Abstract
Background:
This study investigated the potential clinical utility of circulating free DNA (cfDNA) as a source of BRAF mutation detection in patients enrolled into a phase II study of AZD6244, a specific MEK1/2 inhibitor, in patients with advanced melanoma.
Methods:
BRAF mutations were detected using Amplification Refractory Mutation System allele-specific PCR. BRAF mutation status was assessed in serum-derived cfDNA from 126 patients enrolled into the study and from 94 matched tumour samples.
Results:
Of 94 tumour samples, 45 (47.9%) were found to be BRAF mutation positive (BRAF+). Serum-derived cfDNA was BRAF+ in 33 of 126 (26.2%) samples, including in five samples for which tumour data were unavailable. Of BRAF+ tumours, 25 of 45 (55.6%) were BRAF+ in cfDNA. In three cases in which the tumour was negative, cfDNA was BRAF+. Progression-free survival (PFS) of patients with BRAF+ tumour and cfDNA was not significantly different compared with tumour BRAF+ but cfDNA BRAF-negative patients, indicating that cfDNA BRAF detection is not associated with poorer prognosis on PFS in stage III/IV advanced melanoma.
Conclusions:
These data demonstrate the feasibility of BRAF mutation detection in cfDNA of patients with advanced melanoma. Future studies should aim to incorporate BRAF mutation testing in cfDNA to further validate this biomarker for patient selection.
Keywords: AZD6244, amplification refractory mutation system, BRAF , circulating free DNA, cutaneous melanoma
Somatic mutations in the BRAF oncogene have been documented at a high frequency in cutaneous melanomas, occurring in up to 60% of cell lines and tumour samples (Brose et al, 2002; Davies et al, 2002). The most documented BRAF mutation is a thymidine-to-adenine switch at nucleotide position 1799 in exon 15 (Davies et al, 2002). This encodes for a valine-to-glutamic acid substitution at amino acid 600 in the kinase domain of the protein (p.V600E, previously known as p.V599E; Smalley and Herlyn, 2004). The resulting structural change mimics phosphorylated BRAF and has an elevated basal kinase activity substantially higher than that for wild-type BRAF, causing constitutive activation of downstream signalling in the Ras–Raf–MEK–extracellular ligand-regulated kinase pathway, which can lead to malignant transformation (Mansour et al, 1994).
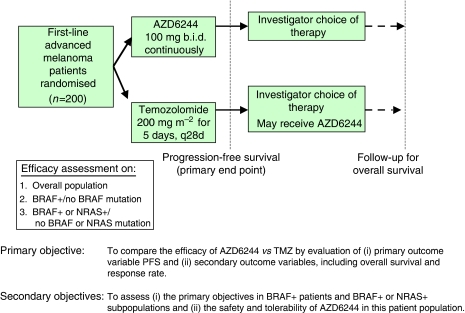
AZD6244 is an orally available, potent, selective, ATP-uncompetitive inhibitor of MEK1/2, with pre-clinical activity in BRAF mutation-positive (BRAF+) and RAS mutation-positive tumour models (Yeh et al, 2007). Clinical responses were observed in a recent 200-patient phase II study of AZD6244 vs temozolomide in patients with advanced melanoma (study D1532C00003, Figure 1; Dummer et al, 2008). Six patients randomised to AZD6244 had a confirmed partial response, of whom five had BRAF+ tumours. Nine patients randomised to temozolomide had a confirmed partial response, three of whom had BRAF+ tumours. In subsequent clinical trials of this drug, patients will be screened and selected on the basis of the mutational status of their tumour. In particular, future clinical trials of AZD6244 in advanced melanoma will evaluate patient selection on the basis of tumour mutation status.
Figure 1.
Melanoma phase II study design. Abbreviations: BRAF+, BRAF mutation positive; NRAS+, NRAS mutation positive; PFS, progression-free survival; TMZ, temozolomide.
Mutation status is traditionally assessed by analysis of DNA extracted from archival tumour tissue samples. However, biopsy material is not always readily available, even in patients with metastatic melanoma. Furthermore, limited and degraded amounts of DNA extracted from tumour biopsies and formalin-fixed paraffin-embedded (FFPE) tissues present an inherent technical challenge in mutation detection. If the response to a therapeutic agent depends on the presence or absence of a particular DNA mutation, then the availability of tumour-derived DNA for mutation analysis becomes critically important.
