Abstract
Nearly identical cells can exhibit substantially different responses to the same stimulus. We monitored the nuclear localization dynamics of nuclear factor κB (NF-κB) in single cells stimulated with tumor necrosis factor-α (TNF-α) and lipopolysaccharide (LPS). Cells stimulated with TNF-α have quantitative differences in NF-κB nuclear localization, whereas LPS-stimulated cells can be clustered into transient or persistent responders, representing two qualitatively different groups based on the NF-κB response. These distinct behaviors can be linked to a secondary paracrine signal secreted at low concentrations, such that not all cells undergo a second round of NF-κB activation. From our single-cell data, we built a computational model that captures cell variability, as well as population behaviors. Our findings demonstrate that mammalian cells can create “noisy” environments in order to produce diversified responses to stimuli.
Introduction
How can virtually identical cells in the same environment exhibit such diverse phenotypes? This phenomenon has been observed in various systems, examples include differentiation to competence (1-3) and antibiotic persistence (4) in bacteria, retinal development in Drosophila (5), and galactose utilization in yeast (6). In many cases, the causative factor in creating the diverse phenotypes was a “noisy” environment in which a key factor was present at low concentrations (7, 8).
Nuclear factor κB (NF-κB) is a transcription factor family that regulates the expression of hundreds of genes (9). Although primarily involved in the innate immune response, NF-κB has been identified as an important protein in such diverse processes as tumor progression in cancer (10), learning (11), epigenetic regulation of gene expression (12), and aging (13). Because of its importance, the NF-κB-related signaling network has been relatively well studied, which, in turn, has made it the primary test bed for systems biology approaches in mammalian cells. For example, high-throughput and systems approaches have been used to reconstruct the NF-κB signaling network (14) and to determine target genes (15-17) and transcription factor binding sites (18, 19) at the global level. NF-κB activation has also been the subject of several computational modeling studies, beginning with a detailed model of the effects of various inhibitor of κB (IκB) proteins on NF-κB activation mediated by tumor necrosis factor-α (TNF-α) (20), which characterized the oscillatory shuttling of NF-κB across the nuclear membrane, which is characteristic of TNF-α-dependent NF-κB activation, and predicted the effects of transient TNF-α signals on gene expression and NF-κB activation. The Hoffmann-Levchenko (HL) model has since been expanded to include certain other components, such as the activity of inhibitor of κB kinase (IKK) (21), feedback regulation by the ubiquitin-modifying enzyme A20 (22), and the effects of varying TNF-α concentration (23). These and other studies (24-27) have enhanced the understanding of NF-κB signaling pathway dynamics, particularly in response to TNF-α stimulation.
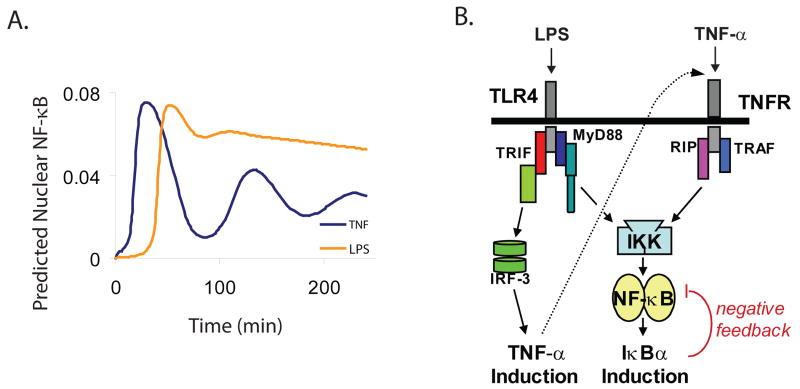
We were originally intrigued by the observation that NF-κB activation undergoes damped oscillations in cells that are stimulated by TNF-α but activation is stable when lipopolysaccharide (LPS) is the stimulus (28) (Fig. 1A). TNF-α stimulates the TNF receptor (TNFR), which interacts with the cytoplasmic adaptors RIP and TRAF, and LPS stimulates the Toll-like receptor TLR4, which interacts with the cytoplasmic adaptors TRIF and MyD88 (Fig. 1B). The oscillatory behavior observed with TNF-α stimulation depends on a negative feedback circuit due in large part to the NF-κB induced expression of the gene encoding inhibitory protein IκBα (29-31). The stable activation observed with LPS stimulation depends on the coordination of two independent pathways upstream of NF-κB mediated through two different adaptor proteins, a MyD88-dependent and a TRIF-dependent pathway (29, 32-35). By expanding the HL model to include mathematical representations of the MyD88- and TRIF-dependent pathways, we predicted that the stable activation of NF-κB occurring in response to LPS stimulation depended on the contributions of two pathways that oscillated out of phase with each other. The model simulations further indicated that oscillations were set out of phase by a time delay in the TRIF-dependent pathway, which we identified experimentally as a gene expression event whereby the transcription factor interferon regulatory factor 3 (IRF-3) was activated and caused expression of the gene encoding TNF-α. Secretion of TNF-α re-stimulated NF-κB through the pathway mediated downstream of the TNFR, its own pathway (Fig. 1B).
Fig. 1. LPS and TNF-α signal through separate receptors and pathways to trigger NF-κB activation with different dynamics.
