Abstract
Protein cleavage is a common feature in human neurodegenerative disease. Ataxin-3 protein with an expanded polyglutamine (polyQ) repeat causes spinocerebellar ataxia type-3 (SCA3), also called Machado–Joseph disease, and is cleaved in mammalian cells, transgenic mice and SCA3 patient brain tissue. However, the pathological significance of Ataxin-3 cleavage has not been carefully examined. To gain insight into the significance of Ataxin-3 cleavage, we developed a Drosophila SL2 cell-based model as well as transgenic fly models. Our data indicate that Ataxin-3 protein cleavage is conserved in the fly and may be caspase-dependent as reported previously. Importantly, comparison of flies expressing either wild-type or caspase-site mutant proteins indicates that Ataxin-3 cleavage enhances neuronal loss in vivo. This genetic in vivo confirmation of the pathological role of Ataxin-3 cleavage indicates that therapies targeting Ataxin-3 cleavage might slow disease progression in SCA3 patients.
INTRODUCTION
Protein cleavage plays a critical role in a number of different neurodegenerative disease situations. Cleavage of amyloid precursor protein (APP) by a series of proteases leads to the production of beta-amyloid (Aβ) peptide and the relative abundance of Aβ 42 versus Aβ 40 is thought to play an important role in the development of Alzheimer's disease (1). Increasing evidence also indicates that caspase-mediated cleavage of APP plays important roles in Aβ production (2). Tau protein, found in Alzheimer's disease and other disease lesions, is also subject to cleavage by caspases, which may contribute to disease progression in Alzheimer's disease and tauopathies (3,4). Huntingtin protein, which is responsible for Huntington's disease (HD), is also cleaved by proteases and its cleavage fragments are detected in HD patient brains (5). Site-directed mutagenesis targeting a caspase recognition site of the Huntingtin protein was shown to drastically reduce HD-like neurodegeneration in mice (6).
HD is one of a larger class of diseases known as polyglutamine (polyQ) diseases, of which there are numerous ataxias, including the most common dominantly inherited ataxia spinocerebellar ataxia type-3 [SCA3, also known as Machado–Joseph disease (MJD)]. Data from mouse transgenic models and human disease tissue indicate that the pathogenic Ataxin-3 protein, encoded by the ATXN3 gene, is proteolyzed in vivo (7). In mice, proteolysis is seen in transgenic lines with high protein expression levels that show degeneration, and is stronger in animals that are sick, and milder in those without symptoms. A normal protein cleavage product is reported, although it is unclear whether protein expression levels are comparable with that of disease animals. Epitope mapping of the cleavage site indicated that it occurs N-terminal to aa221, more recently narrowed to N-terminal to aa190, and the resulting cleavage fragment includes the polyQ repeat (7,8). A similar proteolytic fragment was seen in western samples from three human patients (7). These studies suggest that proteolysis of the pathogenic protein may occur, may contribute to disease, and that the stable fragment contains the pathogenic polyQ repeat.
In a second study, proteolysis of the normal and pathogenic Ataxin-3 proteins was seen in Cos-7 cells upon staurosporine-induced apoptosis (9). Endogenous Ataxin-3 with a normal polyQ length repeat also underwent cleavage with staurosporine treatment. The cleavage in these situations was inhibited by caspase inhibitor zVAD-fmk. The cleavage was mapped to a similar location as in the Goti et al. (7) study, and was shown to be largely caspase-1 dependent. Examination of patient brain tissue for proteolytic fragments failed to reveal a clear fragment.
Drosophila models expressing human Ataxin-3 proteins have provided novel insights into SCA3 disease (10–13). The Ataxin-3 protein has an N-terminal ubiquitin protease domain, with the polyQ repeat near the C-terminal region of the protein (14). The ubiquitin protease domain has been shown to have de-ubiquitination activity, and is speculated to be a ubiquitin chain editing enzyme (15,16). In the fly, the activity of the normal protein associated with the ubiquitin protease domain substantially mitigates toxicity of the pathogenic protein (13). When this domain is deleted (as with an N-terminally truncated protein), or if the ubiquitin protease domain is mutated to make it inactive (C14A mutation), the protein with a pathogenic repeat is substantially more toxic in both degree and tissue range of toxicity. These data indicate that the N-terminal domain has activity to mitigate toxicity of the pathogenic protein, and that, should N-terminal proteolysis occur, it is predicted to substantially enhance toxicity of the protein. N-terminal truncation of the protein also affects the ability of the protein to interact with P97/VCP, which also enhances toxicity (17).
Previous reports about Ataxin-3 protein cleavage are in cell-based studies or correlative studies in transgenic mouse models or patient brain samples (7,9,18,19). In order to gain experimental evidence of the pathological significance of Ataxin-3 protein cleavage in vivo, we used the fruit fly model system to define potential cleavage of the Ataxin-3 protein in the nervous system in vivo, and the consequences of preventing that cleavage pattern. Our data suggest that Ataxin-3 cleavage enhances the progression of neurodegeneration in the fly model for SCA3, indicating that such cleavage in the human situation may enhance disease progression.
