Abstract
Background. Current advances in genetic technology continue to expand the list of medical conditions associated with autism. Clinicians have to identify specific autistic-related syndromes, and to provide tailored counseling. The aim of this study is to elucidate recent advances in autism research that offer important clues into pathogenetic mechanisms of syndromic autism and relevant implications for clinical practice. Data Sources. The PubMed database was searched with the keywords “autism” and “chromosomal abnormalities,” “metabolic diseases,” “susceptibility loci.” Results. Defined mutations, genetic syndromes, and metabolic diseases account for up to 20% of autistic patients. Metabolic and mitochondrial defects may have toxic effects on the brain cells, causing neuronal loss and altered modulation of neurotransmission systems. Alterations of the neocortical excitatory/inhibitory balance and perturbations of interneurons' development represent the most probable pathogenetic mechanisms underlying the autistic phenotype in Fragile X-Syndrome and Tuberous Sclerosis Complex. Chromosomal abnormalities and potential candidate genes are strongly implicated in the disruption of neural connections, brain growth, and synaptic/dendritic morphology. Conclusion. Metabolic testing may be appropriate if specific symptoms are present. High-resolution chromosome analysis may be recommended if a specific diagnosis is suspected because of obvious dysmorphisms. Identifying cryptic chromosomal abnormalities by whole genome microarray analysis can increase the understanding of the neurobiological pathways to autism.
1. Introduction
Autism and related autism spectrum disorders (ASDs) are heterogeneous neurodevelopmental disorders behaviorally defined by significant deficits in social interaction and communication and by the presence of restricted interests and repetitive behaviors [1]. Autism Disorder (AD) comorbidy with seizures and mental retardation occurs in up to 30% and in 80% of autistic patients, respectively [2]. Its pathogenetic substrates are still largely unknown. Despite strong familial components, clinical complexity has posed a major challenge to our understanding of autism pathogenesis. Genetically, the picture is complicated by significant interindividual heterogeneity, numerous contributing loci, and multiple genes and gene-environment interactions [3]. Several lines of evidences strongly support a prenatal onset for developmental abnormalities later leading to autism [4]. Autism in its very broad spectrum of severity is known to have many different etiologies. In the last few years, significant progresses have been made in comprehending the causes of autism, and their multiple impacts on the developing brain [5]. The primary goals of treatment are optimizing the quality of life and minimizing the impairment due to the core symptoms of autism [6].
In this article we discuss current understanding of the pathogenesis of syndromic autism and the multiple pathways responsible for the ASD phenotype.
2. Metabolic Diseases
Several inborn errors of metabolism, including phenylketonuria, biotinidase deficiency, disorders of cerebrospinal fluid (CSF) neurotransmitters such as deficiencies of folic acid, Smith-Lemli-Opitz syndrome (SLOS), creatine deficiency syndromes, metabolic purine disorders, have an autistic phenotype [7]. A better understanding of some of them has implications both for discovery of the pathophysiologic underpinnings of the disorder and for the development of effective interventions.
In untreated children affected by phenylketonuria, the high levels of phenylalanine may have toxic effects on the brain cells, causing reduction of myelin, neuronal loss, and decreased levels of interneuronal connections [8]. Hyperphenylalaninemia also competes with the absorbition of other amino acids and consequently lower tyrosine and tryptophan concentrations can result in a low production of dopamine and serotonin in the prefrontal cortex [9].
In some of the most severe metabolic diseases, like adenylosuccinase deficiency or creatine deficiency syndromes, neurological and behavioral symptoms are probably not caused by deficiency of metabolites, but are more likely due to the toxic effects of the accumulating substances on the brain [8]. A direct role in modulation of dopaminergic and serotoninergic neurotransmission systems and axonal guidance has been hypothesized for the adenosine deaminase deficiency as pathologic mechanisms for the development of altered pathways involved in autistic symptoms [10]. The role of mitochondrial disorders has been revitalized by the association between autism and variants of the SLC25A12 gene, which encodes the predominant isoform of the mitochondrial aspartate (asp)/glutamate (glu) carrier (AGC) in brain [11]. Altered Ca2+ homeostasis is responsible for boosting AGC activity, mitochondrial metabolism, and, to a more variable degree, oxidative stress in autistic brains [12]. Based on our clinical experience, routine metabolic screening studies should be used on a case by-case basis, in the presence of the autistic regression, or suggestive clinical findings, such as lethargy, cyclic vomiting, early onset seizures, dysmorphic features, mental retardation with neurologic deficits, unexplained immune deficiency or unexplained hemolytic anemia, hyper- or hypotonia, self-mutilation, and muscle weakness [13]. Table 1 summarizes the main clinical features, diagnostic test and therapeutic options of the metabolic diseases most frequently associated with ASD.
Table 1.
