Abstract
In dogs hip joint laxity that can lead to degenerative joint disease (DJD) is frequent and heritable, providing a genetic model for some aspects of the human disease. We have used Portuguese water dogs (PWDs) to identify Quantitative trait loci (QTLs) that regulate laxity in the hip joint.A population of 286 PWDs, each characterized by ca. 500 molecular genetic markers, was analyzed for subluxation of the hip joint as measured by the Norberg angle, a quantitative radiographic measure of laxity. A significant directed asymmetry was observed, such that greater laxity was observed in the left than the right hip. This asymmetry was not heritable. However, the average Norberg angle was highly heritable as were the Norberg angles of either the right or left hips. After correction for pedigree effects, two QTLs were identified using the metrics of the left and right hips as separate data sets. Both are on canine chromosome 1 (CFA1), separated by about 95 Mb. One QTL, associated with the SSR marker FH2524 was significant for the left, but not the right hip. The other, associated with FH2598, was significant for the right but not the left hip. For both QTLs, some extreme phenotypes were best explained by specific interactions between haplotypes.
Keywords: quantitative trait loci (QTLs), dog, hip laxity, bilateral asymmetry, Norberg angle, Canine genetics, hip dysplasia
INTRODUCTION
About 1% of humans have hip “dysplasia,” with higher percentages in certain groups or isolates [Henricson et al., 1966; Brier, 1999; Evans, 2001]. Moreover, there have been reports that congenital hip dysplasia in infants occurs with a higher incidence in the left than in the right hip [Smith et al., 1963]. Because a high proportion of larger dogs (>20 kg) are afflicted with the disorder, canine hip dysplasia may serve as a genetic model for some aspects of the human condition [Smith et al., 1963; Riser, 1975]. In dogs, it is a developmental trait that is inherited quantitatively, and expressed morphologically and clinically in response to heritable and environmental influences [Henricson et al., 1966; Hutt, 1967; Hedhammar et al., 1979]. Heritability estimates vary in different studies from 0.2 to 0.6, and both morphological and clinical phenotypes occur along a continuum, from nearly normal to severely abnormal. [Henricson et al., 1966; Leighton et al., 1977; Hedhammar et al., 1979].
Affected dogs appear to have normal hip joint conformation at birth, with development of the initial joint laxity becoming apparent as early as a few weeks of age or as late as mid-life [Mansson and Norberg, 1961; Henricson et al., 1966; Riser, 1973; Smith et al., 2002]. The phenotypic expression of joint laxity is recognized radiographically as subluxation of the femoral head from the acetabulum of the affected hip joint [Olsson, 1971; Riser, 1973]. In many affected joints, the ultimate outcome of the resulting chronically abnormal loading and remodeling is observed histologically as synovitis, erosion of articular cartilage [Farquhar et al., 1997], hyperplasia, and fibrosis of the joint capsule, edema, and elongation of the round ligament, and osteophyte formation [Olsson, 1971; Riser, 1973]. Changes that involve pathological remodeling of bone, altered joint conformation, and osteophyte formation are visible radio-graphically as osteoarthritis [Olsson, 1971; Riser, 1973]. Collectively, these changes often are described as degenerative joint disease (DJD) [Alexander, 1979; Olsewski et al., 1983; Lust, 1997].
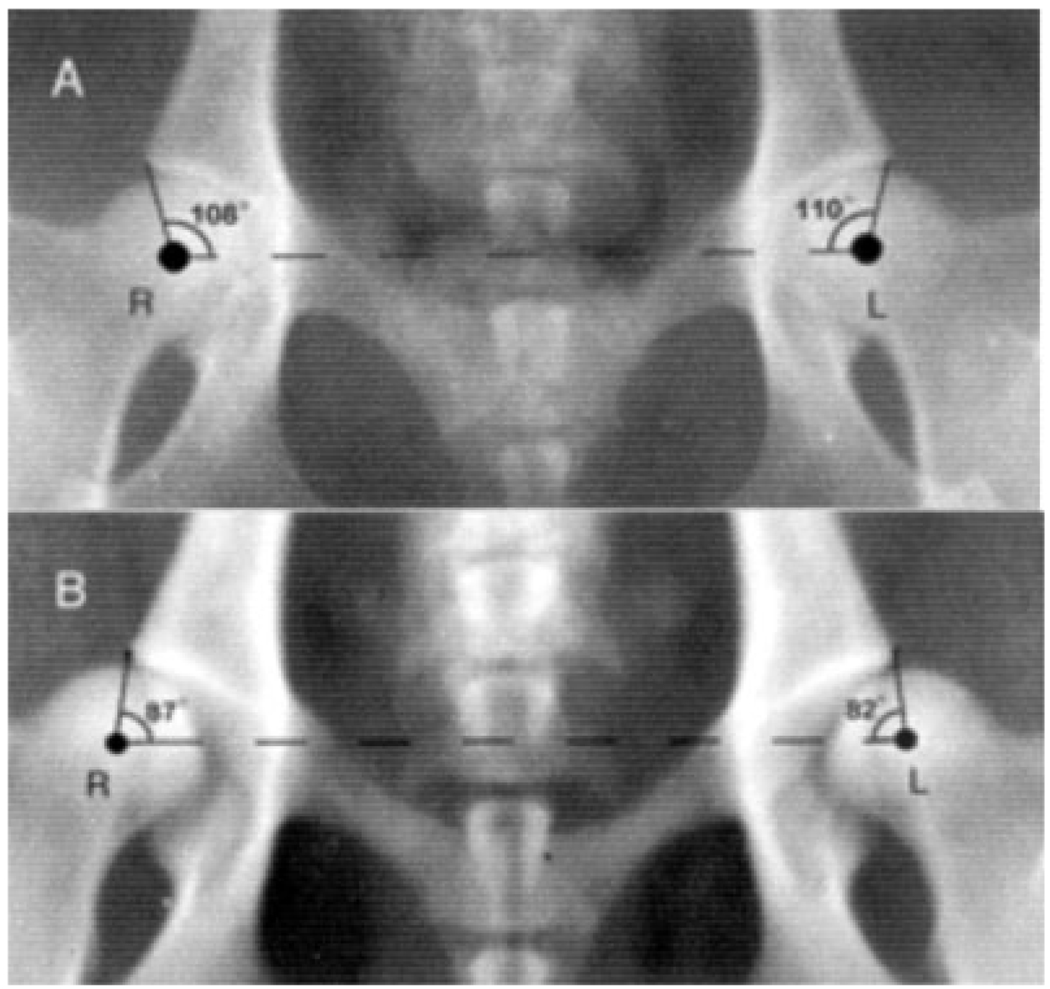
Various radiographic methods have been devised to estimate joint laxity and femoral head subluxation [Federation Cynologique Internationale, 1984; Orthopedic Foundation for Animals, Inc., 1989]. The Norberg angle is a quantitative measure of joint laxity (femoral head subluxation) [Kealy et al., 1992] and is measured from radiographs of the pelvis with the subject in ventrodorsal position, hind legs extended parallel in a posterior direction (Fig. 1); the smaller the angle the greater the subluxation [Olsson, 1961; Hickman, 1964; Kealy et al., 1997]. Previous data established an inverse correlation between the value of the Norberg angle and the development of DJD [Kealy et al., 1992, 1997].
