Abstract
Background and Aims
Gene flow and genetic variability within and among alpine plant populations can be greatly influenced by the steep environmental gradients and heterogeneous topography of alpine landscapes. In this study, the effects are examined of natural isolation of alpine habitats on genetic diversity and geographic structure in populations of C. thyrsoides, a rare and isolated European Alpine monocarpic perennial with limited seed dispersal capacity.
Methods
Molecular diversity was analysed for 736 individuals from 32 populations in the Swiss Alps and adjacent Jura mountains using five polymorphic microsatellite loci. Pollen flow was estimated using pollen grain-sized fluorescent powder. In addition, individual-based Bayesian approaches were applied to examine population structure.
Key Results
High within-population genetic diversity (HE = 0·76) and a relatively low inbreeding coefficient (FIS = 0·022) were found. Genetic differentiation among populations measured with a standardized measure was considerable (G′ST = 0·53). A significant isolation-by-distance relationship was found (r = 0·62, P < 0·001) and a significant geographic sub-structure, coinciding with proposed postglacial migration patterns. Altitudinal location and size of populations did not influence molecular variation. Direct measures of pollen flow revealed that insect-mediated pollen dispersal was restricted to short distances within a population.
Conclusions
The natural isolation of suitable habitats for C. thyrsoides restricts gene flow among the populations as expected for a monocarpic species with very limited seed dispersal capacities. The observed high within-population genetic diversity in this rare monocarpic perennial is best explained by its outcrossing behaviour, long-lived individuals and overlapping generations. Despite the high within-population genetic diversity, the considerable genetic differentiation and the clear western–eastern differentiation in this species merits consideration in future conservation efforts.
Key words: Alpine plant, Campanula thyrsoides, genetic diversity, gene flow, genetic differentiation, glacial history, G′ST, habitat isolation, microsatellites, monocarpy, SSR
INTRODUCTION
Alpine landscapes are characterized by pronounced environmental gradients and heterogenous topography (Körner, 2003). Steep valleys and high mountain ridges demarcate the plants' habitats and might consequently limit gene flow among plant populations and colonization opportunities of new sites (Cain et al., 2000; Theurillat and Guisan, 2001). Limited gene flow due to habitat isolation could lead to a stronger genetic differentiation among populations compared with plants found in less-isolated habitats and/or lower altitudes (Till-Bottraud and Gaudeul, 2002). Genetic diversity and differentiation can also be influenced by several other factors such as population size, founder effects and mating system. For example, genetic drift can be enhanced in small populations, which could lead to extinction of alleles and loss of genetic variability (Barrett and Kohn, 1991; Ellstrand and Elam, 1993; Young et al., 1996; Lowe et al., 2004). Genetic diversity may be reduced as a consequence of a bottleneck during colonization by a small number of colonists and subsequent inbreeding (Hartl and Clark, 1997). The impact of reproductive systems on genetic diversity in plant populations can also be large. Outcrossing species usually have a high within-population diversity and low population differentiation, whereas selfing species often have low within population diversity and high differentiation among populations (Loveless and Hamrick, 1984; Hamrick and Godt, 1996, 1997; Booy et al., 2000; Till-Bottraud and Gaudeul, 2002; Nybom, 2004).
Molecular studies can also provide valuable information on past biogeographical processes, especially when macrofossils of herbaceous alpine plants are scarce or completely absent (Schönswetter et al., 2005, and references therein). Numerous molecular phylogeographic studies with a focus on potential glacial refugia for Alpine plants in the European Alps have been published over the last years (e.g. Stehlik, 2003; Tribsch and Schönswetter, 2003; Hewitt, 2004), and a recent synthesis demonstrated that glacial refugia were located along the rim of the European Alps as well as in some unglaciated central Alpine areas (Schönswetter et al., 2005). Molecular data showing genetic subdivisions in species' ranges might thus provide signals of past biogeographical processes, such as postglacial migration (Comes and Kadereit, 1998; Stehlik, 2003; Tribsch and Schönswetter, 2003).
To explore the effect of natural isolation of alpine habitats on gene flow within and among alpine plant populations, the genetic variability and geographical structure of the Alpine plant species Campanula thyrsoides were studied. Campanula thyrsoides was selected as the study species because this rare plant species only grows on carbonate-bearing soils in the Alps and adjacent mountain ranges in habitats usually isolated by large distances from each other (Aeschimann et al., 2005; Kuss et al., 2007). Furthermore, seed dispersal capacity of this plant is limited because its seeds lack morphological adaptations for dispersal (Tackenberg and Stöcklin 2008). Seeds of C. thyrsoides are only dispersed when strong winds, rain or animals shake the seeds out of the cup-like flowering bracts because of the basal capsule-opening mechanism (Kuss et al., 2007). Consequently, gene flow among populations was expected to be low and genetic differentiation to be high. The accompanying study on the breeding system of C. thyrsoides showed that the species is predominantly outcrossing (Ægisdóttir et al., 2007a). Another important characteristic of C. thyrsoides is that it is monocarpic. Monocarpy reduces flowering population density and consequently limits mating possibilities. The shorter generation time of monocarpic species in combination with a lower population density could lead to a reduction in the effective population size, which should promote population differentiation (Loveless and Hamrick, 1984; Vitalis et al., 2004). In the present study, a large geographical and altitudinal range was covered, and a combination of microsatellite markers and a field-based estimator of pollen dispersal distances was used. Furthermore, tests were carried out for signs of recent bottlenecks and a model-based clustering method implemented in structure 1·0 (Pritchard et al., 2000) was used to determine the optimal number of genetic clusters present in a population.
Previously, the same leaf material system had been screened using randomly amplified polymorphic DNA (RAPD) markers and a moderately high within-population diversity that was not influenced by population size and altitude was found. Population relatedness showed a clear geographic structuring between populations in the western part of Switzerland and the central and eastern part of Switzerland (Kuss et al., 2008a). In contrast to RAPD markers, which are dominant, microsatellites are co-dominant and this enables a straightforward calculation of allele frequencies, Hardy–Weinberg proportion and the inbreeding coefficient (FIS). Another advantage to the use of microsatellites is their selectively neutral and highly polymorphic nature, which, together with co-dominance, makes them very suitable for studying gene flow and population structure (Ouborg et al., 1999; Scribner and Pearce, 2000; Lowe et al., 2004).