Diagnosis and potential therapeutic approaches in some metabolic diseases associated with autism.
| Metabolic diseases | Potential patogenetic mechanisms | Clinical features | Diagnosis | Therapetic options | Refs |
|---|---|---|---|---|---|
| Phenylketonuria | Low production of dopamine and serotonin. Toxic effects on the brain cells. Reduction of myelin. | Neonatal onset Autism, seizures, severe mental retardation, hyperactivity, EEG abnormalities and seizures, microcephaly, albinism (excessively fair hair and skin) or a tendency to hypopigmentation and eczema, “musty or mousy” odor of skin, hair, sweat, and urine. | Quantitative plasma amino acids analysis. Dosage of phenylpyruvic acid in urine. | Restricted diet + aminoacids administration. | [28, 29] |
| Adenylosuccinase deficit | Toxic effects of the accumulating succinyl purines on the brain. | Onset in the first year. Autistic phenotype, profound psychomotor retardation, epilepsy, hypotonia, peripheral hypertonia, failure to thrive. No dismorphic features. | Succinyl aminoimidazole, carboxamide riboside and succinyl adenosine in urine and cerebrospinal fluid. | Therapy with D-ribose. | [7, 30] |
| Smith-Lemli-Opitz syndrome | Neurosteroid deficiency. Alteration of neuroendocrine functions and disruption of the growth and development of many body systems. | Onset in infancy. Autism, mental retardation, sensory hyperreactivity, irritability, language impairment, sleep cycle disturbance, self-injurious behavior, microchepaly, hypotonia, syndactyly, hypogenitalism, malformations of the brain, lung, heart, and gastrointestinal tract. | Abnormal sterol pattern (low plasma and tissue cholesterol concentrations, and increased plasma and tissue 7-dehydrocholesterol reductase and its metabolite). | Cholesterol replacement therapy. | [31, 32] |
| Creatine deficiency syndromes | Neurotoxic effect of guanidinoacetate or other guanidine compounds. | Autistic phenotype, mental retardation, speech delay, epilepsy, extrapyramidal symptoms, progressive encephalopathy with muscular hypotonia, dyskinetic movements, developmental arrest/regression. | Blood and urinary concentration on creatine and guanidinoacetate, Brain magnetic resonance spectroscopy. | Oral creatine supplementation. Restriction of arginine and substitution of ornithine. | [33] |
3. Epilepsy and Regressive Autism
The relationship among epilepsy, autism, and regression is a poorly understood and controversial subject. There are several epilepsy syndromes in which regression of language, cognition, and behavior may lead to clinical manifestations that overlap with the behavioral syndrome of autism, such as infantile spasms, slow spike-wave discharges during sleep, and focal centrotemporal spikes. An epileptic disorder must be considered in all children with a low functioning ASD, especially when a history of regression and electroencephalogram (EEG) epileptic abnormalities is present [14]. Severe epileptiform abnormalities may permanently alter the critical synaptogenesis by strengthening synaptic contacts that should have been naturally pruned [15]. Cognitive functions decline in those patients who have early-onset EEG abnormalities and a prolonged active phase of continuous spike-and-wave discharges during sleep [16]. In children with autistic regression, there is no evidence that treatment of the seizures or of the interictal epileptiform activity makes a difference in regard to the outcome of the language and social deficits. Because the relationship between autism, epilepsy, and regression is complex, the clinician's index of suspicion for epilepsy should be high, and treatment of the epilepsy should be pursued when necessary.
Although the pathogenetic link between autism and epilepsy is poorly understood, the existence of altered Ca2+ signaling in ASD and the bioelectrical instability resulting from mutations of the L-type voltage-gated Ca2+ channels associated to autism may account for the high prevalence of seizures and/or EEG abnormalities present among autistic individuals [17].
4. Genetic Diseases Associated with Autism
Single gene defects and chromosomal abnormalities may account for approximately up to 10% [18] of individuals with autism, and the fraction is likely to be higher when microarray comparative genome hybridization is used [19]. Table 2 summarizes the most frequent genetic syndromes and cytogenetic abnormalities associated with autism.
Table 2.
Genetic syndromes associated with autism.
