Summary
During breast cancer development, increased presence of leukocytes in neoplastic stroma parallels disease progression; however, the functional significance of leukocytes in regulating protumor versus antitumor immunity in the breast remains poorly understood. Utilizing the MMTV-PyMT model of mammary carcinogenesis, we demonstrate that IL-4-expressing CD4+ T lymphocytes indirectly promote invasion and subsequent metastasis of mammary adenocarcinomas by directly regulating the phenotype and effector function of tumor-associated CD11b+Gr1-F4/80+ macrophages that in turn enhance metastasis through activation of epidermal growth factor receptor signaling in malignant mammary epithelial cells. Together, these data indicate that antitumor acquired immune programs can be usurped in protumor microenvironments and instead promote malignancy by engaging cellular components of the innate immune system functionally involved in regulating epithelial cell behavior.
Introduction
Clinical and experimental studies have established that chronic infiltration of neoplastic tissue by leukocytes, i.e., chronic inflammation, promotes development and/or progression of various epithelial tumors (de Visser et al., 2006; Mantovani et al., 2008); however, the organ-specific cellular and molecular programs that favor protumor, as opposed to antitumor, immunity are incompletely understood. Although some subsets of leukocytes certainly exhibit antitumor activity, including cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells (Dunn et al., 2006), other leukocytes, most notably mast cells, B cells, dendritic cells, granulocytes, and macrophages, exhibit more bipolar roles, by virtue of their capacity to either hinder or potentiate tumor progression (de Visser et al., 2005; Mantovani et al., 2008).
Breast cancer development is characterized by significant increases in the presence of both innate and adaptive immune cells, with B cells, T cells, and macrophages representing the most abundant leukocytes present in neoplastic stroma (DeNardo and Coussens, 2007). Retrospective clinical studies in human breast cancer have revealed that high immunoglobulin (Ig) levels in tumor stoma (and serum), and increased presence of extra follicular B cells, T regulatory (Treg) cells, and high ratios of CD4/CD8 or TH2/TH1 T lymphocytes in primary tumors or in draining lymph nodes (LNs) correlate with tumor grade, stage, and overall patient survival (Bates et al., 2006; Coronella-Wood and Hersh, 2003; Kohrt et al., 2005); thus, some facets of adaptive immunity might indeed be involved in fostering cancer development in the breast.
However, experimental studies have demonstrated that macrophages in primary mammary adenocarcinomas regulate late-stage carcinogenesis by virtue of their proangiogenic properties (Lin and Pollard, 2007), as well as foster pulmonary metastasis by providing epidermal growth factor (EGF) to malignant mammary epithelial cells (MECs) and thereby enhancing their invasive (and metastatic) behavior (Pollard, 2004). Based on these seemingly disparate observations, we sought to determine whether adaptive immunity also fosters malignancy in the breast by regulating the phenotype or effector functions of tumor-associated macrophages (TAMs) and either activating their protumor properties or alternatively by suppressing their antitumor capabilities. To address this, we utilized an aggressive transgenic mouse model of murine mammary adenocarcinoma development (MMTV-PyMT mice) (Guy et al., 1992) where late-stage carcinogenesis and pulmonary metastasis are regulated by colony stimulating factor (CSF)-1 and tissue macrophages (Lin et al., 2001). We evaluated MMTV-PyMT mice harboring homozygous null mutations in genes regulating development of specific lymphocyte subtypes and found that CD4+ T cells potentiate pulmonary metastasis of mammary adenocarcinomas indirectly by enhancing aspects of protumor immunity mediated by TAMs.
Results
CD4+ T Cells Regulate Pulmonary Metastasis of Mammary Adenocarcinomas
As observed in several types of solid tumors, human breast adenocarcinomas are characterized by infiltration of both innate and adaptive immune cells (Figure 1A). Immunohistochemical (IHC) detection of CD68+ myeloid cells (macrophages), CD4+ and CD8+ T cells and CD20+ B cells in human breast cancer reveals an increase in each cell type paralleling cancer development (Figure 1A). Given the critical role of adaptive immunity in regulating innate immune cell effector function in chronic inflammatory diseases, and in some mouse models of cancer development (de Visser et al., 2005), we hypothesized that B and/or T lymphocytes might exert a functional role in regulating protumor properties of myeloid cells during mammary carcinogenesis. Because infiltration of CD4+ T cells and F4/80+ macrophages increases progressively during mammary carcinogenesis in MMTV-polyoma middle T (PyMT) mice (Figures 1B and 1C), similar to human breast cancer development (Figure 1A), we addressed this hypothesis by generating PyMT mice harboring homozygous null mutations in the recombinase activating gene-1 (RAG1) functionally impairing development of B and T cells, i.e., PyMT/RAG1−/−, and compared them for characteristics of neoplastic progression to PyMT mice lacking B cells, i.e., PyMT/JH−/−, versus selective subsets of T cells, i.e., PyMT/CD4−/−, PyMT/CD8−/− and PyMT/CD4−/−/CD8−/− mice. Strikingly, we found no gross histopathological or quantitative differences between these cohorts when evaluated for primary tumor latency, tumor burden, or tumor angiogenesis as a function of complete or selective lymphocyte deficiency (Figures 2A–2D; see Figure S1 available online). In contrast, selective loss of CD4+ T cells in either PyMT/RAG1−/−, PyMT/CD4−/−/CD8−/−, or PyMT/CD4−/−/CD8+/− mice resulted in significant attenuation of pulmonary metastasis including reduced numbers of metastatic foci, decreased tumor burden in lungs (Figures 2E and2F), decreased presence of circulating CD45-cytokeratin+ carcinoma cells (Figure 2G), and cells expressing PyMT mRNA (Figure 2H) in peripheral blood (PB). To verify that the attenuated metastatic phenotype of CD4-deficient/PyMT mice was specifically due to lack of CD4+ T cells, as opposed to a genetic anomaly in homozygous null mice, we depleted 85-day-old PyMT mice of CD4+ T cells for 25 days and again found reduced pulmonary metastasis (Figure 2I) and presence of circulating carcinoma cells in PB (Figure 2G). In contrast, adoptive transfer of naive CD4+ T lymphocytes into PyMT/CD4−/−/CD8−/− mice significantly enhanced pulmonary metastasis (Figure 2I), together indicating that CD4+ T lymphocytes mediate metastasis of late-stage malignant mammary epithelial cells in a CD8+ CTL-independent manner.
Figure 1. Concomitant Recruitment of Adaptive and Innate Immune Cells in Breast Cancers.
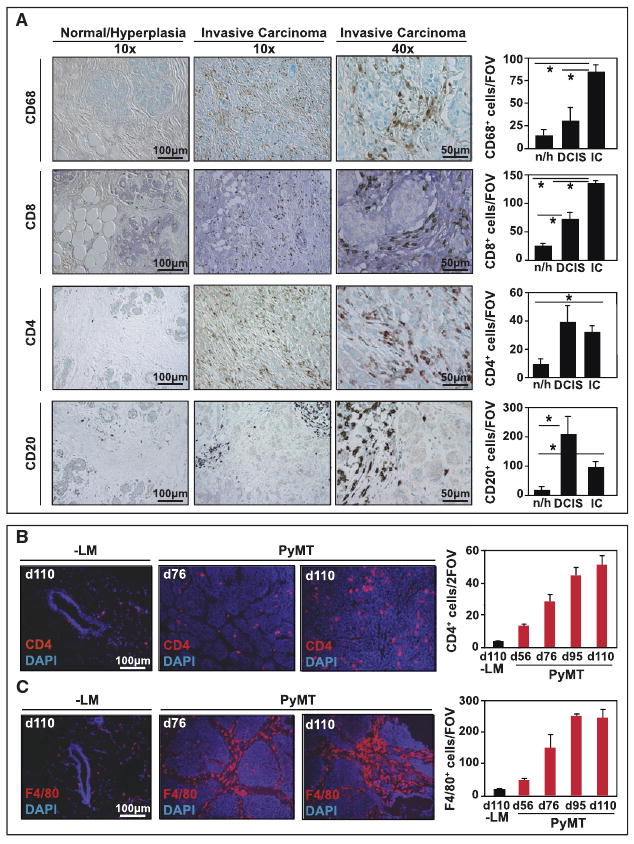
(A) The number of CD68+, CD20+, CD4+, and CD8+ cells was analyzed in patient samples of normal/hyperplastic breast tissue (n/h; n = 9), ductal carcinoma in situ (DCIS; n = 6), and invasive ductal carcinomas (IC; n = 150) using tissue microarrays. Representative 10× and 40× images are shown and the average number of positive cells as depicted reflects the mean number of cells in each disease stage, evaluated by counting all high power fields (20×) per tissue section (1.1 mm)/two sections/patient. *p < 0.05 by Mann-Whitney. (B, C) CD4+ and F4/80+ cell presence was evaluated during MMTV-PyMT mammary tumor development and is depicted by representative images in normal mammary tissue (-LM) and tumors from 76 and 110-day-old PyMT mice. CD4+ or F4/80+ cells were quantitatively assessed and data reflects the mean number of positive cell evaluated in 10 high-power fields (20×) per tumor, n = 4 mice per group.
Graphs are depicted as mean values and standard error of the mean (SEM) in all panels.
Figure 2. CD4+ T Cells Promote Metastasis, but Not Primary Tumor Development.
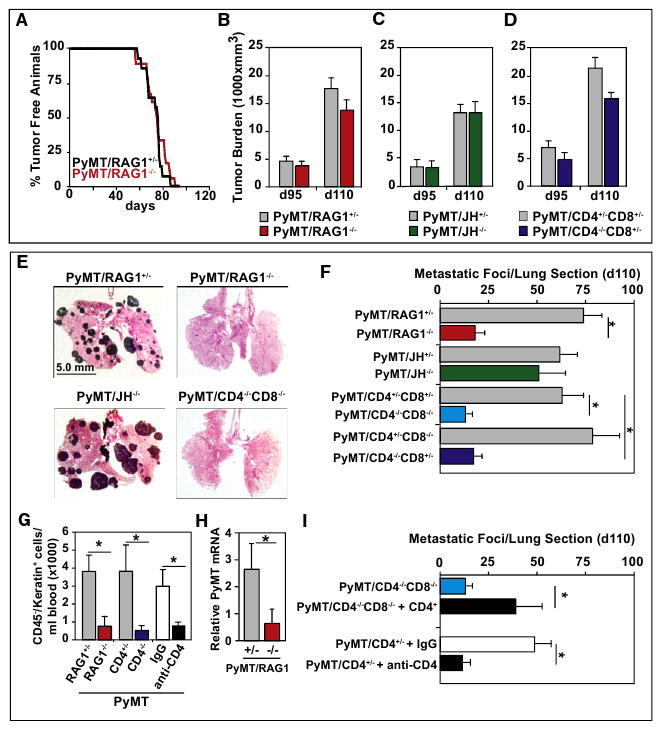
(A) Mammary adenocarcinoma incidence in PyMT/RAG1+/− and PyMT/RAG1−/− mice (n = 15 and 18 mice/group, respectively) is depicted as percentage of tumor-free mice. Mice were considered to be tumor free until a palpable mass (>4.0 mm) persisted for longer then 4 days. No statistical differences between cohorts were observed as evaluated by Wilcoxon test. (B–D) Total tumor burden of PyMT/RAG1 (B), PyMT/JH (C), and PyMT/CD4/CD8 (D) cohorts evaluated at both 95 and 110 days of age, shown as mm3 (n = > 20 mice per cohort). Tumor size was determined by caliper measurement and multiple tumors in one animal were added together. No statistical differences between groups were found as evaluated by Mann-Whitney test.
