Abstract
Mental retardation in Down syndrome (DS), the most frequent trisomy in humans, varies from moderate to severe. Several studies both in human and based on mouse models identified some regions of human chromosome 21 (Hsa21) as linked to cognitive deficits. However, other intervals such as the telomeric region of Hsa21 may contribute to the DS phenotype but their role has not yet been investigated in detail. Here we show that the trisomy of the 12 genes, found in the 0.59 Mb (Abcg1–U2af1) Hsa21 sub-telomeric region, in mice (Ts1Yah) produced defects in novel object recognition, open-field and Y-maze tests, similar to other DS models, but induces an improvement of the hippocampal-dependent spatial memory in the Morris water maze along with enhanced and longer lasting long-term potentiation in vivo in the hippocampus. Overall, we demonstrate the contribution of the Abcg1–U2af1 genetic region to cognitive defect in working and short-term recognition memory in DS models. Increase in copy number of the Abcg1–U2af1 interval leads to an unexpected gain of cognitive function in spatial learning. Expression analysis pinpoints several genes, such as Ndufv3, Wdr4, Pknox1 and Cbs, as candidates whose overexpression in the hippocampus might facilitate learning and memory in Ts1Yah mice. Our work unravels the complexity of combinatorial genetic code modulating different aspect of mental retardation in DS patients. It establishes definitely the contribution of the Abcg1–U2af1 orthologous region to the DS etiology and suggests new modulatory pathways for learning and memory.
INTRODUCTION
Down syndrome (DS; OMIN #190685) is a complex disorder associated with the trisomy of human chromosome 21 (Hsa21) (1). This condition is a paradigm of human aneuploid disorders with supernumerary copies of a chromosome and could be considered as a prototype of the ‘disorders of the genome’. DS represents one-third of the cases of mental retardation and cognitive impairment in school-aged children (2–4) and is associated with a large panel of dysmorphologies, such as a characteristic facies, skeletal anomalies and brain alterations at the level of the prefrontal cortex, the hippocampus and the cerebellum. DS clinical features also include developmental delay, metabolic defects and other symptoms and associated diseases, but their overall expressivity and penetrance are highly variable.
DS is due to the presence of an additional copy of genetic elements that are dosage sensitive leading to an abnormal gene expression level, and/or to the abnormal number of copies of non-protein coding functional sequences. Furthermore, the effect of some dosage-sensitive genes on the phenotype might be allele specific or could depend on the combination of alleles with qualitative (alleles with amino acid variation) or quantitative (alleles with variation in gene expression level) traits (1). It can also be direct or indirect, taking into account the interaction of aneuploid genes or gene products with non-aneuploid genes or gene products. These hypotheses are currently put forward to explain the variability of the DS alterations, including the most frequent one, affecting learning and memory that vary from moderate to severe. The detailed studies of patients carrying partial duplications of Hsa21 clearly show the contribution of several regions to specific DS phenotypes (5–8).
During the past 20 years, the efforts have been directed to understand the pathogenetic mechanisms translating trisomy of specific genes or genetic regions. To this end, several DS mouse models have been generated and investigated (9–12). These include the transchromosomic Tc1 mouse (10), trisomic (Ts) for the entire Hsa21 and four models encompassing different nested regions of Mus Musculus (Mmu) chromosome 16 (Mmu16) homologous to Hsa21: Ts65Dn (13), Ts1Cje (14), Ts1Rhr (15) and Ts1Yu (16). Three of those are Ts for subregions located on Mmu16, including, respectively, 132 genes from Mrpl39 to Zfp295 in Ts65Dn, 85 from Sfrs15 to Zfp295 in Ts1Cje and finally 33 genes on a smaller segment delimited by Cbr and Mx2 (Ts1RhR), whereas Ts1Yu encompasses the complete homologous region on Mmu16. These models show DS-related phenotypes including cognitive and behavioral impairments, and craniofacial and cardiovascular malformations related to DS phenotypes, although their intensity was not related to the length of the Ts region (9–11,17–23). Using such models, the hypothesis on the major role of a critical region inducing the DS alterations was reviewed recently (15,24) confirming data obtained in human (5,7,8). Accordingly, the Tc1 mouse model, containing a nearly complete extra Hsa21, displays additional phenotypes affecting craniofacial morphology, cognitive function and brain physiology, although the cognitive phenotypes are milder and more oriented toward locomotor deficit (10,25,26). Altogether these data enlighten the involvement of regions located upstream the Mrpl39 gene and downstream Zfp295, in the telomeric part of Hsa21 in DS phenotypes.
Mouse models available up to now leave unexplored the contribution of the gene-rich telomeric part of the Hsa21 that maps to the Mmu17 and Mmu10 and do not completely explain the complexity and variability of some phenotypes observed in DS patients. Thus, additional Ts models are needed to evaluate the contribution of genes or other functional elements within the complete Hsa21 to DS phenotypes (1,8). In the present study, we wanted to evaluate the contribution of the Abcg1–U2af1 genetic interval from the telomeric part of Hsa21. To this end, we have generated a new DS mouse model, the Ts1Yah and the corresponding monosomic (Ms) model, the Ms2Yah. Defects in Ts animals were scored in the open-field, the Y-maze and novel object recognition (NOR) tests. But, unexpectedly, an improvement of the spatial reference memory in the Morris water maze (MWM) and a stronger in vivo long-term potentiation (LTP) in the hippocampus were detected in Ts mice. Most of the genes on the Abcg1–U2af1 interval were dosage sensitive and their change in copy number induced changes in expression levels of the encoded proteins along with phenotypic alterations. Overall, our observation definitely changes our concept of trisomy, modifying our view of the consequence of increased gene dosage and will have a strong impact on the identification of pathways to improve learning and memory in patients.
RESULTS
Generating a targeted segmental duplication and deletion of the Abcg1–U2af1 genetic interval
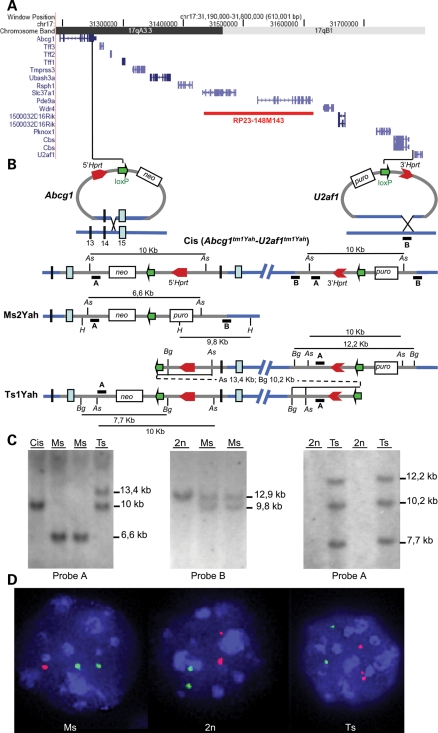
We used chromosomal engineering (reviewed in 27) based on the MICER system (28) to produce in vitro the segmental duplication and the corresponding deletion of the Abcg1–U2af1 0.59 Mb interval located on Mmu17. Using an embryonic stem (ES) cells clone carrying both the 5′- and 3′-Hrpt vectors inserted at the level of the Abcg1 and upstream of the U2af1 gene in a cis configuration [Cis(Abcg1tm1Yah–U2af1tm1Yah)], we were able to generate the segmental duplication and the deletion, after Cre expression in the G2 phase. Derived aneuploid mouse models correspond to the deletion and the reciprocal tandem duplication of the corresponding target region, namely Ts1Yah for the trisomy and Ms2Yah for the monosomy of the Abcg1–U2af1 region (Fig. 1A and B). Confirmation of the genomic rearrangements was obtained using Southern blot analysis (Fig. 1C) and FISH (Fig. 1D). Both Ts and Ms mice were fully viable and fertile after up to seven backcrosses on the C57BL6/J background.
Figure 1.
Generating a 0.59 Mb deletion and duplication between the Abcg1 and U2af1 loci. (A) The 0.59 Mb targeted region, defined by the Abcg1 and U2af1 genes, contains 12 genes as shown from the image captured from the UCSC genome browser (http://genome.ucsc.edu). (B) The targeting vectors containing a loxP site (green arrow), a selectable antibiotic resistance gene (puro, neo), and part of the Hprt gene (3′ or 5′ Hprt, red arrows) were integrated successively in the Abcg1 locus and between the Cbs and U2af1 genes [Cis(Abcg1tm1Yah–U2af1tm1Yah)]. (C) Checking of the new genetic configurations by Southern blot. In embryonic stem cells (left panel), southern analysis with AscI digestion and probe A reveals a 10 kb fragment for the Cis(Abcg1tm1Yah–U2af1tm1Yah) allele, a 6.6 kb for the Ms2Yah allele and two bands at 10 and 13.4 kb for the Ts1Yah locus. In mice (middle), the Ms2Yah allele was checked with probe B, showing an additional HindIII fragment of 9.8 kb compared with the wild-type allele (12.9 kb), whereas the Ts1Yah locus (right panel) was confirmed using the BglII enzyme and probe A. (D) Interphase FISH analysis with BAC probes that map in the Abcg1–U2af1 region (red) and outside (green). The wild-type (2n) showed two red and two green adjacent signals, whereas nuclei from Ms2Yah (Ms) showed two green and only one red signal due to the deletion of the Abcg1–U2af1 region. The Ts1Yah (Ts) nuclei showed two green and three red signal due to the duplication. As: AscI, Bg: BglII, H: HindIII, puro: puromycin, neo: neomycin.
