Abstract
Purpose:
DNA damage recognition and repair play a major role in risk for breast cancer. We investigated 104 single nucleotide polymorphisms in 17 genes whose protein products are involved in double stranded break repair (DSBR).
Experimental Design:
We used a case-control design. Both the case individuals affected with breast cancer or with both breast and ovarian cancer; and the controls had similar familial risk of breast cancer and were participants in a high risk cancer registry.
Results:
We found that 12 of the polymorphisms are associated with breast or breast and ovarian cancer, most notably rs16888927, rs16888997 and rs16889040, found in introns of RAD21, suggesting that SNPs in other genes in the DSBR pathway in addition to BRCA1 and BRCA2 may affect breast cancer risk.
Conclusions:
SNPs within or near a number of Double Strand Break Repair (DSBR) DNA repair pathway genes are associated with breast cancer in individuals from a high-risk population. In addition, our study reemphasizes the unique perspective that recruitment of cases and controls from family cancer registries has for gene discovery studies.
INTRODUCTION
It is well established that alterations of genes involved in DNA damage recognition and repair are associated with cancer risk. Mutations of genes in the DNA repair pathways, such as BRCA1, BRCA2 (1) and ATM (2) confer an increased risk of breast cancer. However, the majority of familial breast cancers have yet to yield single causative mutations. It has been hypothesized that common genetic polymorphisms within DNA damage repair pathway genes may contribute to the development of familial breast cancer.
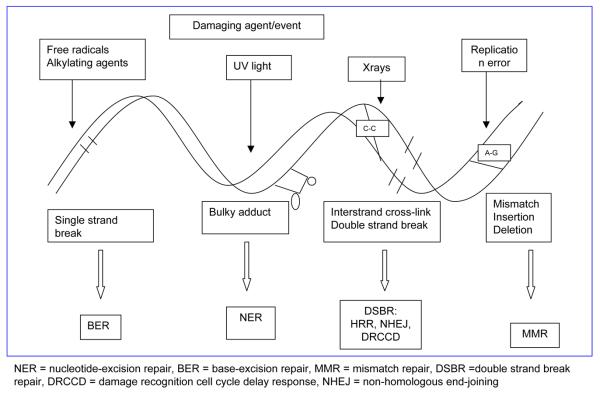
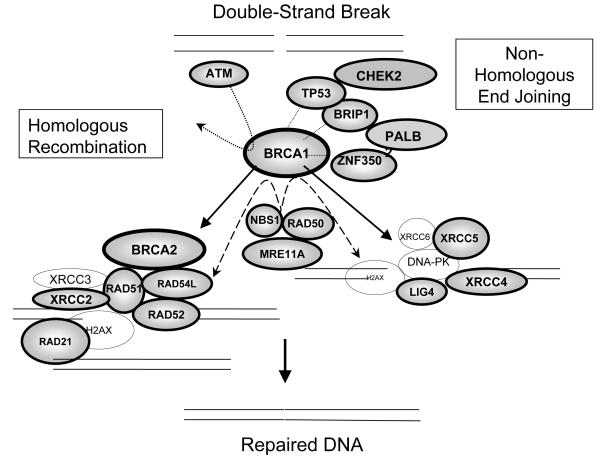
DNA is subject to continuous damage by exogenous chemical or physical agents, or by endogenous byproducts of normal cellular metabolism. DNA damage can trigger cell cycle arrest and DNA repair or, when the damage is too severe for repair, cellular death by apoptosis. Long-term effects of DNA damage may lead to irreversible mutations, contributing to oncogenesis. Thus, compromised DNA repair function likely plays an important role in carcinogenesis and acts as a genetic risk factor for cancers, including breast cancer. Cells from patients with inherited syndromes involving mutations in genes responsible for the integrity of the genome and chromosomal stability, and associated with either cancer predisposition or accelerated senescence, display elevated frequencies of chromosomal aberrations (3-5). Several partially overlapping DNA repair pathways operate in mammals, including nucleotide-excision repair (NER), base-excision repair (BER), mismatch repair (MMR), and double strand break repair (DSBR) (Figure 1). Both BRCA1 and BRCA2 function in the DSBR pathways of the cell (Figure 2).
Figure 1.
Mechanisms of DNA repair
Figure 2.
Roles of the selected genes in the DSBR pathway.
A large number of single nucleotide polymorphisms (SNPs) in DNA-repair genes have been identified. Single nucleotide alterations may influence protein expression or gene function, either by directly leading to amino acid change and modification in protein function or through indirect epigenetic changes in synonymous or non-coding SNPs. Much of the genetic variation in the DNA repair capacity of the general population likely resides in common SNPs in DNA-repair genes. Individual susceptibility to breast cancer may lie in variations in the ability of the cell to respond to DNA damage. Some studies have suggested that specific single nucleotide polymorphisms (SNPs) in genes in the DSBR pathway are associated with breast cancer risk including: XRCC2 (6-8), XRCC4 (9), ZNF350 (10, 11), RAD51 (12), RAD52, LIG4 (8), and ATM (13). A recent candidate gene study using a population based study design (14) systematically examined DSBR genes (164 SNPs in 20 DSBR genes) and found none of these genes was associated with breast cancer after correcting for multiple testing. Our study also systematically examines DSBR genes, (104 SNPs in 17 genes) including 13 genes examined by Pharaoh et al. but in a very differently ascertained sample of cases and controls.
Through the UCLA Familial Cancer Registry, we have access to a highly enriched population of individuals selected for their risk of familial breast cancer. Although some of these individuals carry BRCA1 or BRCA2 (BRCA1/2) mutations, the majority do not. The primary aim of this study is to examine selected SNPs in 17 important DNA-DSBR genes to determine their association with breast cancer in this high risk population.
SUBJECTS AND METHODS
UCLA Familial Cancer Registry
Epidemiological and clinical data and biological specimens for this study were collected from the UCLA Familial Cancer Registry and Genetic Evaluation Program. The program is a shared resource of the Jonsson Comprehensive Cancer Center that was established to facilitate genetic, behavioral, and biological research. To be eligible for the Registry, both affected and unaffected participants must meet the following criteria: age 18 or older, and either (1) their family history must contain at least 2 first- or second-degree relatives with the same primary cancer or cancers that are known to be related (i.e., breast and ovarian), or (2) they have a personal/family history of known cancer genetic susceptibility (e.g., a mutation in BRCA1). The Registry began accepting participants in September of 1998 and recruits new participants on an ongoing basis; the women in the current study were enrolled between April 1999 and December 2005 when this study was initiated. Men with similar medical histories or family backgrounds were excluded from this study due to small numbers in the Registry and their potentially different genetic risks of breast cancer. Further details of Registry recruitment and procedures have been discussed elsewhere (15, 16).
Study Patient population
All study subjects have given signed informed consent to participate in the Registry research protocol, which includes this and other studies and has been approved by the UCLA Institutional Review Board. As a result, specimens and questionnaire data could be used for this study without additional consent. We selected for analysis unrelated Caucasian women who either had breast cancer, breast and ovarian cancer, or were unaffected (controls). See the result section for details.
At enrollment, registry participants completed an initial questionnaire (covering demographics, personal health habits and health status, reproductive history, medication use, and family history of cancer). In addition, a Certified Genetic Counselor (JLS) interviewed participants and prepared a multi-generation pedigree containing all known genetic- and cancer-related information on relatives. Questionnaires and pedigree data are updated annually.
A vial of whole blood was used for the DNA purification; the blood samples were processed according to the specified protocol and stored in the designated Revco −80 C Freezer for the project located at UCLA Immunogenetics Center. Results from BRCA1 and BRCA2 testing were obtained for each participant via commercial clinical testing at Myriad Laboratories or in some cases in outside of the US. In some cases, where testing was limited to a specific familial mutation in either BRCA1 or BRCA2, we inferred that the individual was negative for the other.