(E) Representative lung tissue sections depicting metastatic tumor burden from 110-day-old PyMT/RAG1−/−, PyMT/JH−/−, PyMT/CD4−/−/CD8−/− mice following hematoxylin and eosin (H&E) staining (5× magnification).
(F) Quantification of metastatic foci/lung section/mouse from 110-day-old PyMT/RAG1, PyMT/JH, PyMT/CD4/CD8, PyMT/CD4, PyMT/CD8 cohorts. Each lung was serially sectioned and six sections 100 mm apart were H&E stained and total number of metastatic foci (greater then 5 cells) quantified. Each of the six sections was averaged per mouse (n = > 20 mice per cohort).
(G) Circulating carcinoma cells were analyzed by flow cytometry and counted as the number of cytokeratin+/CD45− cells in blood from 110-day-old PyMT/RAG1+/− (n = 10), PyMT/RAG1−/− (n = 10), PyMT/CD4+/− (n = 20), PyMT/CD4−/− (n = 15), or 110-day old PyMT mice treated with anti-CD4 depleting IgG (n = 8) or IgG control (n = 6) for 18 days. Data are depicted as the mean number of carcinoma cells per milliliter of blood.
(H) PyMT mRNA expression in circulating blood cells. RNA from whole blood cells of 110-day-old PyMT/RAG1+/− and /RAG1−/− mice was evaluated for PyMT mRNA gene expression by RT-PCR (25 cycles) (n = 8 mice/group). Results from ethidium bromide stained gels are depicted following quantification of pixel density using GelDoc software.
(I) Average number of metastatic foci/lung section/mouse from 110-day-old PyMT/CD4+/− mice treated with anti-CD4 depletion antibody (GK1.5) versus IgG control (CD4+/− IgG) or PyMT/CD4−/−CD8−/− mice following adoptive transfer of naive CD4+ T cells (CD4+ rescue). Each lung was serially sectioned and assessed as described above. Twenty mice were used for PyMT/CD4−/−CD8−/−, six mice for CD4+ rescue, and eight mice for CD4+/− IgG or GK1.5 groups. (B–I) SEM is shown and asterisk denotes p < 0.05 by Mann-Whitney.
CD4+ T Cells Regulate Macrophage and Immature Myeloid Cell Phenotype and Effector Bioactivity
Because the attenuated metastatic phenotype of PyMT/CD4−/− mice mirrored tissue macrophage deficiency previously reported by Pollard and colleagues (Lin et al., 2001), we addressed the possibility that CD4 deficiency might result in altered myeloid cell presence and/or function in late-stage carcinomas. Using flow cytometry and IHC analysis, we found no change in CD45+ leukocyte infiltration in primary adenocarcinomas of 95 and 110-day-old PyMT mice resulting from CD4+ T cell-deficiency (Figures 3A and 3B). In addition, using flow cytometry to evaluate the spectrum of leukocytes infiltrating mammary carcinomas, we found no significant variation in leukocyte composition including CD11b+Gr1-F4/80+ macrophages or CD11b+Gr1HiF4/80− immature myeloid cells (IMCs) (Figure 3C).
Figure 3. CD4+ T Lymphocytes Do Not Regulate Leukocyte Infiltration but Instead Regulate Bioeffector Function of Myeloid Cell Subsets.
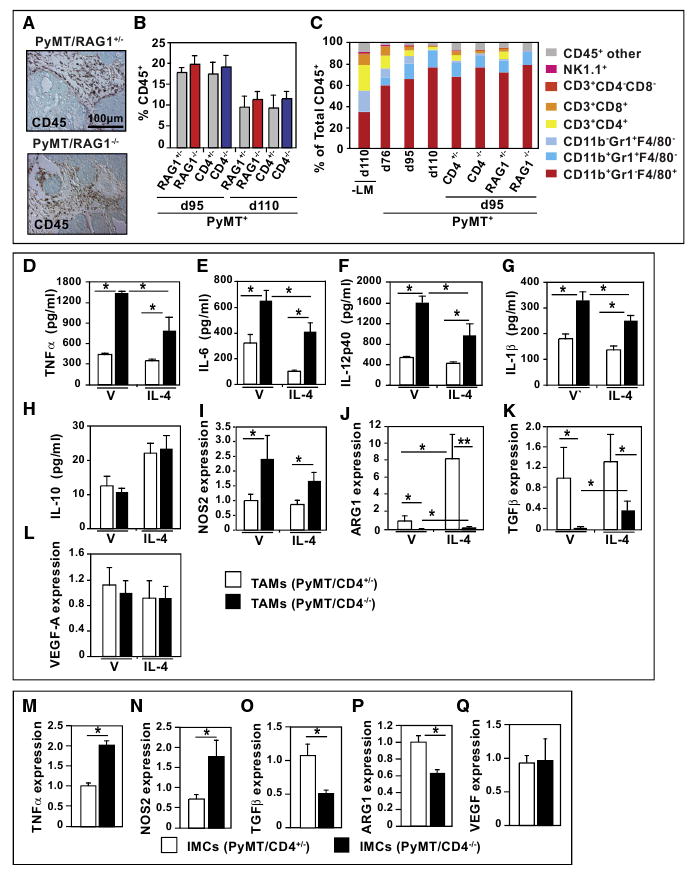
(A) Immunodetection of CD45+ cells in 95-day-old PyMT/RAG1+/− and PyMT/RAG1−/− mammary carcinomas. Representative 20× images are shown.
(B) Flow cytometric analysis of CD45+ cells in tumors from 95 and 110-day-old PyMT/RAG1 and PyMT/CD4 mice. Data are depicted as the mean percent of live cells ± SEM, n = 4 mice per cohort.
(C) Flow cytometric analysis of individual leukocyte populations as a percent of total CD45+ cells in mammary carcinomas of PyMT mice during progression and in day 95 tumors from RAG1-and CD4-deficient/PyMT mice. Data are depicted as the mean value from four mice/cohort ± SEM. No statistical differences were found between groups by Mann-Whitney test.
(D–L) Cytokine expression by TAM. Tumor-associated CD45+F4/80+Gr1− macrophages were isolated by dual magnetic and flow sorting of mammary tumors from 95 day-old PyMT/CD4+/− and PyMT/CD4−/− mice (n = 3/cohort). Cytokine expression (TNF-α, IL-6, IL-12p40, IL-1β, IL-10) was assessed by ELISA of conditioned medium or by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR; Nos2, Arg1, Tgfβ and Vegf-a) from TAMs (50,000) following 18 hr of culture with or without exogenous recombinant IL-4 (10 ng/ml). Representative assays of mammary carcinomas from three or four mice evaluated independently in triplicate and depicted as mean ± SEM.
(M–Q) Analysis of tumor-derived IMC phenotype in PyMT/CD4−/− mice. Tumor-associated CD45+ CD11b+Gr1Hi IMCs were isolated by flow from mammary tumors of 95-day-old PyMT+/CD4+/− and PyMT/CD4−/− mice (n = 3/cohort). Isolated cells were lysed and RNAs assessed by qRT-PCR as described above. Representative assays from two independent cohorts each run at least in triplicate are depicted as mean values ± SEM. Asterisk denotes p < 0.05 by Mann-Whitney in all panels.
Once in tissues, however, the differentiation state, phenotype, and effector functions of myeloid cells, including macrophages and IMCs, can be directly regulated by their immune microenvironment. The bioactive state of macrophages, for example, correlates with classical TH1 and TH2 nomenclature and is often referred to as M1 (classical) or M2 (alternative) activation, respectively (Mantovani et al., 2007). Classically activated M1 macrophages are regulated by TH1 cytokines like IFNγ, TNF-α, and granulocyte-monocyte-colony stimulating factor (GM-CSF) that, in part, enhance macrophage cytotoxic activity. In contrast, tissue macrophages exposed to TH2 cytokines common to tumors, including interleukin (IL)-4, IL-13 or IL-10 manifest an alternative (M2) phenotype that can be potentiated by immune complexes, IL-1, IL-21, transforming growth factor β (TGF-β), and glucocorticoids. Alternatively activated/M2 macrophages are commonly found associated with solid tumors and are thought to possess immunosuppressive, proangiogenic and pro-tissue remodeling bioactivities, as well as expressing high levels of EGF (Leek et al., 2000; Mantovani et al., 2007).
Thus, we evaluated differentiation/maturation and activation status of TAMs (Figures 3D–3L) and IMCs from carcinomas of 95-day-old CD4-proficient versus CD4-deficient/PyMT mice (Figures 3M–3Q). Expression analysis of lineage differentiation markers, including CD45, F4/80, CD11b, and Gr1, in TAMs revealed no significant alteration based on CD4+ T cell presence (Figures S2A and S2B). Although similar percentages of CD45+ CD11b+Gr1-F4/80+ TAMs infiltrated adenocarcinomas in both cohorts (Figure 3C), TAMs of CD4-deficient and RAG1-deficient/PyMT mice expressed significantly elevated levels of type 1 cytokines (e.g., TNF-α, IL-6, IL-12p40, and IL-1β) and Nos2 mRNA, indicative of a prevalent M1 TAM phenotype (Figures 3D–3G, Figure S2C) as compared with TAMs from CD4-proficient/PyMT mice (Figure 3I). Conversely, expression of factors indicative of alternatively activated (M2) TAMs, including arginase-1 (Arg-1) and Tgfβ were significantly reduced in TAMs isolated from mammary tumors of PyMT/CD4−/− mice as compared with CD4-proficient littermates (Figures 3J and 3K). Expression levels of IL-10 and Vegf-a were similar in TAMs from both cohorts (Figures 3H and 3L, Figure S2C). Moreover, cytokine expression of TAMs isolated from PyMT mice where CD4+ T cells had been depleted via neutralizing antibodies evidenced similar profiles as observed in TAMs of PyMT/CD4−/− mice (Figures S2D and S2E).