Fig. 1.
Radiographs (ventrodorsal) illustrating Norberg angles for: (a) a dog with almost no subluxation in the coxofemoral joint, and (b) a dog with very much subluxation in the coxofemoral joint. Because of the ventrodorsal position of the animals, the left hip is on the right side of the radiographs.
We have searched for quantitative trait loci (QTLs) associated with the Norberg angle. To do this, we examined coxofemoral joints in a population of Portuguese water dogs (PWDs). This breed descends from a limited number of founders and is characterized by complete and accurate pedigree records [Molinari, 1993; Chase et al., 1999]. Combining molecular genetic markers with radiographic data, we have begun to examine the genetic basis of mammalian morphological diversity using this breed as a model system. In a previous report, we described QTLs that regulate body size and shape [Chase et al., 2002]. Here we describe two loci that regulate variation of the Norberg angle. A surprising result was that their effects appear to be asymmetric in that one locus affects the right hip and the other the left.
MATERIALS AND METHODS
Materials
Radiographs and blood for DNA were collected from owners of 286PWDs through the Georgie Project (http://www.georgieproject.com, Karen Miller director) [Chase et al., 2002]. The dogs, ranging in age from 1.7 to 17 years (median age, 6 years), comprised 118 males and 168 females and represented a cross-section of the entire PWD population. They trace their ancestry to 31 founders through ca. 24 generations and consanguinities range from 0 to 0.6 with a mean of 0.2 [Chase et al., 1999]. We have associated a founder with specific marker alleles using the consanguinity between that founder and all dogs known to carry the allele. Permutation tests are used to establish the significance of each association [Alroy et al., 2000].
Norberg angles (Fig. 1) were measured on ventrodorsal roentgengrams of the pelvis as illustrated in Figure 2 of Chase et al. [2002]. There was no significant effect of age or sex on the Norberg angle. DNA was isolated from each X-rayed dog and characterized by PCR amplification and electrophoretic identification of the alleles of ca. 500, largely tetranucleotide based, simple sequence repeat genetic markers [Francisco et al., 1996; Mellersh et al., 2000; Chase et al., 2002].
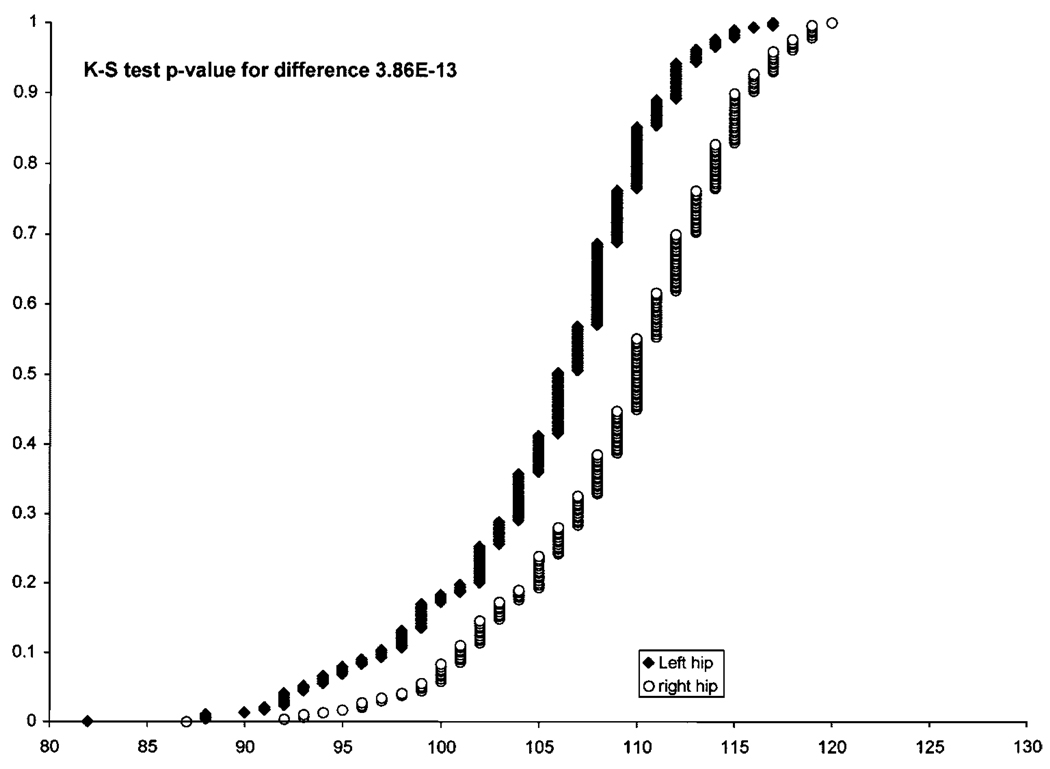
Fig. 2.