RESULTS
Ataxin-3 protein is cleaved in Drosophila in a pattern similar to mammals
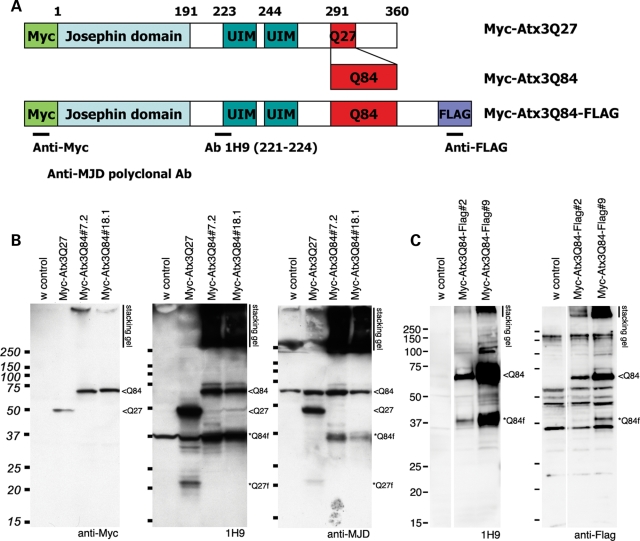
Studies using various antibodies suggest that the major cleavage product of Ataxin-3 includes the polyQ domain, and that expanded pathogenic Ataxin-3 protein from the brain tissues of transgenic mouse models or SCA3 patients generates a ∼36 kDa fragment, with the size fragment dependent on the length of the polyQ repeat (7,9). We examined whether similar protein cleavage products were generated in the nervous system of Drosophila expressing Ataxin-3 with either normal length (Atx3Q27) or expanded polyQ domain (Atx3Q84). Polyclonal antibody to Ataxin-3 and the monoclonal antibody 1H9, which recognizes aa221–224, as well as anti-Myc and anti-Flag antibodies, were chosen to compare our data with previous studies (Fig. 1A) (7,9,18). Previously reported Myc-tagged Ataxin-3 proteins of normal or expanded polyQ length were used along with newly generated double-tagged (N-terminal Myc and C-terminal Flag) proteins.
Figure 1.
Ataxin-3 protein cleavage products are generated in Drosophila. (A) Schematic representation of Ataxin-3 proteins used. Site of the epitopes for the antibodies used are also indicated. (B) and (C) Western immunoblot analysis of proteins expressed in the brain of 7d fly heads. (B) Initial studies performed with Myc-Atx3Q27 and Myc-Atx3Q84 proteins. Left, Anti-Myc antibody detects the full-length proteins. Middle, the mouse monoclonal antibody 1H9 and right, the polyclonal anti-MJD antibody detect Ataxin-3 fragments migrating at ∼22 kDa for Atx3Q27 and ∼37 kDa for Atx3Q84. Each antibody also detects non-specific bands when compared with the control. Strong signals are detected as SDS-insoluble fractions in the stacking gel. (C) Left, transgenic Drosophila head extracts expressing Myc-Atx3Q84-Flag show a similar cleavage pattern by 1H9, of the full-length protein and ∼37 kDa fragments. Right, anti-Flag antibody detects the full-length protein and a ∼37 kDa fragment. These data suggest that ∼37 kDa fragments, also reported in transgenic mouse and some MJD patient samples, are likely to contain the C-terminal portion of MJD protein, as well as the polyQ domain of the protein. Genotypes: (B) elav-gal4/+; UAS-Myc-Atx3Q27/+ or UAS-Myc-Atx3Q84/+ and (C) elav-gal4/+; UAS-Myc-Atx3Q84-Flag/+. Full-length proteins, <Q84 or <Q27, fragments indicated as *Q27f and *Q84f.
Analysis of the cleavage patterns of the normal and pathogenic protein with these different antibodies revealed that the Ataxin-3 protein is cleaved in the nervous system in vivo to generate smaller stable fragments that include the polyQ repeat (Fig. 1B). With the 1H9 and anti-MJD antibodies, significant amounts of cleavage products were readily detected. The pathogenic Atx3Q84 protein runs at ∼73 kDa, and is cleaved to generate fragments running at ∼37 kDa. Depending on the antibody, the cleavage product appears as a doublet or single band. The normal length Atx3Q27 protein generates a smaller fragment of ∼22 kDa, suggesting that the major cleavage product includes the polyQ domain (Fig. 1B). The presence of ∼37 kDa fragments, similar in size to the fragment seen in mouse and human (7), with expression of pathogenic Ataxin-3 suggests that Ataxin-3 cleavage is conserved in Drosophila. Interestingly, our initial studies showed that cleavage of Ataxin-3 was much more prominent when the protein was expressed in the nervous system than in the eye, which has predominantly accessory cells along with neuronal cells (Supplementary Material, Fig. S1). The full-length Ataxin-3 protein is also not very toxic to non-neural cells of the eye (1), suggesting that the cleavage of the protein may be more particular in vivo to the nervous system, and that this may relate to the toxicity of the protein.
The anti-Myc antibody which recognizes the N-terminal end of tagged forms of Ataxin-3 protein highlighted the full-length protein, which migrated just below the 75 kDa marker, and no significant smaller products (Fig. 1B). This indicated that cleavage products do not include the N-terminal end of the protein. With the double-tagged protein, we observed a similar cleavage pattern of the Atx3Q84 protein indicating C-terminal cleavage. Notably, antibodies to the C-terminal FLAG tag detect a cleavage product migrating just above ∼37 kDa, further confirming that stable cleavage products are derived from the C-terminus of the protein (Fig. 1C).
Caspase inhibitor zVAD-fmk inhibits Ataxin-3 protein cleavage in S2 cells
Previous reports suggest that Ataxin-3 protein cleavage is dependent on caspase activities and is blocked by caspase inhibitors (9,19). Calpain proteases and the proteasome have also been implicated in studies in vitro using neuroblastoma cell extracts (18). To further define potential proteases that may function for protein cleavage in the fly, we developed a Drosophila SL2 cell-based system to determine whether the Ataxin-3 protein is proteolyzed by any of the proteases previously implicated using various chemical inhibitors (Fig. 2A).