| Syndrome | Gene(s) associated with the syndrome | Proportion of patients with an ASD that have the syndrome | Proportion of patients with the syndrome that have an ASD | Clinical signs | Refs |
|---|---|---|---|---|---|
| Fragile-X syndrome | FMR1 | 2–5% | 20–40% | Mental retardation, long face with prominent ears, macroorchidism, social anxiety, sensory hypersensitivity, stereotypies, poor motor coordination, delayed speech development. | [56, 57] |
| Tuberous sclerosis | TSC1, TSC2 | 3–4% | 43–86% | Epilepsy, mental retardation, specific learning disabilities, ADHD disorder, autistic spectrum disorders. | [58, 59] |
| 15q duplication Angelman/Prader Will syndrome | UBE3A GABAr cluster | 1–2% | >40% | Ataxia, language delay, epilepsy, mental retardation, repetitive movements, obsessive-compulsive symptoms. | [60] |
| 16p11 deletion | PCKB1 | 1% | High | Developmental delay, distinct facial appearance, autism. | [46, 61] |
| 22q deletion | SHANK3 | 1% | High | Speech and language disability, social impairment. | [62, 63] |
| 2q37 deletion | KIF1A, GBX2 | Unknown | 50% | Developmental delay, mental retardation, hypotonia, hyperactivity, autistic traits, dysmorphic features (cleft palate, temporal bone abnormalities, hypoplastic lungs). | [64] |
| Joubert syndrome | AHI1 | Unknown | 40% | Partial/complete agenesis of the cerebellar vermis, ataxia, abnormalities of ocular movements, cognitive, and behavioral dysfunction. | [65] |
| Timothy syndrome | CACNA1C | Unknown | 60–70% | Cardiac arrhythmia, long QT syndrome, mental retardation, and ASD. | [66] |
| Cortical dysplasia-focal epilepsy syndrome | CNTNAP2 | Rare | 70% | Seizures and language regression. | [67, 68] |
4.1. Fragile X Syndrome
Abnormalities in long-term synaptic plasticity of excitatory synapses and in baseline synaptic connectivity may be the underlying neurological substrate of autism associated with FXS [20, 21]. Alterations in the neocortical excitatory/inhibitory balance as well as abnormal neural synchronization have been also reported in mouse model of FXS [22], resulting in hyperexcitability of neocortical circuits. An immature dendritic morphology may also increase susceptibility to epilepsy and anxiety in FXS patients [23].
4.2. Tuberous Sclerosis Complex
Tuberous sclerosis complex (TSC) is an inherited disorder resulting from mutations in one of two genes, TSC1 (Hamartin) and TSC2 (Tuberin), characterized by benign hamartomatous tumors that involve multiple organ systems. It is commonly associated with neuropsychiatric complications like epilepsy, mental retardation, autism, and other behavioral problems. Seizures can be present in the first year of life and up to one third of children develop infantile spasms. Neurobehavioral phenotypes in TSC may arise from perturbations of interneurons development, which can selectively impact frontal and parietal areas [24]. TSC2 gene localized on 16p13.3 locus encodes for tuberin, a protein highly expressed in frontal regions [25]. Furthermore, several studies have described the TSC 1 locus 9q34 as an important region of vulnerability for the developmental of autism. A loss of a single TSC1 gene copy in mice is sufficient to perturb cytoskeletal dynamics and dendritic spine structure, highlighting generalized neurotrophic roles for these genes, in addition to cell growth regulation. Circuitry alterations are the possible biological substrate of autism associated with TSC.
5. Chromosomal Abnormalities
A wide number of cytogenetic abnormalities have been described [26], particularly in the low functioning autistic population with dysmorphic features [27].
5.1. Chromosome 15
Chromosomal rearrangements in 15q11-15q13 region might be the most frequent cytogenetic abnormality in ASD [34], accounting for 1–2% of patients. A chromosome 15 phenotype II, characterized by ataxia, language delay, epilepsy, mental retardation, repetitive movement disorders, and facial dysmorphic features, has been described in individuals with chromosome 15 duplications [35]. Within the 15q11–15q13 locus, gamma-aminobutyric acid A receptor beta 3 (GABRB3), an inhibitory neurotransmitter receptor, are currently thought to be central likely to play a significant role in the development of ASD, due to its role in the neuronal inhibition and its expression in early development [36]. This finding is particularly interesting in light of the high incidence of seizures and EEG abnormalities in autistic patients.
5.2. Chromosome 7
Two of the loci most commonly associated with ASD by genetic linkage studies [37, 38] (7q22 and 7q31 regions) contain several genes implicated in the pathogenesis of autism. The RELN gene, found within the 7q22 region, has a pivotal role in neuronal migration and prenatal development of neural connections, [39, 40] and is potently inhibited by toxic substances, such as organophosphates [41].
Increased risk for autism can be also linked to a functional polymorphism in the MET gene, found within the 7q31 locus [42], which plays a role into development of the cerebral cortex and cerebellum. The Williams-Beuren syndrome (WBS) region (7q11.23) also contains several genes associated with impairment in language and social interaction [43–45], suggesting the existence of a specific subgroup of autistic patients, characterized by dysmorphic features, mental retardation, language delay, congenital heart disease, and hypersensitivity to sound.
5.3. Chromosome 16
An association between a larger microdeletion on 16p11.2 and a syndrome that included developmental delay and distinct facial appearance (hypertelorism, a broad nasal bridge and a broad nasal tip with a prominent columella, a short philtrum, long ears, a large mouth) has been described [46–48]. The chromosomal region 16p11.2 also encompasses the PRKCB1 locus, an interesting gene previously found associated with autism [49], and expressed in the CNS, the immune system, the digestive tract, and the kidney. A recent study has described an association between PRKCB1 and an enhanced urinary peptide excretion rate [50].