Values of the Norberg angles of the 286 dogs in the Portuguese water dog (PWD) population. The cumulative distributions of values (X-axis) for the right (o) and left hips (◆) are shown.
Methods
QTL detection and estimation of genotypic means: We used a mixed model to estimate the QTL means [Kennedy et al., 1992]. We assume that all of the background genetic effects are additive and use the model:
where yij is the phenotype of the jth individual of genotype i, gi is the effect of genotype i, aij is the effect of the additive genetic background, and eij is the environmental deviation. The effects of the genotypes (gi) are treated as fixed effects; the additive genetic background (aij) and environmental deviations (eij) are treated asr andom effects. The best linear unbiased estimate (BLUE) of the genotypic means (Lynch and Walsh, 1998) is given by:
where X is the design matrix of 1s and 0s indicating the genotype of the individual, V is the covariance matrix for the vector of phenotypes, y;
, the additive genetic variation was estimated using the polygenic function of SOLAR [Almasy and Blangero, 1998]; A is the additive genetic relationship matrix where Aij = 2 σij (twice the coefficient of consanguinity [Falconer and Mackay, 1996]; is the environmental variance; and I is the identity matrix.
The ratio of the variance of the estimated genotypic means to the variance of the total phenotype was used as a test statistic for associations between markers and phenotypes. We estimate the significance of this statistic using Monte Carlo simulations in which a random trait of the same heritability was simulated using the known pedigree. The distribution of the test statistic under the null model was estimated as the beta distribution with the best fit to a set of 5,000 simulations. The P-value of each marker is estimated as the cumulative probability of the null distribution exceeding the observed value. The P values (in Table I of the results) were then corrected for the number of trials (traits times markers).
TABLE I.
Phenotypic, Genotypic, and QTL Parameters of the Norberg Angle in Portuguese Water Dogs
| Norberg angle parameters | QTL linked to marker | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Heritability | FH2524 | FH2598 | ||||||
| Trait | Mean | SD | Skew | h2 | h2 · SD | R2 | P-value | R2 | P-value |
| Left | 105.32 | 5.93 | −0.94 | 0.61 | 0.12 | 0.142 | <0.04 | 0.057 | n.s. |
| Right | 109.13 | 5.75 | −0.73 | 0.46 | 0.15 | 0.053 | n.s. | 0.16 | <0.0005 |
| Average | 107.23 | 4.99 | −0.89 | 0.73 | 0.11 | 0.108 | n.s. | 0.12 | <0.08 |
Phenotypic means were calculated from the raw data (see Fig. 2). Heritabilities were calculated from computer program SOLAR (see “Methods”). QTLs were identified and characterized after correction for pedigree effects (see “Methods”). Pedigree correction uses best linear unbiased estimates (BLUE) of the genotypic means taking into account additive pedigree effects. Values for R2 were calculated as Gvar/Tvar. Gvar: variance of the BLUE phenotypic estimates; Tvar: variance of raw phenotypic data. P-values for R2 were established 5,000 Monte Carlo simulations of traits with similar heritabilities (see “Materials and Methods”). The resulting P values were corrected for the number of trials and independent (vis a vis linkage) markers tested. (n.s., not significant.).
We estimated the variance of the pedigree corrected means using the delete-one jackknife [Wu, 1986]. The jackknife estimate of the pedigree corrected mean for each dog j of genotype i is defined as:
where n is the total number of dogs with genotype i, gi is the pedigree corrected mean using the total population, and gi,−j is the estimation of the pedigree corrected mean when dog j is omitted. The mean of these corrected values is reported as the pedigree corrected mean (in Table II of the results). The standard error of these values is defined as where is the sample variance of jackknife estimates for genotype i.
TABLE II.
Norberg Angle Phenotypes Associated With Individual Genotypes of FH2524 and FH2598
| Left hip | Right hip | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Raw data | Pedigree corrected | Raw data | Pedigree corrected | ||||||
| Genotypes | Count | Mean | SE | Mean | SE | Mean | SE | Mean | SE |
| FH2524 | |||||||||
| CF | 10 | 102.2 | 2.65 | 100.2 | 2.6 | 108.0 | 2.01 | 107.9 | 2.2 |
| CE | 15 | 100.5 | 2.13 | 101.4 | 2.1 | 105.9 | 1.84 | 107.2 | 1.9 |
| EF | 32 | 100.4 | 1.05 | 101.9 | 1.2 | 106.2 | 0.94 | 107.6 | 0.9 |
| FF | 9 | 103.1 | 1.81 | 103.9 | 1.8 | 109.8 | 1.60 | 111.7 | 1.2 |
| AF | 6 | 104.7 | 2.42 | 104.8 | 2.1 | 109.2 | 1.54 | 109.2 | 1.7 |
| AD | 5 | 106.8 | 1.36 | 106.1 | 1.5 | 109.6 | 1.36 | 109.4 | 1.5 |