An alternative source of tumour-derived DNA is cell-free or circulating free DNA (cfDNA). This can be extracted from plasma and serum, providing an opportunity to develop a less-invasive and more accessible source of tumour DNA for mutation detection. Previous studies have demonstrated the feasibility of detecting tumour-specific mutations in cfDNA from patients with cancer, including detection of epidermal growth factor receptor mutations in patients with non-small-cell lung cancer (Kimura et al, 2006, 2007) and KRAS mutations in patients with pancreatic and colorectal cancers (Sorenson, 2000). More recently, BRAF mutations have been detected in cfDNA of patients with melanoma (Daniotti et al, 2007; Shinozaki et al, 2007; Yancovitz et al, 2007). However, these initial studies included a small number of patients with only limited numbers of matched tumour samples.
In this study, we investigated the potential utility of cfDNA for the detection of BRAF mutation status in a large group of patients enrolled into the AZD6244 advanced melanoma phase II study to determine whether cfDNA mutations could be used for patient selection as an alternative to tissue biopsies.
Materials and methods
Patients and samples
A total of 200 patients with advanced melanoma were enrolled into study D1532C00003. The study was conducted according to Good Clinical Practice and the Declaration of Helsinki. All patients provided written informed consent before participation in the main study. Consent for analysis of tumour biopsy material was obtained from all patients enrolled in the study and additional voluntary consent for collection of serum samples for genetic analysis was given by 126 patients.
Blood processing for cfDNA extraction
At the time of enrolment, 8.5 ml of peripheral blood was taken in a Becton-Dickinson Vacutainer Serum Collection Tube and gently inverted for a minimum of five times to ensure a thorough mixing of the sample. The blood was allowed to clot for 30 min and was centrifuged at 2000 g for 10 min. The resultant serum supernatant was transferred to a clean tube and stored at −80°C until analysis.
For cfDNA extraction, serum was thawed at room temperature and cfDNA was extracted from 1 ml of serum using a QIAamp MinElute Virus Spin Kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions, with the following modifications: to each 1 ml sample of serum, 3 μg tRNA (Sigma, Stockholm, Sweden), 125 μl kit proteinase and 1 ml kit lysis buffer were added. After a 1-h incubation, 1250 μl of 100% (v/v) ethanol was added. Each sample was filtered through the MinElute column in aliquots until the sample was exhausted. Following standard kit wash procedures (as per the manufacturer's instructions), the DNA was eluted twice in 50 μl elution buffer.
Germ line DNA extraction
A 9-ml blood sample was collected in a polypropylene tube containing EDTA and gently inverted for a minimum of five times to mix the sample thoroughly. Genomic DNA was extracted using the Nucleon DNA extraction procedure (Tepnel Life Sciences, Manchester, UK).
Tumour DNA extraction
For analysis of tumour samples, haemotoxylin- and eosin-stained slides were reviewed by a pathologist to ensure sufficient viable tumour (as defined by the presence of at least 100 melanoma cells). DNA was extracted from 40-μm unstained sections of FFPE samples by digesting the samples in proteinase K (Fluka, Buchs, Switzerland) for 48 h, boiling in 5% chelex (Sigma-Aldrich, Dorset, UK) and phase extracting through chloroform-and-ethanol precipitation (Coombs et al, 1999). The DNA was re-suspended by adding 50 μl of 0.1 M TE buffer and was then stored at –20°C until use.
Mutation detection
BRAF mutations were detected using a new Amplification Refractory Mutation System (ARMS) allele-specific PCR with Taqman probe assay designed at AstraZeneca (Cheshire, UK) using in-house software (Newton et al, 1989). The primer and probe sequences used are shown in Table 1. ARMS primers were designed to detect mutations at amino-acid 600 within the BRAF gene. The assay can detect p.V600E, p.V600K and p.V600D mutations within the BRAF gene, but does not distinguish between them. Control primers were designed to amplify an area of the BRAF gene with no known mutations or single-nucleotide polymorphisms. Primer and probe sequences were modified for the analysis of cfDNA to allow amplification of smaller PCR products.