The following questions were addressed. (a) How much genetic diversity exists within populations of C. thyrsoides and how does it relate to population size and altitude? (b) Does the inbreeding coefficient FIS indicate inbreeding within the populations? (c) Has the strong isolation of habitats restricted gene flow and consequently led to a strong genetic differentiation among C. thyrsoides populations? (d) Is there a geographic subdivision in population differentiation that may be related to postglacial recolonization history?
MATERIALS AND METHODS
Study species
Campanula thyrsoides is a subalpine to alpine plant species, found on carbonate-bearing soils at altitudes between 1000 and 2900 m a.s.l. throughout the European Alps and adjacent mountain ranges (Lauber and Wagner, 2001; Aeschimann et al., 2005). Campanula thyrsoides is a diploid (2n = 34; Rosen, 1931; Larsen, 1954; Gadella, 1964), rosette-forming monocarpic plant species (Kuss et al., 2007). In the year of flowering (mean 7·5 years; range 3–16 years), a 10- to 40-cm-tall inflorescence is formed that carries about 50–200 flowers in a compact spike (Kuss et al., 2007). Campanula thyrsoides is exclusivelly insect pollinated, mainly by bumblebees. It is an outcrossing species, which has protandrous flowers and a gametophytic self-incompatible system, i.e. it is mostly unable to self but can mate with its half-siblings (Ægisdóttir et al., 2007a). It has a limited seed dispersal capacity (Kuss et al., 2007) and the seed dispersal spectra, obtained from simulations with the software pappus (Tackenberg and Stöcklin, 2008) showed that most seeds (99·99 %) are dispersed within 10 m of the mother plant and only about 15 seeds appear to be dispersed over 1 km (given an average population of 100 individuals and an average fecundity of 15 000 seeds per individual; Kuss et al., 2007).
Campanula thyrsoides is considered rare throughout its native range but locally abundant with occasional population sizes exceeding 50 000 individuals. It is predominantly found in pastures and hay-meadows and shows the tendency to ruderalize along road shoulders. In Switzerland it is common in the northern calcareous Alps, whilst in the central siliceous Alps, it is only found on isolated carbonate-bearing outcrops (Kuss et al., 2007). The species is red-listed in different Alpine countries. In Germany, C. thyrsoides is protected at the national, in Austria and Switzerland at regional level (Kuss et al., 2007).
Sampling sites
Leaf material was sampled from 32 populations in Switzerland ranging from the Jura Mountains in the west to the easternmost part of the Swiss Alps (Table 1 and Fig. 1). In each population, a constant number of individual plants (n = 23) was randomly sampled. The altitude of the sampled populations ranged from 1340 to 2430 m a.s.l. The leaf material was dried in silica gel and stored at room temperature. Population size was assessed either by complete census or through extrapolation of average density counts from population subsets (Table 1).
Table 1.
Geographical location, population code, co-ordinates, altitude and estimated population size of 32 populations of Campanula thyrsoides in Switzerland
| Location* | Population code | Co-ordinates† | Altitude (m a.s.l.) | Population size |
|---|---|---|---|---|
| (1) Col du Marchairux, VD | JUM | 508′ 900/156′ 400 | 1440 | 1000 |
| (2) Les Amburnez, VD | JUA | 507′ 480/155′ 100 | 1340 | 1000 |
| (3) Pre du Rolle, VD | JUR | 508′ 983/155′ 652 | 1377 | 150 |
| (4) Pres de Four, VD | JUF | 498′ 400/148′ 450 | 1430 | 10 000 |
| (5) Col du Jamon, VD | JAA | 564′ 830/145′ 050 | 1630 | 100 |
| (6) Col du Jamon, VD | JAC | 564′ 589/144′ 944 | 1670 | 80 |
| (7) Lac du Fully, VS | FUL | 574′ 000/113′ 200 | 2100 | 500 |
| (8) Trient, Les Tseppes, VS | TRI | 564′ 350/099′ 500 | 2020 | 50 |
| (9) Lac du Moiry, VS | MOI | 609′ 932/109′ 638 | 2266 | 50 000 |
| (10) Stockhorn, BE | STO | 607′ 737/171′ 103 | 1980 | 100 |
| (11) Schynige Platte, BE | SPO | 636′ 225/167′ 625 | 1990 | 600 |
| (12) Schynige Platte, BE | SPU | 636′ 600/167′ 150 | 1890 | 500 |
| (13) Furka, UR/VS | FUR | 674′ 850/158′ 825 | 2430 | 30 000 |
| (14) Unterschächen, Butzlichöpf, UR | UNB | 702′ 500/193′ 200 | 1900 | 500 |
| (15) Langwies, Listboden, GR | LAL | 776′ 750/191′ 510 | 2000 | 300 |
| (16) Langwies, Strassberg, GR | LAS | 775′ 875/190′ 550 | 1870 | 7000 |
| (17) Langwies, Holzbüel, GR | LAH | 775′ 010/188′ 875 | 1700 | 50 |
| (18) Vals, Peil, GR | VAL | 735′ 375/160′ 425 | 1850 | 100 |
| (19) Safiental, GR | SAF | 742′ 851/174′ 289 | 1857 | 50 |
| (20) Medels, Parjurs, GR | MED | 742′ 800/157′ 700 | 1870 | 50 |
| (21) Monstein, Mäschenboden, GR | MOM | 779′ 668/173′ 708 | 1961 | 45 |
| (22) Monstein, Fanexmeder, GR | MOF | 780′ 750/174′ 910 | 2220 | 250 |
| (23) Parsennmeder, GR | PMA | 784′ 030/191′ 473 | 1995 | 5000 |
| (24) Parsennmeder, GR | PMB | 784′ 478/191′ 548 | 1910 | 100 |
| (25) Churwalden, Joch, GR | CHJ | 762′ 300/185′ 100 | 1890 | 150 |
| (26) St. Antönien, GR | STA | 782′ 203/201′ 989 | 1943 | 250 |
| (27) Alp Laret, GR | LAR | 784′ 234/153′ 944 | 2180 | 300 |
| (28) Albula Pass, Naz, GR | NAZ | 778′ 193/162′ 751 | 1755 | 150 |
| (29) Schuol, La Motta, GR | SCM | 816′ 400/188′ 400 | 2142 | 2000 |
| (30) Ftan, Prui, GR | FTA | 812′ 505/187′ 750 | 2100 | 150 |
| (31) Tschlin, Alp Tea, GR | TEA | 828′ 250/198′ 250 | 2200 | 150 |
| (32) Tschlin, Alp Tea, GR | TEB | 827′ 800/198′ 000 | 2150 | 200 |
A constant number of 23 C. thyrsoides individuals was sampled at each location.