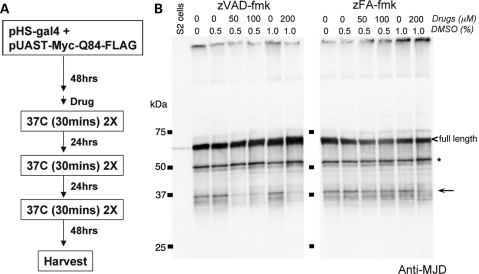
Figure 2.
zVAD, a broad-acting caspase inhibitor, can suppress Ataxin-3 protein cleavage in an SL2-cell-based system. (A) Experimental procedure to detect the effect of various protease inhibitors on Ataxin-3 protein cleavage in SL2 cells. Expression of the Ataxin-3 protein was driven by heat shock using a Gal4 (pHS-gal4) construct. The construct in all cases was pUAST-Myc-Atx3Q84. Drug was added 1 h prior to the first heat shock and was then continuously present until harvesting of the cells. (B) Ataxin-3 protein cleavage pattern in SL2 cells. The full-length protein is marked with a carrot (<) and the ∼37 kDa fragments highlighted with an arrow. In addition to the ∼37 kDa fragments seen in the fly in vivo, additional Ataxin-3 fragments were detected in SL2 cells, most prominently a fragments at ∼50 kDa (*). zVAD, a general caspase inhibitor, inhibited the production of the ∼37 kDa fragments, without significantly altering the generation of the ∼50 kDa fragment. zFA, a control peptide, had no effect. All drugs were dissolved in DMSO, with final percentage of DMSO indicated.
As observed in the fly in vivo, we detected the ∼37 kDa cleavage products when the pathogenic Atx3Q84 protein was transiently expressed in SL2 cells (Fig. 2B). The cleavage pattern of the protein in SL2 cells was similar to the cleavage pattern of the protein in the fly in vivo (Supplementary Material, Fig. S2). Additional cleavage products were also detected in cultured cells, most prominently a fragment of ∼50 KDa (* in Fig. 2B, ** in Supplementary Material, Fig. S3). We then used inhibitors to define potential cleaving proteases. zVAD-fmk, a broad caspase inhibitor, consistently reduced the level of ∼37 kDa fragment, while the control peptide zFA-fmk had no effect (Fig. 2B, Supplementary Material, Figs S3 and S4). Interestingly, with zVAD-fmk, there appeared to be an accumulation of a higher molecular weight fragment (**in Supplementary Material, Fig. S3), suggesting the possibility of sequential processing of the Ataxin-3 protein.
Although the proteasome inhibitor MG-132 and the calpain inhibitor ALLN have been previously shown to inhibit Ataxin-3 cleavage in vitro (18), in SL2 cells, only MG-132 inhibited the appearance of a minor band (*** in Supplementary Material, Fig. S3), but it did not inhibit generation of the ∼37 kDa fragments. Furthermore, when zVAD-fmk and MG-132 were combined, the results were additive rather than synergistic, suggesting that the proteasome and caspases may act independently.
Site-directed mutagenesis of caspase-recognition sites significantly modify Ataxin-3 protein cleavage in cells and in the fly
Goti et al. (7) used a series of monoclonal antibodies to identify the potential Ataxin-3 protein cleavage site. Those studies indicated that the cleavage site may be near or just anterior to the ubiquitin-interacting motifs (UIMs) N-terminal to the polyQ repeat. Berke et al. (9) reported several putative caspase-recognition sites and showed that the modification of six or nine caspase sites by site-directed mutagenesis eliminated the appearance of cleavage products in Cos-7 cells. The data with zVAD-fmk indicated that Ataxin-3 is a target of caspases also in Drosophila cells. To test the significance of caspase cleavage, we modified the aspartic acid residues to asparagines at select caspase sites. Because of our interest in the ∼37 kDa fragments, we focused on the six putative caspase sites at amino acids 171, 208, 217, 223, 225 and 228 (Fig. 3A).
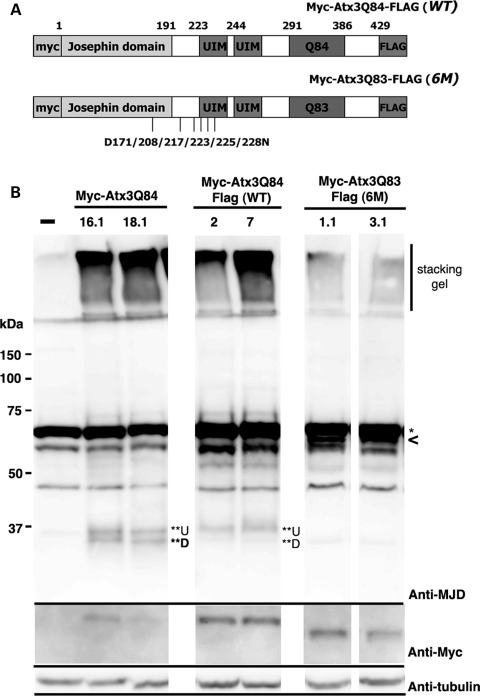
Figure 3.