Table 1. Primer and probe sequences.
| ARMS assay | ARMS primer (5′-3′) | Common primer (5′-3′) | TaqMan probe (5′-3′) | Size (bp) |
|---|---|---|---|---|
| BRAF for FFPE 1799T>A | AAAAATAGGTGATTTTGGTCTA GCTACATA | TAGTTGAGACCTTCAATGACTTT CTAGTAA | Yakima Yellow –AATCTCGATGGAGT GGGTCCCATCAGTTTGAACA-Bhq | 179 |
| Control for FFPE | AGGACACCGAGGAAGAG GACTT | GGAATCACCTTCTGTCTTCATTT | Cy – CCATCTTCTTCCTGCCTGATGA GGGGAAA-Elle | 252 |
| BRAF for cfDNA | AAAAATAGGTGATTTTGGTCTA GCTACATA | CATCCACAAAATGGATCC AGACAA | Yakima Yellow – GATGGA+GTGGGTC+ CCATC+AG-Bhq | 91 |
| Control for cfDNA | CTCCAGATCTCAGTAAGG TACGG | GGGAAAGAGTGGTCTCTCATC | Cy5 – CATGA+AG+AGATTAAT GGCAG+AGTG+CC-Elle | 101 |
Abbreviations: ARMS=Amplification Refractory Mutation System; cfDNA=circulating free DNA; FFPE=formalin-fixed paraffin embedded tissues.
Each reaction was carried out in a 25-μl reaction volume containing 1 × Brilliant II PCR mix (Stratagene, Cedar Creek, TX, USA), 2 μM each of BRAF ARMS primer and reverse primer, 0.5 μm BRAF probe, 0.1 μm each of control forward and reverse primers, 0.2 μM control probe and 0.8 mg ml−1 bovine serum albumin. A 5-μl aliquot of DNA template was added to each reaction. The reactions were amplified on a Stratagene Mx3000P under the following conditions: 95°C for 10 min, followed by 50 cycles of 94°C for 45 s, 60°C for 1 min and 72°C for 45 s. In all cases, samples were assessed in duplicate.
Data were interpreted as follows: if only the control reaction occurred with no mutant reaction, the sample was classified as wild type; if neither reaction occurred, then the sample was classified as unknown, as the concentration of DNA was below the limit of detection; if the mutant reaction occurred, the sample was classified as mutant only if the reaction ΔCt between control and mutant reaction was smaller than the ΔCt for each of the control wild-type standards on the run to ensure that the mutant reaction was not simply a nonspecific signal (where ΔCt is defined as the difference between threshold cycles (mutation Ct−control Ct)). If there was discordance between the replicates or if the ΔCt was within 1 Ct of the ΔCt cutoff, then the experiment for the sample was repeated in triplicate, and the sample was considered positive only if all three replicates were positive.
Positive cell line controls were created using DNA extracted from the HT29 cell line, known to be heterozygous for the p.V600E mutation. Human genomic DNA (Roche, Basel, Switzerland) was used as a non-mutant-DNA-containing negative control and appropriate reagent control was used in all PCR runs.
All FFPE-extracted DNA samples found to be positive for the BRAF mutant by ARMS were sequenced to determine the exact nucleic acid change.
Sequencing reactions for tumour DNA
Tumour DNA was added to duplicate PCR assays containing primers that amplified BRAF exon 15 (forward primer: 5′-TTTCCTTTACTTACTACACCTC-3′; reverse primer: 5′-CTTTCTAGTAACTCAGCAGCATC-3′). The resulting PCR products were sequenced in forward and reverse directions using ABI BigDye sequencing and analysed using SeqScape (Foster City, CA, USA). A mutation result was accepted if it was present in both forward and reverse sequencing traces, and in duplicate PCRs (to eliminate false-positive mutations occurring because of sample fixation artefacts).
Cloning and sequencing for BRAF mutations
To confirm the presence of BRAF mutations in cfDNA from samples in which cfDNA was BRAF+ but the matched tumour sample was negative for a mutation by ARMS, cfDNA was extracted from 1 ml of serum and cloned and sequenced for the presence of BRAF mutations. Cloning was performed using the TOPO TA Cloning kit (Invitrogen, Paisley, UK) (containing pCR 2.1-TOPO) with chemically competent Escherichia coli strain TOP 10F’ (Invitrogen, Paisley, UK). PCR products containing the BRAF sequence were obtained using the same primer sequences and conditions as those used for exon 15 BRAF sequencing as described above. A mutation result was accepted if it was present in both forward and reverse sequencing traces.
Reproducibility of BRAF detection in cfDNA over 1 year
The reproducibility of BRAF detection in cfDNA was tested in 24 serum samples stored at –80°C for 6 months, 13 of which were positive for BRAF mutations on initial sampling. A separate set of 24 serum samples stored at −80°C for 12 months were re-tested for BRAF mutations, 17 of which were positive for BRAF mutations on initial sampling.