Evaluation of motor activity, exploratory behavior, anxiety, learning and memory in Ts1Yah mice
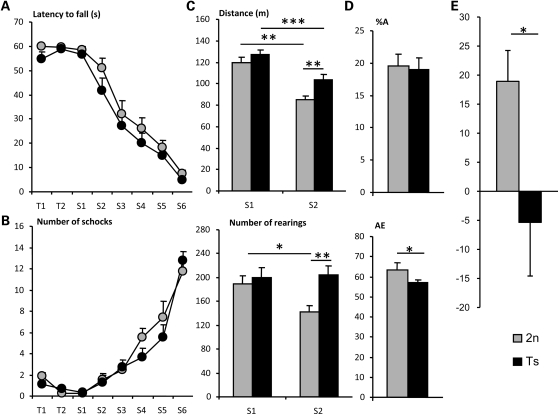
No significant differences in motor coordination, motor learning, balance or muscular tonus were detected between Ts1Yah and diploid mice on the rotarod and the treadmill tests (Fig. 2A and B), thus suggesting that the Abcg1–U2af1 genetic interval does not contribute to the DS locomotor defects. Activity pattern and exploratory behavior were evaluated in the open-field test. The absence of significant differences in distance traveled and number of rears between Ts1Yah and their wild-type littermates in the first session (S1) indicates that the aneuploid mice have a normal motor and exploratory activity and no anxiety-induced locomotor activity (Fig. 2C). No additional abnormal behavior was noticed. Twenty-four hours later in the second session (S2), a reduction in activity due to habituation was observed in both genotype, but the Ts1Yah were significantly more active than wild-types, as shown by the higher levels of horizontal and vertical activities [21% increase in travelled distance (P < 0.01, Student's t-test) and 44% increase in rearing (P < 0.01, Student's t-test) with respect to wild-types]. This observation suggested reduced inter-session habituation in Ts animals. Habituation is a sign of non-associative learning in which there is a progressive diminution of behavioral response probability with repetition of a stimulus or to an environment, which is thus partly altered in Ts1Yah mice. Since anxiety behavior or emotional reactivity could be implicated, the Ts1Yah mice were tested for anxiety-related behavior on the elevated plus maze. No significant differences between Ts and wild-types were found (Supplementary Material, Fig. S1A) in anxiety-related behavior.
Figure 2.
Motor activity and behavioral analysis of Ts1Yah mice. Rotarod (A) Mean ± SEM of the latencies to fall from the rotarod and (B) number of shock received on the treadmill by Ts1Yah (black circles) and wild-type males (gray circles) during training sessions (T1 and T2) and along test sessions (S1–S6) with increasing fixed rotational speeds (4, 10, 14, 19, 24, and 34 r.p.m.). Ts1Yah mice do not show significant motor coordination, nor motor learning impairment [Rotarod: F(1,28) = 0.222 P = 0.641; Treadmill: F(1,28) = 0.347 P = 0.561, repeated measures ANOVA]. (C) Mean ± SEM of the traveled distance (in meters (m) upper panel) and number of rears (lower panel) performed by Ts1Yah mice (black bars) and wild-type mice (gray bars) in an open-field during two sessions of 30 min (S1 and S2) with 24 h inter-session interval. During the first session (S1), similar locomotor activity was observed in Ts1Yah mice, but in the second session (S2), a reduced habituation was detected as shown by the 21% increase in travelled distance (***P < 0.001, **P < 0.01, Student's t-test) and 44% increased rearing activity (**P < 0.01, Student's t-test) for the Ts1Yah mice compared with their wild-type peers. (D) Mean ± SEM percentage of alternation (A; upper panel) and number of arm entries (AE; lower panel) of Ts1Yah (black bars) and wild-type mice (gray bars) during a single 5 min session in a Y-maze. Normal motor activity with lower percentage of alternation was found for Ts1Yah mice (*P < 0.05, Student's t-test). In the novel object recognition test (E), working memory is altered in Ts1Yah at 1 h retention. The discrimination index (N−F/N+F) reveals a significant deficit in short-term recognition for Ts1Yah mice (black) compared with the wild-type mice (gray) (Kruskal–Wallis one-way analysis *P < 0.05).
Short-term memory was assessed by recording spontaneous alternation in the Y-maze test (Fig. 2D). The Y-maze test is based on the innate preference of animals to explore an arm that has not been previously explored, a behavior that, if occurred with a frequency greater than 50%, is called spontaneous alternation. In our experiments, the total number of arm entries recorded during a single 5 min session did not differ significantly between aneuploid and diploid mice, confirming that the level of activity is unaffected in Ts1Yah mice (Fig. 2D). However, the percentage of alternation between the three arms was significantly lower in the Ts1Yah mice compared with their diploid littermates (P < 0.05, Student's t-test). This low alternation indicates a deficit in short-term memory in Ts1Yah mice.
On the basis of the previous studies in other DS mouse models, we analyzed the hippocampal cognitive traits in Ts1Yah using the NOR designed for the assessment of short-term recognition memory and the MWM tests that serve to analyze recognition and visuo-spatial learning and memory (17,29). The NOR test is based on the natural tendency of rodents to investigate a novel object instead of a familiar one. The choice to explore the novel object as well as the reactivation of exploration after object displacement reflects the use of learning and (recognition) memory processes. No significant difference in the time spent to explore two identical objects in the NOR was observed during the training session between Ts1Yah and euploid (Eu) mice, and all the mice had the same curiosity and motivation for this task (data not shown). However, Ts1Yah mice spent less time in exploring the novel object in the second session as indicated by the discrimination index (Fig. 2E; Supplementary Material, Fig. S1B), suggesting that Ts mice have impaired short-term recognition memory.
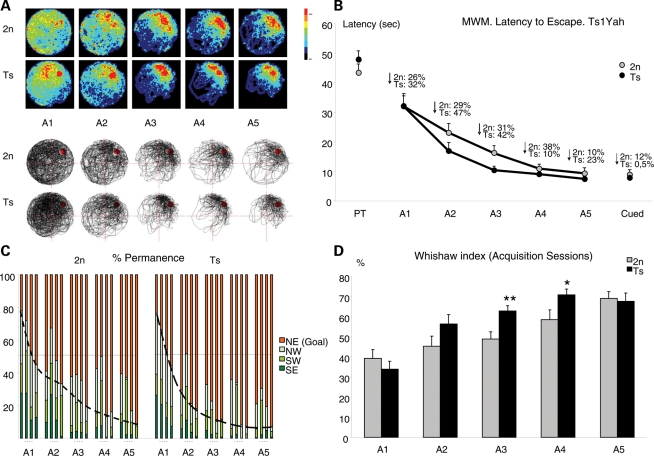
Visual–spatial learning and memory was explored using the hidden-platform MWM. In this task, animals learned to locate the position of the submerged platform by using extra-maze spatial cues. During the pre-training phase of the test, Ts1Yah mice learned to locate the visible platform, equally to wild-types, indicating no procedural learning impairment. However, during the learning phase, with the hidden platform, Ts1Yah mice displayed enhanced ability to locate its position, as indicated by the increased permanence in the target quadrant along sessions and the more focused search (Fig. 3A and B), which resulted in a significant shift to the left of the acquisition curve (P < 0.001, repeated measures ANOVA; Fig. 3C), thus indicating more efficient learning. Moreover, estimation of other more specific learning indexes such as the Whishaw index (defined as percentage path inside the optimal corridor to reach the platform) indicated the use of more efficient learning strategies in Ts mice (Fig. 3D). No changes in swimming speed or thigmotaxic behavior were observed (Supplementary Material, Fig. S2A and B), thus discarding any anxiety-related effects in the phenotypes observed. In the removal session, we found a tendency to increase the distance travelled in the trained quadrant in Ts mice (Supplementary Material, Fig. S2C), although it did not reach statistical significance. Accordingly, the learning process was similar between both groups in the reversal session. As a conclusion, we found that the increased ability of the Ts1Yah mice is specific for visual–spatial learning with almost no impact on spatial memory.
Figure 3.
Analysis of hippocampal-dependent explicit spatial learning and memory. (A) Learning strategies are shown in the graphical representation of swimming paths of wild-type versus Ts1Yah mice along MWM acquisition sessions (A1–A5). Upper panel: Color-coded histograms representing occupancy of wild-type (upper panel) and trisomic (Ts) (lower panel) mice during acquisition sessions of the MWM task. Color scale is given on the right of the histograms. The Ts1Yah mice focused their search in the trained location earlier and more efficiently than wild-type. Lower panel: representative swim paths of a wild-type and a Ts mouse illustrating that controls swam more irregularly than the Ts. (B) Ts1Yah mice learned the platform position more quickly than wild-types, as shown by the steeper acquisition curve (latency to find the platform) although significant differences in escape latency were observed in session 3 [F(1,28) = 7.018 P = 0.014, ANOVA]. As a consequence, Ts1Yah mice showed a significantly higher improvement along sessions (F(1,28) = 5.671 P = 0.025, Repeated Measures ANOVA). (C) Permanence time in quadrants, trial by trial, during acquisition sessions. Ts1Yah mice show a steeper slope than 2n mice, indicating a better performance in learning task. (D) The Whishaw index, defined as percentage path inside an optimal corridor to reach the platform, confirmed better performance of Ts1Yah than control mice [F(1,28) = 8.513 P = 0.007, Repeated Measures ANOVA].