Mutation of putative caspase recognition sites in Ataxin-3 dramatically reduces the amount of cleavage product in vivo. (A) Schematic diagram of Atx3Q83 protein with the positions noted of the six caspase sites targeted in the site-directed mutagenesis (D->N). (B) Western immunoblots. Top panel, Ataxin-3 protein cleavage pattern probed with anti-MJD polyclonal antibody. Head extracts from 7d adult flies were used. Sextuplet mutation strongly suppresses Ataxin-3 protein cleavage. A non-specific band (*, ∼70 kDa) masks the full-length band. SDS-insoluble fractions are within the stacking gel, and the ∼37 kDa cleavage products (**U and **D) are marked. Genotypes: elav-gal4/UAS-Myc-Atx3Q84, elav-gal4/ UAS-Myc-Atx3Q84-Flag (WT) and elav-gal4/UAS-Myc-Atx3Q83-Flag (6M). Two transgenic lines of each genotype are shown. Lower panels are the same blot, probed with Anti-Myc for the full-length protein, and with Anti-tubulin, for a loading control.
Upon generation of the caspase-resistant forms, we found that caspase cleavage of Ataxin-3 is a dynamic process. That is, single site (D208N or D217N) or even triple mutation (D171/208/217N) of the putative caspase sites did not fully eliminate cleavage, but rather lead to an alteration in the cleavage pattern in SL2 cells (Supplementary Material, Fig. S5 and data not shown). Strikingly, however, elimination of all six putative caspase-sites (6M) eliminated the ∼37 kDa fragments in SL2 cells (Supplementary Material, Fig. S5A). Such mutation of the protein also had an effect on the overall mobility of the protein in gels. This may be due to a conformational change caused by the mutations or could result from additional cleavage at the C-terminal end of the protein (9,19).
To extend these findings in vivo in order to understand the pathological impact of modified Ataxin-3 protein cleavage, we generated transgenic animals expressing either wild-type protein (Myc-Atx3Q84-Flag (WT)) or the sextuplet mutant protein (Myc-Atx3Q83-Flag (6M)). Analysis of flies expressing equal levels of these proteins revealed that the 6M mutant protein had dramatically reduced levels of cleavage product in vivo (Fig. 3). Whereas the Myc-Atx3-Q84 protein showed two cleavage products at the ∼37 kDa region (**U and **D in Fig. 3B), the upper fragment of the double-tagged Myc-SCA3Q84-Flag protein was typically more prominent; thus, the C-terminal Flag tag may influence the Ataxin-3 protein cleavage process. The 6M mutant protein, however, which also contained the C-terminal Flag tag, completely lacked the upper ∼37 kDa band, and only faint signals were detected for the lower molecular weight fragment.
The amount of SDS-insoluble material in the stacking gel in the 6M caspase-mutant form of the protein was also significantly reduced, as detected by the anti-MJD antibody (Fig. 3B). Interestingly, the amount of SDS-insoluble material trapped in the stacking gel appeared to correlate with the amount of the upper ∼37 kDa product (**U). In the 6M transgenic lines, lines with the lowest amount of cleavage product also showed the lowest amount of SDS-insoluble material with the anti-MJD antibody (data not shown).
Sextuplet caspase-site mutation (6M) does not significantly modify nuclear inclusion formation
The correlation between the amount of SDS-insoluble material by western analysis and the amount of the upper ∼37 kDa fragment raised the interesting hypothesis that this fragment may act as a seed for the formation of nuclear inclusions in tissues expressing the pathogenic Ataxin-3 protein. To test this hypothesis, we conducted a time course experiment to determine whether there was a difference in onset or size of nuclear inclusion formation by Ataxin-3 in vivo. We used the late-onset rhodopsin (rh1-Gal4) driver that allows sensitive analysis of inclusion formation (11,13).
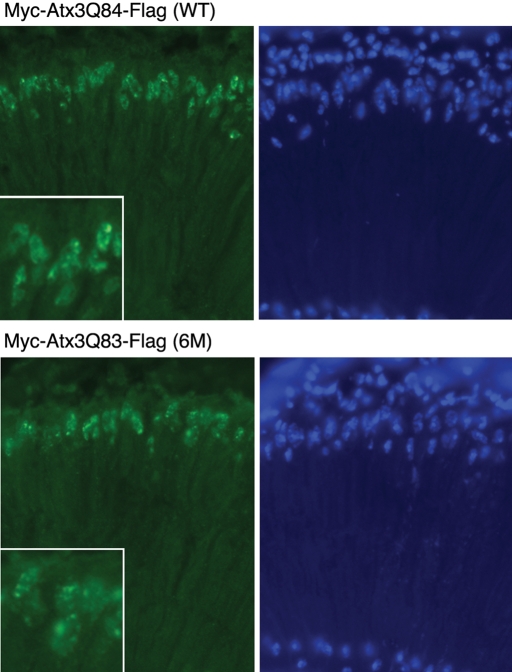
These studies revealed no difference in protein accumulation between the WT and 6M form of pathogenic Ataxin-3 with 7d animals, showing a similar level and size of nuclear inclusions between WT and 6M flies (Fig. 4). Analysis at earlier time points also indicated that the rate of inclusion formation was similar between WT and 6M flies (data not shown). Therefore, mutation of the caspase cleavage sites and reduced proteolysis of the pathogenic Ataxin-3 protein does not affect protein accumulation. This finding also suggests that protein misfolding and clearance are not significantly affected by the presence of these caspase cleavage products, in agreement with MG-132 data from SL2 cells (see Supplementary Material, Fig. S3).
Figure 4.
Elimination of putative caspase cleavage sites in Ataxin-3 protein has little impact on protein accumulations. Cryosections immunostained for Ataxin-3 protein with Myc (green, left panels) and for nuclei with Hoechst (blue, right panels) of retinal sections from 7d adult flies. Protein was expressed with an adult onset driver (rh1-gal4), which is a sensitive method to detect subtle differences in nuclear inclusion formation. Flies expressing either the WT protein (top) or the 6M protein (bottom) showed inclusions typical of the full-length Ataxin-3 protein, being of irregular shape when assayed with this late-onset driver, and had the same onset and size. Higher magnification insets are also included in the Myc panels. Genotypes: rh1-gal4/ UAS-Myc-Atx3Q84-Flag (WT) and rh1-gal4/ UAS-Myc-Atx3Q83-Flag (6M).