(A) Population-based computational models of NF-κB nuclear localization (20, 28). (B) A schematic of NF-κB activation by TNF-α and LPS. Dashed line from TNF-α induction to TNFR represents secretion of TNF-α. The red and green proteins associated with TLR4 are TRAM and TRIF, respectively. The dark blue and teal blue proteins associated with TLR4 are Mal and Myd88, respectively.
The nuclear translocation dynamics of transcription factor NF-κB have been observed at the single-cell level with fluorescent protein fusions (36-39). These studies indicated that NF-κB nuclear localization in single cells differs from the average localization observed in populations of cells. Theoretical analyses of NF-κB dynamics using the Gillespie algorithm to model fluctuations in chemical kinetics (40) and noise in gene transcription (41, 42) have postulated that the average population-level behavior can be explained from single-cell variation due to stochastic effects. In this paper, we combined live-cell imaging and computational modeling to characterize NF-κB dynamics at the single-cell level in response to LPS and TNF-α.
Results
LPS and TNF-α trigger dynamically distinct activation profiles for NF-κB
To compare TNF-α- and LPS-dependent NF-κB activation at the single-cell level, we created a system to monitor near-endogenous amounts of NF-κB in primary cells (fig. S1A) (37). The most common form of NF-κB is a heterodimer comprised of the transcription factors p50 and p65. We cloned a p65-fluorescent protein (FP) fusion, with DsRed or enhanced green fluorescent protein (EGFP), into a lentiviral system to stably infect cells (43). Because researchers have disagreed about the effect of p65 overexpression upon NF-κB dynamics (36, 44, 45), we mimicked the endogenous regulation of NF-κB by cloning the 1.5 kb upstream of the relA gene (encoding p65) into the lentiviral construct to control expression of p65-FP and then infected relA-knockout 3T3 cells. The response of p65-FP reconstituted cells to TNF-α at the population level was the similar to the response of wild-type 3T3 cells in terms of the degradation kinetics and NF-κB-dependent expression of IκBα (fig. S1B). As in other studies (20, 28), we focused on the first four hours of stimulation in order to minimize the extensive downstream effects that follow NF-κB activation (16).
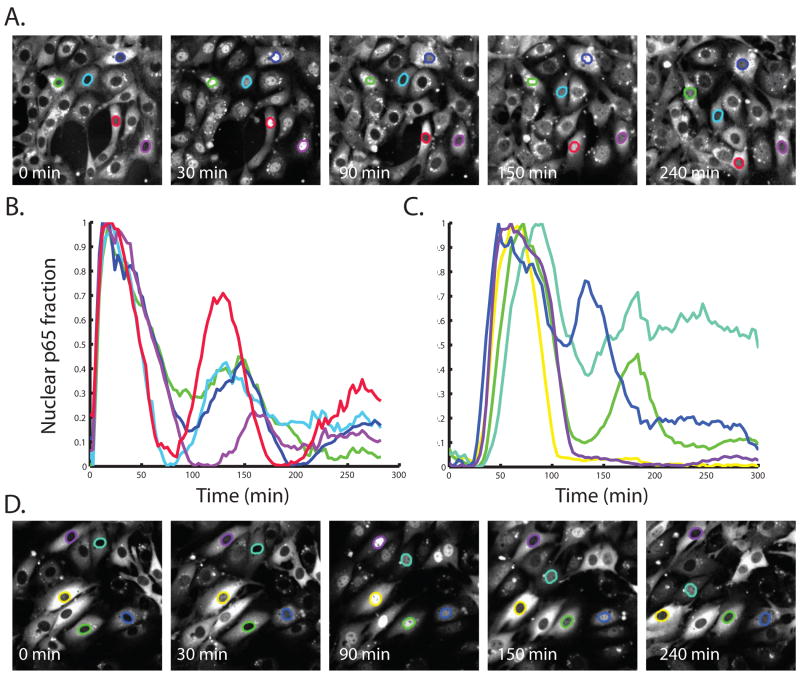
We monitored the movement of NF-κB to and from the nucleus by time-lapse confocal microscopy of the p65-FP-expressing cells over 4 or more hours in response to TNF-α or LPS (Fig. 2A, D). Nuclear images were analyzed to quantify the amount of nuclear NF-κB at any given time (Fig. 2B, C). This analysis revealed a key difference between cells stimulated with TNF-α versus LPS. In the case of TNF-α, we saw that the single-cell traces looked qualitatively similar with most of the cells exhibiting some oscillatory NF-κB behavior in agreement with fluorescence microscopy studies with cells transfected with p65-FP (36, 37) and cells isolated from p65-FP knock-in mice (46) (Fig. 2A, B). In contrast, the LPS-stimulated cells exhibited qualitative differences in NF-κB nuclear localization at late times (>2 hours) after stimulation (Fig. 2C, D).
Fig. 2. NF-κB nuclear localization exhibits oscillatory dynamics when cells are stimulated with TNF-α but stable nuclear accumulation when cells are stimulated with LPS.
(A and D) Single-cell images of EGFP-p65 transduced relA-/- 3T3 cells exposed to TNF-α (A) or LPS (D) for the indicated times. (B) Time course showing the NF-κB localization in the cells in A. (C) Time course showing the NF-κB localization in the cells in B. The colors used to highlight nuclei in (A) and (D) correspond to traces in (B) and (C). Time course data were normalized by the minimum and maximum value of nuclear NF-κB during the time course to account for the varying overall intensities in different cells. All scale bars represent 25 μm.