| DE | 42 | 105.9 | 0.69 | 106.3 | 0.8 | 109.3 | 0.91 | 109.5 | 1.0 |
| DD | 24 | 106.0 | 1.12 | 106.5 | 1.0 | 109.7 | 1.24 | 110.7 | 1.3 |
| BE | 13 | 106.3 | 1.03 | 106.6 | 1.1 | 108.5 | 1.55 | 107.8 | 1.6 |
| DF | 40 | 106.3 | 0.99 | 106.7 | 1.1 | 110.0 | 0.76 | 109.8 | 1.0 |
| CD | 20 | 108.1 | 1.29 | 107.4 | 1.4 | 110.9 | 1.41 | 110.2 | 1.3 |
| BF | 11 | 107.5 | 1.28 | 107.5 | 1.4 | 109.4 | 2.17 | 109.5 | 2.0 |
| EE | 16 | 105.8 | 1.20 | 107.5 | 1.1 | 108.9 | 1.74 | 110.1 | 1.5 |
| BD | 17 | 108.5 | 0.78 | 108.2 | 0.9 | 110.1 | 1.47 | 110.2 | 1.5 |
| AE | 5 | 111.4 | 1.12 | 111.6 | 1.5 | 113.4 | 1.50 | 113.0 | 2.0 |
| FH2598 | |||||||||
| EE | 83 | 104.3 | 0.71 | 105.1 | 0.8 | 106.8 | 0.71 | 106.1 | 0.8 |
| AE | 12 | 107.5 | 1.29 | 105.7 | 1.3 | 110.3 | 2.24 | 107.5 | 2.1 |
| EH | 79 | 104.6 | 0.70 | 106.0 | 0.6 | 108.6 | 0.58 | 107.8 | 0.6 |
| HH | 24 | 106.3 | 1.20 | 107.7 | 1.0 | 109.8 | 0.99 | 109.1 | 1.1 |
| EF | 7 | 107.7 | 0.75 | 108.0 | 1.0 | 110.3 | 2.10 | 109.4 | 1.7 |
| NG | 13 | 104.1 | 1.42 | 104.7 | 1.4 | 109.5 | 1.05 | 110.2 | 1.1 |
| CH | 14 | 106.2 | 1.03 | 106.3 | 1.0 | 111.4 | 1.33 | 110.9 | 1.5 |
| CE | 19 | 106.5 | 1.05 | 106.1 | 1.1 | 111.9 | 1.01 | 110.9 | 1.0 |
| DE | 7 | 110.0 | 1.70 | 110.1 | 1.7 | 112.6 | 1.07 | 112.7 | 0.9 |
| BE | 5 | 102.2 | 4.27 | 101.1 | 5.2 | 113.0 | 1.38 | 113.2 | 1.2 |
| EG | 8 | 107.0 | 1.22 | 109.6 | 1.2 | 115.5 | 1.04 | 116.0 | 0.9 |
Genotypes were characterized by alleles of SSR markers FH2524 and FH2598. Pedigree corrected means were established using jacknife resampling as described in “Methods.”
We conducted an extensive review of these methods using simulations to validate the precision and found that BLUE estimates of the genotypic means were much better than ordinary least square estimates. We also found that the jackknife estimates of the standard errors were more precise than sub-sampling estimates. In 95%of the simulations, the true genotypic mean was within 2.1 jacknife standard errors of the BLUE.
To explain the genotypic means, we invoked additive models. We tested these using a weighted multiple regression. Dummy variables of the allele counts were used to predict the pedigree corrected genotypic means, weighted by the jacknife variance estimates. For marker FH2524, we also tested an additive plus dominance model by including dominance variables for the D, E, and F alleles to indicate the presence or absence of an allele in each genotype.
RESULTS
We measured the Norberg angle in 286 PWDs and calculated the heritability of this trait (Table I) finding a value close to that reported for coxofemoral joint laxity in German Shepherd dogs in an earlier study [Leighton et al., 1977]. Significant heritability also was obtained from data for only the right or only the left joints, although the values were somewhat lower. The Norberg angle was independent of age and gender. There was no significant correlation between inbreeding coefficient and the Norberg angle. Thus, none of the variation in the Norberg angle could be attributed to inbreeding depression. Finally, as in previous studies of other breeds, the average Norberg angle was inversely correlated with radiographic evidence of osteoarthritis. Because most of the PW dogs in our study were young, the correlation was relatively weak (−0.24, P = 0.00004). Radiological examination of these dogs as they age may lead to a stronger correlation.
A striking characteristic of our measurements is consistently greater laxity in the left hip (Fig. 2). This directional asymmetry is highly significant (Kolmogorov–Smirnov non parametric test: P < 10−12) and the polarity (left Norberg angle smaller than right) is observed in 80% of the dogs. However, this difference between the Norberg angles of the right and left joints did not show significant heritablilty. Thus, the asymmetry is probably the result of interaction with the environment. If specific genes are involved, they already are either fixed in the PWD population, or segregation is restricted to very few individuals.
In view of this polarity, we searched for QTLs using the right and left hips as separate data sets. Two significant QTLs were identified after correcting for pedigree effects (Table I). Both are located on chromosome CFA 1 (Fig. 3), but are separated by about 94.6 Mb on the canine radiation hybrid genetic map [Guyon et al., 2003].One of these, linked to FH2524, affected variation in the left hip and accounted for 14% of the heritable variation in that hip. It had no significant effect on the right hip. The other, linked to FH2598, was responsible for 16% of the heritable variation in the right hip but had no significant effect on the left hip.
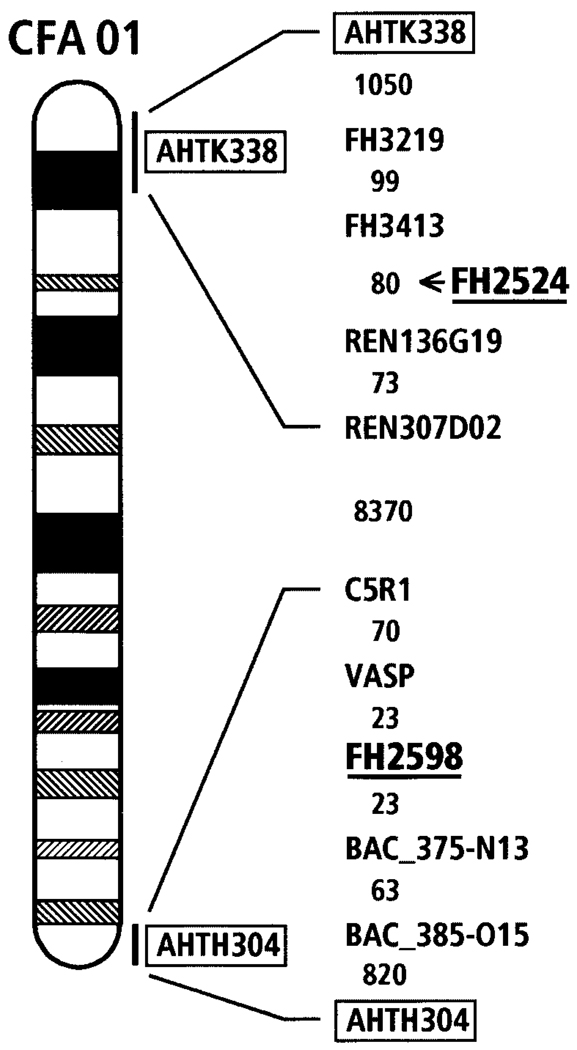
Fig. 3.