The reproducibility of BRAF detection in cfDNA stored at −20°C for 6 months was tested on 26 samples, 17 of which had tested positive for BRAF mutations at the initial analysis. The reproducibility of BRAF detection in cfDNA stored at −20°C for 12 months was tested on a further set of 24 samples, 16 of which had tested positive for BRAF mutations at the initial analysis.
Biostatistical analysis
The primary end point in study D1532C00003 was PFS and, similar to the primary analysis for this study, a multivariate analysis of PFS was carried out for those patients with serum results, using the Cox proportional hazards model allowing for the effect of treatment and adjusting for the following covariates: lactate dehydrogenase (LDH; ⩾2 × upper limit of normal (ULN) vs <2 × ULN); BRAF mutational status by cfDNA; World Health Organization (WHO) performance status (WHO PS; 0 vs 1, 2) and tumour type (uveal vs non-uveal).
To assess whether allowing cfDNA-detected BRAF+ patients into a selected trial would result in the study population being enriched for patients with differing prognoses from the main study population, analyses were carried out using patients who were BRAF+ by tumour but with serum results also available. A univariate analysis was carried out to compare PFS between patients who were BRAF+ in serum and patients in whom BRAF mutations were not detected. In addition, summary 2 × 2 tables were produced to assess a potential correlation between BRAF+, as detected by cfDNA, and the known prognostic factor LDH. LDH levels were available for 190 of the 200 patients enrolled into the study.
Results
Assessment of BRAF assay sensitivity
Using the cell line HT29 (known to be heterozygous for the p.V600E mutation), several serial dilution studies of HT29 DNA in human genomic DNA were performed to determine the sensitivity of the BRAF ARMS assay. The BRAF mutant could be detected at a level as low as 5 copies of HT29 DNA in a background of 5000 copies of wild-type DNA (0.1% sensitivity, data not shown).
BRAF p.V600 mutation detection in clinical samples
Of the 200 patients enrolled in the trial, 176 tumour samples were obtained; 163 samples were FFPE and the remaining 13 were fresh frozen specimens. Of the 176 tumour samples analysed, 158 (90%) generated acceptable ARMS results. DNA sequence data for BRAF were obtained for 147 (84%) tumour samples. In total, 70 BRAF mutations in tumour DNA were identified by ARMS (70 of 158 (44%)). Of the BRAF mutations detected by ARMS, five were determined by sequencing to be complex g.1798-1799GT>AA changes resulting in a BRAF p.V600K alteration, rather than the more common p.V600E (g.1799T>A). Sequencing detected two samples with additional mutation types that could not be captured using the specific ARMS assays in this study: BRAF g.1742A>T (p.N581S) and g.1801A>G (p.K601E). Eighteen mutations were detected by ARMS but failed DNA sequencing because of low DNA yields, indicating that ARMS is the more robust method, particularly for analysis of DNA extracted from FFPE specimens; although confined to detecting known mutations.
Of the 96 tumour samples available from patients with cfDNA data, 45 (47%) were detected to be BRAF+ by ARMS. Sequencing had confirmed these mutations to be p.V600E in 42 cases and p.V600K in 3 cases. A further tumour sample was shown to harbour a p.K601E mutation, which was not detectable by the ARMS assay design.
Serum samples were available for 126 of the 200 patients enrolled in study D1532C00003; cfDNA was extracted from samples as described and analysed for the presence of a BRAF mutation (results summarised in Table 2).
Table 2. Summary of BRAF mutation analysis of tumour and cfDNA in n=126 patients.
|
Tumour DNA
|
|||||
|---|---|---|---|---|---|
| BRAF positive | BRAF negative | BRAF unknown | Total | ||
| cfDNA | BRAF positive | 25 | 3 | 5 | 33 |
| BRAF negative | 20 | 46 | 27 | 93 | |
| Total | 45 | 49 | 32 | 126 | |
Abbreviation: cfDNA=circulating free DNA.
In total, 33 (26%) BRAF mutations were detected in cfDNA by ARMS. Of the 126 patients with serum samples, 96 had matched tumour data available. For the remaining 32 patients, tumour data were unavailable either because there was no available tumour sample (n=20) or because analysis had failed because of insufficient DNA extracted from the tumour sample (n=10).