Cluster analysis reveals three discrete populations of cells, one for the TNF-α response phenotype and two for LPS response phenotype
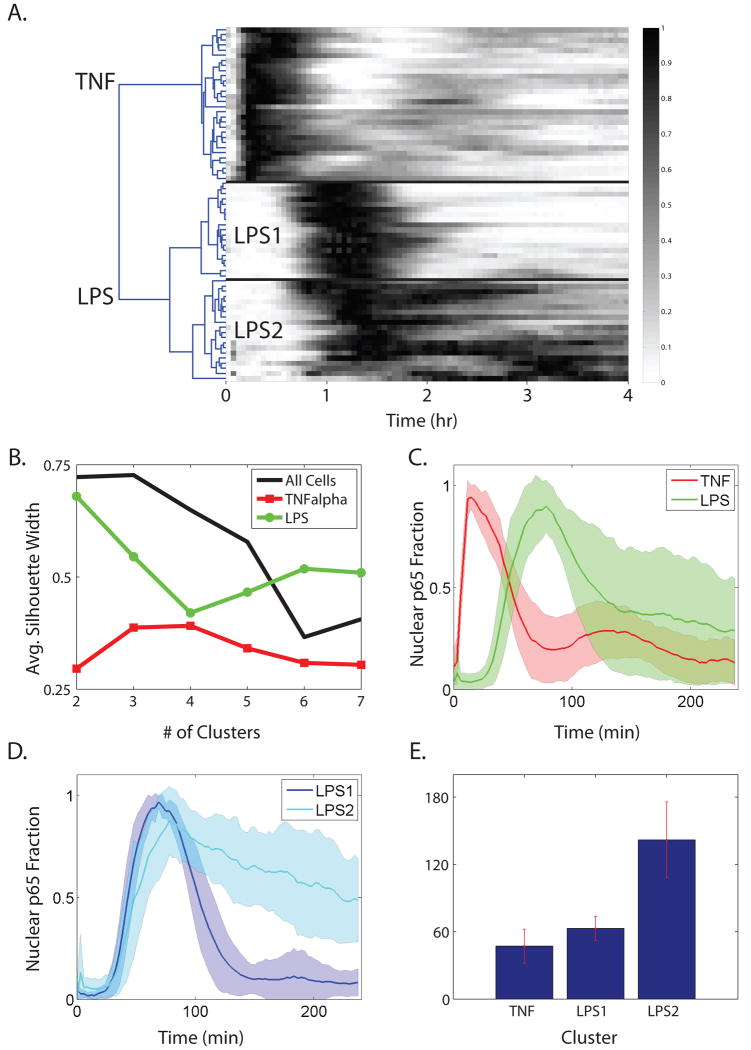
In order to quantify the variation in cellular response for cells stimulated with TNF-α or LPS, we grouped the NF-κB activation profiles using hierarchical clustering and silhouette analysis (Fig. 3A, B). The average silhouette width is a metric of how well the data can be separated into a given number of clusters and is commonly used for microarray analysis, among other applications (47). Silhouette analysis (Fig. 3B) of the single-cell profiles indicated that the data could be most appropriately divided into three major clusters. The first division in the dendrogram was between the cells responding to TNF-α and cells responding to LPS. The primary feature dividing the TNF-α and LPS responses is the substantially later initial NF-κB nuclear localization in LPS-stimulated cells (Fig. 3C). The second division involved only LPS-stimulated cells. All cells stimulated with LPS showed initial NF-κB nuclear localization at roughly the same time, then two populations emerged. For the LPS1 group NF-κB left the nucleus quickly, whereas for the LPS2 group NF-κB persisted in the nucleus for hours (Fig. 3D), more than double the time of either the TNF-α-stimulated cells or cells exhibiting the LPS1 phenotype (Fig. 3E).
Fig. 3. NF-κB nuclear localization time courses can be clustered into distinct groups.
(A) Cluster diagram for 69 cells observed in 8 different experiments, clustered hierarchically using angle cosine as a distance metric between time courses. The diagram is organized and shaded to resemble a series of gel shift assay results stacked on top of each other. (B) Average silhouette widths for different subsets of the dataset calculated for variable number of clusters. (C) TNF-α- and LPS-stimulated cells can be separated into two separate groups on the basis of the dynamics of the initial NF-κB activiation. (D) LPS-stimulated cells can be further separated into two clusters. The shaded areas in C and D correspond to the standard deviation of the cluster around the cluster average. (E) Duration of time where nuclear p65-EGFP was greater than 50% of the maximal value for each cluster.
We found that variation in cellular NF-κB dynamics is much greater between clusters than the variation within a cluster. (Fig.3 A, B). For example, the TNF-α-stimulated cells all exhibited similar, oscillatory behavior with only relatively minor differences in dynamics (fig. S2). The first peak of NF-κB localization has little variation in timing, whereas the second peak is more variable. In addition, the relative timing and amplitudes between the first and second peak also vary between individual cells.