5.4. Chromosome 2
Deletions involving 2q37 have been observed in more than 70 individuals with autism, mental retardation, and dysmorphic features (prominent forehead, depressed nasal bridge, dysmorphic ears and nose, short stature, and short hands and feet) [51, 52]. Three different breakpoints of 2q37 (2q37.1, 2q37.2, 2q37.3) have been analyzed to clarify the genotype-phenotype relationships associated with different terminal deletions [53], and several candidate genes for autism have been identified in 2q37.3 band [54]. Furthermore, a correlation between autism and a de novo cryptic deletion of chromosome 2p25.2 has been described [55]. The interaction between potential candidate genes that are expressed on these loci may explain the phenotypical heterogeneity and the spectrum of neuropsychological deficits associated with 2q37 and 2p25.2 deletion syndromes.
Other regions implicated in the ASDs with possible candidate genes are summarizes in Table 3.
Table 3.
Candidate genes associated with autism.
| Genes | Chromosomes | Proteins | Proteins' functions | Neurobiological abnormalities | Clinical phenotypes | Refs |
|---|---|---|---|---|---|---|
| NGL3 NGL4 |
Xq13.1 Xp22.3 |
Neuroligin 3/4. | Synaptic transmission, differentiation of synaptic contacts. | Synaptic or dendritic changes. | Autism with motor tics, Mild to severe autism, PDD-NOS, “regression” at disease onset, with a loss of initially-acquired social and verbal milestones, no dysmorphic features. | [79, 80] |
| SHANK3 | 22q13 | Shank scaffolding proteins. | Master organizer of postsynaptic glutamatergic density. | Synaptic or dendritic changes. | Multiple developmental delays, dysmorphic features, autism with severe language/social deficits. | [81, 82] |
| MET/HGF | 7q31 | MET receptortyrosine kinase/hepatocyte growth factor. | Regulation of dendritic morphology and promoting neurite outgrowth. | Abnormalities in development of the cerebral cortex and cerebellum. | Autism, increased anxiety, seizures, immune, and gastrointestinal problems. | [83] |
| MECP2 | Xq28 | Methyl-CpG-binding protein 2. | Synapse maintenance and remodeling. | Synaptic or dendritic changes. | Rett syndrome with regression, mental retardation, microcephaly, stereotyped behaviors, epilepsy and breathing problems; verbal Rett variants. | [84] |
| HOXA1 | 7p15.3 | Homeobox protein. | Regulation of brain growth. | Abnormalities of numbers of neurons or glia in the brain. | Mental retardation, autism and distinct clinical features (horizontal gaze abnormalities, focal weakness, hypoventilation, vascular malformations). | [85] |
| PTEN | 10q23 | phosphatase and tensin homologue. | Regulation of cells proliferation/differentiation. | Abnormalities in brain growth. | Macrocephaly, macrosomia, autism and developmental delay, increased risk of developing a variety of PTEN-related cancers in adulthood. | [86, 87] |
6. Pathogenetic Pathways
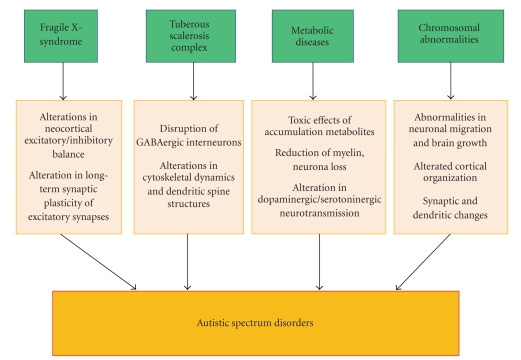
Several molecular pathways potentially involved in the disruption of neurodevelopmental trajectories during intrauterine or postnatal brain development may be associated with abnormal developmental processes, from neuronal migration and cortical organization to synaptic and dendritic conformation [69]. Furthermore, environmental factors, including maternal/intrauterine infections, exposure to toxins, and ossidative stress, may modify the underlying genetic substrate and leading to abnormalities in neuronal organization and cortical network development [70]. Figure 1 summarizes both genetic background and epigenetic factors involved in the pathogenesis of ASD, and explains how their multifactorial influence may be necessary for full expression of the autistic phenotype. Defined medical syndromes, chromosomal abnormalities and de novo copy number variations (CNVs) may account for 10% of ASD cases [71]. Figure 2 illustrates many different types of potential pathogenetic mechanisms responsible for the ASD phenotype in the most common medical conditions associated with autism. Widespread genetic testing would be expensive, time-consuming, and generally inappropriate due to the etiological complexity, while the appropriate use of genetic testing in subgroups of autistic patients showing particular clinical features is relevant to good clinical practice and may allow the identifications of new susceptibility variants. The advent of fluorescent in situ hybridization (FISH) techniques has expanded the list of chromosomal hot spots in autism. Individual FISH studies may be indicated in the confirmation of a clinically suspected condition [72], and in the evaluation of low functioning patients with an IQ <50 [73]. When dysmorphic features are present, it is reasonable to suspect chromosomal rearrangements even if the karyotype appears normal, and oligo-array-based CGH analysis is highly advisable in these cases [74]. Whole genome-scanning by array-based technology has detected copy-number variations (CNVs), which are copy-number changes involving a DNA fragment, and represent submicroscopic deletions or duplications that are undetectable at the routine cytogenetic analysis [75–78]. In conclusion, as etiologies of ASD are progressively discovered, the number of individuals with idiopathic autism will progressively shrink. The role of the neuropediatrician will be to understand the neurological basis of autism, and to identify more homogenous subgroups with specific biologic markers. Because autism represents an extremely heterogeneous group of disorders, a better understanding of underlying biological processes will lead to more targeted intervention approaches that can be designed for specific subtypes of autism.