SNP Selection
Seventeen genes involved in the DNA-DSBR pathway were systematically selected for evaluation based on evidence for their role in either the homologous recombination repair (HR) or the non-homologous end joining (NHEJ) pathways as shown in figure 2. When possible, we included known functional SNPs within the DNA double strand break repair pathway and potentially functional SNPs such as amino-acid-changing (nonsynonymous) SNPs (nsSNPs). All the SNPs selected have a minor allele frequency (MAF) greater than 5% in the NCI SNP 500 project (http://snp500cancer.nci.nih.gov/start.cfm), NCBI's dbSNP http://www.ncbi.nlm.nih.gov/projects/SNP/), NIEHS's GeneSNPs http://egp.gs.washington.edu/B.html or http://www.genome.utah.edu/genesnps/), as well as in the published literature.
The final set of SNPs used in the analysis were selected based on (1) successful genotyping on at least 75% of the Caucasian participants (see Genotyping QC for details), (2) MAFs greater than 0.10 in the unaffected Caucasian participant sample, (3) p-values from HWE testing that are greater than 0.0050 in the unaffected Caucasian participant sample, (4) limited linkage disequilibrium (LD) among the SNPs in the unaffected Caucasian participant sample as defined by r2 <=0.95 with all the other SNPs, or selection as a tag SNP using Haploview Tagger algorithm (haploview 4.0, 17, 18), and (5) the availability of coding information through Applied Biosystems SNPlex technology. The resulting 104 SNPs that met these requirements are in or near one of 17 DNA-DSBR genes (Table 1). SNPs were grouped into haplotype blocks using Haploview version 4.1 (denoted by brackets in Table 1). Haplotype blocks were defined by evidence of linkage disequilibrium greater than D'of 0.95 as described by Gabriel et al. (Science, 2002).
Table 1.
SNPs analyzed, organized by gene
| Chromosome 1 |
| RAD54L: rs104877 |
| Chromosome 2 |
| XRCC5: *[rs3770521, rs3815855], rs2303400, rs3821107, [rs9288518, rs2440], rs207887 |
| Chromosome 5 |
|
XRCC4: rs2075685, rs10474081, rs1120476, rs10514249, rs2731858, rs1193695, rs301279, [rs1056503, rs9293337] |
| RAD50: rs10520114, rs2240032 |
| Chromosome 7 |
| XRCC2: rs3218536, [rs3218499, rs3218408, rs3218374], rs3218373, rs6464268 |
| Chromosome 8 |
|
NBS1: [rs1063054, rs1063053], rs6470522, [rs709816, rs1805786, rs7010210, rs2234744, rs741778, rs1805794, rs1063045] |
| RAD21: [rs16888887, rs2289938, rs1050838], [rs16888927, rs3816342, rs16888997, rs16889040] |
| Chromosome 11 |
| MRE11A: rs499952, rs654718, rs533984 |
| ATM: [rs228591, rs623860, rs228592, rs1801516, rs170548] |
| Chromosome 12 |
|
RAD52: rs7310449, [rs11571476, rs10744729], rs9634161, rs10849594, rs1833095, rs7311151, rs2887532, rs2887531, rs3748523 |
| Chromosome 13 |
|
BRCA2: rs206143, rs1799943, [rs3752451, rs2126042, rs144848, rs1801406, rs3764792], rs542551, [rs206340, rs1012129, rs15869] |
| LIG4: rs1805388 |
| Chromosome 15 |
| RAD51, rs2619681, [rs12593359, rs11855560, rs11852786] |
| Chromosome 17 |
| TP53: rs2909430 |
| BRCA1: [rs12516, rs2236762], rs3737559, [rs799917, rs16940, rs1799949, rs799923] |
|
BRIP1: rs4986763, [rs4986764, rs4986765, rs6504063, rs2158005, rs2191249], rs9897928, rs1978244, [rs6504072, rs12453935], rs2378908, [rs4988344, rs4968451], rs12451910 |
| Chromosome 19 |
| ZNF350: rs2278414, [rs4988334, rs2278416, rs2278418, rs2278419], rs8112515 |
Brackets are used to denote those SNPs located within the same haplotype block, where haplotype blocks were defined using the method described by Gabriel et al. (49).
Whole Genome Amplification
Genomic DNA was isolated from peripheral blood mononuclear cells using the Qiagen extraction method (Qiagen, Valencia, CA). The REPLI-g Midi Kit (Cat. No. 150045) from Qiagen, was used for the Whole Genome Amplification (WGA) according to the manufacturer's specifications. Briefly, 2.5 ul of each DNA sample was denatured by adding 2.5 ul of denaturation buffer. After denaturation was stopped by the addition of 5.0 ul neutralization buffer, a master mix of 40 ul containing REPLI-g Midi Reaction buffer and DNA polymerase was added to each reaction tube. The reaction tubes were incubated at 30° C for 16 hours and 65° C for 3 minutes and 4° C on GeneAmp 9700, Applied Biosystems. DNA concentration and yield was determined by Quant-iT PicoGreen dsDNA reagent kit (Invitrogen, Cat. No. P7581), and the fluorescence was measured in a BioTek FLX800 fluorescence microplate reader with excitation set at 480 nm and emission at 520 nm. The DNA concentration values were obtained by plotting the concentration of DNA standards (ug/ml) (X-axis) against the fluorescence reading generated by the microplate reader (y-axis). The average product length was typically greater than 10 kb (range 2 kb-100 kb) and yielded approximately 40 ug of amplified DNA per 50 ul reaction. As a verification of whole genome amplification we assayed the PON1 rs662 polymorphism using 14 pairs of original and amplified DNA samples. We found 100% concordance between the original and the amplified DNA.
Genotyping assays
Genotypes were determined using the Applied Biosystems SNPlex™ assay (19). This method assays SNP genotypes in 48 SNP PCR multiplexes using an oligonucleotide ligation process. Each SNP allele was ligated to an allele specific oligonucleotide, and then hybridized with a Zipchute™ probe with a mobility modifier and a fluorescent label, allowing the probe to be separated and detected by capillary electrophoresis. Detection was performed on an Applied Biosystems 3730 DNA Analyzer, and data interpretation was performed with the Applied Biosystems Genemapper software v4.0, which uses a clustering algorithm with stringent quality checks to call genotypes. Only genotypes with a Genemapper Quality Score of greater than 95% were passed. With the SNPlex genotyping assay the UCLA Genotyping Core achieves an average call rate of 96%, a reproducibility rate of 99.7%, and a concordance rate of 99.8% (personal communication JCP). Genotype concordance is validated by allelic discrimination assay on an Applied Biosystems 7900HT Fast Real-Time PCR System.
Genotyping QC
Prior to starting the genotyping assays, each whole genome amplified DNA sample was run on an agarose gel to check DNA quality. Every SNP primer set was checked for performance by running it against an Applied Biosystems standard DNA set: SNPlex System Dried gDNA Plates Kit (PN 4362637).
All genotyping was performed blind to family structure and disease status (affected or unaffected by cancer). Genemapper genotyping was performed by two different individuals: one individual doing the first scoring, and the second individual checking the results obtained by the first scorer.
After genotype scoring using the Genemapper software, the data were exported to the SNP Core's Integrated Genetic Database (IGDB). This database holds all the genotypes generated in the Core as well as pedigree information, phenotype data, and marker information such as heterozygosity, ethnic allele frequencies, and genetic and physical maps. This information is used in the data quality checks. All genotypes were checked for HWE and divergence from published allele frequencies. These checks were performed across the entire sample, and also on individual genotyping runs, in order to identify any possible bad runs. Any markers, sample sets, or genotyping runs that gave suspect results on HWE or on any quality control validation tests were manually rescored. If the problem persisted upon manual rescoring, the assays were repeated. Questionable data that could not be resolved by reevaluation or rerunning were discarded.
When several members of the same family were genotyped, an additional QC check using multipoint mistyping analysis was performed using Mendel 7.0 (20, 21). These mistyping procedures identify both Mendelian errors and unlikely double recombinants. All genotypes flagged as potential errors were checked in the raw data. If the errors could not be resolved by reevaluation of the genotype calls, all genotypes that lead to the Mendelian errors, or unlikely double recombinants, were removed if the probability was greater than 0.6.
