Abstract
Summary: We present a web-based service, SimCT, which allows to graphically display the relationships between biological objects (e.g. genes or proteins) based on their annotations to a biomedical ontology. The result is presented as a tree of these objects, which can be viewed and explored through a specific java applet designed to highlight relevant features. Unlike the numerous tools that search for overrepresented terms, SimCT draws a simplified representation of biological terms present in the set of objects, and can be applied to any ontology for which annotation data is available. Being web-based, it does not require prior installation, and provides an intuitive, easy-to-use service.
Availability: http://tagc.univ-mrs.fr/SimCT
Contact: carl.herrmann@univmed.fr
Supplementary information: Supplementary data are available at Bioinformatics online
1 INTRODUCTION
The wealth of data available from large-scale experiments in recent years has made the development of efficient tools to visualize, analyze, interpret and share post-genomic data a crucial endeavor. Among these, biomedical ontologies have been increasingly developed to annotate genome-wide features, and numerous fields of biology are now covered by a dedicated ontology. If Gene Ontology (GO) appears to be the pioneering project, many other projects are actively pursued (e.g. Robinson et al., 2008; see http://www.obofoundry.org/ for available biomedical ontologies) and their adoption by genome databases such as MGI, Wormbase and Flybase, will ensure that they will be increasingly used in the community. In this note, we present a generic, web-based tool called SimCT (Similarity Clustering Tool) which allows the visualization of the relations between biological objects (e.g. genes, proteins, etc.) based on their annotations to an ontology, in the form of a clustering tree. Our clustering procedure is a way to turn the ontology into a simplified tree (which is a subgraph of the ontology), which better represents the terms associated to a list of objects, therefore highlighting their relationships. This representation could neither be obtained by mapping the annotations onto the ontology, due to its complexity, nor by searching for overrepresented terms, which by definition overlooks terms that are not statistically relevant. The visualization is done using a dedicated java applet. Although many tools have been developed for GO, very few comparable tools exist for other biomedical ontologies yet.
2 METHODS
To measure the specificity of a term t in an ontology O, we have introduced the notion of precision as follows (see Supplementary Material for details, in particular the glossary for definition of terms used):
| (1) |
where Nd(t) represents the number of descendant terms of t, Na(t) the number of ancestor terms of t, N the total number of terms in O and Namax the maximal number of ancestors a term can have in O. Interestingly, our definition of precision only depends on the structure of the ontology and not on annotation statistics like in Lord et al. (2003). Therefore, it can be applied to any existing ontology. Additionally, precision differs from information content, which gives equal specificity to all leaves of the ontology (Resnik, 1999; Schlicker et al., 2006; Wang et al., 2007). Based on precision, we define the similarity of two terms as the precision of their most precise common ancestor.
Given a list of objects annotated to ontology, we consider the set of (object| ontology term) pairs. If an object has several annotations, it generates several (object|ontology term) pairs. We have implemented an aggregative clustering algorithm that builds the clustering tree based on the similarity between terms. The leaves of the resulting tree are the (object|ontology term) pairs and the internal nodes are ontology terms. We attach to each internal node a numerical index called Subtree Relevance Index (SRI):
| (2) |
where T represents a subtree, t the ontology term attached to it, p(t) its precision and N(T) is the number of leaves of the subtree. It measures the relevance of each term for the list of objects submitted (Supplementary Material). The topology of the tree respects that of the underlying ontology (i.e. it is included in the directed acyclic graph (DAG) of the ontology).
3 IMPLEMENTATION
SimCT can be used in two different ways, depending on the ontology the user is interested in:
With GO, the user can input a list of genes/proteins, select the corresponding organism (29 are currently available) and the GO sub-ontologies. The system retrieves available annotations.
For other ontologies, the user must provide a two-columns list of objects associated to their annotations, and select the corresponding ontology among the 25 currently available. The user can also provide custom GO annotations.
In both the cases, the (object|ontology term) list is processed by the clustering algorithm. Once done, the user can open a java applet which displays the tree(s). As an example, the clustering of 300 leaves takes ∼40 s.