Computational modeling reveals potential mechanisms underlying single-cell variation in TNF-α-dependent NF-κB activation
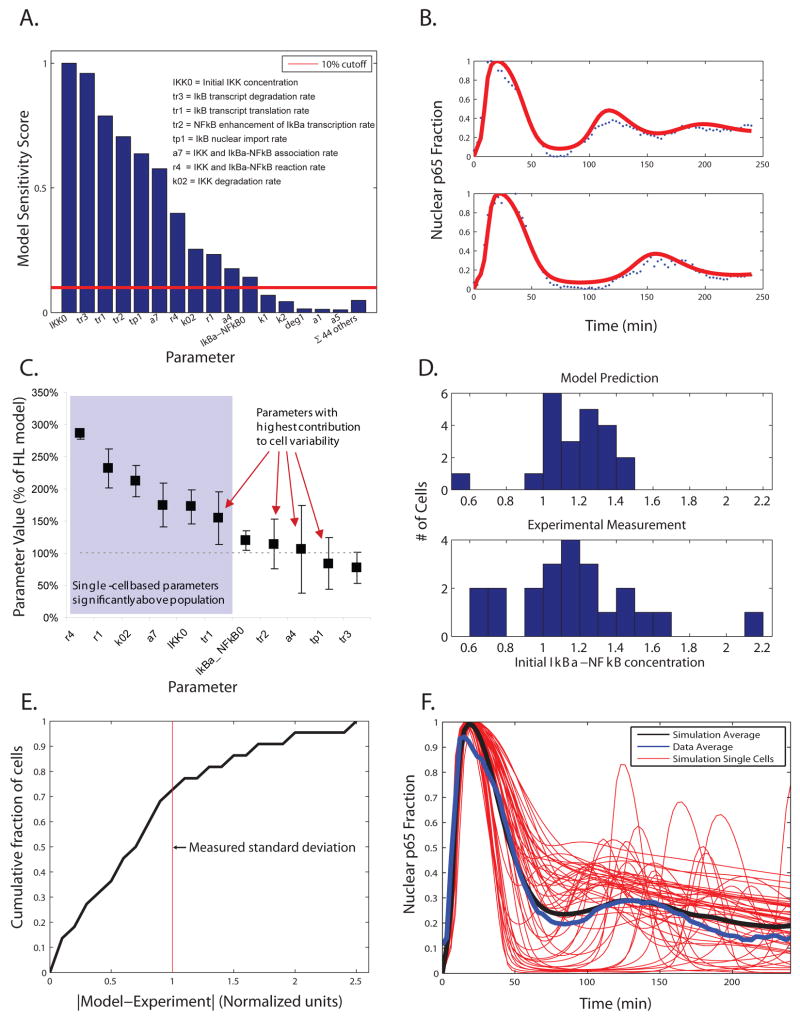
We used computational modeling to determine the cause of the single-cell variation for cellular response to TNF-α and LPS. The HL computational model was based on population-level analysis and cannot predict the variant behavior of individual cells. Because the qualitative behaviors exhibited by single cells agreed generally with the population-level response, we postulated that we could capture single-cell behavior simply by varying certain parameters in the population model. To pursue this hypothesis, we performed a sensitivity analysis to see in which cases a small parameter change would lead to a high degree of variation in the NF-κB localization response (Fig. 4A). Our analysis highlighted eleven parameters, including two initial conditions, as suitable candidates to vary. The two initial conditions were the resting concentrations of IKK and IκBα bound to NF-κB. These parameters all related to the upstream IKK signal or the negative feedback by IκBα, as expected (25).
Fig. 4. Characterizing and modeling the quantitative differences in NF-κB localization dynamics for TNF-α-stimulated cells.
(A) Sensitivity analysis of the base HL model. Each parameter in the model was varied +/- 50% and the distance between the resulting simulations and the base model were calculated and added as shown. We chose parameters for which the score was 10% or higher of the maximum value. (B) Fitting the parameters to single-cell data. The eleven parameters identified in A were fit to cellular NF-κB activation time courses as described in Materials and Methods. Two representative fits are shown and the remaining fits can be found in fig. S3. (C) The resulting distributions for all sets, shown as an average and standard deviation as a percentage of the base HL model (100%, dashed line) value. The distributions most closely fit a mixture of Gaussians, with parameters as shown in table S1. (D) Distribution of fitted and experimental values for the parameter IκBα_NFκB0 (the initial concentration of the IκBα and NF-κB complex). (E) Correlation between model fit and experimental measurement plotted as the cumulative fraction of cells versus error. (F) Representative and average results from a set of ten thousand model simulations where the values of the eleven parameters were chosen randomly from each distribution.
We assumed that the values of these eleven parameters could vary somewhat between cells and used our single-cell NF-κB time-course data to identify possible distributions for these values. Each single-cell time course was used to fit the set of eleven parameters using a gradient descent method where fits were constrained to about one order of magnitude of original parameter value (roughly a three-fold increase or decrease). For 23 out of 30 cells, we identified parameter sets that gave a good model fit of the data (Fig. 4B, fig. S3).
We hypothesized that, taken together, the distribution of these fit parameter values should approximate the distribution across the cell population. It is less likely that each individual fit parameter value corresponds specifically to the physical properties of a given cell. Our parameter distributions highlighted two main differences between the single-cell and population data (Fig. 4C). First, there was a set of 6 parameter distributions with mean values that were higher for single cells than the values of the same parameters that were observed for the population. This would imply that deriving these parameters from population data could be misleading, for example because of single cells exhibiting asynchronous oscillations (44).