Statistical Analysis
Unconditional logistic regression analyses were used to calculate adjusted odds ratios (aORs) and 95% confidence intervals (CIs) to examine associations between each SNP and breast cancer while adjusting for other confounding factors (covariates). The potential confounding factors included in our analysis were: age, education, Ashkenazi Jewish ancestry, BRCA1 and BRCA2 mutational status, smoking and alcohol history, body mass index (BMI) (current and change from age 18), past use of oral contraceptives and hormone replacement therapy, parity, and menopausal status. Age, BMI (kg/m2), and changes in BMI from age to age 18 to present (kg/m2) were coded as continuous variables, while tobacco history (≥100 vs. >100 cigarettes in lifetime), alcohol history (<12 vs. ≥12 drinks in last 12 months), oral contraceptive use and hormone therapy history (never vs. ever), pregnancy (never vs. ever), and menopausal status (menstrual period in last 12 months: no/yes) were coded as binary 0 or 1 variables. Education was grouped into three levels and treated as a categorical variable ((1) grade school to associate degree, (2) college graduate to some college or professional school after college graduation, (3) masters degree and higher). BRCA1 and BRCA2 mutational status were coded as binary variables; BRCA1/2 variants that confer risk were coded as a 1, negative results and polymorphisms were coded as 0, and variants of uncertain significance (VUS) were treated as missing data. The dependent variable, breast cancer status, was coded as a binary outcome, 0 for unaffected, and 1 for those participants with either breast cancer or both breast and ovarian cancer.
Model selection was performed on these covariates using a backwards and forwards stepwise logistic regression using the statistical package R: A Language and Environment for Statistical Computing (R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria, 2006, http://www.R-project.org). Putative confounding factors found to significantly improve the prediction of breast cancer status were then included as covariates in the final models in which SNPs were introduced individually. Each SNP was analyzed assuming log-additive and dominant genetic effects and adjusting for the significant covariates.
A haplotype analysis was performed on SNPs of individual genes for which there were more than one significant SNP. The haplotype frequencies were estimated using the Combining Loci option of Mendel version 8.0 (20-22) and haplotype counts for each individual were calculated using the QTL association option of Mendel (23). Using the most frequent haplotype as a reference group, an additive model was used to introduce haplotype counts for a given gene into the logistic regression. A likelihood ratio test was used to investigate whether the haplotypes were associated with breast cancer.
RESULTS
Participant Selection
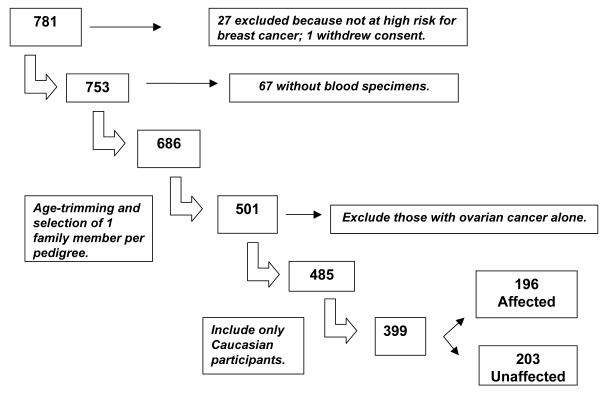
Our maximum sample size was 781, representing the total number of female participants enrolled in the Registry as of December 1, 2005. Twenty-seven women were excluded because they were not at high risk of breast cancer, but rather were enrolled in the Registry because of high risk for other types of cancers (e.g. colorectal). Additionally, one participant withdrew consent from the Registry. Biological specimen collection was initiated in April 2002 (N = 753). Registry participants who had enrolled before that date were asked retrospectively to provide a blood specimen, and only 67 individuals did not participate. As a result there were samples available from 686 individuals who were genotyped and used in the genotype QC procedures. In our statistical analyses the sample was further reduced for the following reasons. To avoid problems due to statistical dependencies among family members, we selected only 1 family member per pedigree. Our sample was then age-trimmed so that the age range of unaffected participants matched that of the breast cancer patients, limiting our sample size to 501 individuals. We additionally excluded 16 participants who were affected with ovarian cancer alone, and limited our analysis to those who either had breast cancer, breast and ovarian cancer, or were unaffected (controls) for a sample size of 485. Finally to avoid population stratification and because the number of non-Caucasians was relatively small (86) and highly varied in ethnicity, we limited our analysis to those who were Caucasian, for a final sample size of 399 individuals (see Figure 3).
Figure 3.
Participant selection.
Participant characteristics
Analyses were performed using all Caucasian participants selected (Data set 1 (DS1), maximum sample size = 399), those not known to have BRCA1 or BRCA2 risk conferring variants (Data set 2 (DS2), maximum sample size = 333) and using all Caucasian individuals with known or inferred BRCA1 and BRCA2 test results (Data set 3 (DS3) maximum sample size =306). Clinical and demographic characteristics of Caucasian breast cancer patients (N=196) and controls (N=203) are listed in Tables 2A and 2B.
Table 2A.
Demographic characteristics of caucasian breast cancer patients and age-matched control participants.
| Affected (N=196) | Unaffected (N=203) | p-value† | ||
|---|---|---|---|---|
| Age, mean ± SD (range) | 51.1 ± 10.6 (26-81) | 46.6 ± 10.6 (26-78) | 2.0E-05 | |
| Ashkenazi Jewish, No. (%) | 80 (41) | 86 (42) | 0.83 | |
| Education | ||||
| High school graduate | 8 (4) | 5 (2) | ||
| Vocational/training school | 5 (2) | 5 (2) | ||
| Some college | 22 (11) | 36 (18) | ||
| Associate degree | 15 (8) | 13 (6) | ||
| College graduate | 48 (24) | 43 (21) | ||
| Some professional school | 23 (12) | 36 (18) | ||
| Master's | 47 (24) | 47 (23) | ||
| Doctoral | 28 (14) | 18 (9) | 0.22 | |
| Income | ||||
| Missing/Declined to state | 7 (3) | 3 (1) | 0.31 | |
| <$15,000 | 4 (2) | 3 (2) | ||
| $15,000-30,000 | 10 (5) | 6 (3) | ||
| $30,000-60,000 | 28 (15) | 36 (18) | ||
| $60-100,000 | 49 (26) | 49 (25) | ||
| > $100,000 | 98 (52) | 106 (53) | 0.71 | |
| Smoking History | ||||
| No. reporting > 100 cigarettes/lifetime (%) | 90 (46) | 89 (44) | 0.75 | |
| Alcohol History | ||||
| Missing/Declined to state | 9 (4) | 9 (4) | 0.87 | |
| No. reporting >= 12 drinks/last year (%) | 128 (68) | 137 (70) | 0.72 | |
|
Total number of female first degree relatives, mean ± SD (range) |
2.99 ± 1.7 (1-12) | 2.89 ± 1.5 (0-8) | 0.55 | |
|
Total number of affected first degree relatives, mean ± SD (range) |
0.77 ± 0.50 (1-4) | 0.96 ± 0.50 (1-4) | 0.01 | |
p-values calculated using Chi-square tests for categorical variables and two-sided t-tests for continuous variables.
Table 2B.