Yang and colleagues recently reported that CD11b+Gr1Hi IMCs are recruited into mammary carcinomas and regulate pulmonary metastasis in PyMT mice via activation of TGF-β-regulated signaling pathways (Yang et al., 2008). In order to determine whether CD4+ T cells were also regulating the bioactivity of IMCs, we analyzed their cytokine profile in mammary carcinomas of CD4-proficient versus deficient PyMT mice (Figures 3M–3Q, Figure S2F) and found significantly elevated expression of factors indicative of an M1 activation state (e.g., TNF-α and Nos2) with parallel reduction in M2-type factors (e.g., Arg-1 and Tgfβ). Thus, CD4+ T lymphocytes significantly regulate cytokine and mediator expression in both IMCs and TAMs in mammary adenocarcinomas.
TAM Phenotype in Mammary Carcinomas Is Treg Independent
In vitro, both CD4+ T effector and Treg cells have the capacity to modulate macrophage cytokine expression (Tiemessen et al., 2007). In order to determine which of these populations were regulating TAM bioactivity in vivo, we immune-depleted CD25+ Tregs by treating cohorts of 80-day-old PyMT mice with anti-CD25 IgG (PC61), versus isotype control Ig, for 20 days and found no differences in expression of M1-type cytokines (e.g., TNFα, IL-6, IL-12p40, IL-12p35, Nos2) or M2-induced genes Arg1, IL-10, or Tgfβ (Figures S3A–S3C). Thus, CD4+ T effector lymphocytes, but not CD25Hi Treg cells, significantly regulate TAM phenotype and bioeffector function.
CD4+ T Lymphocytes in Mammary Adenocarcinomas Express TH2 Cytokines
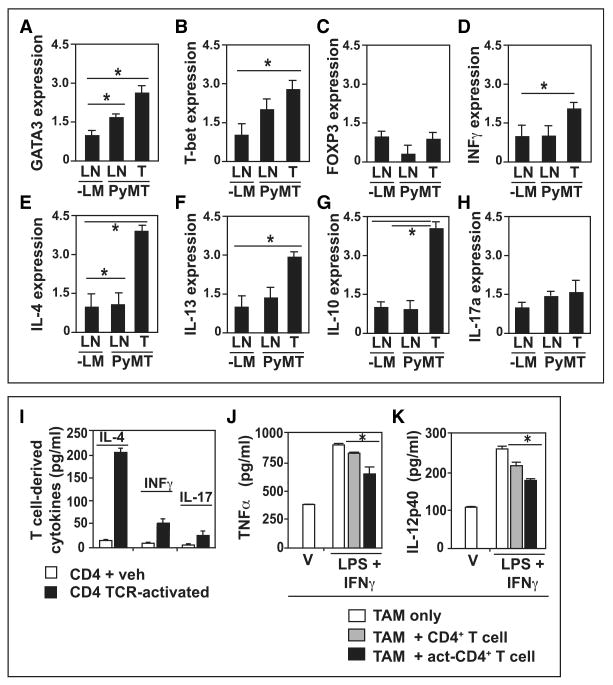
To determine whether CD4+ T cells regulated TAM phenotype by a TH2 cytokine-mediated pathway, we evaluated mRNA expression of CD4+ T cells isolated from LNs and mammary carcinomas of 95-day-old PyMT mice (Figures S4A and S4B) for transcription factors and effector molecules indicative of Treg, TH1, TH2, or TH17-type responses. CD4+ T lymphocytes isolated from draining LNs (LNs) and mammary carcinomas of PyMT mice exhibited elevated expression of GATA3 (TH2) and T-bet (TH1) mRNA, but not FOXP3 (Treg), when compared with LNs of wild-type littermates (Figures 4A–4C) indicating that both TH1 and TH2 effector lineages were expanded in LNs and in tumors. In order to assess the functional consequences of these, we assessed the cytokine expression profile of CD4+ cells and found significant induction in TH2 cytokines including IL-4, IL-13, and IL-10 and to a lesser extent the TH1 cytokine IFNγ, and by contrast, IL-17a was not significantly expressed (Figures 4D–4H). These results were further confirmed by ex vivo activation of CD4+ T lymphocytes (isolated from spleen, draining LNs, and tumors of PyMT mice) with anti-CD3/CD28 Ig. Analysis of expression of IL-4, IFNγ, and IL-17 by enzyme-linked immunosorbent assay (ELISA), and IFNγ and IL-4 by intracellular flow cytometry, and found that activated CD4+ T cells expressed higher levels of IL-4 than IFNγ or IL-17 (Figure 4I), and that IL-4-expressing CD4+ T cells represented a larger fraction of the total CD4+ T cells present in mammary tumors in vivo (Figure S5A).
Figure 4. CD4+ T Lymphocytes are TH2 Cells in Primary Mammary Carcinomas.
(A–H) Analysis of cytokine expression by tumor-associated CD4+ T cells. CD4+ T cells were isolated by flow sorting from LNs and tumors of 95-day-old PyMT mice and corresponding negative littermates (n = 4/cohort). Sorted cells were lysed and RNAs were assessed by qRT-PCR for GATA3, T-bet, FOXP3, IFNγ, IL-4, IL-13, IL-10, and IL-17a expression. Data are depicted as the mean fold change from the standardized sample (-LM LN).
(I) Cytokine analysis in CD4+ T cells ex vivo. Tumor-associated CD4+ lymphocytes were isolated by flow sorting from mammary tumors of 95-day-old PyMT/CD4+/− mice (n = 3) and IL-4, IFNγ, and IL-17 expression assessed by ELISA after 18 hr of culture prior to (veh, white bars) or following TCR activation (black bars). Data are represented as the mean of three replicates.
(J, K) CD4+ T cells repress TAM M1 phenotype. Tumor-associated CD45+CD3+CD4+ T lymphocytes and CD45+F4/80+Gr1− TAMs were isolated by flow sorting from mammary tumors of 95-day-old PyMT/CD4+/− mice. TAMs were untreated (V, white bars), or cultured with IFNγ (5 ng/ml) and LPS (50 ng/ml) in the presence of control CD4+ T cells (treated with control IgGs, gray bars), or activated CD4+ T cells (black bar, Act-CD4). TNF-α and IL-12p40 expression in conditioned medium evaluated by ELISA after 18 hr of coculture. Representative data from two independent experiments are depicted.
(A–K) SEM is shown and asterisk denotes p < 0.05 by Mann-Whitney test.
To determine whether IL-4 produced by CD4+ T cells was involved in differentially regulating macrophage effector functions, we assessed M1/M2 cytokine profile of TAMs isolated from CD4-deficient and CD4-proficient/PyMT mice using an ex vivo assay. We found that brief exposure of primary TAMs (isolated from PyMT/CD4−/− or PyMT/RAG1−/− adenocarcinomas) to exogenous IL-4 resulted in significantly reduced M1-type cytokine expression, simultaneous with enhanced expression of M2-type factors and mirroring cytokine expression of TAMs from CD4-proficient/PyMT mice (Figures 3D–3L, Figures S2C–S2F), thus indicating a dual role for IL-4 and perhaps CD4+ T cells in regulating macrophage polarity.
We next determined if tumor-associated CD4+ T lymphocytes by virtue of their expression of IL-4 directly repressed TAM M1 phenotype ex vivo. Whereas IFNγ/LPS treatment of TAMs led to increased TNFα and IL-12 expression (indicative of M1 activation), this effect was repressed in the presence of activated CD4+CD25-T effector cells (Figures 4J and 4K), thus indicating that tumor-associated CD4+ T lymphocytes actively repress M1 TAM effector function, while simultaneously fostering a protumor alternative/M2 TAM phenotype via expression of cytokines like IL-4.
CD4+ T Cells Regulate Macrophage-Induced MEC Invasive Behavior
To reveal whether CD4+ T cell activation of TAMs translated into enhanced invasive behavior of MECs, a requirement for metastasis in vivo, we utilized an ex vivo three-dimensional (3D) organotypic coculture model with primary murine cells. Primary MECs were isolated from either 76-day-old PyMT mice (pMECs) or 12-week-old virgin negative littermates (nMECs), placed in 3D overlay culture and allowed to form stable noninvasive organoids (Figure 5A) as previously described (Debnath et al., 2003). Following formation of stable organoids (2–3 weeks), CD45+ CD11b+Gr1-F4/80+ TAMs isolated from mammary carcinomas of 95-day-old PyMT mice (Figures S2A and S2B) were added, resulting in elaboration of an invasive MEC phenotype in a significant percentage of organoids (Figure 5A). When invasive pMEC organoids formed, TAMs were typically localized at the “invasive fronts” of invading structures (Figure 5A, panels c–f). In addition, when TAMs were cocultured with pMEC organoids in the presence of TH2-type cytokines (IL-4 or IL-13), organoid disruption and formation of invasive structures was significantly enhanced (Figure 5B) in a TAM and IL-4 dose-dependent manner (Figures S6A and S6B). In contrast, when IL-4 or IL-13 cytokines were added to organoids alone (without TAMs), no significant change in organoid stability or invasive behavior was observed (Figure 5B) indicating that cytokine stimulation of pMEC invasion was mediated by TAMs. Analogous results were found utilizing IL-4-activated TAMs to induce pMEC invasion in standard Boyden Chamber migration assays (Figure S6C). In contrast, when TAMs were cocultured with pMEC organoids in the presence of M1-type cytokines (IFNγ or LPS) or the immunosuppressive cytokine IL-10, the invasive pMEC phenotype was significantly inhibited and instead additional stability of organoids was observed (Figure 5B). Moreover, to evaluate if tumor-associated CD4+ T cells were involved or perhaps directly regulating TAM-induced MEC invasion, TAMs and pMEC organoids were “tricultured” with CD4+ T cells isolated from mammary carcinomas of 95-day-old PyMT mice (Figure S4A), resulting in a significant enhancement of pMEC invasive organoids in an IL-4 dependent manner (Figure 5C). Taken together with data from the in vivo analysis of TAM phenotype, these data indicate that tumor-associated TAMs are alternatively (M2) activated by IL-4-expressing CD4+ T cells, that together induce invasive behavior of MECs, a bioactivity that is not supported by TAMs isolated from CD4+ T-cell-deficient adenocarcinomas, or when TAMs are “classically” activated by factors like IFNγ or LPS, or engaged in immunosuppressive programs regulated by IL-10.
Figure 5. M2-Activated TAMs Induce Invasive Behavior in 3D Mammary Epithelial Organoids.