* The two-letter code after the localities indicates the canton in Switzerland.
† Co-ordinates according to the Swiss topographical maps (Bundesamt für Landestopopgraphie, Wabern, Switzerland).
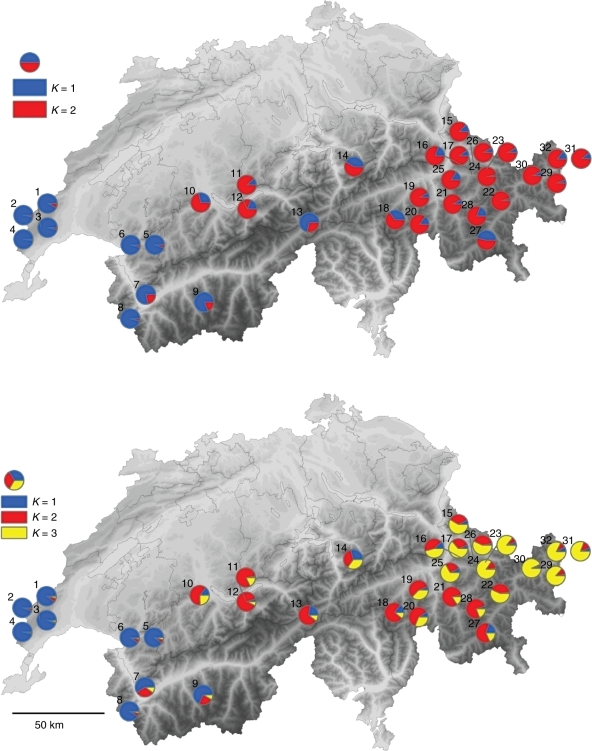
Fig. 1.
Geographical structure of 32 populations of Campanula thyrsoides in Switzerland based on the clustering method implemented in structure 1·0. The figure shows the distribution for K = 2 and K = 3, where K = genetic clusters. The numbers indicate the location in Table 1. The lightest grey colour is at 190 m a.s.l. and the altitude increases with darker colour up to about 4800 m a.s.l.
DNA extraction and PCR amplification
DNA was extracted from 10 mg of silica-gel dried and milled (Retsch MM300) leaf tissue using a DNeasy Plant 96 Kit (Qiagen, Hombrechtikon, Switzerland). To remove polyphenols, which may adversely affect PCR amplification, the first extraction step of the manufacturer's protocol was modified by adding 25 mg of polyvinylpyrrolidone (Fluka, Buchs, Switzerland) to the 600 µL of lysis buffer of each sample. The DNA was quantified using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA) spectrophotometer. Samples were screened using seven polymorphic microsatellite loci of which five were used for the final analyses: Campthy 1, Campthy 3, Campthy 5, Campthy 13 and Campthy 15 (Ægisdóttir et al., 2007b). That was done in order to minimize error rate and avoid pseudoreplication in the event of linkage disequilibrium [as recommended by Bonin et al. (2004) and Selkoe and Toonen (2006)].
Polymerase chain reaction (PCR) amplification was performed in a 10-μL reaction volume containing 15 ng of genomic DNA, 0·125 µm each of forward and reverse primers, 1 µL of 10× PCR buffer, 150 µm dNTP and 0·5 U HotstarTaq (Qiagen, Hombrechtikon, Switzerland). After a denaturation step at 95 °C for 15 min, PCR was performed for 30 cycles: 30 s annealing at a locus-specific temperature (56 °C for Campthy 1, 3, 5, 6, 9 and 12; 60°C for Campthy 13 and 15), 30 s at 72 °C, and 30 s at 95 °C. The PCR ended with 1 min at a locus-specific temperature, followed by a final 30-min extension step at 72 °C. Horizontal gel electrophoresis of PCR products was performed using Spreadex® gels with a resolution of 2 bp in a SEA-2000™ submerged gel electrophoresis system (Elchrom Scientific AG, Cham, Switzerland). Ethidium bromide (1 µg/mL)-stained banding patterns were observed under UV light and analysed by careful manual verification of each gel at least twice.
All ambiguous genotyping results were repeated to minimize genotyping errors, and low quality or unreliable DNA samples and markers discarded. The genotyping error rate was calculated by re-extracting and re-amplifying 39 randomly chosen plants (5·3 % of the dataset). Subsequently allelic differences were calculated between those 39 plants and the original datasets. The error rate was estimated to be 6·1 % and was due to genotyping and scoring errors. Eventually, some individuals had to be excluded from the original dataset. The excluded individuals had microsatellite profiles with only one scored allele at a specific locus. Such loci cannot be included in analyses that require information of two alleles per locus (such as HE, Hardy–Weinberg equilibrium, and calculation in the program structure). The reason for excluding such individuals was due to stuttering or smearing of bands (e.g. the ‘second’ allele could not be scored correctly) or due to evidence of null alleles (i.e. false positive homozygous genotypes). The total percentage of excluded data for each locus was as follows: 1, 3·9 % of all individuals; 3, 0·9 %; 5, 4 %; 6, 4·8 %; 15, 10·5 %; 13, 7·5 %. Nevertheless, individual profiles with only one allele can still be incorporated in population genetic programs, e.g. for estimation of population differentiation (FST, GST and G′ST; see below).