Medical and gynecological history of caucasian breast cancer patients and age-matched control participants.
| Affected (N=196) | Unaffected (N=203) | p-value† | |||
|---|---|---|---|---|---|
| BRCA1 testing | |||||
| Not tested, No. (%) | 27 (14) | 77 (38) | 7.4E-08 | ||
| Inferred negative, No. (%) | 2 (7) | 9 (12) | |||
| Unknown, No. (%) | 25 (92) | 68 (88) | |||
| Of tested, | |||||
| No deleterious mutation, No. (%) | 138 (82) | 109 (87) | |||
| Positive for deleterious mutation, No. (%) | 25 (15) | 17 (13) | |||
| VUS, No. (%) | 6 (3) | 0 (0) | 0.09 | ||
| BRCA2 testing | |||||
| Not tested, No. (%) | 29 (15) | 88 (43) | 7.6E-10 | ||
| Inferred negative, No. (%) | 4 (14) | 20 (23) | |||
| Unknown, No. (%) | 25 (86) | 68 (78) | |||
| Of tested, | |||||
| No deleterious mutation, No. (%) | 148 (89) | 99 (86) | |||
| Positive for deleterious mutation, No. (%) | 13 (8) | 11 (10) | |||
| VUS, No. (%) | 6 (3) | 5 (4) | 0.82 | ||
| BMI, mean ± SD (range) | 25.2 ± 5.1 (15.0-46.6) | 24.5 ± 4.8 (16.2-41.2) | 0.17 | ||
| BMI18, mean ± SD (range) | 20.7 ± 2.4 (16.3-30.1) | 20.7 ± 2.7 (16.1-32) | 0.98 | ||
| Pregnancy | |||||
| Missing/Declined to state | 0 (0) | 1 (1) | 0.99 | ||
| No. Nulliparous (%) | 37 (19) | 51 (25) | |||
| No. reporting >=3 pregnancies (%) | 76 (39) | 78 (39) | 0.25 | ||
| Age at first live birth, mean ± SD (range) | 28.0 ± 6.2 (18-46) | 28 ± 5.6 (18-43) | 0.98 | ||
| Menstruation in last 12 months, No. reporting Yes (%) | 82 (42) | 127 (63) | 5.3E-05 | ||
| Age at menopause, mean ± SD (range) | 46.7 ± 5.9 (22-58) | 46.1 ± 7.2 (23-56) | 0.59 | ||
| Reason menstrual periods stopped (%*) | |||||
| Natural menopause | 44 (38) | 41 (56) | |||
| Hysterectomy alone | 17 (15) | 12 (16) | |||
| Bilateral oophorectomy alone | 0 (0) | 2 (3) | |||
| RT or chemotherapy alone | 1 (1) | 0 (0) | |||
| Consequence of multiple therapies | 45 (39) | 13 (18) | |||
| Other | 7 (6) | 5 (7) | 0.02 | ||
| Prophylactic surgeries | |||||
| Mastectomy | 21 (11) | 2 (1) | 7.7E-05 | ||
| Oophorectomy | 12 (6) | 17 (8) | 0.50 | ||
| Oral contraceptives | |||||
| Ever used OCPs | 121 (62) | 142 (70) | 0.10 | ||
| Hormone replacement | |||||
| Ever used HRT | 48 (24) | 47 (23) | 0.84 | ||
% of those reporting no menstruation in last 12 months.
p-values calculated using Chi-square tests for categorical variables and two-sided t-tests for continuous variables.
As shown in Tables 2A and 2B, the affected and unaffected populations were very similar with respect to almost all variables, with non-significant p-values when comparing education, income, tobacco and alcohol history, body mass index, pregnancy, age of menopause, oral contraceptive use, and hormone replacement use. The mean age in the affected group was significantly greater (p<0.0001), but ranges were similar. While it is striking that a significantly high percentage of individuals were of Ashkenazi Jewish heritage in our study (41% of affected and 42% of unaffected participants), the percent of the population with Ashkenazi Jewish ancestry was similar in the two groups (p=0.83).
Of note, the percentage of individuals not tested for BRCA1 and BRCA2 was significantly higher in the unaffected group (p<0.0001). We suspect that this difference is related to an increased exposure to counseling regarding mutation testing in a person recently diagnosed with cancer. For those who were tested, the percentage of women who tested positive was not significantly different between cases and controls for either mutation.
Also of note are the significant differences between menstruation history in the last 12 months (p<0.0001), reasons menstrual periods stopped, with increased likelihood of multiple therapies in the affected group (p<0.0001), and increased prevalence of prophylactic mastectomies in the affected group (p<0.0001). The fact that fewer affected women had menstrual periods than the unaffected women can be explained by the fact that the affected women are 4.5 years older on average. In addition, it is likely that more women diagnosed with cancer underwent combined radiation and chemotherapy as part of a treatment regimen for their cancer, effecting early menopause. None of these covariates were significant in the logistic regression (see below). Likewise it is understandable why women diagnosed with cancer might be more likely to undergo prophylactic mastectomy. Clearly prophylactic mastectomy is a consequence of cancer diagnosis not a cause. We do not expect these differences to have a significant impact on our analysis.
Although our study was not designed to directly match cases to controls, we did trim the sample to include only Caucasian participants, and unaffected participants of the same age-range as the affected group. All of the participants were enrolled in the UCLA Family Cancer Registry and thus were based in the same geographic region, Southern California. The high enrollment of Ashkenazi Jewish women is the result of this, as the Los Angeles metropolitan area has the second largest Jewish population in the United States. We did not compare Gail scores in our study because individuals in the affected group are known to have cancer. Likewise we do not report BRCAPRO scores, but we do note that the total number of female first degree relatives for the two groups is not significantly different (pvalue = 0.55, Table 2A), while the total number of affected 1st degree female relatives is higher in the unaffected group (p-value = 0.01, Table 2A), which is likely to be related to the criteria for entry in the study.
Model selection for hereditary and environmental covariates
Results from the backwards and forwards stepwise logistic regression with alpha = 0.20 as the criterion for their inclusion revealed the following covariates: age (aOR 1.045 per year, p=0.00021, 95% CI = 1.02-1.07), Ashkenazi Jewish ancestry (aOR = 0.633, p = 0.067, 95% CI = 0.37-0.90), and education (aOR 1.464, p = 0.14, 95% CI = 0.84-2.09, for masters or greater). All other potential covariates were excluded. Although menopausal status (no menstrual period within the last 12 months) was significantly different in the unadjusted comparison (Table 2B), it was not significant as a covariate when age is also included in the model. All of the subsequent models described below in which SNPs were introduced individually were adjusted for age, education and Ashkenazi Jewish ancestry. Education was used even though it was not significant at a 0.05 level because of the important association of education and socioeconomic status with cancer incidence reported previously (24). Likewise Ashkenazi Jewish ancestry was included because it is a well-known risk factor for breast cancer, though it is unclear whether there is an increased risk of breast cancer in individuals with Ashkenazi Jewish ancestry who do not carry BRCA1 and BRCA2 mutations (25). When using data set 3 (DS3), we also included BRCA1 and BRCA2 status, even though these covariates are not significant in the logistic regression for our sample, because of their importance in DNA double strand repair.
Association of SNPs: Additive and dominant models adjusted for covariates
Table 3 lists the odds ratios (OR) and p-values for 12 SNPs where the p-value was less than 0.05 in one or more of the data sets assuming either a log-additive or dominant genetic effect. These 12 SNPs were located within or near 8 genes: XRCC4 (intronic SNPs rs2075685, rs10474081, rs1120476), XRCC2 (5′ of gene, rs6464268), NBS1 (3′UTR SNP rs1063054), RAD21/hHR21 (intronic SNPs rs16888927, rs16888997, and rs16889040), RAD51/RAD51a (intronic SNP rs2619681), P53 (intronic SNP rs2909430), BRIP (synonymous coding SNP rs4986763), and ZNF350 (5′ of gene, SNP rs8112515). We show the results for three data sets to help determine the effects of the SNPs adjusted for BRCA1 and BRCA2 mutation status. We also present results using a model where the allelic effects are additive on the log scale (additive model) as well as a dominant genetic model. In general, the results for these two models are similar, reflecting the fact that in general there are relatively few individuals for a homozygous for the minor allele,
Table 3A.