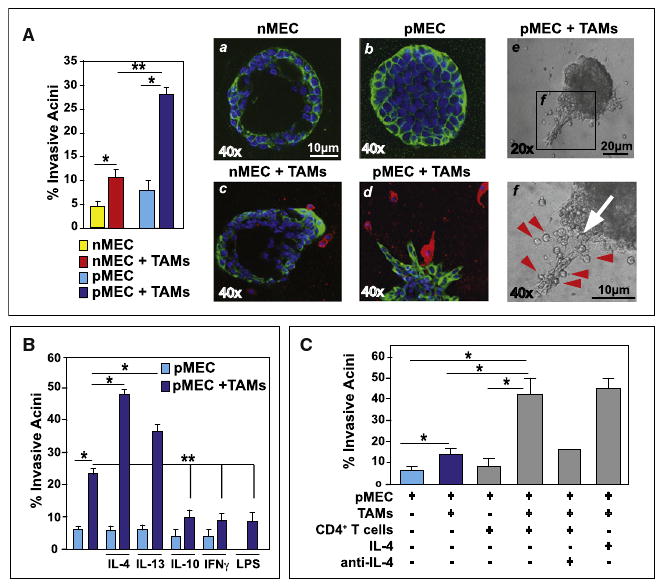
(A) Quantitation of invasive organoids following coculture of TAMs. Stable wild-type MEC (nMEC) or PyMT-derived MEC (pMEC) organoids were allowed to form over 14–20 days and then cocultured with TAMs (48 hr). Representative immunofluorescent images of nMEC (a, c) and pMEC (b, d) organoids in the presence or absence of TAMs evaluated for cytokeratin 7 (green), F4/80 (red), and DAPI (blue) are shown. Representative bright field images of invasive pMEC organoids in coculture with TAMs are depicted at 20× magnification and 40× inset. TAMs are denoted by red arrows.
(B) Quantification of organoid disruption following coculture of TAM with pMEC spheroids in the presence of IL-4 (20 ng/ml), IL-13 (20 ng/ml), IL-10 (10 ng/ml), IFNγ (5 ng/ml), or LPS (50 ng/ml).
(C) Quantification of pMEC organoid disruption (formed over 14 days) following coculture (48 hr) with TAMs and/or tumor-associated CD3+CD4+ T cells. Cocultures were also exposed to exogenous recombinant mouse IL-4 (10 ng/ml) and/or an anti-mouse IL-4 neutralizing antibody (0.5 mg/ml; clone OP06) added 12 hr prior to leukocytes.
IL-4 Regulates TAM Phenotype and Pulmonary Metastasis of Mammary Adenocarcinomas
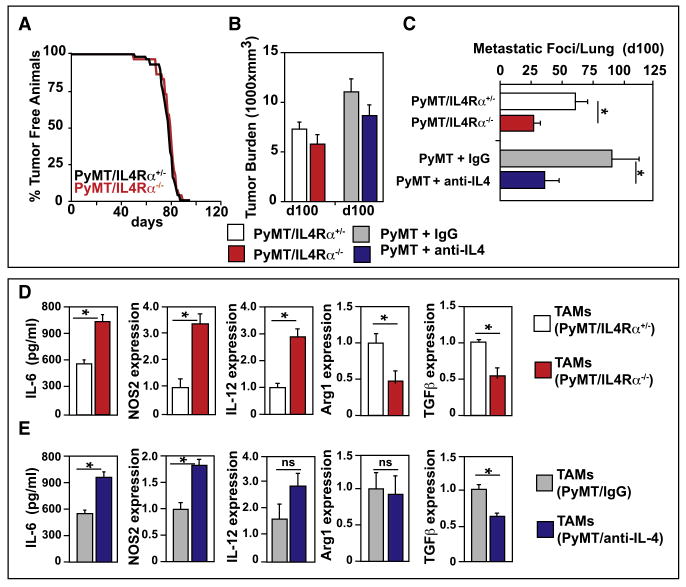
Because the capacity of tumor-associated CD4+ T lymphocytes to regulate TAM phenotype and pMEC invasion was dependent on TH2-type cytokines, we hypothesized that neutralization of IL-4 or its receptors in vivo would mirror the phenotype of CD4-deficient/PyMT mice and limit pulmonary metastasis. To address this, we generated PyMT mice either harboring a homozygous inactivating mutation in the IL-4 receptor alpha (IL4Rα) gene (FVB/n, N6), or treated PyMT mice with a neutralizing antibody to IL-4. Similar to CD4+ T-cell-deficient/PyMT mice, both cohorts of PyMT/IL4Rα−/− and IL-4-neutralized/PyMT mice exhibited no significant change in primary tumor latency or burden as compared with controls (Figure 6A and 6B). However, loss of either IL-4 activity or expression of IL4Rα resulted in significantly reduced numbers of metastatic foci in lungs and overall attenuation of total pulmonary metastasis (Figure 6C). Moreover, cytokine analysis of TAMs isolated from both PyMT/IL4Rα−/− and IL-4-depleted/PyMT mice revealed increased expression of M1-type factors (IL-6, Nos2, IL-12p35) and reduced expression of M2-type genes (Arg1 and Tgfβ) as compared with TAMs from control mice (Figures 6D and 6E) and thus phenocopied the characteristics of TAMs isolated from CD4+ T cell-deficient/PyMT mice (Figure 3).
Figure 6. IL-4 Signaling Promotes Metastasis, but Not Primary Tumor Development.
(A) Kaplan Meyer analysis of tumor incidence in PyMT/IL-4Rα+/− and PyMT/IL-4Rα −/− mice (n = 15/group) depicted as percentage of tumor-free animals. No statistical differences between cohorts by generalized Wilcoxon test were found.
(B) Total mammary tumor burden of PyMT/IL-4Rα+/− and −/− mice, and PyMT mice treated for 20 days with either IL-4 neutralizing Ig (11B11) or control IgG and evaluated at day 100, shown as mm3 (n = > 20 mice per cohort). Tumor size was determined by caliper measurement and multiple tumors in one animal were added together.
(C) Quantification of average number of metastatic foci/lung/mouse of 100-day-old PyMT/IL-4Rα and PyMT mice treated with either IL-4 neutralizing or control IgG. Each lung was serially sectioned, and six sections 100 μm apart were stained by H&E and total number of metastatic foci (greater then 5 cells) quantified. Each of the six sections was summed and each bar represents n ≥ 23 mice for all cohorts and ≥ 12 for Ig-treated groups.
(D and E) Tumor-associated CD45+F4/80+Gr1− macrophages were isolated by flow sorting of mammary carcinomas from (D) PyMT/IL-4Rα or (E) PyMT mice treated with either IL-4 neutralizing IgG (11B11) or control IgG. ELISA was performed on conditioned medium from TAMs (50,000) after 18 hr of culture. Quantitative RT-PCR analysis was performed using the comparative threshold cycle method to calculate fold change in gene expression normalized to GAPDH as reference gene. Representative assays from three independent cohorts each run at least in triplicate are depicted as the mean fold change from the standardized sample. (B–E) Data are represented as mean ± SEM, and asterisk denotes p < 0.05 by Mann-Whitney test.
IL-4 Signaling Induces Macrophage EGF mRNA Expression and EGFR-Dependent Invasion and Metastasis
Because elaboration of the invasion MEC phenotype certainly involved activation of intracellular MEC signal transduction programs, we next sought to identify the soluble mediators released by TAMs following their activation by IL-4 or CD4+ T cells. Thus, we assessed expression of several growth factors associated with epithelial cell invasion and found that TAMs isolated from CD4-proficient/PyMT mice exhibited elevated levels of EGF and Tgfβ mRNA expression, as compared with TAMs from CD4+ T-cell-deficient/PyMT mice (Figures 3K and 7A, and data not shown). In addition, TAMs represent the most abundant cellular source of EGF mRNA in mammary carcinomas (Figure S6E). To determine whether enhanced EGF mRNA expression by TAMs was directly due to IL-4 exposure, we evaluated EGF mRNA expression of TAMs following brief exposure to IL-4, CSF-1, IL-4 plus CSF-1, as compared with pMEC conditioned medium alone, and found that EGF mRNA expression was significantly enhanced by IL-4, but only in the presence of CSF-1 or pMEC conditioned medium (Figures 7A and S6D).
Figure 7. IL-4 and CSF-1 Signaling Intersect and Regulate Macrophage EGF Expression, pMEC Invasion, and Metastasis.
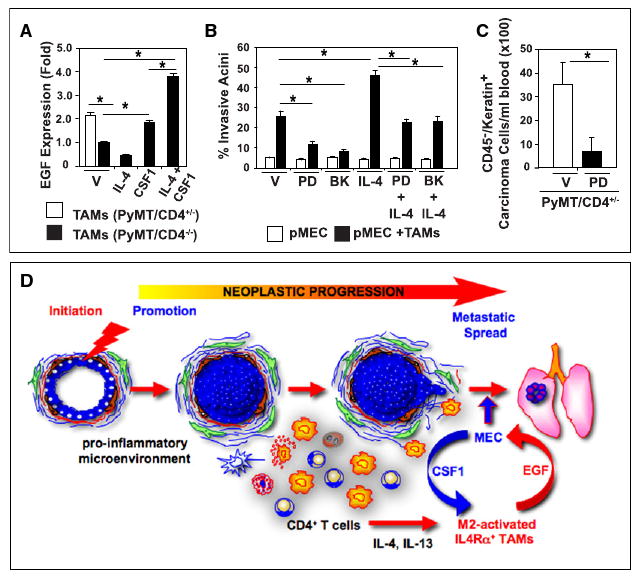
(A) EGF mRNA expression analysis from CSF-1- and IL-4-activated TAMs. TAMs were isolated from mammary tumors of 95-day-old PyMT/CD4+/− or PyMT/CD4−/− mice, and placed into culture with CSF-1 (10 ng/ml) and/or IL-4 (20 ng/ml) for 16 hr. Quantitative RT-PCR analysis of EGF mRNA expression is depicted as fold change from vehicle (CD4−/− no treatment is set to 1.0) assessed by the comparative threshold cycle method normalized to reference gene expression.
(B) Quantification of pMEC organoid disruption following coculture of TAMs (48 hr) +/− IL-4 (10 ng/ml) or EFGR small molecule inhibitors PD153035 (0.1 μM) or BIBX1382 (10 nM). Invasive organoids were counted and data represented as a percentage of the total organoids (>100 replicate). Representative data from two independent experiments performed in quadruplicate are depicted.
(C) EGFR signaling regulates metastasis in PyMT mice. The 110 day-old PyMT mice were treated with the EGFR small molecule inhibitor PD153035 (25 mg/kg) versus vehicle (DMSO) by i.p. injection 5 hr prior to analysis (six mice/group). Presence of circulating carcinoma cells (cytokeratin+CD45−) was assessed by FACS evaluation of live cells in PB.
(A–C) Data are represented as mean ± SEM, and the asterisk denotes p < 0.05 by Mann-Whitney test.