Five cfDNA samples were positive for a BRAF mutation in which no tumour data were available. Of the BRAF+ tumours, 25 out of 45 (56%) had BRAF mutations detected in the serum. In three samples, BRAF mutations were identified in the serum but the tumour was BRAF mutation negative. For each of these samples, cfDNA was extracted from a further 1 ml of serum for repeat analysis; in all samples, a BRAF mutation was confirmed. In two of these cases, sufficient serum was available to allow extraction of cfDNA from a further 1 ml of serum, and the resultant cfDNA was cloned and sequenced for the presence of BRAF mutations. In both these cases, BRAF mutations were confirmed in these samples with 13 and 7% of clones positive for a mutation.
In total, of the 96 cases with matched tumour and cfDNA data, the concordance in BRAF mutation detection was 76% (95% confidence interval 66–84%). If a BRAF mutation was present in tumour DNA, the pick up rate in cfDNA was 56% (95% confidence interval 40–70%).
Importantly, in all samples, analysis of germ line DNA by ARMS was negative for BRAF mutations, confirming that any BRAF mutations detected were tumour derived.
Reproducibility
The reproducibility of BRAF detection in cfDNA was tested in 24 serum samples stored at −80°C for 6 months and a further 24 serum samples stored at −80°C for 12 months. All serum samples analysed after 6 months storage yielded BRAF mutation results identical to the initial analysis. After storage for 12 months, 21 of the 24 serum samples yielded BRAF mutation results identical to the initial analysis. In two samples, the BRAF mutation was no longer detected and, in one sample, a BRAF mutation was detected when initial analysis had been negative. In all of these cases, the tumour sample had been positive for a BRAF mutation.
The reproducibility of BRAF detection in cfDNA stored at −20°C for 6 months was tested on 26 samples, 17 of which had tested positive for BRAF mutations at the initial analysis. At repeat analysis, 16 of the 17 samples that had previously been found to be positive were still BRAF+. The one negative sample had previously been positive with a high ΔCt, suggesting low levels of BRAF mutations within the sample. This patient was known to have a BRAF+ tumour. A further sample tested positive for a BRAF mutation when previously it had tested negative. Again, the ΔCt of this sample was high, suggesting low levels of mutant BRAF within the sample.
A similar result was observed after analysis of 24 DNA samples stored for 12 months at −20°C. Of the 16 samples previously BRAF+, all were BRAF+ after 12 months. A further sample was positive for a BRAF mutation in which initial analysis had been negative with a high ΔCt; this sample was from a patient known to have a BRAF+ tumour. These data imply that in some samples the level of BRAF mutations is very low and sampling differences during analysis could explain the discordant results.
cfDNA as a prognostic indicator
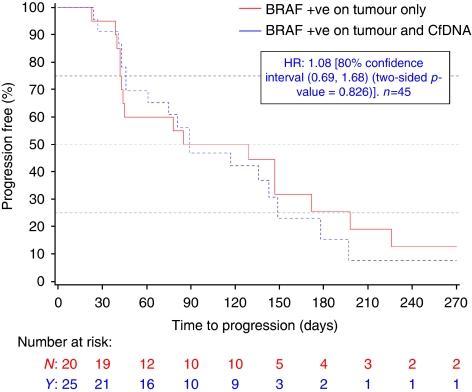
The PFS of the 126 patients with cfDNA results did not differ significantly from the PFS of the study D1532C00003 population as a whole (data not shown). BRAF status by tumour sample (in the primary analysis of 200 patients) or cfDNA (n=126) was not shown to be a prognostic factor for PFS (data not shown). In addition, in those patients with BRAF+ tumours, in whom serum was available for analysis, there was no difference in PFS between patients in whom BRAF mutations could be detected in the serum compared with those patients in whom cfDNA BRAF mutations were not detected (n=45, HR 1.08, 80% confidence interval 0.69, 1.68, two-sided p value=0.826, Figure 2). This suggests that the presence of detectable mutant BRAF in serum of patients with BRAF+ tumours is not associated with a poorer prognosis on the basis of PFS compared with patients with BRAF+ tumours in whom BRAF mutations are not detected in cfDNA .
Figure 2.
The presence of circulating BRAF mutations does not effect progression-free survival. There was no difference in PFS between patients in whom BRAF mutations could be detected in the serum compared with those patients in whom cfDNA BRAF mutations were not detected. HR calculated using an unadjusted Cox proportional hazards model. Abbreviations: BRAF+, BRAF mutation positive; cfDNA, cell-free DNA; HR, hazard ratio; CI, confidence interval.