An ideogram of canine chromosome 1 (CFA01) highlighting locations of FH2524 and FH2598. Mapping data is based on a radiation hybrid map of CFA01 (Guyon et al., 2003, submitted) using the Multimap [Matise et al., 1994] and TSP Concorde Software programs [Agarwala et al., 2000]. Excerpts of markers neighboring FH2524 and FH2598 (underlined) are shown with inter-marker distances in TSP units between the markers. For CFA01, one unit corresponds to approximately 11 kb. The total length of CFA01 is 12,353 U or 137 Mb. Markers (AHTK338 and AHTH304) assigned to chromosomes by previous FISH mapping studies link the DAPI-banded ideogram to the RH map and are shown within a box [Breen et al., 2001]. FH2524 was mapped subsequent to other markers and is located between REN136G19 and FH3413 (as indicated).
This genetic asymmetry can be seen in the raw data and after correction for pedigree effects. Table II presents the phenotypic means of the different marker genotypes associated with each QTL. The data for FH2524 have been sorted according to increasing values for the left hip after pedigree correction. The data for FH2598 were sorted in the same manner using values for the right hip. The maximum pedigree correction was about 2.0° (left hip) and 1.9° (right hip) for FH2524 and FH2598 genotypes respectively (the average corrections were 0.7° and 0.95°, respectively). Most of the corrections tended to bring phenotypic values closer to the mean, increasing values for the left hip of FH2524 genotypes and decreasing values of the right hip for FH2598 genotypes.
The frequency of homozygous genotypes for both FH2524 and FH2598 was consistent with the frequencies predicted from allele frequencies on the basis of random mating. Ten genotypes of the marker FH2524 and 13 genotypes of FH2598 were represented by fewer than five individuals. Among these were homozygotes of FH2524 (two dogs each of genotypes BB & CC and one of II); and homozygotes of FH2598 (one dog each of genotypes BB, CC, & GG).
These differences are seen graphically in Figure 4. Cumulative distributions of phenotypic values associated with some of the genotypes are shown. For both markers, the non-heritable difference between the left and right hips (Fig. 4, upper versus lower panels) is apparent. However, the extent of phenotypic variation between genotypes is different for the two QTLs with greater variation in the left hip associated with marker FH2524, and more in the right hip associated with FH2598. We corrected for pedigree effects assuming that an additive mode of inheritance was responsible for most of the heritability of this phenotype. The range of corrected phenotypic means (Table II) associated with FH2524 genotypes is greater in the left hip (100°–111°) than in the right (107°–113°) and the opposite differential is observed for the phenotypes associated with FH2598 genotypes (left, 105°–109° <right, 106°–116°). These differences in variation are responsible for the differences in QTL significance (P-values, Table I) between the right and left hips.
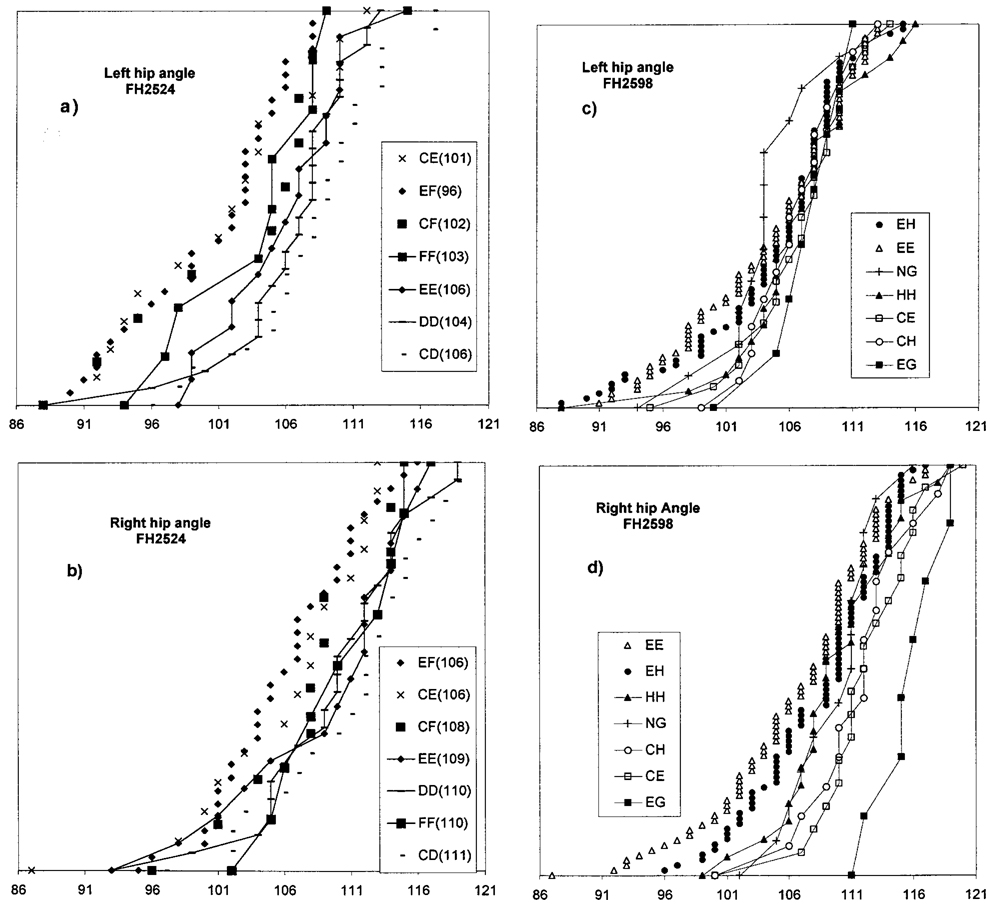
Fig. 4.