Association of specific SNPs with breast cancer.*
|
SNP |
Distance Between SNPS (bp) |
Gene |
Chr. |
MAF** |
Data Set 1. All participants, N=399 |
Data Set 2. Excluding Individual who are BRCA1or 2 Positive, N=333 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additive | Dominant | Additive | Dominant | |||||||||||
| N | OR | p | OR | p | N | OR | p | OR | p | |||||
| rs2075685 | XRCC4 | 5 | 0.39 | 358 | 1.33 | 0.060 | 1.75 | 0.015 | 300 | 1.43 | 0.033 | 1.94 | 0.010 | |
| rs10474081 | 18218 | XRCC4 | 5 | 0.41 | 347 | 1.32 | 0.088 | 1.77 | 0.020 | 290 | 1.25 | 0.215 | 1.75 | 0.036 |
| rs1120476 | 31760 | XRCC4 | 5 | 0.37 | 321 | 1.46 | 0.025 | 2.04 | 0.004 | 274 | 1.41 | 0.059 | 2.09 | 0.006 |
| rs6464268 | 7016 | XRCC2 | 7 | 0.16 | 358 | 1.20 | 0.340 | 1.30 | 0.268 | 298 | 1.20 | 0.375 | 1.30 | 0.309 |
| rs1063054 | NBS1 | 8 | 0.28 | 320 | 1.47 | 0.038 | 1.51 | 0.082 | 273 | 1.45 | 0.066 | 1.53 | 0.100 | |
| rs16888927 | RAD21/hHR21 | 8 | 0.31 | 339 | 0.64 | 0.014 | 0.51 | 0.004 | 289 | 0.62 | 0.014 | 0.48 | 0.004 | |
| rs16888997 | 6948 | RAD21/hHR21 | 8 | 0.29 | 347 | 0.56 | 0.003 | 0.48 | 0.002 | 294 | 0.53 | 0.003 | 0.45 | 0.002 |
| rs16889040 | 1215 | RAD21/hHR21 | 8 | 0.29 | 356 | 0.63 | 0.019 | 0.57 | 0.013 | 298 | 0.58 | 0.010 | 0.50 | 0.006 |
| rs2619681 | RAD51/RAD51a | 15 | 0.18 | 339 | 0.71 | 0.132 | 0.62 | 0.051 | 289 | 0.74 | 0.221 | 0.66 | 0.126 | |
| rs2909430 | TP53 | 17 | 0.11 | 301 | 1.71 | 0.034 | 2.01 | 0.011 | 257 | 1.58 | 0.085 | 1.86 | 0.035 | |
| rs4986763 | BRIP1 | 17 | 0.40 | 330 | 0.64 | 0.014 | 0.56 | 0.013 | 280 | 0.57 | 0.002 | 0.46 | 0.003 | |
| rs8112515 | ZNF350 | 19 | 0.21 | 318 | 0.63 | 0.042 | 0.62 | 0.060 | 271 | 0.75 | 0.252 | 0.72 | 0.219 | |
| Table 3B. Association of specific SNPs with breast cancer, adjusting for BRCA1 and BRCA2 status. | ||||||||||||||
|
SNP |
Distance between SNPS (bp) |
Gene |
Chr. |
MAF** |
Data Set 3. Including only individuals with known BRCA1 and BRCA2 status (N=306). |
|||||||||
| Additive | Dominant | |||||||||||||
| N | OR | p | OR | p | ||||||||||
| rs2075685 | XRCC4 | 5 | 0.39 | 266 | 1.13 | 0.499 | 1.34 | 0.291 | ||||||
| rs10474081 | 18218 | XRCC4 | 5 | 0.41 | 255 | 1.16 | 0.480 | 1.26 | 0.443 | |||||
| rs1120476 | 31760 | XRCC4 | 5 | 0.37 | 241 | 1.41 | 0.087 | 1.75 | 0.053 | |||||
| rs6464268 | 7016 | XRCC2 | 7 | 0.16 | 260 | 1.66 | 0.048 | 2.23 | 0.010 | |||||
| rs1063054 | NBS1 | 8 | 0.32 | 240 | 1.37 | 0.146 | 1.37 | 0.252 | ||||||
| rs16888927 | RAD21/hHR21 | 8 | 0.31 | 247 | 0.62 | 0.027 | 0.45 | 0.005 | ||||||
| rs16888997 | 6948 | RAD21/hHR21 | 8 | 0.29 | 255 | 0.53 | 0.007 | 0.43 | 0.003 | |||||
| rs16889040 | 1215 | RAD21/hHR21 | 8 | 0.29 | 259 | 0.54 | 0.009 | 0.47 | 0.006 | |||||
| rs2619681 | RAD51/RAD51a | 15 | 0.18 | 248 | 0.68 | 0.138 | 0.56 | 0.043 | ||||||
| rs2909430 | TP53 | 17 | 0.11 | 224 | 1.42 | 0.213 | 1.71 | 0.086 | ||||||
| rs4986763 | BRIP1 | 17 | 0.40 | 240 | 0.68 | 0.046 | 0.64 | 0.101 | ||||||
| rs8112515 | ZNF350 | 19 | 0.21 | 238 | 0.62 | 0.067 | 0.61 | 0.100 | ||||||
Models adjusted for age, education, and Ashkenazi Jewish ancestry.
Minor allele frequency (MAF)calculated using the unaffected individuals
Models adjusted for age, education, Ashkenazi Jewish ancestry, and BRCA1 and BRCA2 status.
Minor allele frequency (MAF) calculated using the unaffected individuals
The three intronic SNPs of RAD21/hHR21 have the strongest associations and gave consistent results with all three data sets as well as both genetic models. The minor allele of each of these three SNPs was inversely associated with breast cancer. The significance of the other SNPs depends on the model or data set. The minor allele of SNP rs4986763 in BRIP1 was inversely associated with an increase in risk in all three data sets assuming an additive genetic model. Interestingly, using dominant models, the three SNPs in XRCC4, rs1120476 (aOR=2.04, p=0.004 using DS1, aOR=2.09, p=0.006 using DS2) , rs2075685 (aOR=1.75, p=0.015 using DS1, aOR=1.94, p=0.010 for DS2) and rs10474081 (aOR=1.77, p=0.020 using DS1, aOR=1.75, p=0.036 using DS2) are significantly associated with an increase in risk in Data Sets 1 and 2, but show reduced effect sizes and diminished significance when BRCA1 status and BRCA2 status are included as covariates (aOR=1.75, p=0.053 for rs1120476, aOR=1.34, p=0.291 for rs2075685, aOR=1.26, p=0.443 for rs10474081, using DS3). The same trend is seen using log-additive models for these SNPs. On the other hand, in XRCC2, SNP rs646426 shows increased effects and smaller p-values when BRCA1 status and BRCA2 status are included as covariates (log-additive models aOR=1.20, p=0.340 using DS1, aOR=1.20, p=0.375 using DS3, and aOR=1.66, p=0.048; dominant models aOR=1.30, p=0.268 using DS1, aOR=1.30, p=0.039 using DS2, and aOR=2.23, p=0.010 using DS3). The p-values for the SNP rs2909430 in P53 increased dramatically and the adjusted odds ratios decreased when BRCA1 status and BRCA2 status were included as covariates (log-additive aOR=1.71, p=0.034 using DS1, aOR=1.58, p = 0.85 using DS2, and aOR=1.42, p=0.213; dominant aOR= 2.01, p = 0.011 using DS1, aOR=1.86, p=0.035 using DS2, and aOR=1.71, p = 0.086 using DS3). SNP rs2619681 in RAD51/RAD51a was significantly associated only in a dominant genetic model when BRCA1 status and BRCA 2 status were included as covariates (aOR=0.56, p=0.043 using DS3). SNP rs81112515 5′ of ZNF350 was only significantly associated under a log-additive model when all data from all the Caucasian participants were used (aOR=0.63, p=0.042 using DS1).