A second set of parameter distributions had relatively large standard deviations, so these parameters may be thought of as the key contributors to cellular variation and noise in the single-cell response (Fig. 4C). In general, noise-related effects on phenotype may be classified as intrinsic (attributable to stochastic events inherent to gene expression) or extrinsic (dependent on fluctuations in cellular environment or regulatory factors) (48, 49). The model parameters with the largest coefficients of variation include IκBα translation, transcription, association with NF-κB, and nuclear import rates, which correspond to sources of intrinsic noise (49).
We tested our predicted parameter distributions by investigating the initial concentration of the IκBα-NF-κB complex. We measured the average cytoplasmic fluorophore intensity in each cell for each dataset. The distribution of initial IκBα-NF-κB predicted by the fitted parameters is similar to that detected experimentally in cells (Fig. 4D). In individual single cells, the parameter fits were within one standard deviation of the measured value in ∼70% of the cells (Fig. 4E).
It should be noted that these parameter distributions do not necessarily represent biologically relevant kinetic rates, given the stochastic nature of reactions at the single-cell level (40) and loose parameter sensitivities in large models of biological systems (50). Furthermore, other models exist that explicitly model stochastic gene expression (37, 42), albeit without all of the IκB isoforms that contribute to the response to TNF-α (20). However, these findings underscore the importance of determining certain parameter values at the cellular level, and suggest that not all parameters contribute equally to single-cell variability. Furthermore, the cellular variation in initial concentrations of the IκBα-NF-κB complex represents an independent validation of our single cell-based computational approach.
We wanted to see if the parameter distributions that we determined from our single-cell time courses could be used to adapt the population-based computational model to single-cell studies. We ran the model 10,000 times, with a different set of eleven parameter values randomly chosen from the distributions each time, and found that the new set of simulations predicted both the variation in single-cell behavior and the population-level response (Fig. 4F). Thus, a computational model that accounts for variation in cell behavior may be a more accurate and comprehensive representation of TNF-α-dependent NF-κB activation.
A paracrine TNF-α signal produces the late phase of LPS-dependent NF-κB activation
As mentioned above, the cells stimulated with LPS exhibited more qualitative differences in their NF-κB localization dynamics, clustering into a transient and a persistent group with respect to NF-κB nuclear localization (Fig. 3A, D). These distinct clusters are present even when the stimulated cells are clonal (fig. S4). LPS-dependent activation of NF-κB occurs through two independent pathways, one that depends on MyD88 and another that is depends on TRIF (51). On the basis of our earlier work, it seemed possible that the difference in behavior depended on which of these pathways were activated. Therefore, we infected primary mouse embryo fibroblasts (MEFs) that were deficient in MyD88 or TRIF with our p65-FP construct. The Trif-/- MEFs showed a peak activation of NF-κB (nuclear fraction > 0.5) between 20 and 75 minutes whereas the MyD88-/- MEFs had peak activity between 60 and 115 minutes (Fig. 5A). Neither of these knockout MEFs had nuclear NF-κB > 0.5 after 130 minutes, either individually or summed together, suggesting that either a synergy between the two pathways exists, or that interactions from additional pathways are needed to recapitulate the full persistent response.
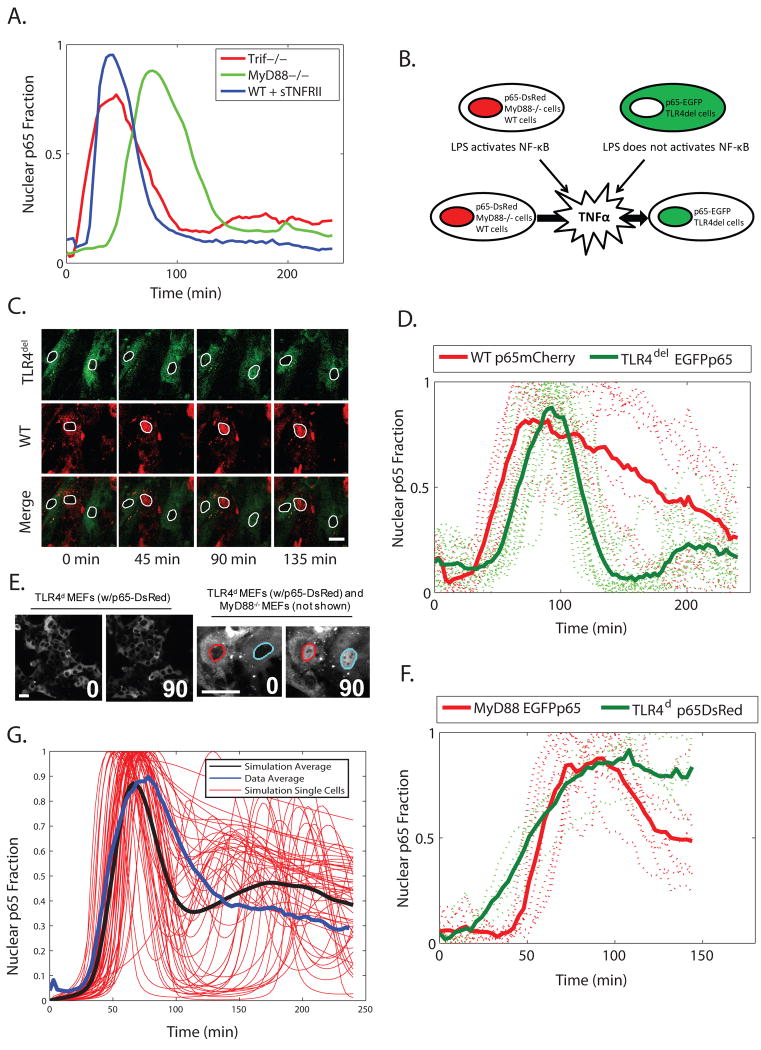
Fig. 5. A noisy paracrine signal determines the cellular NF-κB response to LPS.