(D) Schematic representation of TH2 CD4+ T lymphocytes and their role in breast cancer metastasis. During early breast cancer development, increased presence of leukocytes in neoplastic stroma indicates establishment of a proinflammatory microenvironment. When immune cell infiltrates include high numbers of TH2 CD4+ T lymphocytes that produce IL-4 and IL-13, M2-type TAMs and IMCs are activated and in turn produce EGF, thus resulting in activation of a paracrine-mediated enhancement of malignant cell invasion and dissemination into PB and pulmonary metastasis.
To establish whether activation of EGF receptor (EGFR)-mediated signaling was necessary for TAM-induced pMEC invasion, we evaluated effects of EGFR blockade using the 3D coculture assay, and found that IL-4-regulated TAM-dependent pMEC invasion was significantly diminished in the presence of EGFR tyrosine kinase inhibitors (Figure 7B). To determine whether this translated to a diminishment in metastasis in vivo, late-stage (day 110) PyMT mice were treated with PD153035 (25 mg/kg) for 5 hr and the presence of circulating malignant cells quantitatively determined. This brief treatment resulted in a significant decrease in the number of circulating carcinoma cells present in PB (Figure 7C), similar to observations by Wyckoff et al. (2007). Taken together, these data indicate that in response to CD4+ T cell-derived IL-4, M2 effector bioactivity is enhanced in TAMs (and IMCs) that in turn activate invasive and metastatic potential of MECs in mammary adenocarcinomas through their production of proinvasive/metastatic factors such as EGF (Figure 7D).
Discussion
We revealed a provocative and functional role for CD4+ T effector cells as potentiators of PB dissemination and pulmonary metastasis of mammary adenocarcinomas through their ability to regulate protumor properties of TAMs. Specifically, TH2-polarized CD4+ T lymphocytes regulate M1 and M2-type TAM bioactivity by their expression of IL-4. M2-TAMs in turn promote invasive behavior of malignant MECs by high-level production of EGF that subsequently activates MEC EGFR signaling programs, an activity essential for entry into PB, dissemination and outgrowth in the lung. These findings indicate that when CD4+ T lymphocytes are present in a TH2-type tumor microenvironment, they can promote metastasis by regulating the protumor properties of TAMs, as opposed to limiting or eradicating malignant cells by engaging cytotoxic mechanisms. This realization provides rational for development of anticancer therapeutics that neutralize the protumor properties of both adaptive and innate immune cells in the tumor microenvironment, that when delivered in combination with cytotoxic drugs that bolster antitumor immunity, might thereby extend survival of breast cancer patients with advanced disease.
Effector Function of CD4+ T Cells in Solid Tumors Is Context Dependent
Although our studies have revealed that CD4+ T cells potentiate dissemination and metastasis of mammary adenocarcinomas, it is clear that CD4+ T lymphocytes in other contexts exhibit other bioactivities. Schreiber and colleagues demonstrated that CD4+ T cell deficiency in methylcholanthrene-initiated sarcomas enhanced tumor development (Koebel et al., 2007). By contrast, following two-stage skin carcinogenesis (dimethylbenzanthracene plus tetradecanoylphorbol-acetate), CD4+ T cell deficiency was associated with diminished tumor development (Girardi et al., 2004). Thus, tumor etiology, in combination with the tumor microenvironment, regulates CD4+ T cell phenotype, and in part determines whether a protumor, as opposed to an antitumor immune program, is favored. In agreement with the tissue context-dependent nature of CD4+ T cells, in a mouse model of skin and cervical carcinoma development where oncogenes from human papilloma virus type 16 are expressed behind the keratin 14 promoter/enhancer, skin carcinoma formation is modestly attenuated by CD4+ T cell deficiency, whereas cervical carcinoma development is significantly enhanced (Daniel et al., 2005; Daniel et al., 2003), again demonstrating that immune responses accompanying tumor development are organ dependent as opposed to oncogene dependent, and, based on the neoplastic and immune microenvironment, can engage either pro- or antitumor immune regulatory programs.
CD4+ T lymphocytes have been traditionally classified as either tumor suppressive, such as TH1 effector cells that repress tumor growth by secretion of IFNγ (among other soluble mediators) and support of CTL function, or alternatively tumor-promoting cells, including Tregs, that foster tumor expansion by suppressing CD8+ CTLs and NK cells (Trzonkowski et al., 2006). The interplay between TH1 and Tregs in regulating tumor immunity is likely critical for the etiology of some malignancies, such as sarcomas (Dunn et al., 2006; Koebel et al., 2007) or lung adenocarcinomas (Woo et al., 2002). In addition to these, a new class of CD4+ T cells expressing IL-17 have been identified (i.e., TH17 cells) that might also regulate chronic inflammation and promote tumor development when activated in the presence of TGF-β and IL-6, or IL-23 (Dong, 2008). In our studies, we found that pulmonary metastasis and M2-bioactivity of TAMs was potentiated by CD4+ T effector cells that express high levels of IL-4, IL-13 and IL-10, as compared with expression of IFNγ or IL-17 (Figures 3 and 4), whereas TAM bioactivities were unaffected by immune depletion of CD25+ Treg cells. Moreover, CD4+ T cells exerted these affects independent of the presence or absence of CD8+ T cells, indicating that protumor functionality does not involve suppression of CTL activity.
In addition to indirectly potentiating cancer development by regulating protumor properties of myeloid cells, research from several laboratories has revealed that IL-4 and IL-13 regulate tumor growth through activation of IL-4/13 receptors on epithelial cells. In some human breast carcinoma cell lines, particularly those that express the estrogen receptor a, IL-4 and IL-13 inhibit basal and estrogen-induced cell proliferation in vitro and in xenograph models in vivo (Gooch et al., 2002; Nagai and Toi, 2000). However, in other breast carcinoma cell lines, IL-4 regulates tumor cell survival by conferring resistance to apoptosis (in vitro) that translates to chemoresistance in xenographs (Todaro et al., 2008). Palucka and colleagues reported that CD4+ T cells directly enhance early tumor development by their production of IL-13 (Aspord et al., 2007). In contrast to these, we found no change in the latency or development of primary mammary adenocarcinomas due to either CD4+ T cell or IL4Rα deficiency, indicating that in this model system, CD4+ T cells and IL-4 likely do not provide a survival or proliferative advantage directly to neoplastic cells. Moreover, using the ex vivo cell-based assay, we also found no change in MEC proliferation, acinar morphology, or organoid stability when cocultured with CD4+ T cells alone, or when MECs were given IL-4 or IL-13. Instead, when cultured together with MECs in the presence of TAMs, either tumor-derived CD4+ T cells, IL-4, or IL-13 induced significant changes in organoid morphology consistent with invasive growth (Figure 5). Taken together, these data indicate that the effects of CD4 T cell-derived TH2 cytokines on tumor development and progression is likely regulated by the organ microenvi-ronment or IL-4/13 receptor status on cell in the tumor microenvironment.
Clinical evaluation of human breast cancers has revealed that presence of CD4+ TH2 and Treg cells increase during cancer development. High percentages of CD4+ T cells positively correlate with tumor stage, including metastatic spread to sentinel LNs and increased primary tumor size (Kohrt et al., 2005). Perhaps more significant, the ratio of CD4+ to CD8+ T cells or TH2 to TH1 cells in primary tumors, where CD4+ or TH2 cells are more frequent than CD8+ or TH1 cells, correlates with LN metastasis and reduced overall patient survival (Kohrt et al., 2005). More recently, unsupervised expression profiling from breast-cancer-associated stroma revealed a gene signature predictive of good prognostic outcome (>98%, 5 year survival) that was functionally enriched for elements of a TH1-type immune response, including genes suggestive of CTL and NK cell activity (Finak et al., 2008). Conversely, high levels of FOXP3+ Treg cells predict diminished relapse-free and overall survival (Bates et al., 2006). The interpretation based upon these clinical studies is that the type of CD4+ effector T cell response elicited in an emergent breast cancer might in part determine malignant and metastatic potential. Our data provide some clarity to these profiles wherein we report that TH2-CD4+ T cells promote metastasis, not by altering CTL responses, but instead by enhancing the protumor bioactivities of myeloid cells, and enhancing intracellular signaling cascades (EGF) required for dissemination and metastasis.
Macrophage-Mediated Pro- versus Antitumor Immunity
Macrophages promote metastasis in several contexts, i.e., by supporting tumor-associated angiogenesis, inducing local immunosuppression, or by promoting malignant cell invasion and entry into circulation (Condeelis and Pollard, 2006); however, the molecular mechanisms regulating each of these “hallmark” protumor TAM properties have yet to be elucidated. Macrophages are implicated in tumor angiogenesis (a prerequisite for metastasis) by virtue of their capacity to express proangiogenic factors including VEGF and matrix metalloproteinase (MMP)-9 (Giraudo et al., 2004), and by clinical data in human breast cancers demonstrating their presence correlates with increased microvessel density (Uzzan et al., 2004). Accordingly, tissue macrophage deficiency in PyMT mice leads to reduced angiogenesis, delayed onset of late-stage carcinomas, and greatly diminished pulmonary metastasis (Lin and Pollard, 2007). By comparison, loss of CD4+ T lymphocytes, similar to loss of tissue TAMs, results in reduced presence of circulating carcinoma cells and diminished pulmonary metastasis, but did not impact microvessel density, character of angiogenic vasculature, or expression of Vegf-a or MMP-9 by TAMs or IMCs. These distinctions reflect the fact that CD4+ T-cell-derived factors, including IL-4, regulate only some aspects of TAM bioactivity, in particular invasive and metastatic properties of MECs that are EGF dependent; thus, proangiogenic TAMs are likely regulated by other factors such as hypoxia.
TAMs exhibit immunosuppressive activity via their expression of arginase, IL-10, and TGF-β (Mantovani et al., 2007). Our data demonstrate that CD4+ T cells and IL-4 induce some but not all of their immunosuppressive properties, specifically expression of Arginase-1, TGF-β, and IL-12, but not IL-10, MHCII, or CD86 (Figure 3 and data not shown). Mantovani and colleagues reported that some aspects of TAM-mediated immunosuppression are regulated by intracellular NF-κB signaling (Saccani et al., 2006). Moreover, Balkwill and colleagues revealed that ovarian-cancer-associated TAMs, due to IL-1R and MyD88, maintain an immunosuppressive M2 phenotype dependent on IKKβ (Hagemann et al., 2008). In vivo, IKKβ-deficient TAMs instead exhibit tumor cell cytotoxicity and switch to a classically activated M1 phenotype (e.g., IL-12high, major histocompatibility complex IIhigh, IL-10low, Arginase-1low) that promotes regression of advanced ovarian carcinomas by induction of TAM tumoricidal activity and activation of IL-12-dependent NK cell recruitment (Hagemann et al., 2008). These experimental findings imply that reprogramming TAM phenotype and/or altering the immune microenvironment to foster antitumor activity could diminish tumorigenicity and improve clinical outcome.