Analysis of overall genetic variation
Molecular diversity was calculated within populations with (a) mean number of alleles per locus (Na), (b) HO (observed heterozygosity), (c) HE (expected heterozygosity under Hardy–Weinberg equilibrium) and (d) inbreeding coefficient (FIS). Moreover, genetic diversity was calculated among populations with (e) FST using the method of Weir and Cockerham (1984) and (f) Nei's GST (Nei, 1973). Both FST and GST were calculated in genetix version 4·05 (Belkhir et al., 1996–2002). Since the standardized measurement of G′ST allows more appropriate measures of genetic differentiation than GST or FST (Hedrick, 2005; Heller and Siegismund, 2009), G′ST was calculated (see eqn 4b in Hedrick, 2005). Confidence intervals (95 %) were calculated by bootstrapping 1000 times over all loci (for FIS within each population, for FST within each population and for FST globally; Weir and Cockerham, 1984; Belkhir et al., 1996–2002).
Using genepop on the web, version 3·4 (Raymond and Rousset, 1995), the probability for linkage disequilibrium was assessed using 5000 dememorizations, 1000 batches and 5000 iterations per batch, and the number of alleles per locus and and deviations from Hardy–Weinberg expectations for each pair of loci calculated across all populations using a Fisher exact test. Confidence intervals were calculated with the Markov chain method (Guo and Thompson, 1992). Bonferroni correction for multiple comparisons was applied in both cases to a significance level of P < 0·05.
Moreover, the effect of population size and altitude on molecular variation (HE, FIS and Na) was assessed by performing single linear regression using the software R 2.0.0 (R Development Core Team, 2004).
For a species that lives in spatially isolated and sometimes small populations within the alpine landscape (like C. thyrsoides), the number of plants colonizing isolated habitats might be limited, which could cause founder effect. Bottlenecks were searched within the populations studied by applying the mode-shift test implemented in the software bottleneck, version 1.2.02 (Piry et al., 1999).
Geographical structure
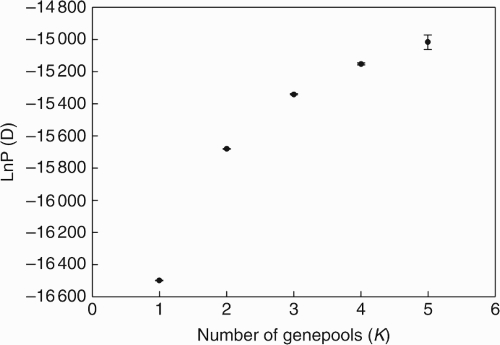
To explore whether subsets of populations were confined to discrete geographical regions (two or three gene pools) as found when using the RAPD markers, a model-based clustering method implemented in structure 1·0 was used (Pritchard et al., 2000). To determine the optimal number of genetic clusters present in a population, structure divides the sampled individuals into a number of clusters (K) based only on multilocus genotypic data. Twenty independent runs were conducted for each K between one and five, and the results from each run plotted with the highest likelihood value (see the Appendix). The admixture model was used and allele frequencies, a burn-in period and Markov chain Monte Carlo (MCMC) iterations of 10 000 each were correlated. The ad hoc statistic ΔK was used to identify the most likely number of clusters in the dataset (Evanno et al., 2005).
Pairwise FST was calculated and differentiation tested between all pairs of populations with a permutation test included in arlequin 3·1 (Excoffier et al., 2005). To test for isolation by distance among populations, the independence between pairwise FST and geographical distances was examined by running 10 000 permutations using a modified Mantel's test (Mantel, 1967) included in the program isolde in genepop (version 3.1c) (Raymond and Rousset, 1995).
Furthermore, tests were carried out for genetic differentiation among populations and between regions (east and west), as observed using structure 1·0 and with analysis of molecular variance (AMOVA) (Excoffier et al., 1992), as applied in arlequin 3·1, using 1000 permutations of the individual (Schneider et al., 2000).
Data on dispersal
Pollen dispersal distance was measured in the field during 1 d in the summer of 2004 and 2 d in the summer of 2005 using fluorescent powder of similar particle size as pollen (Stockhouse, 1976; fluorescent dye from Radiant Colour, Houthalen, Belgium). The study took place on the Furka Pass in the Central Swiss Alps, which holds a big C. thyrsoides population extending over a large area. In both summers, clear sunny days in the middle of the flowering period of C. thyrsoides were chosen for the study. On 6 July 2004, six plants were selected to serve as donors. In 2005, the study took place on 15 July (one donor plant), and 17 July (two donor plants). On each donor plant, the pollen of every open flower (mean number of flowers per plant = 28·33, s.d. = 11·57) was painted with fluorescent powder early in the morning. The population was divided into four parts of equal area and one or two plants were randomly selected within each part of the population. If there were two plants selected within a single part, they were painted with powder of dissimlar colours (pink, yellow, blue and green). Similar colours were only used on plants which were further away (>150 m) from each other to prevent false results. Throughout the day, pollinators frequented the flowers and dispersed the fluorescent powder. After sunset, traces of the fluorescent powder on C. thyrsoides flowers were identified using UV torches. Then the number of plants and flowers visited per plant was counted and the distances to the initially marked plants measured. This method approximates minimum pollen dispersal via insects. However, it is limited in estimating long-distance dispersal due to logistic constraints. For long-distance seed dispersal by wind, modelling work that incorporated turbulences and topographic effects was used (Kuss et al., 2007; Tackenberg and Stöcklin, 2008).
On the same sunny days as the fluorescent powder experiment was done, the number of insects that frequented the C. thyrsoides flowers were monitored and counted. The observations took place in the middle of the day and the observed insect groups were bumblebees, small Diptera spp., beetles and ants. Eleven 0·5- to 1·0-m2 plots were observed during 5·5 h of observation altogether. If flower density was high (>10 flowering C. thyrsoides plants within 1 m2), 0·5-m2 plots were used so that it was possible to observe all visits by pollinators. However, when <10 flowering C. thyrsoides plants where found within an area of 1 m2, 1-m2 plots were used for observations.
RESULTS
Overall microsatellite diversity
Five loci were genotyped in each of 736 individuals of C. thyrsoides and 100 alleles in total and 15–28 alleles per locus with an average of 20 alleles detected. The observed heterozygosity (HO) over all populations was high (mean HO 0·763; range 0·645–0·849) and deviation from expected heterozygosity (HE) was small (mean HE 0·762; range 0·666–0·847) (Table 2). There was no significant difference in expected heterozygosity (HE) among populations (F = 1·72; P = 0·20).
Table 2.