Figure 1.
Genetic and epigenetic factors involved in the pathogenesis of autism. Interactions between multiple genes and environmental factors, such as intrauterine infections, alcohol/toxins exposure, and obstetrical suboptimality, can influence intrauterine and early postnatal brain development and disrupt crucial neurobiological pathways, from neuronal migration and cortical organization to synaptic and dendritic conformation, resulting in alterations of neurobehavioral trajectories that are involved in the pathogenesis of ASD.
Figure 2.
Potential pathogenetic mechanisms of syndromic autism. Several medical conditions associated with syndromic autism appear to influence and potentially disrupt neurodevelopmental processes, including brain growth, cortical connectivity, and neurotransmitters pathways. These neurobiological alterations likely affect the developmental trajectory of social behavior and communication during early stages of childhood and determine the different clinical phenotypes of ASD.
Acknowledgments
Arianna Benvenuto (medical doctor) drew the first draft with the assistance and contribution of Barbara Manzi (Medical Doctor). Riccardo Alessandrelli (Medical Doctor) reviewed relevant articles on the literature under the supervision of Cinzia Galasso (Associated Professor of the Department of Pediatric Neuroscience Unit) on genetic aspects. Professor Paolo Curatolo (Director of the Department of Pediatric Neuroscience Unit) proposed and designed the study, revised the final draft and is the guarantor. All authors contribute to the intellectual contents and approve the final version.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 3rd edition. Washington, DC, USA: American Psychiatric Association; 1994. [Google Scholar]
- 2.Berney TP. Autism—an evolving concept. British Journal of Psychiatry. 2000;176:20–25. doi: 10.1192/bjp.176.1.20. [DOI] [PubMed] [Google Scholar]
- 3.Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends in Neurosciences. 2006;29(7):349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Miller MT, Strömland K, Ventura L, Johansson M, Bandim JM, Gillberg C. Autism associated with conditions characterized by developmental errors in early embryogenesis: a mini review. International Journal of Developmental Neuroscience. 2005;23(2-3):201–219. doi: 10.1016/j.ijdevneu.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Benvenuto A, Moavero R, Alessandrelli R, Manzi B, Curatolo P. Syndromic autism: causes and pathogenetic pathways. doi: 10.1007/s12519-009-0033-2. World Journal of Pediatrics. In press. [DOI] [PubMed] [Google Scholar]
- 6.Myers SM, Johnson CP, Lipkin PH, et al. Management of children with autism spectrum disorders. Pediatrics. 2007;120(5):1162–1182. doi: 10.1542/peds.2007-2362. [DOI] [PubMed] [Google Scholar]
- 7.Manzi B, Loizzo AL, Giana Grazia G, Curatolo P. Autism and metabolic diseases. Journal of Child Neurology. 2008;23(3):307–314. doi: 10.1177/0883073807308698. [DOI] [PubMed] [Google Scholar]
- 8.Huttenlocher PR. The neuropathology of phenylketonuria: human and animal studies. European Journal of Pediatrics. 2000;159(2):S102–S106. doi: 10.1007/pl00014371. [DOI] [PubMed] [Google Scholar]
- 9.Diamond A. Evidence for the importance of dopamine for prefrontal cortex functions early in life. Philosophical Transactions of the Royal Society B. 1996;351(1346):1483–1494. doi: 10.1098/rstb.1996.0134. [DOI] [PubMed] [Google Scholar]
- 10.Okada M, Kawata Y, Murakami T, Wada K, Mizuno K, Kaneko S. Interaction between purinoceptor subtypes on hippocampal serotonergic transmission using in vivo microdialysis. Neuropharmacology. 1999;38(5):707–715. doi: 10.1016/s0028-3908(98)00226-3. [DOI] [PubMed] [Google Scholar]
- 11.Segurado R, Conroy J, Meally E, Fitzgerald M, Gill M, Gallagher L. Confirmation of association between autism and the mitochondrial aspartate/glutamate carrier SLC25A12 gene on chromosome 2q31. American Journal of Psychiatry. 2005;162(11):2182–2184. doi: 10.1176/appi.ajp.162.11.2182. [DOI] [PubMed] [Google Scholar]
- 12.Palmieri L, Papaleo V, Porcelli V, et al. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. doi: 10.1038/mp.2008.63. Molecular Psychiatry. In press. [DOI] [PubMed] [Google Scholar]
- 13.Kayser MA. Inherited metabolic diseases in neurodevelopmental and neurobehavioral disorders. Seminars in Pediatric Neurology. 2008;15(3):127–131. doi: 10.1016/j.spen.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Canitano R. Epilepsy in autism spectrum disorders. European Child and Adolescent Psychiatry. 2007;16(1):61–66. doi: 10.1007/s00787-006-0563-2. [DOI] [PubMed] [Google Scholar]
- 15.Holmes GL. Influence of brain development on status epilepticus. Epilepsia. 2007;48(supplement 8):19–20. doi: 10.1111/j.1528-1167.2007.01339.x. [DOI] [PubMed] [Google Scholar]