LDH levels were available for 190 patients enrolled in study D1532C00003. Consistent with previous literature, ULN had a worse prognosis on PFS for patients in the study with an LDH level greater than 2x than it did for patients with lower LDH levels. The proportion of patients with BRAF mutations in the serum was greater in the elevated LDH group compared with that of patients in the study as a whole (24 vs 16%; Table 3) However, if patients with high LDH are excluded from future trials, preselecting on cfDNA BRAF serum status will not enrich for poor prognosis patients (as defined by LDH> 2 × ULN).
Table 3. Correlation of BRAF mutations in cfDNA with poor prognostic factors (lactate dehydrogenase).
|
CfDNA patients (n=126); serum mutation results
|
Total study | ||
|---|---|---|---|
| LDH | BRAF positive (n=33) | BRAF negative (n=93) | population (n=200) |
| LDH<2XULN | 24 (73%) | 82 (88%) | 158 (79%) |
| LDH=2 × ULN | 8 (24%) | 8 (9%) | 32 (16%) |
| LDH unknown | 1 (3%) | 3 (3%) | 10 (5%) |
Abbreviations: cfDNA=circulating free DNA; LDH=lactate dehydrogenase; pts=patients; ULN=upper limit of normal.
Discussion
This study has demonstrated the detection of BRAF mutations in cfDNA extracted from the serum of patients with advanced melanoma enrolled in a phase II study of AZD6244 versus temozolomide. The concordance rate of cfDNA BRAF mutations with tumour BRAF mutations was 56%, which is consistent with that of other reports (Daniotti et al, 2007; Yancovitz et al, 2007).
Although other groups have demonstrated the feasibility of detecting BRAF mutations in serum and plasma of patients with melanoma (Daniotti et al, 2007; Shinozaki et al, 2007; Yancovitz et al, 2007), this is the first study that compares tumour and cfDNA results from a large cohort of patients and demonstrates the potential clinical application of cfDNA mutation detection for patient selection within clinical trials.
Yancovitz et al demonstrated BRAF mutations in cfDNA extracted from plasma of 14 of 26 (54%) stage IV melanoma patients (Yancovitz et al, 2007). Of 17 available tissue samples, the concordance of results was 10 of 17 (59%). Daniotti et al compared cfDNA and tumour BRAF mutations in 20 patients and found that cfDNA was positive for a BRAF mutation in 5 of 13 cases in which the tumour harboured a BRAF mutation (Daniotti et al, 2007). Shinozaki et al demonstrated BRAF mutations in 38 of 103 (37%) patients with melanoma (Shinozaki et al, 2007). However, they do not record any tumour data to assess concordance of their assay.
Our series identified three cases in which cfDNA was positive for a BRAF mutation but the tumour DNA was negative. Yancovitz et al and Daniotti et al both identified two patients in whom BRAF mutations were detected in cfDNA but not in tumour DNA (Daniotti et al, 2007; Yancovitz et al, 2007). Often, tumour BRAF status is derived from a primary lesion that occurred months or years earlier. In our study, the source of tumour material, whether primary or metastatic, was not captured. It is possible that the BRAF status of metastatic tumour is different compared with that of primary tumour. However, BRAF mutations are thought to develop early in the pathogenesis of melanomas, and analyses of a series of paired primary and metastatic lesions from the same patients indicate that BRAF mutations are preserved in metastases (Omholt et al, 2003). Heterogeneity in BRAF mutation status between metastatic sites may exist (Chang et al, 2004), and thus cfDNA might provide a more accurate representation of BRAF mutation status within a patient than a biopsy of any single lesion. In this regard, three samples in this study had BRAF mutations detected in the serum, wherein the tumour was BRAF mutation negative. In two of these samples, the DNA yield from the tumour sample was very low, and in the third sample, histological evaluation of the tumour sample revealed only small amounts of melanoma. Therefore, the difference in mutation results between tumour and cfDNA in these cases may be explained by the fact that the tumour DNA for these samples was not representative of the tumour as a whole. This again highlights the technical challenges in mutation detection in tumour DNA. In two of these three cases, there was sufficient remaining sample to be able to confirm the presence of BRAF mutations in cfDNA by cloning and sequencing. These data increase confidence that the BRAF mutation was indeed present in cfDNA and that the tumour results are either false negative due to sampling error or not reflective of the mutation status of the metastatic disease.