Mean number of alleles per locus (Na), expected heterozygosity HE), observed heterozygosity (HO) and inbreeding coefficent (FIS) for each studied population of Campanula thyrsoides across five loci
| Site | Na | HE | HO | FIS† |
|---|---|---|---|---|
| 1 | 7·4 | 0·756 | 0·682 | 0·018*** |
| 2 | 7·4 | 0·767 | 0·763 | 0·021 |
| 3 | 7·2 | 0·750 | 0·798 | 0·024 |
| 4 | 5·6 | 0·666 | 0·673 | 0·022 |
| 5 | 9·6 | 0·813 | 0·712 | 0·017*** |
| 6 | 9·8 | 0·793 | 0·849 | 0·024*** |
| 7 | 7·6 | 0·767 | 0·821 | 0·024 |
| 8 | 7·8 | 0·741 | 0·773 | 0·023 |
| 9 | 8·8 | 0·832 | 0·817 | 0·021 |
| 10 | 10·2 | 0·847 | 0·841 | 0·021 |
| 11 | 7·8 | 0·778 | 0·690 | 0·018*** |
| 12 | 7·6 | 0·729 | 0·728 | 0·022*** |
| 13 | 8·4 | 0·797 | 0·739 | 0·019*** |
| 14 | 10·0 | 0·834 | 0·813 | 0·021*** |
| 15 | 9·6 | 0·766 | 0·819 | 0·024 |
| 16 | 9·4 | 0·798 | 0·799 | 0·021 |
| 17 | 9·0 | 0·745 | 0·778 | 0·022 |
| 18 | 8·4 | 0·766 | 0·835 | 0·024 |
| 19 | 6·6 | 0·704 | 0·707 | 0·021 |
| 20 | 7·6 | 0·759 | 0·836 | 0·025 |
| 21 | 6·0 | 0·701 | 0·671 | 0·020 |
| 22 | 6·6 | 0·740 | 0·791 | 0·023 |
| 23 | 9·2 | 0·786 | 0·734 | 0·020** |
| 24 | 9·2 | 0·780 | 0·806 | 0·023 |
| 25 | 9·0 | 0·787 | 0·762 | 0·021 |
| 26 | 7·4 | 0·739 | 0·645 | 0·017** |
| 27 | 8·0 | 0·749 | 0·722 | 0·020 |
| 28 | 6·4 | 0·718 | 0·785 | 0·024 |
| 29 | 6·8 | 0·750 | 0·761 | 0·022*** |
| 30 | 7·0 | 0·715 | 0·771 | 0·024 |
| 31 | 7·2 | 0·761 | 0·771 | 0·022 |
| 32 | 7·2 | 0·746 | 0·734 | 0·021** |
| Mean | 8·0 | 0·762 | 0·763 | 0·022 |
† Statistically significant deviations from Hardy–Weinberg expectations by P < 0·01 (**) and P < 0·001 (***).
The average inbreeding coefficient was relatively low (FIS = 0·022), ranging from 0·017 to 0·025 at the population level (Table 2). Eleven out of 32 populations deviated significantly from Hardy–Weinberg equilibrium (Table 2). None of the studied populations showed significant signs of a recent bottleneck. No evidence was found for linkage disequilibrium between the studied loci.
Results from linear regression indicated that altitude (m a.s.l.) had no influence on molecular variation within populations (HE, r = 0·2, P = 0·29; FIS, r = 0·14, P = 0·89; Na, r = 0·06, P = 0·72). Similarly, population size had no influence on the molecular variation (HE, r = 0·1, P = 0·078; FIS, r = 0·17, P = 0·37; Na, r = 0·105, P = 0·56).
Genetic differentiation and geographic structure
Geographic structure based on assignment tests in structure revealed that two genetic clusters (K = 2) have the best ad hoc statistical fit (ΔK; Evanno et al., 2005). Here, a clear west–east division was found with populations numbered 1–9 clustering together in western Switzerland and populations numbered 10–32 clustering in central and eastern Switzerland (Fig. 1, top). For K = 3 and K = 4, the west–east divisions were still very clear but other divisions with new genepools appeared in the east (one regional genepool for K = 3, and two for K = 4; see Fig. 1, bottom, for K = 3).
The average genetic differentiation in the populations studied was comparably low, based either on FST = 0·10 or GST = 0·12 algorithms, though considerably higher in the case of the standardized G′ST = 0·53.
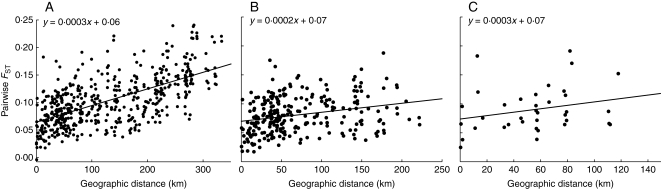
Genetic differentiation between the populations correlated significantly with geographical distances (Mantel test based on pairwise FST; r = 0·62; P < 0·001; Fig. 2A). When the two regions (east and west) were tested seperately, there was still a significant correlation between the geographical distances and the pairwise FST in the eastern region (r = 0·26; P < 0·05; Fig. 2B). In the western region, however, the relationship was only marginally significant (r = 0·263; P = 0·052; Fig. 2C). AMOVA results are listed in Table 3.
Fig. 2.
Pairwise genetic distance (FST) and geographical distances (km) between populations of Campanula thyrsoides: (A) all studied populations, (B) populations in the eastern region (numbers 10–32) and (C) populations in the western region (numbers 1–9).
Table 3.