Odds ratios for the SNPs rs16888927, rs16888997 and rs168889040 from RAD21/hHR21 vary based on the model, but the minor allele for each consistently remains significantly inversely associated with breast cancer in each data set and model. For example, the minor allele from rs16888997 is inversely associated with breast cancer, with an adjusted odds ratio of 0.56 (p=0.003) for the additive model, and 0.48 (p=0.002) for the dominant model after adjusting for age, education and Ashkenazi Jewish heritage in Data Set 1 which includes all participants. This association persists for rs16888997 in Data Set 2, in which individuals with known mutations for either BRCA1 or BRCA2 are excluded, with an aOR of 0.53 (p=0.003) and 0.45 (p=0.002). Thus the inverse association of this SNP with breast cancer is not restricted to individuals who carry risk from a BRCA1 or BRCA2 mutation. Finally, rs16888997 is again inversely associated with breast cancer with aOR of 0.53 (p=0.007) in the additive model, and 0.43 (p=0.003) in the dominant model, after adjusting for BRCA1 and BRCA2 status, in addition to age, education and Ashkenazi Jewish ancestry, using the population of individuals with known BRCA1 and BRCA2 status (Data Set 3). Thus the aOR are not biased by the differences in missing data in test results. By contrast, rs2909430 from TP53 was found to be significantly inversely associated with breast cancer in models adjusting for age, education and Ashkenazi Jewish ancestry, with aOR 1.71 (p = 0.034 additive, p = 0.011 dominant), whereas this association was lost after adjusting for BRCA1 and BRCA2 status (p=0.213 additive, p=0.086 dominant).
We adjusted for multiple testing by setting the tolerated false discovery rate at 15% (26). Using this criterion, we find that the following SNPs are significant after adjusting for multiple testing: in XRCC4, rs1120476 (dominant model, Data Sets 1 & 2), in RAD21, rs16888927 (dominant model, Data Sets 1, 2 & 3), rs16888997 (additive and dominant models, Data Sets 1, 2 & 3), rs16889040 (dominant model, Data Sets 2 & 3), and in BRIP1, rs4986763 (additive and dominant models, Data Set 2). These results are designated in bold type in Table 3.
Cumulative association and haplotype analysis
We examined additive models of cumulative risk for the twelve SNPs found to have an association with breast cancer, after adjusting for age, education and Ashkenazi Jewish ancestry. For SNPs in which the minor allele was found to be inversely associated with breast cancer, we used the major allele as the “high risk” allele. We found a cumulative association with breast cancer for the additive effect of the “high risk” alleles from these twelve SNPs. The trend in the number of risk alleles, with the adjusted odds ratio for presence of each additional risk allele is 1.13 (p = 0.017).
Since several SNPs were implicated in the XRCC4 and RAD21 genes, we tested the association of the haplotypes with breast cancer using Data Set 1 adjusting for age, education and Ashkenazi Jewish heritage. Using an additive model, we found that the C-A-A haplotype of RAD21 (snps rs16888927, rs16888997, rs889040) was positively associated with breast cancer (p-value 0.0068). Likewise the T-G-G and T-G-A haplotypes of XRCC4 (snps rs2075685, rs10474081, rs1120476) were positively associated with breast cancer (p-value 0.0330). However, for both genes the haplotypes did not result in a more significant association than the corresponding allele of the most associated SNP.
DISCUSSION
Our results suggest that SNPs within or near a number of Double Strand Break Repair (DSBR) DNA repair pathway genes are associated with breast cancer in individuals from a high-risk population. After adjustment for multiple testing, 5 SNPs remain significant when the false discovery rate is set at 15%. Although they may be false positives, the results of all 12 SNPs identified with a p-value of less than or equal to 0.05 in any of the models are potentially useful in generating hypotheses regarding the role of the DSBR pathways in breast cancer susceptibility.
Our most significant results are for 3 SNPs found in RAD21. The three SNPs are in linkage disequilibrium and any one of them is sufficient to account for the association. Results from a large-scale case-control association study using > 25,000 SNPs, located within approximately 16,000 genes, lend support for RAD21 as a susceptibility gene for breast cancer (27). RAD21 is implicated in homologous recombination-mediated DSB DNA repair, and plays a role in cell cycle regulation (28). RAD21 is also involved in the mitochondrial apoptosis pathway, and most recently has been shown to play an important role in predicting the outcome in breast cancer patients with supraclavicular lymph node metastases (29).
It is of note that the minor alleles from SNPs within XRCC4, XRCC2, and NBS1 are positively associated with breast cancer, whereas the minor alleles from SNPs within the RAD21/hHR21, RAD51/RAD51a, BRIP1 and ZNF350 are inversely associated with breast cancer. Inverse associations have been previously reported in DNA repair genes. For example, the R194W polymorphism in the base excision repair gene XRCC1 has been shown to be associated with reduced risk of bladder cancer (30), and the codon 399 Arg->Gln polymorphism has been shown to be associated with reduced risk of breast cancer in African Americans who smoke tobacco (31), suggesting that the SNP may modulate effects of environmental exposures. An inverse association may either mean that the minor allele confers protection or that the major allele confers risk. The functional significance of what may be happening is not clear. The minor allele could be involved in a regulatory pathway and actually may confer protection, or the SNP may be in linkage disequilibrium with another SNP in which the minor allele confers risk. It is important to remember that both cases and controls in our study come from a high risk breast cancer population. With this study design, controls are representatives of a resistant population, suggesting the possibility of protective SNPs. These polymorphisms could be responsible for a woman with a number of risk factors for breast cancer to remain cancer free. Alternatively, the major allele may be the more recent polymorphism that has increased in frequency due to random drift.
Several of the SNPs in our study that were shown to be associated with cancer risk in this high-risk population have been previously implicated in breast cancer. The TP53 SNP rs2909430 (32) and the XRCC4 SNP rs2075685 (9) were associated with breast cancer in two independent case-control studies. Some SNPs identified in our study have been implicated in other types of cancer. For example, XRCC2 SNPs rs321834 and rs6464268 have been found to be associated with bladder cancer risk (33).
DNA repair SNPs are also candidate modifiers of highly penetrant genes. For example, SNPs of DSBR genes RAD51 (34-37) and ATM (38), as well as MDM2, a regulator of p53 (39), have been implicated as possible modifiers of BRCA1/2-associated breast cancer risk. The BRCA1 gene induces expression of RAD21 (40). Unfortunately, the number of individuals with BRCA1 risk variants (N=42) or BRCA2 risk variants (N=24) are too small to test for gene-gene interactions in this study. In the future, we plan to examine the question of SNP-BRCA interactions once there are sufficient numbers of individuals in the Registry with BRCA1 or BRCA2 risk variants.
Some of the genes studied here have previously been shown to be associated with breast cancer, but with different SNPs than those described here. These genes and SNPs include XRCC4: rs1805377 (9), ZNF350: rs4986771 (11), TP53: rs9894946, rs1625895, rs1614984 (32), MRE11A: rs601341 (41), and XRCC2: rs1799794 (7, 8, 41, 42). SNP rs1614984 from the TP53 gene is in LD with rs2909430 one of the SNPs found to be associated with breast cancer in our study, with an R2 value of 0.803, based on analyses of the CEPH HapMap data. In addition, SNPs from genes XRCC2 have also been associated with increased risk in pancreatic cancer (43).
Some other SNPs previously observed to be associated were tested in our study and were not significant. These SNPs include BRCA1 rs799917 (44), BRCA2: rs144848 (11), and XRCC4: rs2075686 (9). A candidate gene study examined 710 tag SNPs in 120 candidate genes in 4,400 breast cancer cases and 4,400 controls and found several SNPs associated with breast cancer (14). SNPs significantly associated with breast cancer arose from different pathways, including the cell-cycle control pathway, steroid hormone metabolism and signaling and DNA repair pathways. The following SNPs were in both our study and theirs: rs799917 and rs3737559 from BRCA1, rs144848, rs542551 and rs2126042 from BRCA2, rs2191249, rs4988344, rs4968451, and rs2378908 from BRIP1, rs2619681 and rs11852786 from RAD51, rs6470522 from NBS1, rs3218536, rs3218499 and rs3218374 from XRCC2 pathways. None of these SNPs were found to be significant in the Pharoah study (14) and only one was found significantly associated with familial breast cancer in our study: rs2619681 from the RAD51 gene. RAD21 and XRCC4, the genes with the strongest support in our study, were not examined in the Pharoah study.