In vivo electrophysiological activity in the CA1 of Ts mice
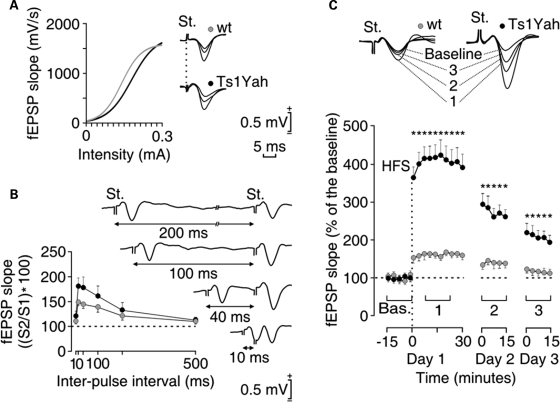
Hippocampal LTP, a form of synaptic plasticity, provides a putative physiological basis for hippocampus-dependent learning and memory (30,31). We hence assessed synaptic function in the Ts1Yah hippocampus by recording the activity-dependent changes at the CA3–CA1 synapse in conscious mice. Both diploid and Ts mice presented similar increases in the slope of field excitatory post-synaptic potentials (fEPSP) evoked at the CA1 area following the presentation of single pulses of increasing intensity at the ipsilateral Schaffer collaterals (Fig. 4A and B). In both genotypes, these relationships were fitted by sigmoid curves suggesting a normal functioning of the CA3–CA1 synapse. It is known that the synaptic facilitation evoked by the presentation of a pair of pulses is a typical presynaptic short-term plastic property of the hippocampal CA3–CA1 synapse, which has been related to neurotransmitter release (32). As illustrated in Figure 4, control and Ts1Yah presented a significant increase of the response to the second pulse at short (20–100 ms) time intervals. However, no genotype-dependent differences were observed at any of the selected intervals, suggesting that short-term plastic processes are not affected in Ts animals. For the LTP study, after presenting the animal with the high-frequency stimulation (HFS) protocol, recording sessions were repeated 24 and 48 h later for 15 min each. Measurement of LTP revealed larger and longer-lasting LTP responses in Ts1Yah mice compared with their respective littermate controls (Fig. 4C), even at the end of the recording time (48 h) being significantly larger in Ts1Yah (≈200%) than in wild types (≈115%) with respect to the baseline values. This result provides a distinct link between certain forms of hippocampal LTP and the enhanced visual–spatial learning detected in Ts1Yah mice in the MWM.
Figure 4.
Change in long-term potentiation in Ts1Yah mice compared with control littermate. (A) Wild-type (gray circles) and trisomic (black circles) mice presented normal input/output curves. To this end, single (100 ms, biphasic) pulse was presented to Schaffer collaterals at increasing intensities (in mA), while recording the evoked fEPSP at the CA1. Some fEPSPs collected from the four types of mouse are illustrated at the right. (B) There were no significant differences in paired-pulse facilitation between wild-type (gray) and transgenic (black) mice. The data shown are mean ± SEM slopes of the second fEPSP expressed as a percentage of the first for six (10, 20, 40, 100, 200, 500) inter-pulse intervals. Some fEPSP paired traces collected from a representative Ts1Yah mouse at different inter-pulse intervals (10–200 ms) are illustrated at the right. (C) At the top are illustrated examples of fEPSPs collected from selected wild-type (gray) and Ts1Yah (black) animals before (baseline) and after (1–3) high-frequency stimulation (HFS) of Schaffer collaterals. The bottom graphs illustrate the time course of LTP evoked in the CA1 area (fEPSP mean ± SEM) following HFS for wild-type and transgenic mice. The HFS was presented after 15 min of baseline recordings, at the time marked by the dashed line. The fEPSP is given as a percentage of the baseline (100%) slope. Although the two groups presented a significant increase (ANOVA, two-tailed) in fEPSP slope following HFS when compared with baseline records, values collected from the Ts1Yah group were significantly (*P < 0.001; F(24,216) = 25.278) larger than those collected from wt mice at the indicated times.
Impact of copy number variation on expression of genes located in the Abcg1–U2af1 genetic interval
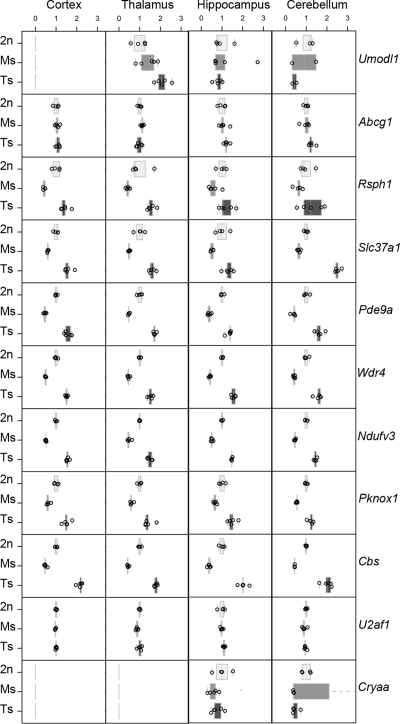
In DS, changes in copy number are associated with alteration of gene expression even though compensation effects have been described (33–36). Thus, we investigated the transcriptional levels of the genes located in the Abcg1–U2af1 interval in both trisomy and monosomy models. Taqman real-time quantitative PCR analysis was performed on samples derived from cortex, thalamus-hypothalamus, hippocampus and cerebellum of Ts1Yah, Ms2Yah (Ms) and euploid (2n) mice. Abcg1 and U2af1 that were at the border of the region were mostly unaffected, with an expression level ∼1, whereas all the other expressed genes within the Abcg1–U2af1 region were globally 0.5-fold under-expressed in Ms2Yah tissues, and their expression was 1.5-fold increased in Ts1Yah tissues (Fig. 5). Ubash3a, Tff2, Tff3 and Tmprss3 were detected only in one sub-region of the mouse brain (hippocampus for Ubash3a; cerebellum for Tff2, Tff3 and Tmprss3) with similar expression pattern in Ts and Ms (data not shown). In addition, we measured expression levels of four genes at the borders of the re-arrangement, namely Umodl1, Abcg1, U2af1 and Cryaa (Fig. 5), three Hsa21 orthologous genes from Mmu10 (Pfkl, Nnp1 and Cstb), eight Hsa21 orthologous genes from Mmu16 (Erg, Pcp4, Dscam, Bace2, Gabpa, Bach1, Runx1 and Sh3bgr; Supplementary Material, Fig. S3), three Hsa21 orthologous genes from Mmu10 (Pfkl, Nnp1 and Cstb) and six housekeeping genes used for input normalization (data not shown). No significant change of gene expression was observed for genes not located in the engineered interval except for the Umodl1 gene whose expression was increased substantially in the Ts thalamus (Fig. 5).
Figure 5.
Expression of genes from the Abcg1–U2af1 region in four different Ts1Yah, euploid (Eu) and Ms2Yah mice brain tissues. Box plots of normalized expression levels of 11 Mmu17 genes expressed in the four tissues analyzed (Cortex, Thalamus-Hypothalamus, Hippocampus and Cerebellum). The X-axis is normalized expression values relative to the mean of Eu mice, the Y-axis is the three genotype groups (2n = Eu, Ms = Ms2Yah and Ts = Ts1Yah mice). Each panel represents a gene (shown on right).
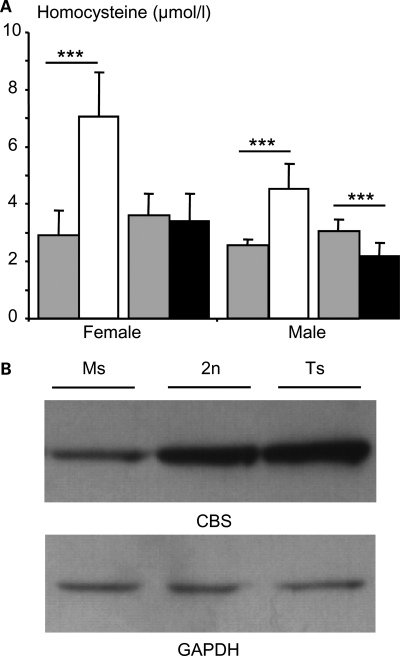
Next, we focused on the Cystathionine beta synthase (Cbs) gene, which is overexpressed in all tested regions of the brain. CBS catalyses the change of homocysteine toward cystathionine and its deficiency causes homocystinuria (OMIN 236250), a disease associated with increased plasma homocysteine and mental retardation. Thus we checked whether the altered gene expression modifies the accumulation or the activity of the encoded protein. We determined the concentration of the plasma homocysteine, the substrate of CBS and showed a significant difference in Ms2Yah mice compared with euploid and Ts1Yah mice (Fig. 6A). Levels of homocysteine were significantly increased in Ms2Yah females and males (P < 0,001, Student's t-test) and decreased in Ts1Yah males (P < 0,001, Student's t-test), but not females (P = 0.628, Student's t-test). These variations correlate with the amount of CBS protein detected in the liver of aneuploid mice (Fig. 6B). When compared with a similar level of GADPH in the extract, level of CBS was decreased in the Ms and increased in the Ts liver. Similarly, overexpression of Cbs in DS patients leads to a decrease in homocysteine levels (37). As expected most of the genes located in the Abcg1–U2af1 genetic interval were sensitive to gene dosage in the brain of both aneuploid models but a few including Rsph1, Slc37a1, Pde9a, Ndufv3, Pknox1 and Cbs were overexpressed in the Ts hippocampus.