Sextuplet caspase site mutation mitigates neurodegeneration
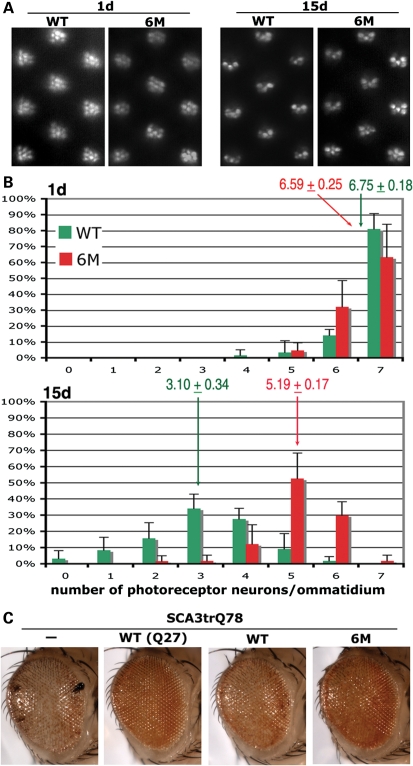
We next asked whether reduction in caspase cleavage modulates Ataxin-3 protein toxicity. Transgenic lines expressing the same level of the WT or 6M proteins were expressed in the nervous system with elav-gal4, and photoreceptor degeneration was assessed. Animals at 1d showed a mild degeneration that was similar with expression of either the WT or 6M protein, with 6–7 photoreceptor neurons per unit eye ommatidial cluster (Fig. 5). Flies expressing the WT pathogenic protein, however, then underwent rapid neural loss, such that 15d animals retained only 3.1 ± 0.34 photoreceptor neurons per ommatidial cluster. In contrast, 15d flies expressing the 6M caspase mutant protein had strikingly less degeneration, with 5.19 ± 0.17 PR neurons per ommatidial unit remaining. To confirm that the 6M mutant protein was properly folded despite the caspase site mutations, we tested whether the ability to suppress the pathogenicity of the truncated SCA3Q78 protein was retained. Normally, the pathogenic full-length Ataxin-3 protein, when co-expressed with the truncated Q78 protein, has activity to suppress toxicity (Fig. 5C; 13). These studies showed that the 6M protein retained suppressor activity comparable with that of the protein without caspase site mutations; this indicates that the 6M protein retains normal Ataxin-3 activity and thus is likely properly folded. Taken together, these data indicate that blocking caspase cleavage of the Ataxin-3 protein dramatically mitigates neural toxicity, despite the fact that nuclear inclusion formation is unaffected.
Figure 5.
Suppression of Ataxin-3 protein cleavage slows down the progression of photoreceptor neuronal degeneration. (A) Images of the retina by pseudopupil technique, showing 1d and 15d representative images of flies expressing Myc-Atx3Q84-Flag (WT) and Myc-Atx3Q83-Flag (6M). (B) Quantitation of the number of photoreceptor neurons in 1d WT (green) and 6M (red) flies. At 1d, flies of both genotypes showed a similar degree of mild photoreceptor neuron degeneration. Average number of photoreceptor neurons indicated ± SD. 15d adult animals show greater degeneration in flies expressing the WT protein than the 6M protein. Genotypes: WT, elav-gal4/+; UAS-Myc-Atx3Q84-Flag (WT)/+ and 6M, elav-gal4/+; pUAS-Myc-Atx3Q83-Flag (6M)/+. (C) The 6M protein retains normal Ataxin-3 activity to suppress pathogenicity of the truncated Q78 protein. External eye pictures. Flies expressing the truncated Q78 protein alone (—) have a degenerate eye. The normal non-pathogenic Ataxin-3 protein [WT(Q27)] suppresses this toxicity when co-expressed; this suppression reflects the normal functional activity of the Ataxin-3 protein (13). The pathogenic protein (WT) also shows suppressor activity, although not as striking as the non-pathogenic protein. The 6M protein (6M) has suppressor activity similar to the WT pathogenic protein. This indicates that the 6M protein is likely properly folded because it retains normal activity of the Ataxin-3 protein to suppress polyQ toxicity. Genotypes: gmr-gal4 UAS-SCA3trQ78(S)/+. gmr-gal4 UAS-SCA3trQ78(S)/UAS-Myc-Atx3Q27-Flag [WT(Q27)]. gmr-gal4 UAS-SCA3trQ78(S)/UAS-Myc-Atx3Q84-Flag (WT). gmr-gal4 UAS-SCA3trQ78(S)/ UAS-Myc-Atx3Q83-Flag (6M).
DISCUSSION
Here we show that the Ataxin-3 protein undergoes cleavage in SL2 cells and in the fly in vivo. Cleavage of the protein has been suggested to be important in the disease process (7). We also show that mutation of six caspase sites in the protein, which prevents cleavage of the protein in vivo, dramatically mitigates neural degeneration. These studies provide in vivo evidence that cleavage of the Ataxin-3 protein in the nervous system may occur and may contribute to disease progression.