Mechanisms of TAMs Induced Epithelial Cell Invasion and Metastasis
Reciprocal interactions between TAMs and MECs together regulate mammary carcinogenesis through activation of a paracrine feed-forward loop involving TAM-expressed EGF and epithelial-expressed CSF-1 (Wyckoff et al., 2004). This paracrine loop is critical for branching morphogenesis (Gouon-Evans et al., 2000), as well as for breast carcinoma cells exhibiting “high-velocity” polarized movement (chemotaxis) along collagen fibers toward blood vessels directed by perivascular macrophages (Wyckoff et al., 2007). We propose that these heterotypic interactions are further regulated by factors derived from CD4+ T lymphocytes including IL-4, IL-13, and possibly IFNγ. Herein, we demonstrated that activation of TAMs by IL-4, in combination with factors derived from malignant MECs such as CSF1, regulate high-level expression of EGF, that in turn stimulates EGFR-induced MEC invasive behavior in vitro and MEC entry into PB and pulmonary metastasis in vivo. As such, a TH2-rich microenvironment likely collaborates with existing genetic mutations in neoplastic cells, and thereby fosters development of highly invasive tumors in vivo.
In addition to EGF, production of TGFβ by M2 TAMs, mesenchymal support cells, and IMCs also enhances invasive and metastatic programming of malignant cells (Yang et al., 2008). Profiling of human breast carcinomas has revealed that a TGF-β-responsive gene signature predicts lung metastasis (Padua et al., 2008). Similarly, we found that the absence of CD4+ T cells also resulted in decreased Tgfβ expression in TAMs (and IMCs) in mammary adenocarcinomas. The prediction based on these data is that the type of CD4+ effector lymphocyte response elicited by the neoplastic microenvironment functionally modulates critical stromal derived factors, such as EGF and TGFβ, that collaborate with tumor-cell-intrinsic programs to regulate invasive and metastatic potential.
Summary
Taken together with clinical and experimental studies, our data indicate that CD4+ T effector lymphocytes potentiate mammary adenocarcinoma metastasis by modulating the protumor properties of TAMs that in turn enhance the invasive potential of malignant mammary epithelial cells. Because late-stage immune-depletion of CD4+ T cells or IL-4 resulted in a significant diminution in circulating malignant carcinoma cells and reduced outgrowth of pulmonary metastases, these provocative findings indicate that anticancer therapeutic strategies targeting the effector bioactivity of T cells might hold promise for treating late-stage disease. Although ongoing genetic alterations clearly play a role in regulating the malignant behavior of a neoplastic cell, our study in combination with others revealing dominant roles played by the tumor microenvironment in regulating malignancy, support the long-standing hypothesis (Bissell et al., 1982) that the host response and microenvironment in which a neoplastic cell evolves is as critical to its evolution as the genetic changes occurring within its nucleus.
Experimental Procedures
Animal Husbandry
Mice carrying the PyMT gene underthe control of the MMTV promoter in the FVB/n background were obtained from Dr. Zena Werb (University of California, San Francisco [UCSF], San Francisco, CA) and have been previously described (Guy et al., 1992). Generation and characterization of FVB/n mice homozygous null (−/−) for RAG-1, CD4 and CD8 have been described previously (de Visser et al., 2005). Homozygous null JH and IL4Rα mice were obtained from Jackson Laboratories. To generate PyMT mice on the RAG-1−/−, JH−/−, CD4−/−, CD8−/−, and IL4Rα−/− backgrounds, RAG-1+/−, JH+/−, CD4+/−, CD8+/−, and IL4Rα+/− mice were backcrossed into the FVB/n strain to N15, N5, N14, N7, and N6, respectively, and then intercrossed with PyMT mice to generate breeding colonies of −/− and +/− PyMT/RAG-1, PyMT/JH, PyMT/CD4, PyMT/CD8, PyMT/CD4/CD8, and PyMT/IL4Rα mice. Immune depleted mice were injected every 5 days intraperitoneally (i.p.) with either anti-CD4 (400 μg, GK1.5), anti-CD25 (400 μg, PC61), anti-IL-4 (1.0 mg, 11B11), or control rat Ig. All mice were maintained within the UCSF Laboratory for Animal Care barrier facility, and all experiments involving animals were approved by the Institutional Animal Care and Use Committee of UCSF.
Primary and Organoid Cell Culture
Primary nMEC and pMEC pools were established by organoid centrifugation as previously described (Pullan and Strueli, 1996). Briefly, mammary tissue biopsies were harvested from 76-day-old PyMT female or 12-week-old virgin negative littermates and digested with Collagenase A 2.0 mg/ml (Roche) and DNase 2.0 units/ml (Roche) for 2 hr. Organoids were then isolated by differential centrifugation and grown in culture conditions as previously described (Pullan and Strueli, 1996). Primary nMECs were used within two passages and primary pMEC cells were used within ten passages. Three-dimensional organotypic cultures were established as previously described (Debnath et al., 2003; Lee et al., 2007). Cultrex basement membrane extract (BME; R&D Systems) was utilized to limit endotoxin levels. Cocultures with primary leukocytes were established only after stable organoid structures had formed (approximately 3 weeks for nMEC, 2 weeks for pMEC). Leukocytes were overlaid in medium containing 0.5% BME. Formation of invasive acini was assessed every 12 hr for 3 days. The cytokines IL-4 (20 ng/ml), IL-13 (20 ng/ml), IL-10 (10 ng/ml), IFNγ (5.0 ng/ml) (Peprotech), or LPS (1.0 mM/ml) were added to cocultures 12 hr after leukocytes overlay. Inhibitors PD153035 (0.1 mM, Calbiochem) or BIBX1382 (10 nM, Calbiochem) were added 1.0 hr prior to the addition of leukocytes. All experiments were repeated two or three times with separate pMEC pools and individual experiments were run at least in triplicate.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism and/or InStat Software. Specific tests used were Student's t test, Mann-Whitney (unpaired, nonparametric, two-tailed), unpaired t test Welch corrected, generalized Wilcoxon test, and log rank analysis. All p values less than 0.05 were considered statistically significant.
Additional experimental procedures are included in Supplemental Experimental Procedures.
Supplementary Material
Figure S1: The impact of CD4+ T cells on primary mammary tumor development A-B) Kaplan Meyer analysis of tumor incidence in CD4, CD8 and B cell-deficient/PyMT mice. The percent of tumor free animals is depicted for PyMT/CD4+/-CD8+/- (n=10), PyMT/CD4-/-CD8-/- (n=46), PyMT/CD4-/-CD8+/- (n=34), PyMT/CD4+/-CD8-/- (n=31) as well as PyMT/JH+/- (n=23) and PyMT/JH-/- (n=28) mice. Animals were considered to be tumor free until a palpable mass (>4.0 mm) persisted for longer then 4 days. There were no statistical differences between tumor incidence curves by generalized Wilcoxon test.
C) Representative images of H&E stained mammary tumor tissue from 76 and 95-day old PyMT/RAG1+/- and PyMT/RAG1-/- mice. Magnification and scale are shown.
D) Flow cytometric analysis of CD31+ endothelial cells in mammary tumors from 95 and 110 day-old PyMT/RAG1+/-, PyMT/RAG1-/-, PyMT/CD4+/-CD8+/-, PyMT/CD4-/-CD8-/- mice (n=4). Data is depicted as the mean % CD31+ cells of the total live cells/tumor ±SEM. No statistical differences were found between groups by Mann-Whitney test. Representative 20x images of paraffin embedded tumor sections from PyMT/CD4+/-, PyMT/CD4-/- sections stained for CD31+ and vasculature revealed as brown staining. Bar in both = 100 μm.
Figure S2: CD4+ T lymphocytes regulate bioeffector function of myeloid cell subsets A-B) Isolation of TAMs from the mammary tumors of PyMT/CD4+/- and PyMT/CD4-/- mice. The purity of isolated CD45+F4/80+Gr1- macrophages was assessed by flow cytometric analysis of the cell suspension before and after selection. Representative analyses are shown from PyMT/CD4+/-(red) and PyMT/CD4-/- (blue) tumors and relative CD45, CD11b, Gr1 and F4/80 expression depicted.
C) Cytokine expression analysis from TAM-conditioned medium. Tumor-associated CD45+F4/80+Gr1- macrophages were isolated by dual magnetic and flow sorting of mammary tumors from 95 day-old PyMT/RAG1+/- and PyMT/RAG1-/- mice (n=3/cohort). Cytokine expression was assessed by ELISA of conditioned medium from TAMs (25,000) following 18 hours of culture, with or without recombinant IL-4 (10 ng/ml). Representative assays from 2 independent cohorts each performed at least in triplicate are depicted as means ± SEM and * denotes p<0.05 by student t test.
D) Lymphocyte depletion by anti-CD4 IgG (GK1.1) was assessed by flow cytometric analysis 4 days after of IgG injection. Representative analyses from anti-CD4 IgG and control IgG treated mice are shown from blood isolated by left ventricle heart puncture. % CD4+ of total cells is depicted.
E) Analysis of TAM polarization following CD4+ T cells depletion. Tumor-associated CD45+F4/80+Gr1- macrophages were isolated by flow sorting mammary tumors from PyMT mice treated with either anti-CD4 IgG (GK1.1) or control IgG. Tumor bearing 85 day-old mice were treated twice over 10 days with Igs and TAMs were isolated from tumors when mice were 95 day-old. Cytokine expression was assessed by ELISA analysis of conditioned medium from TAMs following 18 hours of culture. Gene expression analysis used the comparative threshold cycle method to calculate fold change in gene expression normalized to GAPDH as reference gene. Representative assays from 2 independent cohorts each run at least in triplicate are depicted as mean values ± SEM. * denotes n p<0.05 by Mann-Whitney.
F) Analysis of IMC polarization following CD4+ T cells depletion. Tumor-associated CD45+CD11b+Gr1Hi IMCs were isolated by flow sorting mammary tumors from PyMT mice treated with either anti-CD4 IgG (GK1.1) or control IgG. Tumor bearing 85 day-old mice were treated twice over 10 days with Igs and TAMs were isolated from tumors when mice were 95 day-old. Cytokine expression was assessed by ELISA and quantitative real-time PCR as described above. Representative assays from 2 independent cohorts each run at least in triplicate are represented as mean values ± SEM. * denotes p<0.05 by Mann-Whitney.
Figure S3: Cytokine profiles of TAMs from CD25 Treg-depleted mice. A) Analysis of CD25+ T cell depletion. Tumor bearing 85 day-old PyMT mice (5 mice/cohort) were treated with either anti-CD25 IgG (PC61) or control IgG three times over 15 days. Peripheral blood was analyzed for expression of FOXP3 and CD25 expression on CD4+ T lymphocytes every 5 days by flow cytometry to affirm depletion. Representative analysis for depleted versus control 100 day-old PyMT mice is depicted as FACS plots gated on CD4+ lymphocytes.