Figure 6.
Detection of plasma homocysteine and CBS protein levels in Ms2Yah and Ts1Yah mice. (A) Mean ± SEM plasma levels of total homocysteine (µmol/l) in females and males Ms2Yah (white box) (n = 15 females and 7 males), Ts1Yah (black box; n = 15 females and 14 males) and their wild-type littermate (gray box; Ms2Yah wild-type littermate: n = 7 females and 9 males and for the Ts1Yah wild-type littermate: n = 15 females and 15 males) at 3 months of age. (B) Western blots of CBS in the liver of Ms2Yah (Ms), control (2n) and Ts1Yah (Ts) adult mice (3 months of age). Equal amounts (20 µg) of protein extracts were loaded in each lane and the same blot was incubated with an antibody against GADPH to normalize loading (***P < 0.001, Student's t-test).
DISCUSSION
The aim of this study was to analyze the contribution of the Abcg1–U2af1 region from the telomeric part of the Hsa21 to the DS phenotype. To this end, we generated two new models by chromosome engineering, a trisomy and a monosomy for the 0.59 Mb Mmu17 Abcg1–U2af1 region that is homologous to a distal part of Hsa21. In this report, we focused our interest on the Ts model. We found that the trisomy of the Abcg1–U2af1 region modifies cognitive performance and the neurophysiology of the hippocampus. Furthermore, expression of the genes located in the Abcg1–U2af1 interval was shown to be sensitive to copy number.
Behavioral and neurological analysis of the Ts1Yah mice has shown a clear but complex cognitive phenotype that combines altered habituation (a form of non-associative learning), and spontaneous alternation, reflecting short-term spatial memory and short-term recognition memory in the NOR. However, trisomy of the interval improved moderately the performance in spatial learning in the spatial version of the MWM. Our model mimics DS patients for their reduced attention and their alterations in processes involved in some forms of explicit memory that require the integrity of hippocampal functions (38–40). DS is characterized by a complex neuropsychological profile where some abilities are more impaired than others. In particular, DS people and DS mouse models such as the Ts65Dn (17,20,41–43) show a specific impairment in hippocampal-based declarative learning and memory. Similar to the Ts65Dn model (17), Ts1Yah mice perform poorly in the NOR task that has been related to a poor hippocampal-based declarative memory function, a hallmark of DS cognitive characteristics. However, the role of the hippocampus in object recognition memory processes is unclear since conflicting results have been found in lesion studies of both primates and rodents. In the current view, even though the hippocampus is involved in object recognition memory, parahippocampal structures (e.g. perirhinal cortex) are sufficient to support object recognition memory over short-retention intervals. Interestingly, object recognition is not altered in Ts1Cje mice (9) but long-lasting NOR is modified in the Ts1Rhr (44). Thus, at least several independent genetic intervals, some located on Mmu16, and one located in the Abcg1–U2af1 region should interact in patients. Interestingly, control of this trait might be even more complex as the Tc1 mouse model displays impaired short-term memory of object, whereas their long-term memory of object in not affected (10,26). Taken together, these results could reflect the region-specificity of hippocampal versus para-hippocampal genetic alteration in the different models. More surprisingly, spatial reference learning was improved in the Ts1Yah mice. The observed differences between NOR and MWM with the Ts1Yah was due to impairment of short-term memory and relative improvement of long-term memory and learning.
The MWM result was unexpected as other models, such as Ts65Dn and Tc1 mice are defective in learning in this test (13,20,26,42,43,45–50); a defect similar to the hippocampal deficit observed in DS patients (38,40,51). Furthermore, the hippocampus of Ts65Dn or Tc1 mice was affected for their structure, plasticity, growth, response to signaling pathways and LTP (19,52–57). In Ts65Dn, defects in LTP in the dentate gyrus has been related to enhanced inhibitory synaptic transmission (53,54,58), whereas Tc1 showed a more reduced early LTP that was restored 24 h after the initial stimulation (26). In contrast to both models, performances in the MWM task and LTP are surprisingly enhanced in the Ts1Yah mice. One seeming contradiction in our experiments would be the reduced performance in the NOR that is also partially dependent on hippocampal function. However, although both NOR and MWM assess hippocampus-dependent functions, each test puts specific cognitive demands on the animal, including spatial exploration and the formation of associations between spatial and non-spatial cues. These data reinforced the idea that separate domains mediate recognition versus spatial memory (59). In this regards, some evidence suggests a local processing deficit in DS construction, in which perceptual impairment in processing local forms, a global attention bias, or even an abnormality in the interaction between global and local levels may be involved (60).
Increased performance in the MWM was supported by higher and long-lasting LTP. Somehow both results suggested a convergence of mechanisms leading to improved hippocampal-dependent spatial memory and LTP, and dissociation of hippocampal function between spatial memory, novelty discrimination and working memory in this model. Such an increase in LTP could results from alterations in signaling pathways or in GABAergic mechanisms rather than an alteration in LTP machinery itself (61). Reduced LTP has been observed in the Ts65Dn and to some extent in the Ts1Cje and Ts1Rhr models (44,58,62–65) but less severely in the Tc1 mouse, a more complete Ts model (26). These results provide evidence for supporting a genetic mechanism affecting the severity of cognitive deficits observed in DS patients. Somehow, allelic variations of the genes from the Abcg1–U2af1 and other regions on Mmu16 or Mmu10 should affect the DS cognitive phenotypes that can be either severe or moderate. In addition, controlling the copy number and the expression of the genes from the Abcg1–U2af1 region could also affect the learning and memory in normal population.
By the use of two new genetic configurations, we were able to determine the consequences of copy number variation on the expression of genes from the genetic interval located at the borders or on other homologous region to the Hsa21. Our current analysis, on four different subregions of the brain, showed that all expressed genes from the engineered interval follow the expected transcription level based on copy number, with a decrease of the expression ratio to 0.5× in Ms and an increase to 1.5× in Ts tissues. Our results are similar to those described in other Ts or Ms mouse models (33,66–73) and in human DS patients (34–36,74–76). Two genes located on each side of the borders were studied. Only the expression of Umodl1 was changed in the thalamus of Ts mice, suggesting an altered regulation of transcription of the gene due to the engineered segmental duplication. Similar changes were described in other diseases depending on large genomic rearrangements (77–79). A correlation between protein level and substrate concentration was established in both Ms and Ts males versus control animals and confirmed the parallel, for Cbs, between gene expression level and protein level. Interestingly, the homocysteine level in Ts females was not affected in our study a result similar to some obtained in human T21 patients at the adult stage (80,81) but controversial to others obtained in younger individuals (37).
The present data suggest that the telomeric part of Hsa21 contain genes involved in the global and local perception, attention and construction deficits in DS. In comparison with the previous DS models, whereas the Ts65Dn mouse shows deficiencies across both domains, Ts1Cje appear to have deficiencies restricted to only the spatially oriented domain (9,17,41). Thus, the Abcg1–U2af1 region should contain at least one gene that is implicated in hippocampus-dependent spatial learning and LTP and whose overexpression leads to a gain-of-function phenotype. Most of the genes found within Abcg1–U2af1 have not been functionally characterized and no non-coding RNAs, including miRNAs, have been reported so far in the region. A few genes are associated with behavioral changes, such as polymorphisms of Pde9a with major depressive disorder (82) and Cbs deficiency with mental retardation in homocystinuria. Such an allelic variation of genes from the Abcg1–U2af1 region in the human population might alter differently the behavioral deficit of the DS patients and also might help to consider the hypothesis drawn from the human genetics analysis. Further experiments will be necessary to pinpoint that gene(s) might be responsible for the Ts1Yah phenotype, such as rescue experiments with single gene inactivation.