When considering the use of cfDNA mutation detection as an inclusion criterion for clinical trials, we needed to ascertain whether there was a different overall outcome in those patients with mutant cfDNA compared with those patients with tumour mutations but no cfDNA mutations. If this were the case, then enrolling patients on the basis of cfDNA results may enrich trials for patients with a differing prognosis. Our series has demonstrated that the prognosis by PFS of patients with BRAF+ tumours, in whom BRAF mutations can be detected in cfDNA, is not significantly different from that of patients with BRAF+ tumours in whom BRAF mutations cannot be detected in cfDNA. This increases our confidence that enrolling patients into clinical trials on the basis of cfDNA mutation results will not enrich our trial populations for cohorts of patients with an inherently worse prognosis.
Although this study has provided meaningful and interesting results, there are important limitations to this work that require acknowledgement and discussion. First, 191 of the 200 patients enrolled in study D1532C00003 had stage IV melanoma. It is suggested that the detection of cfDNA mutations is tumour-stage dependent, with decreased accuracy in earlier-stage patients. Daniotti et al reported BRAF-positive cfDNA results in 3 of 13 stage IV melanoma patients but in none of 4 stage I/II patients (Daniotti et al, 2007). This ‘stage-dependency’ may restrict cfDNA as a detection method in trials of agents for advanced disease, although more sensitive technologies may alter this in the future.
Second, it should be noted that in the D1532C00003 trial, cfDNA analysis was conducted using serum, which contains high levels of contaminating wild-type DNA sequences after white cell lyses in the clotting process, which may interfere with mutation detection sensitivity. The alternative, plasma, which is processed from blood collected in a tube containing an anticoagulant, contains fewer wild-type DNA sequences and is hence a more sensitive medium for cfDNA mutation detection (Daniotti et al, 2007). The use of serum in our study could have reduced the mutation detection rate in cfDNA and our future trials will collect plasma for this purpose.
Third, concern has been raised with regard to the reliability of cfDNA mutation detection in stored blood samples (Sozzi et al, 2005; Daniotti et al, 2007). Previous reports have demonstrated that cfDNA mutations are no longer detectable in previously positive serum and DNA samples after 12 months of storage (Daniotti et al, 2007). In this series, mutation detection was not found to be significantly affected by 6 months of storage of serum samples. However, after 12 months of storage of serum at −80°C, there were three discordant results. Discordant results were also noted when cfDNA extracted from serum was stored at –20°C for 6 or 12 months. In all of these cases, ΔCt for the mutation assay was high, suggesting that there was a low proportion of mutant DNA within those samples. This might explain the discordant results; simple sampling differences at the time of the BRAF assay may have affected detection. In this series, insufficient sample was available for extensive repeat testing, but these results raise some concern regarding the reliability of mutation detection in cfDNA with low levels of mutant DNA in the sample, especially in stored samples. Future studies will need to focus on repeated testing of initial samples to fully establish the reliability and precision of these tests. The use of plasma or more-sensitive technologies in future trials may reduce the number of samples with discordant results on repeated testing.
Finally, and importantly, not all patients with BRAF+ tumours had BRAF+ cfDNA. As discussed, it is possible that there are some differences in the mutation status between primary tumour and metastatic deposit, but the most likely explanations are that either the tumour cfDNA is shed at such a low level that it is below the sensitivity of the assay or there are true biological differences between tumours in different patients, indicating that cfDNA is shed at a higher level in some patients than in others. Recent reports from a study of cfDNA mutations in patients with operable colorectal cancer using an ultra-sensitive technology (BEAMing) suggest that mutated cfDNA fragments are present at levels as low as 0.18% (Diehl et al, 2008). Development of these increasingly sensitive technologies may improve detection rates in the future.