B) Quantitative real-time PCR analysis of Nos2, Arg1, Tgfβ, IL-12a and IL-10 mRNAs expression in tumor-associated CD45+F4/80+Gr1- macrophages isolated by flow sorting from mammary tumors of 100 day-old PyMT mice treated with either anti-CD25 Ig (PC61) or isotype control IgG. The comparative threshold cycle method was used to calculate fold change in gene expression normalized to GAPDH as reference gene. Cell isolation and gene expression analysis was performed on 2 separate cohorts of animals (3 mice/group) with similar results. Representative data are depicted as means ± SEM. No statistical differences were found between treated and untreated groups by Mann-Whitney.
C) Cytokine expression analysis from TAM conditioned medium. Tumor-associated CD45+F4/80+Gr1- macrophages were isolated by flow sorting from mammary tumors from 100 day-old PyMT mice treated with either anti-CD25 Ig (PC61) or isotype control IgG for 15 days. Cytokine expression was assessed by ELISA analysis of conditioned medium from TAMs following 18 hours of culture. Cell isolation and cytokine analysis was performed on 3 separate cohorts of animals (2 mice/group) with similar results. Representative data are depicted as mean ± SEM. No statistical differences were found between treated and untreated groups by Mann-Whitney.
Figure S4. CD4 T lymphocyte purification and depletion. A-B) Isolation of CD4+ T lymphocytes from tumors and lymph nodes. Selection was done by magnetic selection for CD4 followed by FACS sorting for CD3 and CD4 positive cells. The purity of isolated cells was assessed by flow cytometric analysis of pre-selection and purified cells. Representative analyses are shown from tumor and LN isolations. % CD4+ of total cells is shown.
Figure S5. CD4+ T lymphocytes are TH2 polarized in primary mammary tumors A) Analysis of intracellular cytokine expression by CD4+ T cells. CD4+ T cells were isolated by flow sorting from spleen (S), lymph nodes (LN) or tumors (T) from 95 day-old (-)LN or PyMT mice (n=3/cohort). Isolated CD4+ T cells were activated ex vivo by treatment with anti-mouse CD3 and CD28 IgGs in the presence of Brefeldin A. Cells were fixed and intracellular IL-4 and IFNγ was calculated after 6 hours of activation. The Ratio of IL-4+ to IFNγ+ CD4+ T cells is depicted as mean % of cytokine positive cells from 3 independent isolation experiments and * denotes p<0.05 by Mann-Whitney.
Figure S6: Alternatively activated TAMs induce malignant behavior. A) Quantification of pMEC organoid disruption following co-culture with TAMs (isolated from mammary tumors of 95 day-old PyMT mice). Metalloprotease activity was inhibited by treatment with a broad-spectrum MMP inhibitor GM6001 (10 μM). Disrupted organoids were quantified and data represented as a percentage of total organoids (>100 replicate/4 replicates). Data depicted as mean ± SEM and * denotes p<0.05 by Mann-Whitney.
B) Quantification of organoid disruption following co-culture of TAM (48 hours) with pMEC, in the presence of either vehicle (V) or increasing concentrations of IL-4 (2-20 ng/ml). Disrupted organoids were counted and then expressed as a percentage of the total organoids present. Disrupted organoids were counted and data represented as a percentage of the total organoids (>100 replicate/4 replicates). Data depicted as mean ± SEM and * denotes p<0.05 by Mann-Whitney.
C) Quantification of pMEC migration/chemotaxis in response to TAMs treated with IL-4 (10 ng/ml) and/or CSF-1 (10ng/ml) assessed using a Boyden chamber assay. The number of pMECs that migrated to the opposite side of the membrane was assessed by H&E staining, four 10× fields were quantified per membrane (using Image J) and 4 inserts were used for each condition. Data are represented as mean ± SEM and * denotes p<0.05 by Mann-Whitney.
D) Quantitative real-time PCR analysis for EGF mRNA expression from CD45+F4/80+Gr1- macrophages isolated by flow sorting from mammary tumors from 95 day-old PyMT/CD4+/- mice. TAMs were with treated pMEC conditioned medium (24 hours conditioning) with or without IL-4 (20 ng/ml). The comparative threshold cycle method was used to calculate fold change in gene expression normalized to β-actin as a gene reference. Representative data from 2 independent experiments is depicted as the mean fold change from the standardized sample ± SEM. In all panels, * denotes p<0.05 by Mann-Whitney.
E) Macrophages as a source of EGF in mammary carcinomas. Quantitative real-time PCR analysis of EGF mRNA expression in tumor-associated CD45+F4/80+Gr1- macrophages (TAMs), CD45+CD11b+Gr1HI IMCs, CD4+ and CD8+ T cells as well as MECs from mammary carcinomas of 100 day-old PyMT mice isolated by flow sorting. The comparative threshold cycle method was used to calculate fold change in gene expression normalized to GAPDH as reference gene. Cell isolation and gene expression analysis was performed on 4 separate cohorts of animals (1-2 mice/cohort). Representative data are depicted as mean ± SEM.
Acknowledgments
The authors thank Drs. Zena Werb and Mikala Egeblad for providing breeding colonies of PyMT mice and for assistance with mammary histopathology, Dr. Jayanata Debnath for instruction and advice with the 3D organoid culture model, and Drs. Lewis Lanier and Zena Werb for invaluable discussion and critical reading of the manuscript. L.M.C. acknowledges Drs. Bonnie Sloane, Joe Gray, Zena Werb, Mina Bissell, and Thea Tlsty for encouraging exploration of breast cancer models. D.D. acknowledges Drs. Nesrine Affara and Magnus Johansson for their scientific input, and support from the American Cancer Society (PF-07-264-01) and NCI grants (T32-CA09043 and T32-CA108462). P.A. is supported by a postdoctoral fellowship from the Cancer Research Institute. L.M.C. was supported by grants from the NIH/NCI R01CA130980, R01CA13256, R01CA098075, P01CA72006, and a DOD BCRP Era of Hope Scholar Award (W81XWH-06-1-0416).
Footnotes
Significance: DeNardo and colleagues demonstrate a tumor-promoting role for TH2-CD4+ T lymphocytes that elicit protumor, as opposed to cytotoxic bioactivities of tumor-associated macrophages and enhancement of prometastatic epidermal growth factor receptor signaling programs in malignant mammary epithelial cells. This work reveals a protumor regulatory program involving components of the acquired and cellular immune systems that effectively collaborate to promote pulmonary metastasis of mammary adenocarcinomas, and identifies cellular targets, namely CD4+ T effector cells and IL-4 for anticancer therapy.
Supplemental Data: Supplemental Data include six figures and Supplemental Experimental Procedures and can be found with this article online at http//www.cell.com/ cancer-cell/supplemental/S1535-6108(09)00216-5.
References
- Aspord C, Pedroza-Gonzalez A, Gallegos M, Tindle S, Burton EC, Su D, Marches F, Banchereau J, Palucka AK. Breast cancer instructs dendritic cells to prime interleukin 13-secreting CD4+ T cells that facilitate tumor development. J Exp Med. 2007;204:1037–1047. doi: 10.1084/jem.20061120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, Banham AH. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24:5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Coronella-Wood JA, Hersh EM. Naturally occurring B-cell responses to breast cancer. Cancer Immunol Immunother. 2003;52:715–738. doi: 10.1007/s00262-003-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel D, Chiu C, Giraud E, Inoue M, Mizzen LA, Chu NR, Hanahan D. CD4+ T Cell-mediated antigen-specific immunotherapy in a mouse model of cervical cancer. Cancer Res. 2005;65:2018–2025. doi: 10.1158/0008-5472.CAN-04-3444. [DOI] [PubMed] [Google Scholar]
- Daniel D, Meyer-Morse N, Bergsland EK, Dehne K, Coussens LM, Hanahan D. Immune enhancement of skin carcinogenesis by CD4+ T cells. J Exp Med. 2003;197:1017–1028. doi: 10.1084/jem.20021047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Girardi M, Oppenheim D, Glusac EJ, Filler R, Balmain A, Tigelaar RE, Hayday AC. Characterizing the protective component of the alphabeta T cell response to transplantable squamous cell carcinoma. J Invest Dermatol. 2004;122:699–706. doi: 10.1111/j.0022-202X.2004.22342.x. [DOI] [PubMed] [Google Scholar]
- Giraudo E, Inoue M, Hanahan D. An amino-bisphosphonate targets MMP-9-expressing macrophages and angiogenesis to impair cervical carcinogenesis. J Clin Invest. 2004;114:623–633. doi: 10.1172/JCI22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch JL, Christy B, Yee D. STAT6 mediates interleukin-4 growth inhibition in human breast cancer cells. Neoplasia. 2002;4:324–331. doi: 10.1038/sj.neo.7900248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann T, Lawrence T, McNeish I, Charles KA, Kulbe H, Thompson RG, Robinson SC, Balkwill FR. “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- Kohrt HE, Nouri N, Nowels K, Johnson D, Holmes S, Lee PP. Profile of immune cells in axillary lymph nodes predicts disease-free survival in breast cancer. PLoS Med. 2005;2:e284. doi: 10.1371/journal.pmed.0020284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek RD, Hunt NC, Landers RJ, Lewis CE, Royds JA, Harris AL. Macrophage infiltration is associated with VEGF and EGFR expression in breast cancer. J Pathol. 2000;190:430–436. doi: 10.1002/(SICI)1096-9896(200003)190:4<430::AID-PATH538>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Pollard JW. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007;67:5064–5066. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Locati M. New vistas on macrophage differentiation and activation. Eur J Immunol. 2007;37:14–16. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- Nagai S, Toi M. Interleukin-4 and breast cancer. Breast Cancer. 2000;7:181–186. doi: 10.1007/BF02967457. [DOI] [PubMed] [Google Scholar]
- Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- Pullan SE, Streuli CH. The mammary gland epithelial cell. In: Harris A, editor. Epithelial Cell Culture. Cambridge: Cambridge University Press; 1996. pp. 97–121. [Google Scholar]
- Saccani A, Schioppa T, Porta C, Biswas SK, Nebuloni M, Vago L, Bottazzi B, Colombo MP, Mantovani A, Sica A. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- Tiemessen MM, Jagger AL, Evans HG, van Herwijnen MJ, John S, Taams LS. CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proc Natl Acad Sci USA. 2007;104:19446–19451. doi: 10.1073/pnas.0706832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M, Lombardo Y, Francipane MG, Alea MP, Cammareri P, Iovino F, Di Stefano AB, Di Bernardo C, Agrusa A, Condorelli G, et al. Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4. Cell Death Differ. 2008;15:762–772. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- Trzonkowski P, Szmit E, Mysliwska J, Mysliwski A. CD4+CD25+ T regulatory cells inhibit cytotoxic activity of CTL and NK cells in humans-impact of immunosenescence. Clin Immunol. 2006;119:307–316. doi: 10.1016/j.clim.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Uzzan B, Nicolas P, Cucherat M, Perret G. Microvessel density as a prognostic factor in women with breast cancer: a systematic review of the literature and meta-analysis. Cancer Res. 2004;64:2941–2955. doi: 10.1158/0008-5472.can-03-1957. [DOI] [PubMed] [Google Scholar]
- Woo EY, Yeh H, Chu CS, Schlienger K, Carroll RG, Riley JL, Kaiser LR, June CH. Cutting edge: Regulatory T cells from lung cancer patients directly inhibit autologous T cell proliferation. J Immunol. 2002;168:4272–4276. doi: 10.4049/jimmunol.168.9.4272. [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, Moses HL. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The impact of CD4+ T cells on primary mammary tumor development A-B) Kaplan Meyer analysis of tumor incidence in CD4, CD8 and B cell-deficient/PyMT mice. The percent of tumor free animals is depicted for PyMT/CD4+/-CD8+/- (n=10), PyMT/CD4-/-CD8-/- (n=46), PyMT/CD4-/-CD8+/- (n=34), PyMT/CD4+/-CD8-/- (n=31) as well as PyMT/JH+/- (n=23) and PyMT/JH-/- (n=28) mice. Animals were considered to be tumor free until a palpable mass (>4.0 mm) persisted for longer then 4 days. There were no statistical differences between tumor incidence curves by generalized Wilcoxon test.