In conclusion, we report that the Mmu17 region orthologous to Hsa21, which had not been previously analyzed, contributes to the cognitive phenotypes observed in DS individuals. Mouse models that are Ts for other non-overlapping Hsa21 orthologous regions also show cognitive phenotypes: the Ts65Dn mouse displays deficiencies in both NOR and MWM, whereas the Ts1Cje model appears to have deficiencies restricted to only the spatially oriented domain (17). Learning and memory phenotype in both trisomies is linked to changes in the structure and the circuitry development of their hippocampus and of their cortical pyramidal layer, associated to reduced LTP and long-term depression (19,52–57). In this report, we found that trisomy of the Abcg1–U2af1 interval also impacted the hippocampal function in a more sophisticated way, disturbing short-term memory while facilitating the spatial learning and LTP. All these data combined suggest that the variable DS cognitive phenotypes are likely to result from the complex interactions of several genes or regions which can have negative or even positive contributions to cognitive performance (7,8,17,24,83). The data presented here show that variation in copy number should not only be considered as deleterious or susceptibility factors, but also can have advantageous or protective factors (12). Substantial progress has recently been made concerning several genes and pathways involved in DS pathogenesis (11), but this study highlights the complexity of this disorder. We showed that trisomy of a specific interval can both contribute to decrease in certain cognitive function and to an improvement of another one. Thus the contribution of the different regions of Hsa21 needs to be further evaluated to pinpoint genes modulating DS traits. This will not only decipher the pathophysiology of the disease but also help to characterize pathways controlling hippocampal-dependent learning and memory that could be targeted to rescue DS alterations or to improve normal condition.
MATERIALS AND METHODS
Generating the Ms2Yah and Ts1Yah mouse strains
The Abcg1 and U2af1 targeting vectors were isolated, respectively, from the 5′Hprt and 3′Hprt libraries (84) and inserted by homologous recombination in HM-1 Hprt-deficient ES cells (85). Subsequently, a transient expression of the Cre recombinase provided ES cells with three different configurations (Fig. 1). We obtained ES cell clones carrying two loxP sites in the Abcg1 and U2af1 loci in a cis configuration, which were named Cis(Abcg1tm1Yah–U2af1tm1Yah), ES cells with a deletion of the Abcg1–U2af1 region, the Ms2Yah clone and the Ts1Yah clone which provide a duplication of this region. Ms2Yah and Ts1Yah clones were injected into C57BL/6J blastocysts to generate chimera. These animals were crossed with C57BL/6J mice to obtain the corresponding mouse line. The Ms2Yah [B6.129Del(17Abcg1–U2af1)] and the Ts1Yah [B6.129Dup(17Abcg1–U2af1)] were generated on 129/Ola and backcrossed on the C57BL/6J genetic background up to N7 in this study.
Southern blot and fluorescence in situ hybridization (FISH) analysis
Between 5 and 10 µg of genomic tail or ES cell DNA extracts were digested with the appropriate enzyme and separated by electrophoresis through 0.7% agarose gel. Southern blot analysis was performed as described by Besson et al. (73). Interphase nuclei were recovered by affixing of a frozen/defrosted sample of kidney on a slide. Mouse BAC clones were chosen to be located inside (RP23-148M13) or outside (RP23-50P6) the Abcg1–U2af1 region; and 1 µg of mouse BAC DNA was used to generate DNA probes labeled by nick translation with DIG-dUTP (for 148M13) and biotin-dUTP (for 50P6). Detection was achieved by the use of both antidigoxigenin–rhodamine and avidin–fluorescein antibodies (Roche, Mannheim, Germany). The slide was mounted with vectashield medium containing DAPI 4′,6 diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA, USA). Images were analyzed by SmartCapture2F (Digital Scientific, Cambridge, UK) using a Zeiss AxioPlanII microscope equipped with a cooled camera (Photometrics, Tuscon, USA).
Behavioral analysis
Experiments were divided into two series. Groups of 15 Ts1Yah and 15 wild-type male mice at N4 backcross level on C57BL6/J (3 months of age) were tested in a first level behavioral analysis: SHIRPA, open-field, Y- and elevated plus mazes. A second groups of 15 Ts1Yah and 14 wild-type male mice at N5 backcross level (5 months of age; 30–35 g) were involved in a more specific analysis: Rotarod, treadmill, NOR and MWM. All the behavioral testing was conducted by the same experimenter in an isolated room and at the same time of the day (between 8:30 a.m. and 3:30 p.m.). Behavioral experimenters were blinded as to the genetic status of the animals. Mice were group housed (3–4 littermate mice per cage) in standard macrolon cages under a 12 h light–dark schedule (lights on at 8:00 a.m.) in controlled environmental conditions of humidity (60%) and temperature (22 ± 2°C) with free access to food and water. All experimental protocols involving the use of animals were performed in accordance with recommendations for the proper care and use of laboratory animals and were performed under authorization of the local Ethics Committee for Animal Experimentation (PRBB 1998/031 OV/2004/184 PNT) through the regulations and policies governing the Care and Use of Laboratory Animals (Law 5/1995 of the Catalonian Government, EU directive no. 86/609 and Council of Europe Convention ETS123, EU decree 2001-486 and regulations of the National Institutes of Health No. A5388-01) (USA). All investigators involved in animal experimentation have the appropriate training (Department de Medi Ambient i Habitatge, decret 214/1997). The CRG is authorized to work with genetically modified organisms (Operacions d'Utilització Confinada d'OGM A/ES/05/I-13 and A/ES/05/14). The experiments followed the recommendations from the standard operating procedures developed by the Eumorphia network (http://www.eumorphia.org). Detailed protocols are listed in Supplementary Materials and Methods.
LTP induction
Animals were anesthetized with 0.8–3% halothane delivered from a calibrated Fluotec 5 (Fluotec-Ohmeda, Tewksbury, MA, USA) vaporizer at a flow rate of 1–4 l/min oxygen. Animals were implanted with bipolar stimulating electrodes aimed at the right Schaffer collateral-commissural pathway of the dorsal hippocampus [2 mm lateral and 1.5 mm posterior to Bregma; depth from brain surface, 1.0–1.5 mm (86)] and with two recording electrodes aimed at the ipsilateral stratum radiatum underneath the CA1 area (1.2 mm lateral and 2.2 mm posterior to Bregma; depth from brain surface, 1.0–1.5 mm). These electrodes were made of 50 µm Teflon-coated tungsten wire (Advent Research Materials Ltd, Eynsham, England). The final position of hippocampal electrodes was determined using as a guide the field potential depth profile evoked by paired (40 ms of interval) pulses presented at the Schaffer collateral pathway (87). A bare silver wire (0.1 mm) was affixed to the skull as a ground. The four wires were connected to a four-pin socket and the socket was fixed to the skull with the help of two small screws and dental cement [see Ref. (87) for details]. A total of ten successful animals at N5 backcross level (about 8 months of age) were prepared for each experimental group.
fEPSPs were recorded with Grass P511 differential amplifiers through a high-impedance probe (2 × 1012 Ω, 10 pF). Electrical stimulus presented to Schaffer collaterals consisted of 100 µs, square, biphasic pulses presented alone, paired or in trains. Stimulus intensities ranged from 0.02 to 500 µA for the construction of the input/output curves. For paired pulse facilitation, the stimulus intensity was set well below the threshold for evoking a population spike, usually 35% of the intensity necessary for evoking a maximum fEPSP response (88). Paired pulses were presented at six different pulse intervals (10, 20, 40, 100, 200, and 500). The stimulus intensity was also set at 35% of its asymptotic value for LTP induction. An additional criterion for selecting stimulus intensity for LTP induction was that a second stimulus, presented 40 ms after a conditioning pulse, evoked a larger (>20%) synaptic field potential than the first (89). For LTP induction, each animal was presented with a HFS protocol consisting of five trains (200 Hz, 100 ms) of pulses at a rate of 1/s. This protocol was presented six times in total, at intervals of 1 min. The 100 µs, square, biphasic pulses used to evoke LTP were applied at the same intensity used for the single pulse presented following CS presentation. Further details of this chronic preparation can be found elsewhere (87). Statistical analyses were carried out using two-way repeated measure ANOVA. Factors were time versus fEPSP slopes, being time a repeated measure. Contrast analysis was added to further study significant differences.
Gene expression analysis
Cerebellum, thalamus-hypothalamus, hippocampus and cortex were isolated from 15 male mice at 3 month of age (six Ts; five Ms and four Eu) and flash frozen. Total RNA was prepared using Trizol (Invitrogen) according to the manufacturer's instructions. The quality of all RNA samples was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was converted to cDNA using Superscript II (Invitrogen) primed with poly d(T). For each sample in the study, 1 µg of total RNA was converted to cDNA. On the basis of Ensembl annotation (http://www.ensembl.org/index.html), we considered for study 14 genes mapping on Mmu17, between the Abcg1 and U2af1 genes where the loxP sites were inserted. Assays for the Umodl1, Abcg1, U2af1 and Cryaa genes were also designed to check the expression, the genes mapping to the borders of the insertions/deletions. In addition, we included assays for 17 non-Mmu17 genes. Among these, three genes for each tissue were selected for normalization and the remaining genes were used as additional controls. Selection of normalization genes was performed with GeNorm software (90). The list of the primers used is available on request. Taqman assays were designed using the program PrimerExpress v 2.0 (Applied Biosystems) with default parameters. Where possible, assays were designed to span an intron, and for the majority of the cases this was possible. Non intron-spanning assay (Pknox1) was tested in standard +/− RT reactions of RNA samples for genomic contamination; and no amplification was observed. Assay efficiencies were calculated using five four-fold serial dilutions of a pool of mouse brain, liver and testis (BLT) cDNAs as described (36). When transcripts were not expressed in BLT, efficiencies were tested in heart, thalamus, cerebellum and total embryo. Two genes (Pknox1 and Pde9a) did not pass our efficiency criteria threshold (between 0.95 and 1.05) and were re-designed successfully, whereas Tff1 was excluded from the analysis because the expression was not detected in any of the tissues studied giving a total of 15 Mmu17 genes and 17 non-Mmu17 genes. Amplicon sequences were checked by BLAST against the mouse genome to ensure that they were specific for the gene being assayed. HPLC-purified FAM-TAMRA-labeled double-dye Taqman probes were obtained from Sigma-Genosys (UK). All reactions used qPCR mastermix from ABgene (Epsom, UK). All PCRs were set up using a Biomek 2000 robot (Beckman), in a 10 µl volume in 384-well plates with six replicates per sample and ran in an ABI 7900 Sequence Detection System (Applied Biosystems) with the following conditions: 50°C for 2 min, 95°C for 10 min and 50 cycles of 95°C 15 s/60°C for 1 min. In total, 11 520 qRT–PCR reactions were performed. Raw Cycle threshold (CT) values were obtained using SDS 2.1 software (Applied Biosystems). Baseline values were automatically determined and threshold values were manually adjusted for each gene. Values with a deviation of ± 0.25 CT with respect to the median were considered outliers and excluded. Transcripts that amplified with a CT value greater than 37.9 were not included in the analysis. Each gene was rescaled using the mean expression value of control samples to give a relative normalized value. Data handling and normalizations were performed using Excel (Microsoft Corporation) and R (R Foundation, see Web Resources). To assess the differences in gene expression values between Ts, Ms and Eu samples, we performed the Kruskal–Wallis (KW) test.