(A) Average NF-κB nuclear localization in Trif-/- MEFs and in MyD88-/- MEFs, as well as wild-type 3T3 cells (WT) pre-treated with soluble TNF-α receptor (sTNFRII), over time. (B) Schematic of the approach to assess paracrine signaling by TNF-α. Wild-type or MyD88-/- and TLR4-deficient (TLRdel or TLRd) cells are grown together, each labeled with a different color of fluorescent protein. Activation of NF-κB in TLRdel would occur through the paracrine pathway (Fig. 1B). (C) TLR4del and wild-type MEFs cultured together and stimulated with LPS (5 μg/ml). Nuclei are outlined for clarity. (D) Single-cell traces for the experiments presented in C. Bold lines correspond to average behavior. (E) TLR4d and MyD88-/- MEFs cultured together and stimulated with LPS (0.5 μg/ml). TLR4d cells do not respond to LPS (left), but do respond in the presence of MyD88-/- MEFs (right). Although not present in the field imaged, MyD88-/- MEFs were present in the culture. (F) Single-cell traces for the experiments presented in E. Bold lines correspond to average behavior. (G) Representative and average results from a set of ten thousand model simulations where activation of the TRIF-dependent pathway occurs as a random event. All scale bars represent 25 μm.
Late LPS-dependent activation of NF-κB through the TRIF pathway depends on the induction of the gene encoding TNF-α by IRF-3 (28). Consistent with this, by treating the p65-FP 3T3s with a soluble TNFR protein to block the TNF-α signal, we abolished the late response of NF-κB (Fig. 5A). Previously, we reported that TNF-α is secreted in very low concentrations in response to signaling through the TRIF pathway (< 30 pg/mL, roughly corresponding to a ratio on the order of 1 TNF-α receptor to 1 TNF-α ligand) (28). Here, we found that a greater number of MyD88-/- MEFs failed translocate NF-κB to the nucleus in response to LPS than was observed for the p65-FP 3T3s and the Trif-/- MEFs (fig. S5). Based on these observations, we thought the NF-κB persistence in the nucleus, but not the initial response, could depend on whether a given cell is stimulated by an adequate amount of TNF-α.
There are multiple possibilities for the propagation of the TNF-α signal. With such low concentrations, it may be TNF-α acts as a strictly autocrine signal: The secreted TNF-α may never or rarely pass the receptors located on the secreting cell, in which case the variation in cell response is determined by whether or not TNF-α is produced by a given cell. Alternatively, if the signal is paracrine then the variation is more likely due to the low TNF-α concentration outside the cell, making the secondary TNF-α activation of NF-κB a stochastic event subject to extrinsic noise.
We applied multiple methods to differentiate between an autocrine and a paracrine TNF-α signal. First, we looked for coupling of activation dynamics between neighboring cells. Although we found no correlation between spatial distance and clustering distance in cells stimulated with either TNF-α or LPS (fig. S6), this is not sufficient to rule out paracrine signaling, because the range of a paracrine signal in tissue culture experiments, with a relatively high ratio of culture medium to cells, can be on the order of hundreds of cell lengths (52).
Therefore, we constructed a system to monitor the communication between cells through the secondary TNF-α signal (Fig. 5B). We used two different p65-fluorescent protein fusion constructs (DsRed and EGFP), and MEFs of different genotypes: wild-type cells that were responsive to LPS and TLR4 deletion mutant (TLR4del) cells that were unresponsive to LPS but still responsive to TNF-α. We transduced wild-type MEFs with p65-DsRed and TLR4del MEFs with EGFP-p65 and stimulated the cells with LPS. Individually, the TLR4del mutant cells show no activation of NF-κB in response to LPS (fig. S7) and normal activation of NF-κB in response to TNF-α (fig. S8). By growing both types of cells together, we observed the transmission of the TNF-α signal from the LPS-responsive cells to the LPS-unresponsive cells. In both cell types, NF-κB had localized to the nucleus (Fig. 5C, D). Cotreatment with soluble TNF receptor abolished NF-κB activation in TLR4del cells when cultured with wild-type MEFs (fig. S9). A decreased number of TLR4del cells showed nuclear NF-κB in response to LPS when plating density was decreased (fig. S10). To determine whether the TNF-α secretion mediated by signaling through the TRIF pathway is sufficient for paracrine signaling, we cultured MyD88-/- and TLR4-deficient MEFs individually and together, and stimulated the cells with LPS. When cultured together and then stimulated, both cell types showed similar late NF-κB activation kinetics (Fig. 5F), suggesting that they are responding to the same TNF-α signal. From this evidence of direct transmission of the TNF signal, we conclude that the TNF-α signal is paracrine, and that persistent NF-κB activation is determined by the noisy event of mediated by low concentrations of TNF-α.