Ataxin-3 protein cleavage is conserved
Ataxin-3 protein cleavage appears to be conserved as we observed cleavage products from Drosophila SL2 cells and the fly nervous system similar to those reported from Cos-7 cells, transgenic mice and SCA3 patient brain tissue (7,9). Western blot analysis, using polyclonal antibodies and the 1H9 antibody, indicates that the major cleavage product from transgenic flies contains the polyQ domain, because the size of cleavage fragment varied depending upon the polyQ length. Moreover, the ∼37 kDa fragments were greatly reduced upon zVAD-fmk treatment that inhibits caspase activity, similar to the findings in Cos-7 cells (9). Overall conservation of the Ataxin-3 cleavage pattern suggests that Drosophila can be used as a model to study the mechanism and significance of the protein cleavage.
One hypothesis we had initially was that Ataxin-3 cleavage fragments may act as seeds or facilitators of protein aggregation. According to that hypothesis, we expected to see a delay in nuclear inclusion formation or reduced inclusions in flies expressing the 6M mutant protein. However, there was no obvious difference in inclusion onset, size or accumulation. Pozzi et al. (19) identified multiple Ataxin-3 protein cleavage products in Cos-7 cells and concluded that polyQ expansion in the Ataxin-3 protein partially inhibited its proteolysis. Additionally, it was suggested that the reduced proteolytic degradation may enhance accumulation of full-length Ataxin-3 protein, and thereby contribute to disease. Our data showed no obvious difference in the dynamics of protein accumulation with or without Ataxin-3 cleavage, and suggested that the ubiquitin proteasome system is not differentially affected by caspase site mutations. Despite this, we did see a significant reduction in the rate of photoreceptor neural degeneration when Ataxin-3 protein cleavage was inhibited by mutation.
The UIM domains have been found to modulate toxicity of the pathogenic Ataxin-3 protein (13). Because the 6M mutations potentially affect residues in the first UIM domain, it is possible that the resulting changes in UIM function may change the overall toxicity. However, loss of UIM function has been shown to increase, rather than decrease, Ataxin-3 toxicity as observed here. We also used an assay in vivo, the ability to suppress polyQ toxicity, to assess Ataxin-3 function of the 6M mutant protein (13). This showed that the 6M protein retains normal Ataxin-3 activity. Thus, we suggest that the 6M mutant protein shows reduced toxicity as a result of reduced protein cleavage. Overall, these observations indicate that the cleavage product per se may contribute to disease progression.
Protein cleavage and neurodegeneration
Cleavage of the protein may enhance neurodegeneration in vivo in multiple ways. Cleavage fragments containing the polyQ domain may interact with other proteins in a more robust manner. Nuclear inclusions in polyQ diseases may have components with differing exchange rates (20), and the presence of short cleavage products may be more potent in sequestering key cellular factors (21). Proteomic studies may help reveal insight into why the WT protein is more toxic than the 6M protein.
Cong et al. (22) made an interesting observation that the short N-terminal fragment of Huntingtin with the expanded polyQ remained predominantly monomeric in cells. Considering the apparent lack of a gross difference in the formation of nuclear inclusions by Ataxin-3 cleavage in fly neurons, it is possible that ∼37 kDa cleavage products may not be readily recruited into macroscopic inclusions, but rather may exert toxic effects as soluble monomers. The apparent increase in the SDS-insoluble aggregates with normal Ataxin-3 protein cleavage, however, also suggests that the cleavage product may be a part of the nuclear inclusions in vivo. Moreover, the apparent decrease in the SDS-insoluble aggregates with cleavage-site mutant protein suggests that cleavage products may alter or trigger changes in the biochemical properties of Ataxin-3 accumulations. One such candidate process is cross linking by transglutaminase. A recent study using chemical inhibitors of transglutaminase showed mitigated SCA3-like disease effects when fed to Drosophila (23). While the mechanism of transglutaminase activation in the nervous system remains to be fully elucidated, Ataxin-3 cleavage fragment might selectively interact with transglutaminase and trigger cross linking of aggregates (24).
Ataxin-3 has a ubiquitin chain-modifying activity, and especially targets Lys63 linkages in mixed linkage ubiquitin chains (16). Lys63-linked ubiquitin chains are thought to play critical roles in the NF-kB signal transduction pathway as well as in DNA repair. While the cellular targets are unknown, it is possible that cleavage of the N-terminal ubiquitin protease domain may compromise key cellular pathways normally regulated by Ataxin-3. Moreover, because the cleavage product is likely to keep the UIM domain (19), it is an interesting possibility that it may act as a dominant-negative protein in the affected pathway.
Proteases involved in cleavage of Ataxin-3
Our data indicate that caspases may be responsible for Ataxin-3 cleavage. Activation of caspases as diseased cells undergo apoptosis pathways may trigger cleavage, as is the case with endogenous Ataxin-3 protein in Cos-7 cells (9). While it is reasonable to hypothesize that expanded Ataxin-3 protein may activate caspases that proteolyze Ataxin-3, the observation that normal Ataxin-3, which does not cause neurodegeneration, is also cleaved, suggests that cleavage is not necessarily a consequence of apoptosis. Caspases have been known to play important roles in cell proliferation and differentiation, beyond their roles as inducers/executioners of apoptosis.