Summary of analysis of molecular variance (AMOVA) of 32 populations of Campanula thyrsoides grouped in two regions (west versus east)
| Variance component |
||||
|---|---|---|---|---|
| Source of variation | d.f. | Absolute | % | P |
| Among regions | 1 | 0·11 | 5·39 | <0·001 |
| Among populations | 30 | 0·16 | 7·91 | <0·001 |
| Within regions, within populations | 1440 | 1·73 | 86·7 | <0·001 |
| Total | 1471 | |||
Dispersal
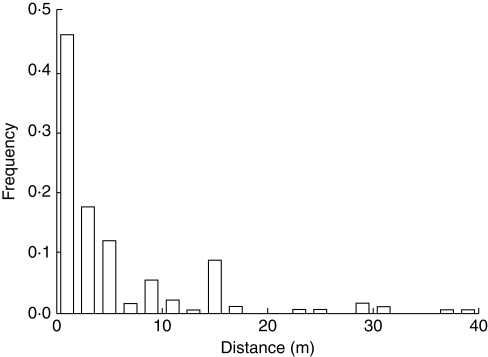
In total, 1102 dye transfers among flowers were recorded on a total of 173 different plants. On average, the powder was found on 6·4 flowers per plant (s.e. = 0·48). The frequency of observed pollen dispersal distances was highest within the first 2 m from the marked plant but decreased markedly with increasing distance. The average dispersal distance was 4·85 m (s.e. = 0·54), the shortest 0·13 m, and the longest observed dispersal distance 39 m (Fig. 3). Bumblebees, which were observed collecting a large amount of pollen from flowers, were the prime flower visitor, accounting for 60 % of the total visits, followed by small Diptera species whose contribution measured about 30 % of the total visits. Other insect groups accounted for a smaller number of visits and are unlikely to be pollinators (beetles, 7 %; ants, 3 %).
Fig. 3.
Frequency of pollen movement distances (m) between Campanula thyrsoides plants on the Furka Pass in the Swiss Alps, as indicated by fluorescent powder traces in summer 2004 and 2005 (n = 173).
DISCUSSION
Genetic diversity within populations
Higher levels of genetic diversity were found within populations of C. thyrsoides (expected heterozygosity; HE = 0·762) than many other microsatellite-based plant studies (e.g. Sun and Salomon, 2003; Mengoni et al., 2000; Galeuchet et al., 2005; Gao, 2005). A review by Nybom (2004) reported, for microsatellite studies, an average HE of 0·55 for short-lived perennials and HE of 0·65 for outcrossing plant species. No indication of genetic erosion or recent bottlenecks were found despite the species rareness and isolation of some of its populations.
Population size did not affect molecular diversity in C. thyrsoides despite the theoretical prediction that small populations might lose genetic variation due to genetic drift, founder effects and population bottlenecks (Barrett and Kohn, 1991; Ellstrand and Elam, 1993; Young et al., 1996; Lowe et al., 2004). Although drift could be expected in some of the C. thyrsoides populations studied here because of their small size (see Table 1), no significant signs of bottlenecks or founder effects were found. Other studies on Alpine plant species that include comparisons of genetic diversity and population sizes of similar size range have either detected (Lloydia serotina, Jones and Gliddon, 1999; Eryngium alpinum, Gaudeul et al., 2000; Trollius europaeus, Despres et al., 2002; Sesleria albicans, Reisch et al., 2002) or have not detected any correlation (Geum reptans, Pluess and Stöcklin, 2004; Epilobium fleischeri, Kuss et al., 2008a). This inconsistent pattern among alpine plants may be a result of different sampling parameters such as sample size and marker system and/or biological traits such as breeding system and life span (Nybom, 2004). In C. thyrsoides, the longevity of individuals despite its monocarpy, the desynchronized flowering of cohorts (generation overlap) are likely factors that prevent negative effects of small size or isolation of populations on genetic diversity (Kuss et al., 2008b).
The high within-population diversity in C. thyrsoides is also likely to be influenced by some life-history traits of the species, especially the type of breeding system, which has been shown to strongly affect the distribution and magnitude of genetic diversity in plant populations. Outcrossing plant species tend to have higher genetic variation within populations, whereas populations of selfing species or species with a mixed mating system are often genetically less variable (Loveless and Hamrick, 1984; Hamrick and Godt, 1996, 1997; Booy et al., 2000; Till-Bottraud and Gaudeul, 2002; Nybom, 2004; Duminil et al., 2007). Since C. thyrsoides is predominantly outbreeding (Ægisdóttir et al., 2007a) with low inbreeding coefficient (FIS = 0·022), we suggest that the breeding system of C. thyrsoides plays an important role in driving the high within-population diversity. Furthermore, the high number of traces of fluorescent powder observed in the field (1102 times on 173 plants) with occasional flight distances of 39 m indicate that pollinators (especially bumblebees) contribute to the high genetic variation within populations.
Heterozygote deficiency
Whilst the inbreeding coefficent (FIS) was low in all populations (average FIS = 0·022), most values were positive and 11 populations out of 32 significantly deviated from Hardy–Weinberg equilibrium. Heterozygote deficiency in a population can occur as a result of inbreeding, selection against heterozygotes, the Wahlund effect (reduction of heterozygosity in a population caused by subpopulation structure) or the presence of null alleles (Sun and Salomon, 2003, and references therein). In our opinion, the most plausible explanation for the observed deviation from Hardy–Weinberg equilibrium in populations of C. thyrsoides is the ability of this species to mate with half-sibs (Ægisdóttir et al., 2007a). This allows some inbreeding which leads to more homozygotes than expected. Yet, other explanations (like the presence of null alleles) may also contribute to the lack of heterozygosity (see Materials and methods).
Genetic differentiation in a landscape context
The genetic differentiation values of C. thyrsoides in this study showed rather low vales of 0·10 for FST and 0·12 for GST. It has, however, been argued that the high levels of within-population heterozygosity usually found with microsatellites yield low differentiation indices among populations (Hedrick, 1999, 2005), and that a standardized measure of genetic differentiation (G′ST) should routinely be reported for microsatellite data (Heller and Siegismund, 2009). Using G′ST, a considerable genetic differentiation (0·53) was observed as could be expected for a rare alpine monocarpic species with limited seed dispersal capacities (Loveless and Hamrick, 1984). The value of G′ST which is about four times higher compared with GST in the present study is in accordance with other comparisons of G′ST and GST (see table S1 in Heller and Siegismund, 2009). Gene flow between the C. thyrsoides populations in the present study is therefore clearly restricted and occurs mostly among close and adjacent populations (see Fig. 2A) as the correlation between geographic distance and genetic distance suggests (Wright, 1943). The restricted gene flow between the studied populations was to be expected. The Alpine landscape with high mountain ridges and steep valleys may limit or even block gene flow among plant populations (Cain et al., 2000; Theurillat and Guisan, 2001). This may result in a stronger genetic differentiation among populations than in more homogenous landscapes (Till-Bottraud and Gaudeul, 2002). Therefore, the considerable population differentiation even among populations that are geographically not very distant (Fig. 2) may relate to the particularities of the landscape in the Alps. In this context, a combination of genetic information with fine-resolution qualitative landscape data appears to be a promising future approach (Holderegger and Wagner, 2008).