- 16.Smith MC, Hoeppner TJ. Epileptic encephalopathy of late childhood: Landau-Kleffner syndrome and the syndrome of continuous spikes and waves during slow-wave sleep. Journal of Clinical Neurophysiology. 2003;20(6):462–472. doi: 10.1097/00004691-200311000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Krey JF, Dolmetsch RE. Molecular mechanisms of autism: a possible role for Ca2+ signaling. Current Opinion in Neurobiology. 2007;17(1):112–119. doi: 10.1016/j.conb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Herman GE, Henninger N, Ratliff-Schaub K, Pastore M, Fitzgerald S, McBride KL. Genetic testing in autism: how much is enough? Genetics in Medicine. 2007;9(5):268–274. doi: 10.1097/gim.0b013e31804d683b. [DOI] [PubMed] [Google Scholar]
- 19.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bureau I, Shepherd GMG, Svoboda K. Circuit and plasticity defects in the developing somatosensory cortex of Fmr1 knock-out mice. Journal of Neuroscience. 2008;28(20):5178–5188. doi: 10.1523/JNEUROSCI.1076-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selby L, Zhang C, Sun Q-Q. Major defects in neocortical GABAergic inhibitory circuits in mice lacking the fragile X mental retardation protein. Neuroscience Letters. 2007;412(3):227–232. doi: 10.1016/j.neulet.2006.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson JR, Bartley AF, Hays SA, Huber KM. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. Journal of Neurophysiology. 2008;100(5):2615–2626. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickett J, London E. The neuropathology of autism: a review. Journal of Neuropathology and Experimental Neurology. 2005;64(11):925–935. doi: 10.1097/01.jnen.0000186921.42592.6c. [DOI] [PubMed] [Google Scholar]
- 24.Napolioni V, Moavero R, Curatolo P. Recent advances in neurobiology of tuberous sclerosis complex. Brain and Development. 2009;31(2):104–113. doi: 10.1016/j.braindev.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nature Neuroscience. 2005;8(12):1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- 26.Vorstman JAS, Staal WG, Van Daalen E, Van Engeland H, Hochstenbach PFR, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Molecular Psychiatry. 2006;11(1):18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 27.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. American Journal of Human Genetics. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baieli S, Pavone L, Meli C, Fiumara A, Coleman M. Autism and phenylketonuria. Journal of Autism and Developmental Disorders. 2003;33(2):201–204. doi: 10.1023/a:1022999712639. [DOI] [PubMed] [Google Scholar]
- 29.Lowe TL, Tanaka K, Seashore M. Detection of phenylketonuria in autistic and psychotic children. Journal of the American Medical Association. 1980;243(2):126–128. doi: 10.1001/jama.1980.03300280024022. [DOI] [PubMed] [Google Scholar]
- 30.Ciardo F, Salerno C, Curatolo P. Neurologic aspects of adenylosuccinate lyase deficiency. Journal of Child Neurology. 2001;16(5):301–308. doi: 10.1177/088307380101600501. [DOI] [PubMed] [Google Scholar]
- 31.Sikora DM, Pettit-Kekel K, Penfield J, Merkens LS, Steiner RD. The near universal presence of autism spectrum disorders in children with Smith-Lemli-Opitz syndrome. American Journal of Medical Genetics Part A. 2006;140(14):1511–1518. doi: 10.1002/ajmg.a.31294. [DOI] [PubMed] [Google Scholar]
- 32.Jira PE, Wevers RA, de Jong J, et al. Simvastatin: a new therapeutic approach for Smith-Lemli-Opitz syndrome. Journal of Lipid Research. 2000;41(8):1339–1346. [PubMed] [Google Scholar]
- 33.Arias-Dimas A, Vilaseca MA, Artuch Iriberri R, Ribes A, Campistol J. Diagnosis and treatment of brain creatine deficiency syndromes. Revista de Neurologia. 2006;43(5):302–308. [PubMed] [Google Scholar]
- 34.Dykens EM, Sutcliffe JS, Levitt P. Autism and 15Q11-Q13 disorders: behavioral, genetic, and pathophysiological issues. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10(4):284–291. doi: 10.1002/mrdd.20042. [DOI] [PubMed] [Google Scholar]
- 35.Shao Y, Cuccaro ML, Hauser ER, et al. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. American Journal of Human Genetics. 2003;72(3):539–548. doi: 10.1086/367846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma DQ, Whitehead PL, Menold MM, et al. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. American Journal of Human Genetics. 2005;77(3):377–388. doi: 10.1086/433195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palferman S, Matthews N, Turner M, et al. Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Human Molecular Genetics. 2001;10(9):973–982. doi: 10.1093/hmg/10.9.973. [DOI] [PubMed] [Google Scholar]