Although the exact source and nature of cfDNA remain unclear, the potential applications of its use are becoming increasingly apparent. It offers significant advantages over tumour DNA analysis: it is less invasive than tissue biopsies and results are quicker to obtain (particularly within clinical trial scenarios when tissue samples are often spread across many sites and countries). In addition, cfDNA can provide us with the opportunity to detect the current or real-time mutation status of a tumour and ultimately could lead to serial sampling to assess tumour progression or the development of resistant mutations. This study has demonstrated that cfDNA analysis could provide an opportunity for the detection of tumour-specific mutations in patients with advanced melanoma, allowing for speedy access to novel agents and enrolment into clinical studies without waiting for tissue procurement. This technology provides patients who have no available tissue samples with the opportunity to be considered for a study without having to undergo further invasive procedures to obtain tissue samples. The study has demonstrated that the detection of BRAF mutations in cfDNA is not significantly prognostic in advanced melanoma, and that, provided high LDH patients are excluded from the study population, entering patients by cfDNA analysis into a BRAF+-selected trial will not enrich for a poor prognostic study population. Further studies on AZD6244 and other targeted agents will focus on improving the tissue/cfDNA concordance rate and will aim to further validate cfDNA as a surrogate marker for tumour DNA mutations and as an inclusion criterion for clinical studies.
Acknowledgments
We thank all the study investigators and patients involved in study D1532C00003, and Dr Miriam Banner, from MediTech Media, who provided editing assistance funded by AstraZeneca. This work was supported by educational grants from AstraZeneca and Cancer Research UK.
Footnotes
Conflict of interest
Gillian Ellison, Maria CM Orr, Karin R Kemsley, Gael McWalter, Laura Y Blockley, Simon P Deardon, Clive Morris, Mireille Cantarini and Andrew Hughes are AstraZeneca employees who hold AstraZeneca shares. All other authors declare no conflict of interest.
References
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, Roth JA, Albelda SM, Davies H, Cox C, Brignell G, Stephens P, Futreal PA, Wooster R, Stratton MR, Weber BL (2002) BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 62: 6997–7000 [PubMed] [Google Scholar]
- Chang DZ, Panageas KS, Osman I, Polsky D, Busam K, Chapman PB (2004) Clinical significance of BRAF mutations in metastatic melanoma. J Transl Med 2: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs NJ, Gough AC, Primrose JN (1999) Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res 27: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniotti M, Vallacchi V, Rivoltini L, Patuzzo R, Santinami M, Arienti F, Cutolo G, Pierotti MA, Parmiani G, Rodolfo M (2007) Detection of mutated BRAFV600E variant in circulating DNA of stage III-IV melanoma patients. Int J Cancer 120: 2439–2444 [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA (2002) Mutations of the BRAF gene in human cancer. Nature 417: 949–954 [DOI] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz Jr LA (2008) Circulating mutant DNA to assess tumor dynamics. Nat Med 14: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dummer R, Robert C, Chapman PB, Sosman JA, Middleton M, Bastholt L, Kemsley K, Cantarini MV, Morris C, Kirkwood JM (2008) AZD6244 (ARRY-142886) vs temozolomide (TMZ) in patients (pts) with advanced melanoma: an open-label, randomized, multicenter, phase II study. J Clin Oncol 26 [Google Scholar]
- Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, Nishio K (2006) Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 12: 3915–3921 [DOI] [PubMed] [Google Scholar]
- Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, Koizumi F, Nishio K, Miyamoto K, Fujimura M, Nakao S (2007) Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 97: 778–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG (1994) Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265: 966–970 [DOI] [PubMed] [Google Scholar]
- Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omholt K, Platz A, Kanter L, Ringborg U, Hansson J (2003) NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res 9: 6483–6488 [PubMed] [Google Scholar]
- Shinozaki M, O′Day SJ, Kitago M, Amersi F, Kuo C, Kim J, Wang HJ, Hoon DS (2007) Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res 13: 2068–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley KS, Herlyn M (2004) Loitering with intent: new evidence for the role of BRAF mutations in the proliferation of melanocytic lesions. J Invest Dermatol 123: xvi–xvii [DOI] [PubMed] [Google Scholar]
- Sorenson GD (2000) Detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Clin Cancer Res 6: 2129–2137 [PubMed] [Google Scholar]
- Sozzi G, Roz L, Conte D, Mariani L, Andriani F, Verderio P, Pastorino U (2005) Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. J Natl Cancer Inst 97: 1848–1850 [DOI] [PubMed] [Google Scholar]
- Yancovitz M, Yoon J, Mikhail M, Gai W, Shapiro RL, Berman RS, Pavlick AC, Chapman PB, Osman I, Polsky D (2007) Detection of mutant BRAF alleles in the plasma of patients with metastatic melanoma. J Mol Diagn 9: 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, Parry J, Smith D, Brandhuber BJ, Gross S, Marlow A, Hurley B, Lyssikatos J, Lee PA, Winkler JD, Koch K, Wallace E (2007) Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res 13: 1576–1583 [DOI] [PubMed] [Google Scholar]