In another candidate gene study examining 12 SNPs from the DNA repair pathways in 1,109 women with breast cancer and 1,177 age-matched healthy controls, SNPs rs1801320 and rs1801321 from the RAD51 gene were not found to be significantly associated with breast cancer (41). It is of note that these SNPs are located approximately 1600 base pairs away from the SNP rs2619681 in RAD51 which was found to be significantly associated in our study. SNPs that were found to be significant in the Loizidou study included rs1799944 from BRCA2 and rs601341 from MRE11A. The MRE11A locus was approximately 20,000 base pairs from the SNPs of MRE11A examined in our study and not found to be significant. The BRCA2 SNP rs1799944 was only 430 base pairs away from rs1801406 which was analysed in our sample and not found to be significantly associated.
Hunter et al. (45) examined the association of a genome-wide panel of polymorphisms with sporadic breast cancer in a population a population of postmenopausal women of European ancestry. The strongest support came from SNPs in the FGFR2 and RELN genes, which are not involved in DNA repair pathways. This study found no evidence of association in regions containing DNA repair genes. Notable differences have been found before when comparing the association of polymorphisms from DNA repair genes with familial breast cancer in a high risk population than the association of these polymorphisms with sporadic breast cancer in the general population. Some of these results may be false positives in one study, but it is also possible that polymorphisms that confer risk in familial breast cancer may be different than the genes that confer breast cancer in sporadic cases (46).
By contrast, some recent genome-wide association studies have examined the association of polymorphisms with familial breast cancer in high risk populations, with the rationale that these individuals are more likely to carry susceptibility alleles. Gold et al. (47) confirmed the association of polymorphisms from FGFR2 in a population of 249 high risk Ashkenazi Jewish breast cancer patients testing negative for both BRCA1 and BRCA2 and 299 cancer-free Ashkenazi Jewish controls, and identified a new locus in chromosome 6q22.33 associated with breast cancer in this population. Easton et al. (48) examined 227,876 tag SNPs from the whole genome in 408 cases of invasive breast cancer, with strong family history (at least 2 affected women first degree relatives), and 400 controls, and found 6 SNPs significantly associated with breast cancer (p < 10−5). These SNPs were from the following genes: FGFR2, TNRC9 (unknown function), lymphocyte specific protein (LSP1), and H19 (associated with insulin growth factor gene regulation). Neither study reported significant associations at or near DSBR genes.
There are a number of reasons that our results differ from some of the results of the studies we mentioned in the preceding paragraphs. It is possible that the associations found in the original studies represent type 1 errors or artifacts of hidden ethnic differences between cases and controls. It is also possible that our lack of support for association of breast cancer with these SNPs is due to an insufficient sample size to detect moderately small risks. However, our results could also be due to the special characteristics of the unaffected participants in our study. Because the controls come from the UCLA Familial Cancer registry, they are individuals at high risk with a family history of breast cancer and they may harbor many of the same risk variants as affected participants.
The characteristics of the study sample warrant additional discussion, as the unique aspects bear both on the interpretation and generalizability of the inferences. The UCLA Family Cancer Registry is a clinic-based repository of information about people who have had cancer or have a strong family history of cancer. This information can include complete pedigree data, survey data on lifestyles, health behaviors, biological specimens such as tissue or blood samples, and results of genetic susceptibility testing. The careful documentation of lifestyles and behaviors as well as demographics allowed us to examine the effects of SNPs adjusted for these covariates. The Family Cancer Registry enrolls individuals who either 1) have a family history of cancer, defined as at least two individuals with the same primary cancer or cancers that are known to be related (e.g., breast and ovarian) and who are at least second-degree relatives of each other, or 2) have a documented, known susceptibility cancer gene (e.g., BRCA1). Registry participants may or may not have had cancer themselves. We chose this clinic-based resource because it allows for investigation of genetic association with breast cancer in a high risk population.
Selecting cases and controls from the Family Cancer Registry leads to a nested study design with specific consequences. It conditions both affected and unaffected women on having a high risk of a familial form of breast cancer. This study design has a disadvantage over using women selected as cases or controls from the general population. The power to detect interactions between the implicated SNPs and other breast cancer risk factors has been reduced. In effect, we have over matched our cases and controls on these other risk factors. Indeed, we find that the affected and unaffected participants in our study had similar prevalence of most of the well-known risk factors for breast cancer. For example, the proportion of women of Ashkenazi Jewish heritage was quite high in our population sample and this prevalence was similar in the unaffected (42%) and affected groups (41%).
There are also advantages to the conditioning on having a high risk of a familial form of breast cancer. This design should increase the likelihood of detecting polymorphisms that may protect against breast cancer. However, with the nested design, we also must guard against direct extrapolation of our findings to the general population. This caution is particularly important for those polymorphisms that may be associated with protection against breast cancer. Because unaffected women in the general population are not “exposed” to the same risks as controls in this study, these protective polymorphisms may not be detected.
In conclusion, we identified several polymorphisms in the RAD21 and XRCC4 genes as well as others involved in double strand DNA repair pathways that modulate breast cancer susceptibility in a high-risk population. Although none of these polymorphisms have adjusted odds ratios close to the same magnitude as BRCA1 and BRCA2, these results are helpful in generating hypotheses about parts of the DNA repair pathway that may play a role in carcinogenesis, and may help explain why some individuals with familial risk of breast cancer get breast cancer why others with the same risk factors do not. Further studies in similar populations with larger numbers of known mutation carriers are warranted.
Acknowledgements
We thank the participants of the UCLA Family Cancer Registry. Research support for this work from the Breast Cancer Research Foundation, NCI Cancer Center Support Grant P30 CA 16042, NCI/NIH R25T CA 87949, a Young Investigator Award from the American Society of Clinical Oncology, from the John A. Hartford Foundation, Center of Excellence: UCLA Hartford/UCLA Program for Advanced Training in Geriatrics Research, and from the United States Public Health Service Grant GM53275.
Footnotes
Statement of Clinical Relevance
It is well known that BRCA1 and BRCA2 mutations are strong risk factors for the development of hereditary breast and ovarian cancer. This study lends support to the notion that, in addition to BRCA1 and BRCA2, other genes involved in the Double Strand DNA Repair Pathway are linked with breast cancer. Most notably, polymorphisms in RAD21 and XRCC4 were found to be associated with breast cancer in a population in which both cases and controls were at high familial risk for breast cancer. Because the sample size is relatively small and the results can not be extrapolated to predict risks in all women, these results must be viewed as preliminary. However, these results warrant validation in larger cohorts, and contribute to the identification of genes associated with breast cancer. Further investigation of these genes may aid in prediction and prevention of breast cancer, and in counseling individuals with a known family history of breast cancer.
Conflicts of Interest: None.