Homocysteine measurement and western blotting
Blood samples were collected on fasting mice in tubes containing lithium heparin. After centrifugation, the plasma was separated and stored at −20°C. Total homocysteine concentration was measured on IMX (Abbott Laboratory) equipment using the commercially available ‘Abbott homocysteine assay’, a fluorescence polarization immunoassay from Abbott Diagnostics according to the manufacturer's instructions. Twenty microgram of total protein from liver extracts were electrophoretically separated in SDS–polyacrylamide gels (12%) and then transferred to nitrocellulose membrane (17 V) over night at room temperature. Non-specific binding sites were blocked with 5% skim milk powder in Tween Tris buffer saline 1 h at room temperature. Immunostaining was carried out with a goat polyclonal anti-Cbs and anti-Gapdh antibodies (Santa Cruz Biotechnology), followed by secondary anti-goat IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology). The immunoreactions were visualized by ECL chemiluminescence system (Amersham Biosciences) and exposure to ECL Hyperfilm (GE healthcare) for 1–5 min.
Statistical analysis
Results were processed for statistical analysis using the Sigma Plot software (Sigma) unless otherwise indicated. Data are represented as the mean ± SEM. All acquired behavioral data were analyzed using a one-way ANOVA test, with genotype as the independent variable. Data collected were also analyzed with general linear model with session as repeated measure. When appropriate, the parametric Fisher–Student t-test was applied otherwise the non-parametric KW test was used. Significant threshold was P < 0.05 or otherwise indicated.
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by grants from CNRS, Fondation Jerome Lejeune, ‘Association Française pour la recherche sur la trisomie 21’, ‘Conseil Général du Loiret’, ‘Région Centre’, Areces Foundation, Marató TV3, SAF2007-31093-E, SAF2007-60827, PI082038 and the AnEUploidy project (LSHG-CT-2006-037627) supported by the European commission under FP6.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the laboratory, of the AnEUploidy consortium for their helpful comments (www.aneuploidy.org). We are grateful to K. Maillard, T. Durand and E. Desale for taking care of the animals and to V. Nalesso for genotyping. We thank the National Centre for Scientific Research, the Foundation Jerome Lejeune, the ‘Association Française pour la recherche sur la trisomie 21’, Dr J. Laneau, the ‘Conseil Général du Loiret’, the ‘Région Centre’ and the European commission with the AnEUploidy project (LSHG-CT-2006-037627) for supports. The mutant mice are available for distribution through the ‘European Mouse Mutant Archive’ (www.emmanet.org).
Conflicts of Interest statement. None declared.
REFERENCES
- 1.Antonarakis S.E., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 2.Nadel L. Down's syndrome: a genetic disorder in biobehavioral perspective. Genes Brain Behav. 2003;2:156–166. doi: 10.1034/j.1601-183x.2003.00026.x. [DOI] [PubMed] [Google Scholar]
- 3.Antonarakis S.E., Epstein C.J. The challenge of Down syndrome. Trends Mol. Med. 2006;12:473–479. doi: 10.1016/j.molmed.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Gibson D., Groeneweg G., Jerry P., Harris A. Age and pattern of intellectual decline among Down syndrome and other mentally retarded adults. Int. J. Rehabil. Res. 1988;11:47–55. doi: 10.1097/00004356-198803000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Korenberg J.R., Chen X.N., Schipper R., Sun Z., Gonsky R., Gerwehr S., Carpenter N., Daumer C., Dignan P., Disteche C., et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl Acad. Sci. USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delabar J.M., Theophile D., Rahmani Z., Chettouh Z., Blouin J.L., Prieur M., Noel B., Sinet P.M. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 7.Lyle R., Béna F., Gagos S., Gehrig C., Lopez G., Schinzel A., Lespinasse J., Bottani A., Dahoun S., Taine L., et al. Genotype–phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 2008;17:454–466. doi: 10.1038/ejhg.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korbel J., Tirosh-Wagner T., Urban A., Chen X., Kasowski M., Dai L., Grubert F., Erdman C., Gao M., Lange K., et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl Acad. Sci. USA. 2009;106:12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez F., Garner C.C. Object recognition memory is conserved in Ts1Cje, a mouse model of Down syndrome. Neurosci. Lett. 2007;421:137–141. doi: 10.1016/j.neulet.2007.04.075. [DOI] [PubMed] [Google Scholar]
- 10.O'Doherty A., Ruf S., Mulligan C., Hildreth V., Errington M.L., Cooke S., Sesay A., Modino S., Vanes L., Hernandez D., et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves R., Garner C. A year of unprecedented progress in Down syndrome basic research. Ment. Retard. Dev. Disabil. Res. Rev. 2007;13:215–220. doi: 10.1002/mrdd.20165. [DOI] [PubMed] [Google Scholar]
- 12.Sussan T.E., Yang A., Li F., Ostrowski M.C., Reeves R.H. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 13.Reeves R.H., Irving N.G., Moran T.H., Wohn A., Kitt C., Sisodia S.S., Schmidt C., Bronson R.T., Davisson M.T. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 14.Sago H., Carlson E.J., Smith D.J., Kilbridge J., Rubin E.M., Mobley W.C., Epstein C.J., Huang T.T. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc. Natl Acad. Sci. USA. 1998;95:6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson L.E., Richtsmeier J.T., Leszl J., Reeves R.H. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Yu T., Morishima M., Pao A., Laduca J., Conroy J., Nowak N., Matsui S., Shiraishi I., Yu Y.E. Duplication of the entire 22.9 Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum. Mol. Genet. 2007;16:1359–1366. doi: 10.1093/hmg/ddm086. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez F., Morishita W., Zuniga E., Nguyen J., Blank M., Malenka R.C., Garner C.C. Pharmacotherapy for cognitive impairment in a mouse model of Down syndrome. Nat. Neurosci. 2007;10:411–413. doi: 10.1038/nn1860. [DOI] [PubMed] [Google Scholar]
- 18.Baxter L.L., Moran T.H., Richtsmeier J.T., Troncoso J., Reeves R.H. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum. Mol. Genet. 2000;9:195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- 19.Belichenko P.V., Masliah E., Kleschevnikov A.M., Villar A.J., Epstein C.J., Salehi A., Mobley W.C. Synaptic structural abnormalities in the Ts65Dn mouse model of Down Syndrome. J. Comp. Neurol. 2004;480:281–298. doi: 10.1002/cne.20337. [DOI] [PubMed] [Google Scholar]
- 20.Holtzman D.M., Santucci D., Kilbridge J., Chua-Couzens J., Fontana D.J., Daniels S.E., Johnson R.M., Chen K., Sun Y., Carlson E., et al. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc. Natl Acad. Sci. USA. 1996;93:13333–13338. doi: 10.1073/pnas.93.23.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenzi H.A., Reeves R.H. Hippocampal hypocellularity in the Ts65Dn mouse originates early in development. Brain Res. 2006;1104:153–159. doi: 10.1016/j.brainres.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Moore C.S. Postnatal lethality and cardiac anomalies in the Ts65Dn Down syndrome mouse model. Mamm. Genome. 2006;17:1005–1012. doi: 10.1007/s00335-006-0032-8. [DOI] [PubMed] [Google Scholar]
- 23.Richtsmeier J.T., Zumwalt A., Carlson E.J., Epstein C.J., Reeves R.H. Craniofacial phenotypes in segmentally trisomic mouse models for Down syndrome. Am. J. Med. Genet. 2002;107:317–324. doi: 10.1002/ajmg.10175. [DOI] [PubMed] [Google Scholar]
- 24.Olson L.E., Roper R.J., Sengstaken C.L., Peterson E.A., Aquino V., Galdzicki Z., Siarey R., Pletnikov M., Moran T.H., Reeves R.H. Trisomy for the Down syndrome ‘critical region’ is necessary but not sufficient for brain phenotypes of trisomic mice. Hum. Mol. Genet. 2007;16:774–782. doi: 10.1093/hmg/ddm022. [DOI] [PubMed] [Google Scholar]
