. Biochemistry. 1999;38:8458–68. doi: 10.1021/bi982870n. [DOI] [PubMed] [Google Scholar]
- 61.Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712–8. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 62.Hindmarsh P, Johnson M, Reeves R, Leis J. Base-pair substitutions in avian sarcoma virus U5 and U3 long terminal repeat sequences alter the process of DNA integration in vitro. J Virol. 2001;75:1132–41. doi: 10.1128/JVI.75.3.1132-1141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brin E, Leis J. Changes in the mechanism of DNA integration in vitro induced by base substitutions in the HIV-1 U5 and U3 terminal sequences. J Biol Chem. 2002;277:10938–48. doi: 10.1074/jbc.M108116200. [DOI] [PubMed] [Google Scholar]
- 64.Brin E, Leis J. HIV-1 integrase interaction with U3 and U5 terminal sequences in vitro defined using substrates with random sequences. J Biol Chem. 2002;277:18357–64. doi: 10.1074/jbc.M201354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–27. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison RW. Stiffness and energy conservation in molecular dynamics: An improved integrator. 1993;14:1112–1122. [Google Scholar]
- 67.Weber IT, Harrison RW. Molecular mechanics calculations on HIV-1 protease with peptide substrates correlate with experimental data. Protein Eng. 1996;9:679–90. doi: 10.1093/protein/9.8.679. [DOI] [PubMed] [Google Scholar]
- 68.Weber IT, Harrison RW. Molecular mechanics calculations on Rous sarcoma virus protease with peptide substrates. Protein Sci. 1997;6:2365–74. doi: 10.1002/pro.5560061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bagossi P, Zahucky G, Tozser J, Weber IT, Harrison RW. Improved Parameters for Generating Partial Charges: Correlation with Observed Dipole Moments. J Mol Model. 1999;5:143–152. [Google Scholar]
- 70.De Lano WL. The PyMol Molecular Graphics System v1.00 edit. 2002 [Google Scholar]
- 71.Heuer TS, Brown PO. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–78. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.