Addition of the paracrine TNF-α signal to the computational model recapitulates single-cell and population responses to LPS
To determine if inclusion of this TNF-α switch more accurately models the LPS-dependent NF-κB response in single cells, we adapted the single cell-based model for TNF-α stimulation with the parameter distributions described above to include the MyD88 and TRIF pathway representations from our previous work (28). We observed that, in response to LPS, the TRIF-deficient MEFs (fig. S11) and the MyD88-deficient MEFs (fig. S12) exhibited variations in activation time, with standard deviations of 7.7 and 11.2 minutes, respectively. Utilizing the parameter distributions we obtained for the response to TNF-α (table S1), the model predicted variations of 4.6 and 8.8 minutes, respectively. Because both the model and the experimental results yield differences in activation time, we assumed that models for the individual pathways include the information necessary to recapitulate the experimentally observed variation in activation time and that we could use the kinetic parameters relating the two pathways from in our previous study in the current model (28).
Therefore, we added one additional stochastic variable and the corresponding parameter to determine whether or not the cells received the secondary TNF-α signal, and ran 10,000 simulations of the new model of NF-κB activation in response to LPS. The resulting model accounted for both transient and persistent cellular behaviors and reproduced the population-level response to LPS (Fig. 5G). The model includes at least some of the synergy between the MyD88 and TRIF pathways, which was encoded in the computational model as a linear addition of the IKK activation profiles created by each pathway.
Discussion
Previously, we found that LPS-dependent activation of NF-κB depended on the integration of two upstream signalling pathways. Through live cell imaging, we observed two distinct NF-κB localization patterns in single cells stimulated with LPS. By co-culturing specifically labelled cells of different genotype together, we could also observe the transmission of a paracrine TNF-α signal between cells. Based on these observations, we present a framework for extending existing population models of NF-κB activation to reflect the diverse responses of individual cells.
Several different sets of parameters and additions to the Hoffmann-Levchenko model have been suggested (21, 23, 25, 27), with different rationales. Other models include explicit representation of the stochastic processes of transcription and translation to account for variations in single cell responses (40-42). Of these, only the original HL model has been extended to capture the behaviour of LPS-stimulated cells (28). We therefore modified the existing LPS-dependent population model to capture the behaviour of single cells. This choice had the additional advantage of allowing us to determine how the fit values of various parameters varies at the single cell and population levels.
This multicolor co-culture system is a novel method using genetic mutants modified to fluoresce in different channels to restrict the emission and monitor the detection of secreted signals. This enables us to experimentally interrogate paracrine and autocrine signalling models. Given the spatial information that we also obtain using this method, it should be possible to quantitatively analyze autocrine and paracrine signalling. This should complement existing computational studies of intercellular signalling (52).
Our computational and experimental approach shows that cells of identical genotype responding to the same stimulus can create a diverse response consisting of qualitatively different behaviours. It remains to be seen whether varied NF-κB activation in response to LPS occurs in vivo. However, one could imagine that multifaceted responses would be extremely useful to the innate immune response, allowing for an extra level of cellular control that does not require differentiation – in this case, an orchestrated and robust response to infection.
Materials and Methods
Cell culture and imaging
We imaged primary MEFs and the 3T3 relA-/- cell line using time-lapse confocal microscopy. TLR4d and TLR4del MEFs were obtained from pregnant mice (Jackson Labs) at day 13 to 14 using standard procedures, all other MEFs were generously provided by S. Akira and M. Yamamoto. MyD88-/- mice can also be purchased through Oriental BioScience (http://myv.ne.jp/obs/index.files/tlr_eng.htm).
To create clonal cell populations, 3T3 relA-/- p65-DsRed cells were additionally transduced with H2B-GFP to assist in nuclear segmentation. Cells were then seeded into 96-well tissue culture plates such that each well received 0.25 cells on average. Clonal populations were isolated from the well plate one week after seeding and were characterized by confocal microscopy. In response to LPS, these cells show both persistent and transient NF-κB response phenotypes (fig. S4) within the sample clonal population.
Co-culture experiments were performed with MEFs of different genotypes. TLR4-mutant MEFs harvested from Jackson Labs mice were infected with the p65 fluorescent protein constructs. Wild-type MEFs harvested from the C57BL/6 background were infected with a different colored p65 construct. Cells of different phenotype were seeded at a 1:1 ratio on gelatin-coated Labtek slides.
Cells were cultured on 4 or 8 chambered coverslips (Labtek) at densities of 75,000 and 35,000 cells per wells, respectively with DMEM supplemented with 10% fetal bovine serum, 100I U/ml penicillin, 100 μg/ml streptomycin, and 2mM L-glutamine. Twenty four hours after seeding, cells were then imaged after stimulation with TNF-α (10 ng/ml, Roche), LPS (0.5 μg/ml, Sigma), sTNFRII (R&D systems), or ultrapure LPS (5 μg/ml, Invivogen, used for TLR4 deletion mutants) with a Zeiss 510 confocal microscope (40× oil or 20× air objective) every 3 to 6 minutes for several hours at 37±1 degrees C with 5% CO2. For primary cell culture, plates and chambers coverslips were coated with gelatin (0.2%, Sigma G1393) or fibronectin (human 25 μg/ml, Millipore).
Nuclear NF-κB monitoring system
DNA constructs encoding EGFP-p65 and p65-DsRed fusion proteins (generous gifts from M. Meffert and M. White, respectively) were cloned into the FUW lentiviral vector (1), under the control of the first 1.5 kb before the relA gene (fig. S1A). Vectors were then used to infect cells using established protocols (43), thereby reconstituting p65 to endogenous amounts and apparently normal p65 activation (Fig. 2D) and IκBα expression (fig. S1B) (22).