- 25.Galante M., Jani H., Vanes L., Daniel H., Fisher E., Tybulewicz V., Bliss T., Morice E. Impairments in motor coordination without major changes in cerebellar plasticity in the Tc1 mouse model of Down syndrome. Hum. Mol. Genet. 2009;18:1449–1463. doi: 10.1093/hmg/ddp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morice E., Andreae L., Cooke S., Vanes L., Fisher E., Tybulewicz V., Bliss T. Preservation of long-term memory and synaptic plasticity despite short-term impairments in the Tc1 mouse model of Down syndrome. Learn. Mem. 2008;15:492–500. doi: 10.1101/lm.969608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brault V., Pereira P., Duchon A., Herault Y. Modeling chromosomes in mouse to explore the function of genes, genomic disorders, and chromosomal organization. PLoS Genet. 2006;2:e86. doi: 10.1371/journal.pgen.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams D.J., Biggs P.J., Cox T., Davies R., van der Weyden L., Jonkers J., Smith J., Plumb B., Taylor R., Nishijima I., et al. Mutagenic insertion and chromosome engineering resource (MICER) Nat. Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 29.Squire L.R., Stark C.E., Clark R.E. The medial temporal lobe. Annu. Rev. Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- 30.Bliss T.V., Collingridge G.L. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 31.Madronal N., Delgado-Garcia J.M., Gruart A. Differential effects of long-term Potentiation evoked at the CA3–CA1 synapse before, during, and after the acquisition of classical eyeblink conditioning in behaving mice. J. Neurosci. 2007;27:12139–12146. doi: 10.1523/JNEUROSCI.3397-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zucker R.S. Short-term synaptic plasticity. Annu. Rev. Neurosci. 1989;12:13–31. doi: 10.1146/annurev.ne.12.030189.000305. [DOI] [PubMed] [Google Scholar]
- 33.Lyle R., Gehrig C., Neergaard-Henrichsen C., Deutsch S., Antonarakis S.E. Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res. 2004;14:1268–1274. doi: 10.1101/gr.2090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sultan M., Piccini I., Balzereit D., Herwig R., Saran N.G., Lehrach H., Reeves R.H., Yaspo M.L. Gene expression variation in ‘Down syndrome’ mice allows to prioritize candidate genes. Genome Biol. 2007;8:R91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aït Yahya-Graison E., Aubert J., Dauphinot L., Rivals I., Prieur M., Golfier G., Rossier J., Personnaz L., Creau N., Bléhaut H., et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am. J. Hum. Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prandini P., Deutsch S., Lyle R., Gagnebin M., Delucinge Vivier C., Delorenzi M., Gehrig C., Descombes P., Sherman S., Dagna Bricarelli F., et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am. J. Hum. Genet. 2007;81:252–263. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogribna M., Melnyk S., Pogribny I., Chango A., Yi P., James S.J. Homocysteine metabolism in children with Down syndrome: in vitro modulation. Am. J. Hum. Genet. 2001;69:88–95. doi: 10.1086/321262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlesimo G.A., Marotta L., Vicari S. Long-term memory in mental retardation: evidence for a specific impairment in subjects with Down's syndrome. Neuropsychologia. 1997;35:71–79. doi: 10.1016/s0028-3932(96)00055-3. [DOI] [PubMed] [Google Scholar]
- 39.Vicari S., Bellucci S., Carlesimo G. Implicit and explicit memory: a functional dissociation in persons with Down syndrome. Neuropsychologia. 2000;38:240–251. doi: 10.1016/s0028-3932(99)00081-0. [DOI] [PubMed] [Google Scholar]
- 40.Pennington B.F., Moon J., Edgin J., Stedron J., Nadel L. The neuropsychology of Down syndrome: evidence for hippocampal dysfunction. Child. Dev. 2003;74:75–93. doi: 10.1111/1467-8624.00522. [DOI] [PubMed] [Google Scholar]
- 41.Fernandez F., Garner C.C. Episodic-like memory in Ts65Dn, a mouse model of Down syndrome. Behav. Brain Res. 2008;188:233–237. doi: 10.1016/j.bbr.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escorihuela R.M., Fernandez-Teruel A., Vallina I.F., Baamonde C., Lumbreras M.A., Dierssen M., Tobena A., Florez J. A behavioral assessment of Ts65Dn mice: a putative Down syndrome model. Neurosci. Lett. 1995;199:143–146. doi: 10.1016/0304-3940(95)12052-6. [DOI] [PubMed] [Google Scholar]
- 43.Escorihuela R.M., Vallina I.F., Martinez-Cue C., Baamonde C., Dierssen M., Tobena A., Florez J., Fernandez-Teruel A. Impaired short- and long-term memory in Ts65Dn mice, a model for Down syndrome. Neurosci. Lett. 1998;247:171–174. doi: 10.1016/s0304-3940(98)00317-6. [DOI] [PubMed] [Google Scholar]
- 44.Belichenko N., Belichenko P., Kleschevnikov A., Salehi A., Reeves R., Mobley W. The ‘Down syndrome critical region’ is sufficient in the mouse model to confer behavioral, neurophysiological, and synaptic phenotypes characteristic of Down syndrome. J. Neurosci. 2009;29:5938–5948. doi: 10.1523/JNEUROSCI.1547-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran T.H., Capone G.T., Knipp S., Davisson M.T., Reeves R.H., Gearhart J.D. The effects of piracetam on cognitive performance in a mouse model of Down's syndrome. Physiol. Behav. 2002;77:403–409. doi: 10.1016/s0031-9384(02)00873-9. [DOI] [PubMed] [Google Scholar]
- 46.Hyde L.A., Frisone D.F., Crnic L.S. Ts65Dn mice, a model for Down syndrome, have deficits in context discrimination learning suggesting impaired hippocampal function. Behav. Brain Res. 2001;118:53–60. doi: 10.1016/s0166-4328(00)00313-2. [DOI] [PubMed] [Google Scholar]
- 47.Hyde L.A., Crnic L.S., Pollock A., Bickford P.C. Motor learning in Ts65Dn mice, a model for Down syndrome. Dev. Psychobiol. 2001;38:33–45. doi: 10.1002/1098-2302(2001)38:1<33::aid-dev3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 48.Granholm A.C., Ford K.A., Hyde L.A., Bimonte H.A., Hunter C.L., Nelson M., Albeck D., Sanders L.A., Mufson E.J., Crnic L.S. Estrogen restores cognition and cholinergic phenotype in an animal model of Down syndrome. Physiol. Behav. 2002;77:371–385. doi: 10.1016/s0031-9384(02)00884-3. [DOI] [PubMed] [Google Scholar]
- 49.Hunter C.L., Bimonte H.A., Granholm A.C. Behavioral comparison of 4 and 6 month-old Ts65Dn mice: age-related impairments in working and reference memory. Behav. Brain Res. 2003;138:121–131. doi: 10.1016/s0166-4328(02)00275-9. [DOI] [PubMed] [Google Scholar]
- 50.Stasko M.R., Costa A.C. Experimental parameters affecting the Morris water maze performance of a mouse model of Down syndrome. Behav. Brain Res. 2004;154:1–17. doi: 10.1016/j.bbr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 51.Vicari S., Bates E., Caselli M.C., Pasqualetti P., Gagliardi C., Tonucci F., Volterra V. Neuropsychological profile of Italians with Williams syndrome: an example of a dissociation between language and cognition? J. Int. Neuropsychol. Soc. 2004;10:862–876. doi: 10.1017/s1355617704106073. [DOI] [PubMed] [Google Scholar]
- 52.Dierssen M., Benavides-Piccione R., Martinez-Cue C., Estivill X., Florez J., Elston G.N., DeFelipe J. Alterations of neocortical pyramidal cell phenotype in the Ts65Dn mouse model of Down syndrome: effects of environmental enrichment. Cereb. Cortex. 2003;13:758–764. doi: 10.1093/cercor/13.7.758. [DOI] [PubMed] [Google Scholar]
- 53.Siarey R.J., Kline-Burgess A., Cho M., Balbo A., Best T.K., Harashima C., Klann E., Galdzicki Z. Altered signaling pathways underlying abnormal hippocampal synaptic plasticity in the Ts65Dn mouse model of Down syndrome. J. Neurochem. 2006;98:1266–1277. doi: 10.1111/j.1471-4159.2006.03971.x. [DOI] [PubMed] [Google Scholar]
- 54.Best T.K., Siarey R.J., Galdzicki Z. Ts65Dn, a mouse model of Down syndrome, exhibits increased GABAB-induced potassium current. J. Neurophysiol. 2007;97:892–900. doi: 10.1152/jn.00626.2006. [DOI] [PubMed] [Google Scholar]
- 55.Hanson J.E., Blank M., Valenzuela R.A., Garner C.C., Madison D.V. The functional nature of synaptic circuitry is altered in area CA3 of the hippocampus in a mouse model of Down's syndrome. J. Physiol. 2007;579:53–67. doi: 10.1113/jphysiol.2006.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aldridge K., Reeves R.H., Olson L.E., Richtsmeier J.T. Differential effects of trisomy on brain shape and volume in related aneuploid mouse models. Am. J. Med. Genet. A. 2007;143:1060–1070. doi: 10.1002/ajmg.a.31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Contestabile A., Fila T., Ceccarelli C., Bonasoni P., Bonapace L., Santini D., Bartesaghi R., Ciani E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with down syndrome and in Ts65Dn mice. Hippocampus. 2007;17:665–678. doi: 10.1002/hipo.20308. [DOI] [PubMed] [Google Scholar]