- 38.Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. International Journal of Developmental Neuroscience. 2007;25(2):69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Hong SE, Shugart YY, Huang DT, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nature Genetics. 2000;26(1):93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 40.Fatemi SH, Snow AV, Stary JM, et al. Reelin signaling is impaired in autism. Biological Psychiatry. 2005;57(7):777–787. doi: 10.1016/j.biopsych.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Quattrocchi CC, Wannenes F, Persico AM, et al. Reelin is a serine protease of the extracellular matrix. The Journal of Biological Chemistry. 2002;277(1):303–309. doi: 10.1074/jbc.M106996200. [DOI] [PubMed] [Google Scholar]
- 42.Campbell DB, Sutcliffe JS, Ebert PJ, et al. A genetic variant that disrupts MET transcription is associated with autism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(45):16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer-Lindenberg A, Mervis CB, Faith Berman K. Neural mechanisms in Williams syndrome: a unique window to genetic influences on cognition and behaviour. Nature Reviews Neuroscience. 2006;7(5):380–393. doi: 10.1038/nrn1906. [DOI] [PubMed] [Google Scholar]
- 44.Kirchhoff M, Bisgaard A-M, Bryndorf T, Gerdes T. MLPA analysis for a panel of syndromes with mental retardation reveals imbalances in 5.8% of patients with mental retardation and dysmorphic features, including duplications of the Sotos syndrome and Williams-Beuren syndrome regions. European Journal of Medical Genetics. 2007;50(1):33–42. doi: 10.1016/j.ejmg.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Edelmann L, Prosnitz A, Pardo S, et al. An atypical deletion of the Williams-Beuren syndrome interval implicates genes associated with defective visuospatial processing and autism. Journal of Medical Genetics. 2007;44(2):136–143. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England Journal of Medicine. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 47.Ballif BC, Hornor SA, Jenkins E, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nature Genetics. 2007;39(9):1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 48.Finelli P, Natacci F, Bonati MT, et al. FISH characterisation of an identical (16)(p11.2p12.2) tandem duplication in two unrelated patients with autistic behaviour. Journal of Medical Genetics. 2004;41(7):p. e90. doi: 10.1136/jmg.2003.016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philippi A, Roschmann E, Tores F, et al. Haplotypes in the gene encoding protein kinase c-beta (PRKCB1) on chromosome 16 are associated with autism. Molecular Psychiatry. 2005;10(10):950–960. doi: 10.1038/sj.mp.4001704. [DOI] [PubMed] [Google Scholar]
- 50.Lintas C, Sacco R, Garbett K, et al. Involvement of the PRKCB1 gene in autistic disorder: significant genetic association and reduced neocortical gene expression. Molecular Psychiatry. 2008:1–14. doi: 10.1038/mp.2008.21. [DOI] [PubMed] [Google Scholar]
- 51.Casas KA, Mononen TK, Mikail CN, et al. Chromosome 2q terminal deletion: report of 6 new patients and review of phenotype-breakpoint correlations in 66 individuals. American Journal of Medical Genetics Part A. 2004;130(4):331–339. doi: 10.1002/ajmg.a.30156. [DOI] [PubMed] [Google Scholar]
- 52.Gorski JL, Cox BA, Kyine M, Uhlmann W, Glover TW. Terminal deletion of the long arm of chromosome 2 in a mildly dysmorphic hypotonic infant with karyotype 46,XY,del(2)(q37) American Journal of Medical Genetics. 1989;32(3):350–352. doi: 10.1002/ajmg.1320320315. [DOI] [PubMed] [Google Scholar]
- 53.Galasso C, Lo-Castro A, Lalli C, Nardone AM, Gullotta F, Curatolo P. Deletion 2q37: an identifiable clinical syndrome with mental retardation and autism. Journal of Child Neurology. 2008;23(7):802–806. doi: 10.1177/0883073808314150. [DOI] [PubMed] [Google Scholar]
- 54.Wassink TH, Piven J, Vieland VJ, et al. Evaluation of the chromosome 2q37.3 Gene CENTG2 as an autism susceptibility gene. American Journal of Medical Genetics Part B. 2005;136(1):36–44. doi: 10.1002/ajmg.b.30180. [DOI] [PubMed] [Google Scholar]
- 55.Lo-Castro A, Giana G, Fichera M, et al. Deletion 2p25.2: a cryptic chromosome abnormality in a patient with autism and mental retardation detected using aCGH. European Journal of Medical Genetics. 2009;52(1):67–70. doi: 10.1016/j.ejmg.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Kaufmann WE, Cortell R, Kau ASM, et al. Autism spectrum disorder in fragile X syndrome: communication, social interaction, and specific behaviors. American Journal of Medical Genetics Part A. 2004;129(3):225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- 57.Hagerman RJ, Ono MY, Hagerman PJ. Recent advances in fragile X: a model for autism and neurodegeneration. Current Opinion in Psychiatry. 2005;18(5):490–496. doi: 10.1097/01.yco.0000179485.39520.b0. [DOI] [PubMed] [Google Scholar]