4 APPLICATIONS
4.1 Disease ontology
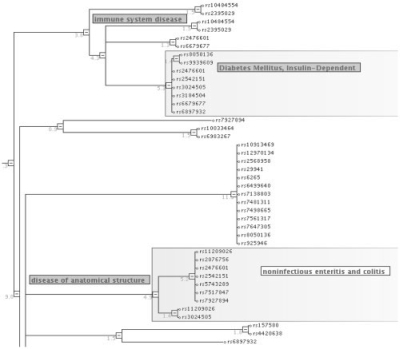
To illustrate the use of SimCT, we have extracted from http://www.genome.gov/gwastudies/ a list of 79 single nucleotide polymorphisms (SNP) associated to a disease, described using the Disease Ontology (http://diseaseontology.sourceforge.net/). Figure 1 shows the resulting tree, in which two subtrees are highlighted: Diabetes Mellitus and Noninfectious enteritis and colitis. Taking the intersection of both trees through the applet menu reveals that three SNPs are simultaneously associated to both diseases (rs3024505, rs2542151, rs2476601). Interestingly, the latter two are close to or inside two genes, respectively, PTPN2 and PTPN22. Inspection of the OMIM entries related to both genes shows that only the first one is explicitly associated to both diabetes and enteritis, while PTPN22 is only associated to diabetes. Thus, our result suggests that we could add an additional annotation to PTPN22, namely enteritis.
Fig. 1.
A clustering tree of SNPs associated to diseases. Highlighted are two subtrees, Diabetes Mellitus (top) and Noninfectious enteritis and colitis (bottom).
4.2 Gene Ontology
We chose a set of 69 coregulated genes extracted from Transcriptome Browser (Lopez et al., 2008) around the natural killer (NK) gene NCR3 in human. We compared the P-values of the nodes with SRI ≥ 2.5 with the P-values given by DAVID (Dennis et al., 2003) and GO:TermFinder (Boyle et al., 2004). The differences between the SimCT approach and the search for overrepresented terms are highlighted in Supplementary Table S2. In particular, although no term related to biopolymer synthesis is found by DAVID as overrepresented, SimCT detects that five genes of the list are related to transcriptional (TBX21, CEBPD, TAF6L, GFI1) or translational (EIF5B, RPS8) processes which are child terms of biopolymer synthesis. These are the effectors at the end of the cascade of NK activity, leading for instance to the production of gamma-interferon.
5 CONCLUSION
Our approach can be compared with GOSurfer (Zhong et al., 2004), GOTreePlus (Lee et al., 2008) or GO::TermFinder (Boyle et al., 2004). However, SimCT includes the possibility to work with other biomedical ontologies than GO, and is web-based. Therefore, it provides an intuitive, easy-to-use and immediately available service, which allows to draw a clear picture of the ontological terms represented in a list of biological objects annotated to ontology, for any biomedical ontology. The viewer applet helps easily exploring and annotating the resulting tree to highlight its most relevant features. As more and more ontologies are being developed, we believe that this tool will prove very useful in working with these.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank E. Choron, M. Hermes, P. Bonnaure, L. Baumgaertel and C. Lepoivre for their participation in the developments of the java TreeViewer applet. We thank P. Rihet, C. Brun and B. Jacq for discussions and feedback.
Conflict of Interest: none declared.
REFERENCES
- Boyle EI, et al. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis GJ, et al. David: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [Epub ahead of print, April 3, 2003] [PubMed] [Google Scholar]
- Lee B, et al. Gotreeplus: an interactive gene ontology browser for proteomics projects. Bioinformatics. 2008;24:1026–1028. doi: 10.1093/bioinformatics/btn068. [DOI] [PubMed] [Google Scholar]
- Lopez F, et al. Transcriptomebrowser: a powerful and flexible toolbox to explore productively the transcriptional landscape of the gene expression omnibus database. PLoS ONE. 2008;3:e4001. doi: 10.1371/journal.pone.0004001. [Epub ahead of print, December 23, 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord PW, et al. Investigating semantic similarity measures across the gene ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- Resnik P. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J. Artif. Intell. Res. 1999;11:95–130. [Google Scholar]
- Robinson PN, et al. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker A, et al. A new measure for functional similarity of gene products based on gene ontology. BMC Bioinformatics. 2006;7:302–308. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, et al. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- Zhong S, et al. Gosurfer: a graphical interactive tool for comparative analysis of large gene sets in gene ontology space. Appl. Bioinform. 2004;3:261–264. doi: 10.2165/00822942-200403040-00009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.