C) Representative images of H&E stained mammary tumor tissue from 76 and 95-day old PyMT/RAG1+/- and PyMT/RAG1-/- mice. Magnification and scale are shown.
D) Flow cytometric analysis of CD31+ endothelial cells in mammary tumors from 95 and 110 day-old PyMT/RAG1+/-, PyMT/RAG1-/-, PyMT/CD4+/-CD8+/-, PyMT/CD4-/-CD8-/- mice (n=4). Data is depicted as the mean % CD31+ cells of the total live cells/tumor ±SEM. No statistical differences were found between groups by Mann-Whitney test. Representative 20x images of paraffin embedded tumor sections from PyMT/CD4+/-, PyMT/CD4-/- sections stained for CD31+ and vasculature revealed as brown staining. Bar in both = 100 μm.
Figure S2: CD4+ T lymphocytes regulate bioeffector function of myeloid cell subsets A-B) Isolation of TAMs from the mammary tumors of PyMT/CD4+/- and PyMT/CD4-/- mice. The purity of isolated CD45+F4/80+Gr1- macrophages was assessed by flow cytometric analysis of the cell suspension before and after selection. Representative analyses are shown from PyMT/CD4+/-(red) and PyMT/CD4-/- (blue) tumors and relative CD45, CD11b, Gr1 and F4/80 expression depicted.
C) Cytokine expression analysis from TAM-conditioned medium. Tumor-associated CD45+F4/80+Gr1- macrophages were isolated by dual magnetic and flow sorting of mammary tumors from 95 day-old PyMT/RAG1+/- and PyMT/RAG1-/- mice (n=3/cohort). Cytokine expression was assessed by ELISA of conditioned medium from TAMs (25,000) following 18 hours of culture, with or without recombinant IL-4 (10 ng/ml). Representative assays from 2 independent cohorts each performed at least in triplicate are depicted as means ± SEM and * denotes p<0.05 by student t test.
D) Lymphocyte depletion by anti-CD4 IgG (GK1.1) was assessed by flow cytometric analysis 4 days after of IgG injection. Representative analyses from anti-CD4 IgG and control IgG treated mice are shown from blood isolated by left ventricle heart puncture. % CD4+ of total cells is depicted.
E) Analysis of TAM polarization following CD4+ T cells depletion. Tumor-associated CD45+F4/80+Gr1- macrophages were isolated by flow sorting mammary tumors from PyMT mice treated with either anti-CD4 IgG (GK1.1) or control IgG. Tumor bearing 85 day-old mice were treated twice over 10 days with Igs and TAMs were isolated from tumors when mice were 95 day-old. Cytokine expression was assessed by ELISA analysis of conditioned medium from TAMs following 18 hours of culture. Gene expression analysis used the comparative threshold cycle method to calculate fold change in gene expression normalized to GAPDH as reference gene. Representative assays from 2 independent cohorts each run at least in triplicate are depicted as mean values ± SEM. * denotes n p<0.05 by Mann-Whitney.
F) Analysis of IMC polarization following CD4+ T cells depletion. Tumor-associated CD45+CD11b+Gr1Hi IMCs were isolated by flow sorting mammary tumors from PyMT mice treated with either anti-CD4 IgG (GK1.1) or control IgG. Tumor bearing 85 day-old mice were treated twice over 10 days with Igs and TAMs were isolated from tumors when mice were 95 day-old. Cytokine expression was assessed by ELISA and quantitative real-time PCR as described above. Representative assays from 2 independent cohorts each run at least in triplicate are represented as mean values ± SEM. * denotes p<0.05 by Mann-Whitney.
Figure S3: Cytokine profiles of TAMs from CD25 Treg-depleted mice. A) Analysis of CD25+ T cell depletion. Tumor bearing 85 day-old PyMT mice (5 mice/cohort) were treated with either anti-CD25 IgG (PC61) or control IgG three times over 15 days. Peripheral blood was analyzed for expression of FOXP3 and CD25 expression on CD4+ T lymphocytes every 5 days by flow cytometry to affirm depletion. Representative analysis for depleted versus control 100 day-old PyMT mice is depicted as FACS plots gated on CD4+ lymphocytes.
B) Quantitative real-time PCR analysis of Nos2, Arg1, Tgfβ, IL-12a and IL-10 mRNAs expression in tumor-associated CD45+F4/80+Gr1- macrophages isolated by flow sorting from mammary tumors of 100 day-old PyMT mice treated with either anti-CD25 Ig (PC61) or isotype control IgG. The comparative threshold cycle method was used to calculate fold change in gene expression normalized to GAPDH as reference gene. Cell isolation and gene expression analysis was performed on 2 separate cohorts of animals (3 mice/group) with similar results. Representative data are depicted as means ± SEM. No statistical differences were found between treated and untreated groups by Mann-Whitney.
C) Cytokine expression analysis from TAM conditioned medium. Tumor-associated CD45+F4/80+Gr1- macrophages were isolated by flow sorting from mammary tumors from 100 day-old PyMT mice treated with either anti-CD25 Ig (PC61) or isotype control IgG for 15 days. Cytokine expression was assessed by ELISA analysis of conditioned medium from TAMs following 18 hours of culture. Cell isolation and cytokine analysis was performed on 3 separate cohorts of animals (2 mice/group) with similar results. Representative data are depicted as mean ± SEM. No statistical differences were found between treated and untreated groups by Mann-Whitney.
Figure S4. CD4 T lymphocyte purification and depletion. A-B) Isolation of CD4+ T lymphocytes from tumors and lymph nodes. Selection was done by magnetic selection for CD4 followed by FACS sorting for CD3 and CD4 positive cells. The purity of isolated cells was assessed by flow cytometric analysis of pre-selection and purified cells. Representative analyses are shown from tumor and LN isolations. % CD4+ of total cells is shown.
Figure S5. CD4+ T lymphocytes are TH2 polarized in primary mammary tumors A) Analysis of intracellular cytokine expression by CD4+ T cells. CD4+ T cells were isolated by flow sorting from spleen (S), lymph nodes (LN) or tumors (T) from 95 day-old (-)LN or PyMT mice (n=3/cohort). Isolated CD4+ T cells were activated ex vivo by treatment with anti-mouse CD3 and CD28 IgGs in the presence of Brefeldin A. Cells were fixed and intracellular IL-4 and IFNγ was calculated after 6 hours of activation. The Ratio of IL-4+ to IFNγ+ CD4+ T cells is depicted as mean % of cytokine positive cells from 3 independent isolation experiments and * denotes p<0.05 by Mann-Whitney.
Figure S6: Alternatively activated TAMs induce malignant behavior. A) Quantification of pMEC organoid disruption following co-culture with TAMs (isolated from mammary tumors of 95 day-old PyMT mice). Metalloprotease activity was inhibited by treatment with a broad-spectrum MMP inhibitor GM6001 (10 μM). Disrupted organoids were quantified and data represented as a percentage of total organoids (>100 replicate/4 replicates). Data depicted as mean ± SEM and * denotes p<0.05 by Mann-Whitney.
B) Quantification of organoid disruption following co-culture of TAM (48 hours) with pMEC, in the presence of either vehicle (V) or increasing concentrations of IL-4 (2-20 ng/ml). Disrupted organoids were counted and then expressed as a percentage of the total organoids present. Disrupted organoids were counted and data represented as a percentage of the total organoids (>100 replicate/4 replicates). Data depicted as mean ± SEM and * denotes p<0.05 by Mann-Whitney.
C) Quantification of pMEC migration/chemotaxis in response to TAMs treated with IL-4 (10 ng/ml) and/or CSF-1 (10ng/ml) assessed using a Boyden chamber assay. The number of pMECs that migrated to the opposite side of the membrane was assessed by H&E staining, four 10× fields were quantified per membrane (using Image J) and 4 inserts were used for each condition. Data are represented as mean ± SEM and * denotes p<0.05 by Mann-Whitney.
D) Quantitative real-time PCR analysis for EGF mRNA expression from CD45+F4/80+Gr1- macrophages isolated by flow sorting from mammary tumors from 95 day-old PyMT/CD4+/- mice. TAMs were with treated pMEC conditioned medium (24 hours conditioning) with or without IL-4 (20 ng/ml). The comparative threshold cycle method was used to calculate fold change in gene expression normalized to β-actin as a gene reference. Representative data from 2 independent experiments is depicted as the mean fold change from the standardized sample ± SEM. In all panels, * denotes p<0.05 by Mann-Whitney.
E) Macrophages as a source of EGF in mammary carcinomas. Quantitative real-time PCR analysis of EGF mRNA expression in tumor-associated CD45+F4/80+Gr1- macrophages (TAMs), CD45+CD11b+Gr1HI IMCs, CD4+ and CD8+ T cells as well as MECs from mammary carcinomas of 100 day-old PyMT mice isolated by flow sorting. The comparative threshold cycle method was used to calculate fold change in gene expression normalized to GAPDH as reference gene. Cell isolation and gene expression analysis was performed on 4 separate cohorts of animals (1-2 mice/cohort). Representative data are depicted as mean ± SEM.