Genetic differentiation among regions
The clear geographical boundary existing between populations in western Switzerland and populations in central/eastern Switzerland (K = 2; Fig. 1A), had already been found with rapd profiles (Kuss et al., 2008a), and remained after increasing the number of groups in structure to K = 3 (Fig. 1B) and K = 4 (figure not shown). AMOVA indicated that 5·4 % of the genetic diversity was found among regions. There were no indications that the two groups of populations relate to a particular structure in the landscape. Since the observed west–east division in the populations also did not correspond to the geographical division between the Jura mountains in the west and the Alps in the east, we consider it most likely that this genetic boundary arose as a result of postglacial colonization history, as it corresponds to the glacial refugia map for mountain plants in the European Alps proposed by Schönswetter et al. (2005).
Using structure, another but less-pronounced gene pool was identified in the eastern-most populations (Fig. 1B) using K = 3. Using four gene pools (K = 4), moreover, one more gene pool was identified that was regional in the eastern part of Switzerland (figure not shown). It remains to be explored to what extent the emerging easternmost Swiss gene pools correspond with the split between the eastern and central Alps as outlined by Schönswetter et al. (2005).
Because of the poor dispersal capacity of C. thyrsoides, the isolation of its suitable habitats in the Alpine landscape and the considerable genetic differentiation found in its populations, we find it most likely that the genetic pattern found in C. thyrsoides is at least partly reflecting a historical gene flow over a timescale of several hundred or thousands of years. Possibly, this gene flow has occurred since the end of the last glacial period (10 000 years ago) when the species invaded Switzerland from at least two different glacial refugia as indicated by the geographical pattern observed with structure (Fig. 1).
Conclusions
A high within-population genetic diversity is reported for the rare monocarpic perennial Campanula thyrsoides in the Swiss Alps, which can best be explained by long-lived individuals, overlapping generations and the outbreeding behaviour of the species. Despite living in isolated populations of different sizes and at contrasting altitudes in the Alpine landscape, neither isolation of habitats nor population size or altitude affected genetic variability within the studied populations.
The considerable genetic differentiation and the isolation by distance relationship observed in the present study show that gene flow is restricted among the C. thyrsoides populations. This was to be expected in a monocarpic perennial that lives in isolated populations in the Alpine landscape and has poor seed dispersal capacities. In addition, a clear geographical boundary was found among populations in western versus central/eastern regions of Switzerland, which corresponds well with the postglacial colonization history of many other Alpine plants (Schönswetter et al., 2005).
The present molecular study found no indication of genetic depletion, even within small populations of C. thyrsoides, despite the rareness of this species in the alpine landscape. Nevertheless, the considerable genetic differentiation among populations and the clear west–east differentiation between two population groups should be considered in future conservation efforts for this species.
ACKNOWLEDGEMENTS
We thank R. Viti, A. Gilgen and F. Linder for support in the laboratory as well as B. Koller and B. Hefti-Gautschi (Ecogenics, Zürich, Switzerland) for help and support during the development of the microsatellite method in our laboratory. Moreover, we are grateful to G. Armbruster for help at all stages, C. Thiel-Egenter for advice on the program structure and J. Bech for help on using GIS. Last but not least, we would like to thank D. Ehrich and P. Schönswetter for answering our questions and giving valuable comments on the manuscript. Funding for this work was provided by the Swiss National Foundation (grant no. 3100AO-100762 to Jürg Stöcklin) and Freiwillige Akademische Gesellschaft (grant to Hafdís Hanna Ægisdóttir).
APPENDIX
Mean values of 20 runs (±s.d.) of ln probability of data [ln P(D)] for K from 1 to 5 as implemented in structure on Campanula thyrsoides.
LITERATURE CITED
- Ægisdóttir HH, Jespersen D, Kuss P, Stöcklin J. No inbreeding depression in an outcrossing Alpine species: the breeding system of Campanula thyrsoides. Flora. 2007;a 202:218–225. [Google Scholar]
- Ægisdóttir HH, Koller B, Kuss P, Stöcklin J. Development and characterization of microsatellite DNA markers for the Alpine plant species Campanula thyrsoides. Molecular Ecology Notes. 2007;b 7:996–997. [Google Scholar]
- Aeschimann D, Lauber K, Moser DM, Theurillat J-P. Flora Alpina. Bern: Haupt Verlag; 2005. [Google Scholar]
- Barrett SCH, Kohn JK. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KH, editors. Genetics and conservation of rare plants. Oxford: Oxford University Press; 1991. pp. 3–30. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Montpellier, France: Université de Montpellier II; 1996–2002. GENETIX 4·04 Logiciel sous Windows TM, pour la Génétique des Populations. Laboratoire Génome, Populations, Interactions. CNRS UMR 5000. [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Booy G, Hendriks RJJ, Smulders MJM, Van Groenendael JM, Vosman B. Genetic diversity and the survival of populations. Plant Biology. 2000;2:379–395. [Google Scholar]
- Cain ML, Milligan BG, Strand AE. Long-distance dispersal in plant populations. American Journal of Botany. 2000;87:1217–1227. [PubMed] [Google Scholar]
- Comes HP, Kadereit JK. The effects of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Science. 1998;3:432–438. [Google Scholar]
- Despres L, Loriot S, Gaudeul M. Geographic pattern of genetic variation in the European globeflower Trollius europaeus L. (Ranunculaceae) inferred from amplified fragment length polymorphism markers. Molecular Ecology. 2002;11:2337–2347. doi: 10.1046/j.1365-294x.2002.01618.x. [DOI] [PubMed] [Google Scholar]
- Duminil J, Fineschi S, Hampe A, et al. Can population genetic structure be predicted from life-history traits? The American Naturalist. 2007;169:662–672. doi: 10.1086/513490. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review Ecology and Systematic. 1993;24:217–242. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin version 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Gadella TWJ. Cytotaxonomic studies in the genus Campanula. Wentia. 1964;11:1–104. [Google Scholar]