- 58.Folstein SE, Rosen-Sheidley B. Genetics of autism: complex etiology for a heterogeneous disorder. Nature Reviews Genetics. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- 59.Curatolo P, Porfirio MC, Manzi B, Seri S. Autism in tuberous sclerosis. European Journal of Paediatric Neurology. 2004;8(6):327–332. doi: 10.1016/j.ejpn.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 60.Dykens EM, Sutcliffe JS, Levitt P. Autism and 15Q11-Q13 disorders: behavioral, genetic, and pathophysiological issues. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10(4):284–291. doi: 10.1002/mrdd.20042. [DOI] [PubMed] [Google Scholar]
- 61.Kumar RA, Karamohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Human Molecular Genetics. 2008;17(4):628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 62.Manning MA, Cassidy SB, Clericuzio C, et al. Terminal 22q deletion syndrome: a newly recognized cause of speech and language disability in the autism spectrum. Pediatrics. 2004;114(2):451–457. doi: 10.1542/peds.114.2.451. [DOI] [PubMed] [Google Scholar]
- 63.Mukaddes NM, Herguner S. Autistic disorder and 22q11.2 duplication. World Journal of Biological Psychiatry. 2007;8(2):127–130. doi: 10.1080/15622970601026701. [DOI] [PubMed] [Google Scholar]
- 64.Lukusa T, Vermeesch JR, Holvoet M, Fryns JP, Devriendt K. Deletion 2q37.3 and autism: molecular cytogenetic mapping of the candidate region for autistic disorder. Genetic Counseling. 2004;15(3):293–301. [PubMed] [Google Scholar]
- 65.Alvarez Retuerto AI, Cantor RM, Gleeson JG, et al. Association of common variants in the Joubert syndrome gene (AHI1) with autism. Human Molecular Genetics. 2008;17(24):3887–3896. doi: 10.1093/hmg/ddn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Splawski I, et al. Ca (V) 1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 67.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. The New England Journal of Medicine. 2006;354(13):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 68.Alarcón M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. American Journal of Human Genetics. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. International Journal of Developmental Neuroscience. 2005;23(2-3):183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathology. 2007;17(4):434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature Reviews Genetics. 2008;9(5):341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Battaglia A, Bonaglia MC. The yield of subtelomeric FISH analysis in the evaluation of autistic spectrum disorders. American Journal of Medical Genetics Part C. 2006;142(1):8–12. doi: 10.1002/ajmg.c.30077. [DOI] [PubMed] [Google Scholar]
- 73.Schaefer GB, Mendelsohn NJ. Genetics evaluation for the etiologic diagnosis of autism spectrum disorders. Genetics in Medicine. 2008;10(1):4–12. doi: 10.1097/GIM.0b013e31815efdd7. [DOI] [PubMed] [Google Scholar]
- 74.Lintas C, Persico AM. Autistic phenotypes and genetic testing: state-of-the-art for the clinical geneticist. Journal of Medical Genetics. 2009;46(1):1–8. doi: 10.1136/jmg.2008.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christian SL, Brune CW, Sudi J, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biological Psychiatry. 2008;63(12):1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. American Journal of Human Genetics. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genetics. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lawson-Yuen A, Saldivar J-S, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. European Journal of Human Genetics. 2008;16(5):614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- 80.Yan J, Oliveira G, Coutinho A, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Molecular Psychiatry. 2005;10(4):329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- 81.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. American Journal of Human Genetics. 2007;81(6):1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature Genetics. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levitt P, Campbell DB. The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. The Journal of Clinical Investigation. 2009;119(4):747–754. doi: 10.1172/JCI37934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zappella M, Meloni I, Longo I, et al. Study of MECP2 gene in Rett syndrome variants and autistic girls. American Journal of Medical Genetics Part B. 2003;119(1):102–107. doi: 10.1002/ajmg.b.10070. [DOI] [PubMed] [Google Scholar]
- 85.Bosley TM, Salih MA, Alorainy IA, et al. Clinical characterization of the HOXA1 syndrome BSAS variant. Neurology. 2007;69(12):1245–1253. doi: 10.1212/01.wnl.0000276947.59704.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buxbaum JD, Cai G, Chaste P, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. American Journal of Medical Genetics Part B. 2007;144(4):484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herman GE, Butter E, Enrile B, Pastore M, Prior TW, Sommer A. Increasing knowledge of PTEN germline mutations: two additional patients with autism and macrocephaly. American Journal of Medical Genetics Part A. 2007;143(6):589–593. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]