Distributions of Norberg angle metrics associated with particular genotypes of the markers FH2524 (a and b) and FH2598 (c and d). The left (top) and right (bottom) hip joints are shown. See Table II for additional information as well as additional genotypes. The collection of infrequent genotypes containing the FH2598 “G” allele (see text) has been denoted as NG. Genotypes are listed in descending order corresponding to the order from left to right in which their distributions occur.
The order of phenotypic values for the right and left hips corresponds for most of the genotypes. However, the FH2524 genotypes CF, CE, and EF are more clearly separated from DD, EE, and CD in the left hip; whereas FF, which was separate from CE and EF in the right hip is less distinct in the left (Table II). The relative order of FH2598 genotypes EE, EH, and EG remain the same, but the indeterminate order of CH, CE, and EH in the left hip is resolved in the right hip where CH, CE, (and EG) have much larger angles (Table II).
We have estimated the phenotypic means using an additive model of inheritance (Fig. 5). The QTL associated with FH2598 (Fig. 5a) fits this model well. However, there are few constraints on this fit, since we do not have adequate data to define the means of AA, CC, DD, FF, or GG. In Table II, the genotype labeled GN represents a group of 13 dogs belonging to 6 genotypes containing the Gallele (1 AG, 1 CG, 1 FG, 1 GG, 4 DG, and 5 GH). It can be seen (Table II, Fig. 4) that the mean value for the EG phenotype is significantly larger than the means of the GN and EE phenotypes. This suggests that the phenotype of the EG genotype may result from an interaction between specific QTL alleles associated with the “E” and “G” haplotypes.
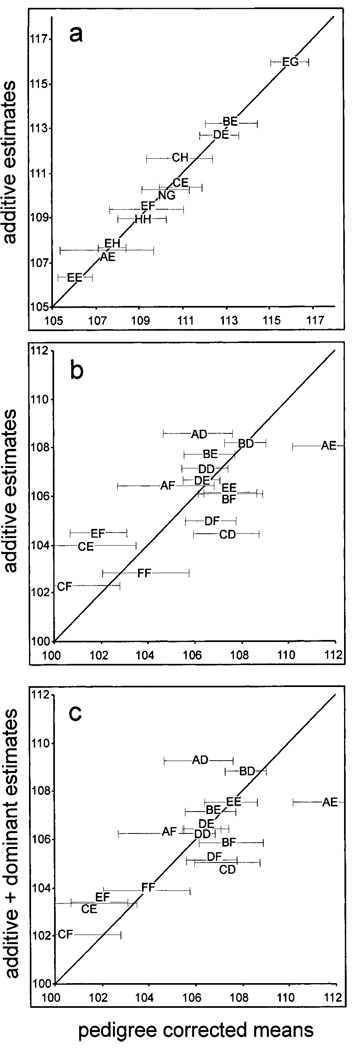
Fig. 5.
Scatter graph of the pedigree corrected estimates of genotypic means (X-axis) graphed against the best fit to an additive model (a and b) for FH2598 and FH2524 (see “Materials and Methods”); or (c) graphed against an additive model with simple dominance for FH2524. Error bars for the standard deviation were estimated using the jackknife procedure (Wu, 1986; see “Materials and Methods”).
The QTL associated with FH2524 (Fig. 5b) does not fit an additive model. A simple dominant model (Fig. 5c), in which one allele is assumed to be dominant to all other alleles, provides a slightly better fit but is not adequate. More complicated models involving specific interactions between alleles might improve the fit of FH2524 genotypes such as CD, CE, and CF as well as AD, AE, and AF or EF, BF and DF. Indeed, the data in Table II suggest that for the QTL associated with this marker, C could be dominant to E but recessive to D; or that F may be dominant to C and E, but not to D or B.
DISCUSSION
We have measured laxity, as defined by the Norberg angle, in 286 genotyped PW dogs. A striking result was the finding of a directed asymmetry in which the left hip is, on the average, significantly more lax than the right. (We have examined Norberg data from several other breeds (Lawler, unpublished observations) and have found a similar significant asymmetry in all of the breeds examined—Labrador Retriever, German Shepherd dog, English Setter, Siberian Husky and Miniature Schnauzer.)
As noted above, a similar asymmetry, left worse than right, has been described for congenital subluxation and luxation in the human [Smith et al., 1963]. In the PWD population, this asymmetry is not heritable. It could have a fixed genetic basis that is not segregating, or be due entirely to environmental effects or assay artifacts. It is difficult to envision an environmental effect (exercise or nutrition) that would produce such an asymmetry in a population that is maintained in an unrestricted manner by a large number of owners. Artifacts such as differential tension when extending the limbs for radiography (technician right hand versus left hand) could conceivably produce such a bias. A fixed genetic basis might be behavioral or physiological. For example, left or right “footedness” (analogous to handedness in humans) might lead to greater stress on one joint than on the other; or organ asymmetry could produce unequal loading, hence differential stress, during running or jumping. Variation in the extent or type of nutrition or exercise might then be translated into variation in the extent of the asymmetry.