REFERENCES
- 1.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–32. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stankovic T, Kidd AML, Sutcliffe A, et al. ATM mutations and phenotypes in ataxia-telangiectasia families in the British Isles: Espression of mutant ATM and the risk of leukemia, lymphoma, and breast cancer. Am J Hum Genet. 1998;62:334–45. doi: 10.1086/301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariyoshi K, Suzuki K, Goto M, et al. Increased chromosome instability and accumulation of DNA double-strand breaks in Werner Syndrome cells. J Radiat Res. 2007;48:219–31. doi: 10.1269/jrr.07017. [DOI] [PubMed] [Google Scholar]
- 4.Mourra N, Zeitoun G, Buecher B, et al. High frequency of chromosome 14 deletion in early-onset colon cancer. Dis Colon Rectum. 2007;50:1881–6. doi: 10.1007/s10350-007-9040-3. [DOI] [PubMed] [Google Scholar]
- 5.Sieber OM, Heinimann K, Tomlinson IPM. Genomic instability – the engine of tumorigenesis? Nat Rev Cancer. 2003;3:701–8. doi: 10.1038/nrc1170. [DOI] [PubMed] [Google Scholar]
- 6.Rafii S, O'Regan P, Xinarianos G, et al. A potential role for the XRCC2 R186H polymorphic site in DNA-damage repair and breast cancer. Hum Mol Genet. 2002;11:1433–8. doi: 10.1093/hmg/11.12.1433. [DOI] [PubMed] [Google Scholar]
- 7.Han J, Hankinson SE, Rannu H, De Vivo I, Hunter DJ. Polymorphisms in DNA double-strand break repair genes and breast cancer risk in the Nurses' Health Study. Carcinogenesis. 2004;25:189–95. doi: 10.1093/carcin/bgh002. [DOI] [PubMed] [Google Scholar]
- 8.Kuschel B, Auranen A, McBride S, et al. Variants in DNA double-stranded break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11:1399–1407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- 9.Fu YP, Yu JC, Cheng TC, et al. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–6. [PubMed] [Google Scholar]
- 10.Desjardins S, Belleau P, Labrie Y, et al. Genetic variants and haplotype analyses of the ZBRK1/ZNF350 gene in high-risk non BRCA1/2 French Canadian breast and ovarian cancer families. Int J Cancer. 2008;122:108–16. doi: 10.1002/ijc.23058. [DOI] [PubMed] [Google Scholar]
- 11.García-Closas M, Egan KM, Newcomb PA, et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet. 2006;119:376–88. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 12.Jara L, Acevedo ML, Blanco R. RAD51 135G > C polymorphism and risk of familial breast cancer in a South American population. Cancer Genet Cytogenet. 2007;178:65–9. doi: 10.1016/j.cancergencyto.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Koren M, Kimmel G, Ben-Asher E, et al. ATM haplotypes and breast cancer risk in Jewish high-risk women. Br J Cancer. 2006;94:1537–43. doi: 10.1038/sj.bjc.6603062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pharoah PDP, Tyler J, Dunning AM, et al. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:401–6. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor MF, Irwin MR, Seldon J, Kwan L, Ganz PA. Pro-inflammatory cytokines and depression in a familial cancer registry. Psychooncology. 2007;16:499–501. doi: 10.1002/pon.1108. [DOI] [PubMed] [Google Scholar]
- 16.Low CA, Bower JE, Kwan L, Seldon J. Benefit finding in response to BRCA1/2 testing. Ann Behav Med. 2008;35:61–9. doi: 10.1007/s12160-007-9004-9. [DOI] [PubMed] [Google Scholar]
- 17.de Bakker, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Tobler AR, Short S, Andersen MR, et al. The SNPlex Genotyping System: A Flexible and Scalable Platform for SNP Genotyping. J Biomolecular Techniques. 2005;16:396–404. [PMC free article] [PubMed] [Google Scholar]
- 20.Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Abstract 1886. Am J Hum Genet. 2001;69S:504. [Google Scholar]
- 21.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinsheimer JS, McKenzie CA, Keavney B, Lange K. SNPs and snails and puppy dogs' tails: analysis of SNP haplotype data using the gamete competition model. Ann Hum Genet. 2001;65:483–90. doi: 10.1017/S0003480001008843. [DOI] [PubMed] [Google Scholar]
- 23.Lange K, Sinsheimer JS, Sobel E. Association Testing with Mendel. Genet Epidemiol. 2005;29:36–50. doi: 10.1002/gepi.20073. [DOI] [PubMed] [Google Scholar]
- 24.Faggiano F, Partanen T, Kogevinas M, Boffetta P. Socioeconomic differences in cancer incidence and mortality. In: Kogevinas M, Pearce N, Susser M, Boffetta P, editors. Social inequalities and cancer. International Agency for Research on Cancer; Lyon (France): 1997. (IARC Scientific Publications. No. 138). [PubMed] [Google Scholar]
- 25.Sagatopan JM, Offit K, Foulkes W, et al. The lifetime risks of breast cancer in Ahskenazi Jewish carriers of BRCA1 and BRCA2 mutations. Cancer Epidemiol Biomark Prev. 2001;10:467–73. [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc. 1995;57:289–300. [Google Scholar]
- 27.Kammerer S, Roth RB, Reneland R, et al. Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Research. 2004;64:8906–10. doi: 10.1158/0008-5472.CAN-04-1788. [DOI] [PubMed] [Google Scholar]
- 28.Atienza JM. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol Cancer Ther. 2005;4:361–8. doi: 10.1158/1535-7163.MCT-04-0241. [DOI] [PubMed] [Google Scholar]
- 29.Oishi Y, Nagasaki K, Miyata S, et al. Functional pathway characterized by gene expression analysis of supraclavicular lymph node metastasis-positive breast cancer. J Hum Genet. 2007;52:271–9. doi: 10.1007/s10038-007-0111-z. [DOI] [PubMed] [Google Scholar]
- 30.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA Repair genes and associations with cancer risk. 2002;11:1513–30. [PubMed] [Google Scholar]
- 31.Duell EJ, Millikan RC, Pittman GS, et al. Polymorphisms in the DNA Repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomark Prev. 2001;10:217–22. [PubMed] [Google Scholar]
- 32.Sprague BL, Trentham-Dietz A, Garcia-Closas M, et al. Genetic variation in TP53 and risk of breast cancer in a population-based case-control study. Carcinogenesis. 2007;28:1680–86. doi: 10.1093/carcin/bgm097. [DOI] [PubMed] [Google Scholar]
- 33.Figueroa JD, Malats N, Rothman N, et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28:1788–93. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- 34.Antoniou AC, Sinilnikova OM, Leone SJ, et al. RAD51 135G->C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadouri L, Kote-Jarai Z, Hubert A. A single-nucleotide polymorphism in the RAD51 gene modifies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. Br J Cancer. 2004;90:2002–5. doi: 10.1038/sj.bjc.6601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sliwinski T, Krupa R, Majsterek I, et al. Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res Treat. 2005;94:105–9. doi: 10.1007/s10549-005-0672-5. [DOI] [PubMed] [Google Scholar]
- 37.Levy-Lahad E, Lahad A, Eisenberg S, et al. A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc Natl Acad Sci. 2001;98:3232–6. doi: 10.1073/pnas.051624098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Ting NSY, Zheng L, et al. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA response. Nature. 2000;406:210–5. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- 39.Yarden RI, Friedman E, Metsuyanim S, et al. MDM2 SNP309 accelerates breast and ovarian carcinogenesis in BRCA1 and BRCA2 carriers of Jewish-Ashkenazi descent. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9797-z. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 40.Atalay A, Crook T, Ozturk M, et al. Identification of genes induced by BRCA1 in breast cancer cells. Biochem Biophys Res Comm. 2002;299:839–46. doi: 10.1016/s0006-291x(02)02751-1. [DOI] [PubMed] [Google Scholar]
- 41.Loizidou MA, Thalia M, Neuhausen SL, et al. DNA-repair genetic polymorphisms and risk of breast cancer in Cyprus. Breast Cancer Res Treat. 2008 Jun 16; doi: 10.1007/s10549-008-0084-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 42.Loizidou MA, Michael T, Neuhausen SL, et al. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res Treat. 2008 Jan 10; doi: 10.1007/s10549-007-9881-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 43.Jiao L, Hassan MM, Bondy ML, Wolff RA, Evans DB, Abbruzzese JL, Li D. XRCC2 and XRCC3 gene polymorphism and risk of pancreatic cancer. Am J Gastroenterol. 2008;103:360–7. doi: 10.1111/j.1572-0241.2007.01615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox DG, Kraft P, Hankinson SE, Hunter DJ. Haplotype analysis of common variants in the BRCA1 gene and risk of sporadic breast cancer. Breast Cancer Res. 2005;7:R171–R175. doi: 10.1186/bcr973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaag A, Walsh T, Renbaum P, et al. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum Mol Genet. 2005;14:555–63. doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- 47.Gold B, Kirchoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Nat Acad Sci. 2008;105:4340–5. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–95. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]