- Galeuchet D, Perret C, Fischer M. Microsatellite variation and structure of 28 populations of the common wetland plant, Lychnis flos-cuculi L., in a fragmented landscape. Molecular Ecology. 2005;14:991–1000. doi: 10.1111/j.1365-294X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- Gao L-Z. Microsatellite variation within and among population of Oryza officinalis (Poaceae), an endangered wild rice from China. Molecular Ecology. 2005;14:4287–4297. doi: 10.1111/j.1365-294X.2005.02758.x. [DOI] [PubMed] [Google Scholar]
- Gaudeul M, Taberlet P, Till-Bottraud I. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Molecular Ecology. 2000;9:1625–1637. doi: 10.1046/j.1365-294x.2000.01063.x. [DOI] [PubMed] [Google Scholar]
- Guo S, Thompson E. Performing the exact test of Hardy–Weinberg proportion from multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London Biological Sciences. 1996;351:1291–1298. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. In: Silvertown J, Franco M, Harper JL, editors. Plant life histories. Cambridge: Cambridge University Press; 1997. pp. 102–117. [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. 3rd edn. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- Hedrick PW. Perspective: highly variable genetic loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Heller R, Siegismund HR. Relationship between three measures of genetic differentiation GST, DEST and G′ST: how wrong have we been? Molecular Ecology. 2009;18:2080–2083. doi: 10.1111/j.1365-294x.2009.04185.x. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderegger R, Wagner HH. Landscape genetics. Bioscience. 2008;58:199–207. [Google Scholar]
- Jones B, Gliddon C. Reproductive biology and genetic structure in Lloydia serontina. Plant Ecology. 1999;141:151–161. [Google Scholar]
- Körner C. Alpine plant life. 2nd edn. Heidelberg: Springer, Germany; 2003. [Google Scholar]
- Kuss P, Ægisdóttir HH, Stöcklin J. The Biological Flora of Central Europe: Campanula thyrsoides L. Perspectives in Plant Ecology, Evolution and Systematics. 2007;9:37–51. [Google Scholar]
- Kuss P, Pluess AR, Ægisdóttir HH, Stöcklin J. Spatial isolation and genetic differentiation in naturally fragmented plant populations of the Swiss Alps. Journal of Plant Ecology. 2008;a 1:149–159. [Google Scholar]
- Kuss P, Rees M, Ægisdóttir HH, Ellner PS, Stöcklin J. Evolutionary demography of long-lived perennials: a time-lagged integral projection model. Journal of Ecology. 2008;b 96:821–832. [Google Scholar]
- Larsen K. Chromosome numbers of some European flowering plants. Botanisk Tidsskrift. 1954;50:163–174. [Google Scholar]
- Lauber K, Wagner G. Flora Helvetica. 3rd edn 2001. Bern: Verlag Paul Haupt. [Google Scholar]
- Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics. 1984;15:65–95. [Google Scholar]
- Lowe A, Harris S, Ashton P. Ecological genetics: design, analysis, and application. Oxford: Blackwell Publishing; 2004. [Google Scholar]
- Mantel N. Detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Mengoni A, Gori A, Bazzicalupo M. Use of RAPD and microsatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa, Mediago sativa. Plant Breeding. 2000;119:311–317. [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating genetic diversity in plants. Molecular Ecology. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Ouborg NJ, Piquot Y, van Groenendael JM. Population genetics, molecular markers and the study of dispersal in plants. Journal of Ecology. 1999;87:551–568. [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Pluess AR, Stöcklin J. Population genetic diversity of the clonal plant Geum reptans (Rosaceae) in the Swiss Alps. American Journal of Botany. 2004;91:2013–2021. doi: 10.3732/ajb.91.12.2013. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2004. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Reisch C, Poschlod P, Wingender R. Genetic variation of Sesleria albicans Kit. Ex Schultes (Poaceae): lack of evidence for glacial relict endemism in central Europe. Plant Biology. 2002;4:711–719. [Google Scholar]
- Rosen W. Zur Embryologie der Campanulaceen und Lobeliaceen. Meddelanden Fran Gotegorgs botaniska tradgard. 1931;7:31–42. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva; 2000. Arlequin: a software for population genetics data analysis, ver 2·000. [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Scribner KT, Pearce JM. Microsatellites: evolutionary and methodological background and empirical applications at individual, population and phylogenetic levels. In: Baker AJ, editor. Molecular methods in ecology. Oxford: Blackwell Science; 2000. pp. 235–273. [Google Scholar]
- Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecology Letters. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- Stehlik I. Resistance or emigration? Response of alpine plants to the ice ages. Taxon. 2003;52:499–510. [Google Scholar]
- Stockhouse RE. New method for studying pollen dispersal using micronized fluorescent dusts. American Midland Naturalist. 1976;96:241–245. [Google Scholar]
- Sun G, Salomon B. Microsatellite variability and heterozygote deficiency in the arctic-alpine Alaskan wheatgrass (Elymus alaskanus) complex. Genome. 2003;46:729–737. doi: 10.1139/g03-052. [DOI] [PubMed] [Google Scholar]
- Tackenberg O, Stöcklin J. Wind dispersal of alpine plant species: a comparison with lowland species. Journal of Vegetation Science. 2008;19:109–118. [Google Scholar]
- Theurillat J-P, Guisan A. Potential impact of climate change on vegetation in the European Alps: a review. Climate Change. 2001;50:77–109. [Google Scholar]
- Till-Bottraud I, Gaudeul M. Intraspecific genetic diversity in alpine plants. In: Körner C, Spehn EM, editors. Mountain biodiversity: a global assessment. New York, NY: Parthenon Publishing; 2002. pp. 23–34. [Google Scholar]
- Tribsch A, Schönswetter P. In search for Pleistocene refugia for mountain plants: patterns of endemism and comparative phylogeography confirm palaeo-environmental evidence in the Eastern European Alps. Taxon. 2003;52:477–497. [Google Scholar]
- Vitalis R, Glémin S, Olivieri I. When genes go to sleep: the population genetic consequences of seed dormancy and monocarpic perenniality. The American Naturalist. 2004;163:295–311. doi: 10.1086/381041. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown AHD. The population genetic consequences of habitat fragmentation for plants. Trends of Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]