We have described two QTLs for joint laxity measured by the Norberg angle. Each accounts for about 15% of the genotypic variation either in the left (FH2524) or right (FH2598) joint. For both QTLs, extreme phenotypes were associated with heterozygous genotypes (Table II): CF, CE, and EF had the smallest (most lax) phenotypic means for the FH2524 associated QTL and EG had a strikingly large mean for the QTL associated with FH2598 (only 10% of the right hip values were larger). Finally, the data for the FH2598 EG and “NG” genotypes (Table II) suggest the existence of a specific interaction between QTL haplotypes.
Most of these dogs were selected from a breeding population in which we are studying the genetic basis of skeletal morphology [Chase et al., 2002]. As a consequence, most of the measurements reflect the laxity of joints acceptable for breeding by the more subjective Orthopedic Foundation for Animals standard of “good” or “excellent.” Hence, the distribution in Figure 2 contains relatively few extremely low values (extreme laxity, OFA “poor” or “bad”). Nevertheless, the high frequency of FH2598 EE genotypes (lowest Norberg values) together with the very low frequency of EG genotypes (highest Norberg values; Table II) was unexpected. However, selection against the FH2598 EE genotype would not tend to be stringent, since its effect is primarily on the variation in the right hip, which tends more often to be acceptable. Moreover, the value of the EE genotypic mean is not exceptionally low (27% of the right hip values are lower, Fig. 2). Selection in favor of EG phenotypes would be slight, since many other, more frequent, genotypes (e.g., EH) are acceptable under OFA criteria. On the other hand, founder effects may have been important in determining haplotype frequencies. Using the pedigree database to associate marker alleles with particular founders [Alroy et al., 2000], we have traced the “E” allele of FH2598 to the original Algar-biorum kennel that founded the breed in the 1930s [Molinari, 1993]. This kennel contributed a large percentage of the present day gene pool. In contrast, the “G” allele was derived from a group of late founders (referred by Molinari [1993] as “others”) that did not become prominent contributors.
For each QTL, there is significant phenotypic variation associated with genotypes of one hip, but not the other. There are, in principle two explanations for this:
The phenotypic variation is dependent on the asymmetry in laxity. The QTL associated with FH2524 can only express its phenotypic variation under conditions of greater laxity (low values of the Norberg angle); whereas the phenotypic variation derived from genotypes of the QTL associated with FH2598 can only be observed under conditions of low laxity (large Norberg angles).
The phenotypic variation is dependent on some form of bilateral asymmetry and is distinguishing the right from the left side of the animal.
Our existing data, although extensive enough to identify these QTLs, does not have the statistical power to discriminate between these explanations. Enlarging the data set to increase the number of animals with extreme phenotypes could allow us to choose: Does extreme laxity in the right hip allow the detection of right hip variation associated with FH2524 or lack of laxity in the left allow the detection of left hip variation associated with FH2598? Despite our inability to discriminate between these hypotheses, it is important to note that separating the left and right data sets has provided more information than would have been gained from the increase in statistical power produced by combining them. This could be true for other data dealing with bilateral, dorsal ventral, or anterior posterior symmetries.
ACKNOWLEDGMENTS
Technical support from Travis Lorentzen, Tyler Jarvik, and Makiko Uemura is gratefully noted. Special thanks to Francis Galibert and members of his laboratory for sharing data. We thank Karen Miller and David Carrier for their continued input and support. We are indebted to Deborah Broughton and to the “Georgie Project” for organizing the collection of X-rays and blood samples. Without the Georgie Project this research would not have been possible. This research was supported by gifts from the Judith Chiara Family Trust and more than 100 PWD owners and breeders. We also acknowledge support from NIH grants R01 CA92167 and NIH/NCI K05 CA90754 to EAO; and GM 63056 to KGL.
Grant sponsor: NIH; Grant number: R01 CA92167; Grant sponsor: NIH/NCI; Grant number: K05 CA90754; Grant sponsor: GM; Grant number: 63056.
REFERENCES
- Agarwala R, Applegate DL, Maglott D, Schuler GD, Schaffer AA. Afast and scalable radiation hybrid map construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JW. Osteoarthritis (DJD) in the dog. Canine Practice. 1979;6:31–34. [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy J, Rush JE, Freeman L, Amarendhra Kumar MS, Karuri A, Chase K, Sarkar S. Inherited infantile dilated cardiomyopathy in dogs: Genetic, clinical, biochemical, and morphologic findings. Am J Med Genet. 2000;95:57–66. [PubMed] [Google Scholar]
- Breen M, Jouquand S, Renier C, Mellersh CS, Hitte C, Holmes NG, Cheron A, Suter N, Vignaux F, Bristow AE, Priat C, McCann E, Andre C, Boundy S, Gitsham P, Thomas R, Bridge WL, Spriggs HF, Ryder EJ, Curson A, Sampson J, Ostrander EA, Binns MM, Galibert F. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier SR. Primary care orthopedics. Mosby: St. Louis; 1999. p. 478. [Google Scholar]
- Chase K, Adler FR, Miller-Stebbings K, Lark KG. Teaching a new dog old tricks: Identifying quantitative trait loci [in dogs] using lessons from plants. J Hered. 1999;90:43–51. doi: 10.1093/jhered/90.1.43. [DOI] [PubMed] [Google Scholar]