In SL2 cells, there were additional fragments beyond the ∼37 kDa fragment seen in mammalian models or fly brain, suggesting that Ataxin-3 protein cleavage may be a multi-step process like for Huntingtin (25). The ∼37 kDa fragments may have the longest half-life and thereby accumulate in vivo. Furthermore, z-VAD-fmk treatment eliminated the ∼37 kDa fragment selectively in SL2 cell, leaving the ∼50 kDa fragments unaffected. Haacke et al. (18) suggested potential involvement of calpain proteases, but calpain inhibitor ALLN had no effect in Ataxin-3 cleavage in SL2 cells. The apparent discrepancy could be due to the use of different cell lines or culture conditions. MG-132 is widely used as an inhibitor of proteasome function and it was effective in reducing Ataxin-3 protein cleavage in vitro (18). We also observed that MG-132 treatment consistently reduced the appearance of some cleavage products in SL2 cells, but the ∼37 KDa fragments were not affected. When cells were treated with a combination of zVAD-fmk and MG-132, we observed an additive but not synergistic effect. These data suggest that Ataxin-3 protein cleavage is a complex process, involving multiple cellular pathways.
In an attempt to define a specific caspase responsible for the cleavage of the Ataxin-3 protein in vivo, we utilized dsRNA-mediated RNA knockdown in SL2 cells and in the fly. We effectively decreased the levels of Dronc, Strica and Drice in SL2 cells, but this had no effect on the Ataxin-3 cleavage pattern (J. Jung and N. Bonini, unpublished data). We further examined knockdown of caspase genes in the fly in vivo using P35 or dsRNA transgenic lines, but no consistent effect was seen (J. Jung et al., unpublished data). It may be necessary to knock down multiple caspases at once, or knock down the genes to a greater extent than was achieved.
Our data demonstrate that Ataxin-3 protein is cleaved in Drosophila as reported in mammalian model systems and in SCA3 patients. Additionally, we provide experimental evidence that Ataxin-3 cleavage significantly enhances neural degeneration induced by the pathogenic Ataxin-3 protein. Taken together, these data indicate that suppression of Ataxin-3 protein cleavage may significantly slow down the progression of neurodegeneration caused by expanded Ataxin-3 protein expression.
MATERIALS AND METHODS
Drosophila lines
Fly lines used were elav-gal4 (C155) for neural and brain expression, rh1-gal4 for photoreceptor cell-specific expression, and gmr-gal4 for eye expression. The Myc-tagged Ataxin-3 lines have been described (13). Constructs with Flag tags were generated by PCR and sequenced to confirm integrity. The WT double-tagged protein had a polyQ length of Q84. The putative caspase site mutant constructs were generated by site-directed mutagenesis (Cat No 12397-014, Invitrogen). Several transgenic lines were chosen and used for analysis. The polyQ length of the 6M mutant line was Q83. Typically, lines Myc-Atx3Q84-Flag (WT) line #7 and Myc-Atx3Q83-Flag (6M) line #1.1 were chosen for more detailed analysis based on western immunoblot analysis of similar protein expression with anti-Myc antibody. We also analyzed additional lines with similar results, including lines (WT) line #2 and (6M) line #3.1.
Immunoblots and immunohistochemistry
Western immunoblot analysis was performed as described (26). Antibodies used were as follows. Rabbit anti-MJD polyclonal antibody (gift of R. Pittman) (27), at 1:20 000, pre-adsorbed overnight at 4°C at 1:50 against 20–50 paraformaldehyde fixed, dissected larvae. Mouse monoclonal antibody 1H9 at 1:1000 (ab78546, Abcam). Anti-Flag antibody (F3165, Sigma, Company) at 1:1000. Mouse anti-Myc antibody (9E11, Santa Cruz Biotechnology) at 1:250. Mouse monoclonal anti-tubulin at 1:1000 (Developmental Studies Hybridoma Bank) was used as a loading control. Immunostaining of cryosections was performed as described previously (12), using the mouse anti-Myc antibody at 1:100. When appropriate, blots were stripped with Reblot™ (Cat No 2060, Millipore).
Cell culture studies
The pUAST-Myc-Atx3Q84-Flag expression vector (2 µg) was co-transfected with the pHS-gal4 driver construct (1 µg) into SL2 cells using the Effectene reagent (Cat No 11668027, Invitrogen) following the manufacturer's protocol. Transfected cells were incubated for 2 days at 25°C. Cells were then rinsed with PBS and fed fresh Schneider's Drosophila medium (Cat No 11720-034, Invitrogen) followed by a 30 min heat-shock treatment at 37°C to induce Atx3Q84 protein expression. Heat-shocked cells were cooled at 25°C for 30 min and the heat-shock treatment was repeated. This double heat-shock treatment was repeated twice more, with a 24 h interval in-between. Cells were harvested for analysis after 2 additional days of incubation at 25°C. For drug treatments, respective drug compounds were solubilized in DMSO, and then diluted to indicated concentrations in the normal growth medium. One hour prior to the first heat-shock treatment, cells were rinsed once with PBS and fed drug-containing medium, followed by the standard heat-shock treatment. Drugs used were N-benzyloxycarbonyl-val-ala-asp(O-Me) fluoromethyl ketone (zVAD-fmk, Sigma), benzyloxicarbonyl-l-phenylalanyl-ala-fluoromethylketone (Z-FA.FMK, Sigma), Carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG-132, EMD Biosciences) and N-acetyl-Leu-Leu-Nle-CHO (ALLN, Cat No 208719, Calbiochem). Cells were kept in drug-containing medium until harvest.