- 58.Kleschevnikov A.M., Belichenko P.V., Villar A.J., Epstein C.J., Malenka R.C., Mobley W.C. Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J. Neurosci. 2004;24:8153–8160. doi: 10.1523/JNEUROSCI.1766-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vicari S., Bellucci S., Carlesimo G.A. Evidence from two genetic syndromes for the independence of spatial and visual working memory. Dev. Med. Child Neurol. 2006;48:126–131. doi: 10.1017/S0012162206000272. [DOI] [PubMed] [Google Scholar]
- 60.Wang P.P., Doherty S., Rourke S.B., Bellugi U. Unique profile of visuo-perceptual skills in a genetic syndrome. Brain Cogn. 1995;29:54–65. doi: 10.1006/brcg.1995.1267. [DOI] [PubMed] [Google Scholar]
- 61.Nosten-Bertrand M., Errington M., Murphy K., Tokugawa Y., Barboni E., Kozlova E., Michalovich D., Morris R., Silver J., Stewart C., et al. Normal spatial learning despite regional inhibition of LTP in mice lacking Thy-1. Nature. 1996;379:826–829. doi: 10.1038/379826a0. [DOI] [PubMed] [Google Scholar]
- 62.Costa A.C., Grybko M.J. Deficits in hippocampal CA1 LTP induced by TBS but not HFS in the Ts65Dn mouse: a model of Down syndrome. Neurosci. Lett. 2005;382:317–322. doi: 10.1016/j.neulet.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 63.Siarey R.J., Stoll J., Rapoport S.I., Galdzicki Z. Altered long-term potentiation in the young and old Ts65Dn mouse, a model for Down syndrome. Neuropharmacology. 1997;36:1549–1554. doi: 10.1016/s0028-3908(97)00157-3. [DOI] [PubMed] [Google Scholar]
- 64.Siarey R.J., Carlson E.J., Epstein C.J., Balbo A., Rapoport S.I., Galdzicki Z. Increased synaptic depression in the Ts65Dn mouse, a model for mental retardation in Down syndrome. Neuropharmacology. 1999;38:1917–1920. doi: 10.1016/s0028-3908(99)00083-0. [DOI] [PubMed] [Google Scholar]
- 65.Siarey R.J., Villar A.J., Epstein C.J., Galdzicki Z. Abnormal synaptic plasticity in the Ts1Cje segmental trisomy 16 mouse model of Down syndrome. Neuropharmacology. 2005;49:122–128. doi: 10.1016/j.neuropharm.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Saran N.G., Pletcher M.T., Natale J.E., Cheng Y., Reeves R.H. Global disruption of the cerebellar transcriptome in a Down syndrome mouse model. Hum. Mol. Genet. 2003;12:2013–2019. doi: 10.1093/hmg/ddg217. [DOI] [PubMed] [Google Scholar]
- 67.Cairns N.J. Molecular neuropathology of transgenic mouse models of Down syndrome. J. Neural. Transm. Suppl. 2001:289–301. doi: 10.1007/978-3-7091-6262-0_24. [DOI] [PubMed] [Google Scholar]
- 68.Chrast R., Scott H.S., Papasavvas M.P., Rossier C., Antonarakis E.S., Barras C., Davisson M.T., Schmidt C., Estivill X., Dierssen M., et al. The mouse brain transcriptome by SAGE: differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res. 2000;10:2006–2021. doi: 10.1101/gr.10.12.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dauphinot L., Lyle R., Rivals I., Dang M.T., Moldrich R.X., Golfier G., Ettwiller L., Toyama K., Rossier J., Personnaz L., et al. The cerebellar transcriptome during postnatal development of the Ts1Cje mouse, a segmental trisomy model for Down syndrome. Hum. Mol. Genet. 2005;14:373–384. doi: 10.1093/hmg/ddi033. [DOI] [PubMed] [Google Scholar]
- 70.Potier M.C., Rivals I., Mercier G., Ettwiller L., Moldrich R.X., Laffaire J., Personnaz L., Rossier J., Dauphinot L. Transcriptional disruptions in Down syndrome: a case study in the Ts1Cje mouse cerebellum during post-natal development. J. Neurochem. 2006;97(Suppl. 1):104–109. doi: 10.1111/j.1471-4159.2005.03624.x. [DOI] [PubMed] [Google Scholar]
- 71.Kahlem P., Sultan M., Herwig R., Steinfath M., Balzereit D., Eppens B., Saran N.G., Pletcher M.T., South S.T., Stetten G., et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 2004;14:1258–1267. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amano K., Sago H., Uchikawa C., Suzuki T., Kotliarova S.E., Nukina N., Epstein C.J., Yamakawa K. Dosage-dependent over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum. Mol. Genet. 2004;13:1333–1340. doi: 10.1093/hmg/ddh154. [DOI] [PubMed] [Google Scholar]
- 73.Besson V., Brault V., Duchon A., Togbe D., Bizot J.C., Quesniaux V.F., Ryffel B., Herault Y. Modeling the monosomy for the telomeric part of human chromosome 21 reveals haploinsufficient genes modulating the inflammatory and airway responses. Hum. Mol. Genet. 2007;16:2040–2052. doi: 10.1093/hmg/ddm152. [DOI] [PubMed] [Google Scholar]
- 74.Mao R., Zielke C.L., Zielke H.R., Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 75.Mao R., Wang X., Spitznagel E.L., Jr, Frelin L.P., Ting J.C., Ding H., Kim J.W., Ruczinski I., Downey T.J., Pevsner J. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.FitzPatrick D.R., Ramsay J., McGill N.I., Shade M., Carothers A.D., Hastie N.D. Transcriptome analysis of human autosomal trisomy. Hum. Mol. Genet. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 77.Merla G., Howald C., Henrichsen C., Lyle R., Wyss C., Zabot M., Antonarakis S., Reymond A. Submicroscopic deletion in patients with Williams–Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am. J. Hum. Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reymond A., Henrichsen C., Harewood L., Merla G. Side effects of genome structural changes. Curr. Opin. Genet. Dev. 2007;17:381–386. doi: 10.1016/j.gde.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Henrichsen C., Vinckenbosch N., Zöllner S., Chaignat E., Pradervand S., Schütz F., Ruedi M., Kaessmann H., Reymond A. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 80.Varga P., V Oláh A., Oláh E. [Biochemical alterations in patients with Down syndrome] Orv. Hetil. 2008;149:1203–1213. doi: 10.1556/OH.2008.28327. [DOI] [PubMed] [Google Scholar]
- 81.Fillon-Emery N., Chango A., Mircher C., Barbé F., Bléhaut H., Herbeth B., Rosenblatt D., Réthoré M., Lambert D., Nicolas J. Homocysteine concentrations in adults with trisomy 21: effect of B vitamins and genetic polymorphisms. Am. J. Clin. Nutr. 2004;80:1551–1557. doi: 10.1093/ajcn/80.6.1551. [DOI] [PubMed] [Google Scholar]
- 82.Wong M.L., Whelan F., Deloukas P., Whittaker P., Delgado M., Cantor R.M., McCann S.M., Licinio J. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc. Natl Acad. Sci. USA. 2006;103:15124–15129. doi: 10.1073/pnas.0602795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seregaza Z., Roubertoux P.L., Jamon M., Soumireu-Mourat B. Mouse models of cognitive disorders in trisomy 21: a review. Behav. Genet. 2006;36:387–404. doi: 10.1007/s10519-006-9056-9. [DOI] [PubMed] [Google Scholar]
- 84.Zheng B., Mills A.A., Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 1999;27:2354–2360. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Magin T., McWhir J., Melton D. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 1992;20:3795–3796. doi: 10.1093/nar/20.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paxinos G., Franklin K.B.J. New York: Elsevier; 2001. The Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- 87.Gruart A., Muñoz M., Delgado-García J. Involvement of the CA3–CA1 synapse in the acquisition of associative learning in behaving mice. J. Neurosci. 2006;26:1077–1087. doi: 10.1523/JNEUROSCI.2834-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gureviciene I., Ikonen S., Gurevicius K., Sarkaki A., van Groen T., Pussinen R., Ylinen A., Tanila H. Normal induction but accelerated decay of LTP in APP+PS1 transgenic mice. Neurobiol. Dis. 2004;15:188–195. doi: 10.1016/j.nbd.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 89.Bliss T., Gardner-Medwin A. Long-lasting potentiation of synaptic transmission in the dentate area of the unanaestetized rabbit following stimulation of the perforant path. J. Physiol. 1973;232:357–374. doi: 10.1113/jphysiol.1973.sp010274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.