- Chase K, Carrier D, Adler FR, Jarvik T, Ostrander EA, Lorentzen TD, Lark KG. Genetic basis for systems of skeletal quantitative traits: Principal component analysis of the canid skeleton. Proc Natl Acad Sci USA. 2002;99:9930–9935. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC. Illustrated orthopedic physical assessment. Mosby: St. Louis; 2001. p. 1035. [Google Scholar]
- Falconer DS, Mackay T. Introduction to quantitative genetics. 4th Edition. New York: Longman Group Ltd; 1996. pp. 82–88. [Google Scholar]
- Farquhar T, Bertram J, Todhunter RJ, Burton-Wurster N, Lust G. Variations in composition of cartilage from the shoulder joints of young adult dogs at risk for developing canine hip dysplasia. J Am Vet Med Assoc. 1997;210:1483–1485. [PubMed] [Google Scholar]
- Federation cynologique internationale. Scientific committee: Hip dysplasia—International certificate and evaluations of radiographs. Helsinki: Federation cynologique internationale; 1984. pp. 1–25. [Google Scholar]
- Francisco LV, Langston AA, Mellersh CS, Neal CL, Ostrander EA. A class of highly polymorphic tetranucleotide repeats for canine genetic mapping. Mamm Genome. 1996;7:359–362. doi: 10.1007/s003359900104. [DOI] [PubMed] [Google Scholar]
- Guyon R, Lorentzen TD, Hitte C, Kim L, Cadieu E, Parker HG, Quignon P, Lowe JK, Renier C, Gelfenbeyn B, Vignaux G, DeFrance HB, Gloux S, Mahairas GG, André C, Galibert F, Ostrander EA. A 1 Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci USA. 2003;100:5296–5301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedhammar A, Olsson S-E, Andersson SA, Persson L, Pettersson L, Olausson A, Sundgren PE. Canine hip dysplasia: A study of heritability in 401 litters of German Shepherd dogs. J Am Vet Med Assoc. 1979;174:1012–1016. [PubMed] [Google Scholar]
- Henricson B, Norberg I, Olsson S-E. On the etiology and pathogenesis of hip dysplasia: A comparative review. J Sm Anim Pract. 1966;7:673–688. doi: 10.1111/j.1748-5827.1966.tb04393.x. [DOI] [PubMed] [Google Scholar]
- Hickman J. Veterinary orthopedics. Philadelphia: JB Lippincott; 1964. p. 316. [Google Scholar]
- Hutt FB. Genetic selection to reduce the incidence of hip dysplasia in dogs. J Am Vet Med Assoc. 1967;151:1041–1048. [PubMed] [Google Scholar]
- Kealy RD, Olsson SE, Monti KL, Lawler DF, Biery DN, Helms RW, Lust G, Smith GK. Effects of limited food consumption on the incidence of hip dysplasia in growing dogs. J Am Vet Med Assoc. 1992;201:857–863. [PubMed] [Google Scholar]
- Kealy RD, Lawler DF, Ballam JM, Lust G, Smith GK, Biery DN, Olsson SE. Five-year longitudinal study on limited food consumption and development of osteoarthritis in coxofemoral joints of dogs. J Am Vet Med Assoc. 1997;210:222–225. [PubMed] [Google Scholar]
- Kennedy BW, Quinton M, van Arrendonk JAM. Estimation of effects of single genes on quantitative traits. J Anim Sci. 1992;70:2000–2012. doi: 10.2527/1992.7072000x. [DOI] [PubMed] [Google Scholar]
- Leighton EA, Linn JM, Willham RL, Castleberry MW. A genetic study of canine hip dysplasia. Am J Vet Res. 1977;38:241–244. [PubMed] [Google Scholar]
- Lust G. An overview of the pathogenesis of canine hip dysplasia. J Am Vet Med Assoc. 1997;210:1443–1445. [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sinauer Associates, Inc.; 1998. pp. 745–751. [Google Scholar]
- Mansson J, Norberg I. Hoftledsdysplasi hos hund (Hip dysplasia in the dog). (Hip joint laxity and secondary acetabular dysplasia induced by hormone administration) Medlemsblad for Sveriges veterinarforbund (J Swedish Vet Assoc) 1961;12:1–8. [Google Scholar]
- Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): A human genome linkage map. Nature Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Hitte C, Richman M, Vignaux F, Priat C, Jouquand S, Werner P, Andre C, DeRose S, Patterson DF, Ostrander EA, Galibert F. An integrated linkage-radiation hybrid map of the canine genome. Mamm Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- Molinari C. The Portuguese water dog. Portugal: ELO-Publicidade; 1993. pp. 1–156. [Google Scholar]
- Olsewski JM, Lust G, Rendano VT, Summers BA. DJD: Multiple joint involvement in young and mature dogs. Am J Vet Res. 1983;44:1300–1308. [PubMed] [Google Scholar]
- Olsson S-E. Advice and directions for roentgen examination of the hip joints of German Shepherd dogs. Hundsport (the Journal of the Swedish Kennel Club) 1961 Supp. l:1–4. [Google Scholar]
- Olsson S-E. DJD (osteoarthrosis): A review with special reference to the dog. J Small Animal Practice. 1971;12:333–342. doi: 10.1111/j.1748-5827.1971.tb06238.x. [DOI] [PubMed] [Google Scholar]
- Orthopedic Foundation for Animals, Inc. Hip dysplasia, a guide for dog breeders and owners. 2 edition. Columbia, MO: Orthopedic Foundation for Animals, Inc.; 1989. pp. 1–28. [Google Scholar]
- Riser WH. The dysplastic hip joint: Its radiographic and histologic development. J Am Vet Radiol Soc. 1973;14:35–50. [Google Scholar]
- Riser WH. The dog as a model for the study of hip dysplasia. Basel: S Karger; 1975. p. 332. [DOI] [PubMed] [Google Scholar]
- Smith WS, Coleman CR, Olix ML, Slager RF. Etiology of congenital dislocation of the hip. J Bone Joint Surg. 1963;45-A:491–500. [Google Scholar]
- Smith GK, Biery DN, Kealy RD, Lawler DF. Effects of restricted feeding on onset, incidence, and severity of hip dysplasia and osteoarthritis in dogs: Diagnostic, therapeutic, and genetic ramifications; Proceedings, The Purina Pet Institute Symposium; St. Louis: Purina Pet Institute; 2002. pp. 21–26. [Google Scholar]
- Wu CJF. Jacknife, bootstrap, and other resampling methods in regression analysis (with discussion) Ann Stat. 1986;14:1261–1350. [Google Scholar]