SUPPLEMENTARY MATERIAL
FUNDING
This study was funded by National Institutes of Health, National Institutes of Neurological Diseases and Stroke (to N.M.B.). N.M.B. is an Investigator of the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Zhenming Yu for critical comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Beher D. Gamma-secretase modulation and its promise for Alzheimer's disease: a rationale for drug discovery. Curr. Top. Med. Chem. 2008;8:34–37. doi: 10.2174/156802608783334051. [DOI] [PubMed] [Google Scholar]
- 2.Rohn T.T., Head E. Caspases as therapeutic targets in Alzheimer's disease: is it time to ‘cut’ to the chase? Int. J. Clin. Exp. Pathol. 2009;2:108–118. [PMC free article] [PubMed] [Google Scholar]
- 3.Gamblin T.C., Chen F., Zambrano A., Abraha A., Lagalwar S., Guillozet A.L., Lu M., Fu Y., Garcia-Sierra F., LaPointe N., et al. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman J., Rissman R.A., Sarsoza F., Kim R.C., Dick M., Bennett D.A., Cotman C.W., Rohn T.T., Head E. Caspase-cleaved tau accumulation in neurodegenerative diseases associated with tau and alpha-synuclein pathology. Acta Neuropathol. 2005;110:135–144. doi: 10.1007/s00401-005-1027-3. [DOI] [PubMed] [Google Scholar]
- 5.Hoffner G., Island M.L., Djian P. Purification of neuronal inclusions of patients with Huntington's disease reveals a broad range of N-terminal fragments of expanded huntingtin and insoluble polymers. J. Neurochem. 2005;95:125–136. doi: 10.1111/j.1471-4159.2005.03348.x. [DOI] [PubMed] [Google Scholar]
- 6.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 7.Goti D., Katzen S.M., Mez J., Kurtis N., Kiluk J., Ben-Haiem L., Jenkins N.A., Copeland N.G., Kakizuka A., Sharp A.H., et al. A mutant ataxin-3 putative-cleavage fragment in brains of Machado–Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J. Neurosci. 2004;24:10266–10279. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colomer Gould V.F., Goti D., Pearce D., Gonzalez G.A., Gao H., Bermudez de Leon M., Jenkins N.A., Copeland N.G., Ross C.A., Brown D.R. A mutant ataxin-3 fragment results from processing at a site N-terminal to amino acid 190 in brain of Machado–Joseph disease-like transgenic mice. Neurobiol. Dis. 2007;27:362–369. doi: 10.1016/j.nbd.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berke S.J., Schmied F.A., Brunt E.R., Ellerby L.M., Paulson H.L. Caspase-mediated proteolysis of the polyglutamine disease protein ataxin-3. J. Neurochem. 2004;89:908–918. doi: 10.1111/j.1471-4159.2004.02369.x. [DOI] [PubMed] [Google Scholar]
- 10.Jung J., Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 11.Lessing D., Bonini N.M. Polyglutamine genes interact to modulate the severity and progression of neurodegeneration in Drosophila. PLoS Biol. 2008;6:266–274. doi: 10.1371/journal.pbio.0060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L.B., Yu Z., Teng X., Bonini N.M. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1011. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warrick J.M., Morabito L.M., Bilen J., Gordesky-Gold B., Faust L.Z., Paulson H.L., Bonini N.M. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 14.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 15.Burnett B.G., Pittman R.N. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl Acad. Sci. USA. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winborn B.J., Travis S.M., Todi S.V., Scaglione K.M., Xu P., Williams A.J., Cohen R.E., Peng J., Paulson H.L. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeddrich A., Gaumer S., Haacke A., Tzvetkov N., Albrecht M., Evert B.O., Muller E.C., Lurz R., Breuer P., Schugardt N., et al. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 2006;25:1547–1558. doi: 10.1038/sj.emboj.7601043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haacke A., Hartl F.U., Breuer P. Calpain inhibition is sufficient to suppress aggregation of polyglutamine-expanded ataxin-3. J. Biol. Chem. 2007;282:18851–18856. doi: 10.1074/jbc.M611914200. [DOI] [PubMed] [Google Scholar]
- 19.Pozzi C., Valtorta M., Tedeschi G., Galbusera E., Pastori V., Bigi A., Nonnis S., Grassi E., Fusi P. Study of subcellular localization and proteolysis of ataxin-3. Neurobiol. Dis. 2008;30:190–200. doi: 10.1016/j.nbd.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Stenoien D.L., Mielke M., Mancini M.A. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 2002;4:806–810. doi: 10.1038/ncb859. [DOI] [PubMed] [Google Scholar]
- 21.Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E., Thompson L.M. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl Acad. Sci. USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong S.Y., Pepers B.A., Roos R.A., van Ommen G.J., Dorsman J.C. Small N-terminal mutant huntingtin fragments, but not wild type, are mainly present in monomeric form: Implications for pathogenesis. Exp. Neurol. 2006;199:257–264. doi: 10.1016/j.expneurol.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Lai T.S., Liu Y., Tucker T., Daniel K.R., Sane D.C., Toone E., Burke J.R., Strittmatter W.J., Greenberg C.S. Identification of chemical inhibitors to human tissue transglutaminase by screening existing drug libraries. Chem. Biol. 2008;15:969–978. doi: 10.1016/j.chembiol.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun W., Lesort M., Tucholski J., Faber P.W., MacDonald M.E., Ross C.A., Johnson G.V. Tissue transglutaminase selectively modifies proteins associated with truncated mutant huntingtin in intact cells. Neurobiol. Dis. 2001;8:391–404. doi: 10.1006/nbdi.2001.0390. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y.J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K.B., Qin Z.H., Aronin N., DiFiglia M. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl Acad. Sci. USA. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L.B., Xu K., Bonini N.M. Suppression of polyglutamine toxicity by the yeast Sup35 prion domain in Drosophila. J. Biol. Chem. 2007;282:37694–37701. doi: 10.1074/jbc.M705211200. [DOI] [PubMed] [Google Scholar]
- 27.Paulson H.L., Das S.S., Crino P.B., Perez M.K., Patel S.C., Gotsdiner D., Fischbeck K.H., Pittman R.N. Machado–Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann. Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.