Image analysis
The images were analyzed to identify nuclei and quantify nuclear intensity using the Image Processing Toolbox in MATLAB. Images were segmented automatically using one of two methods: (i) Canny edge detection, followed by classification and selection based on region area and eccentricity, or (ii) Marker-controlled Watershed segmentation where markers were derived from previous selections. Nuclear boundaries were marked manually when selection failed by the above methods. A software package that can also achieve this purpose is CellTracker (53). For each set of images all of the cells that were adequately visible over the entire course of the experiment were chosen for quantification of NF-κB motion to and from the nucleus. Cells that divided during the course of the experiment or left the field of view were not quantified. The average nuclear intensity was used as a metric to describe the nuclear NF-κB concentration over time. The sum of nuclear and cytoplasmic intensity over time remained relatively constant through the course of experiment, suggesting that photobleaching was not a major effect (fig. S13). Fluctuations in the focal plane and inhomogenous cytoplasmic intensity across a single cell caused average cytoplasmic intensity to fluctuate greatly between time points. As a result, the nuclear:cytoplasmic ratio showed possibly erroneous fluctuations not present in the average nuclear intensity. Thus, average nuclear intensity was chosen and each time course was normalized to the minimum and maximum average nuclear intensity during the time course. To examine whether coupling between cells was a major effect, we compared the cosine distance used in clustering compared to the spatial distances between nuclei (fig. S6).
Parameter fitting
To model the NF-κB nuclear response to TNF-α, we used the HL model (22), which was generously provided as a Mathematica file by A. Hoffmann. The model was fit to the single-cell data using an algorithm that found the best fit, between 0.3 and 3 times the base model parameter value, for each of the 11 parameters that we identified as most sensitive. Because the parameter values are often coupled(54), we restricted the change in each parameter to one-fourth of the change calculated for the parameters singly. Parameter sets converged for each cell after 200 to 400 steps. Fits for 30 cells were examined (fig. S3) and 23 cells were chosen as having satisfactory best fits. A comparison of the distance between the fit and experimental data and average distances in experimental data showed the error in fits is less than the order of variance between cells (fig. S14). The parameter distributions could be most closely approximated as a mixture of two Gaussians, for which the parameters are given in table S1.
Stochastic LPS model
For the LPS model, we used a slightly modified version of our earlier LPS model (30). Because experimentally we found that the first nuclear NF-κB entry following LPS stimulation was later than occurred in the population-based model, we changed three parameters: The time constants associated with the MyD88 and TRIF signals were both set to τ = 20 minutes, and the time delay that precedes activation of the TRIF pathway was set to 50 minutes. The integrated response of these two pathways is implemented as previously described (28). To represent the stochastic switch, we compared a cutoff parameter in the model (0.55, determined from Fig. 4 (D and F) to a random number between 0 and 1; simulations with randomly-generated values greater than the cutoff received the secondary TNF stimulus. MATLAB files for model simulations are found in hard copy as part of this supplement and are available at www.simtk.org.
Supplementary Material
Fig. S1. Lentiviral expression construct and activation of NF-κB in cells reconstituted with p65-EGFP.
Fig. S2. Metrics of variability in the TNF-α response.
Fig. S3. Parameter fitting to single-cell data.
Fig. S4. Clonal cells stimulated with LPS.
Fig. S5. Cells unresponsive to LPS.
Fig. S6. Lack of correlation between NF-κB dynamics and spatial location.
Fig. S7. TLRdel MEFs (EGFP-p65) treated with LPS.
Fig. S8. TLRdel MEFs (EGFP-p65) treated with TNF-α.
Fig. S9. TLRdel MEFs (EGFP-p65) cultured with wild-type MEFs and treated with LPS and soluble TNFR (sTNFRII).
Fig. S10. Coculture plating density influences the activation of NF-κB in TLRdel cells.
Fig. S11. Trif-/- MEFs exposed to LPS.
Fig. S12. MyD88-/- MEFs exposed to LPS.
Fig. S13. Total cellular fluorescence during a typical experiment.
Fig. S14. Single-cell fit distances compared to global variation between cells.
Table S1. Parameter distributions from single-cell fitting.
MATLAB files for simulations
Acknowledgments
The authors would like to thank D. Baltimore for guidance and support; M. Boldin, T.H. Leung, the Baltimore lab and the Covert lab for helpful discussions; and the Fraser lab for assistance with confocal microscopy. This work was funded by the NIH (GM039458-21 and CA125994-01A1), a Robert Black Fellowship through the Damon Runyon Cancer Research Foundation (DRG-#1835-04) to M.W.C., a Stanford Graduate Fellowship to T.K.L., and a Stanford Bio-X Graduate Fellowship to J.C.S. and J.J.H.
References and notes
- 1.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–9. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–50. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 3.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–9. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 4.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–5. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 5.Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–80. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–5. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 7.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–8. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samoilov MS, Price G, Arkin AP. From fluctuations to phenotypes: the physiology of noise. Sci STKE. 2006;2006:re17. doi: 10.1126/stke.3662006re17. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107:241–6. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meffert MK, Baltimore D. Physiological functions for brain NF-kappaB. Trends Neurosci. 2005;28:37–43. doi: 10.1016/j.tins.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 12.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-{kappa}B activity. Genes Dev. 2007 doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappaB signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell SM, Holm GH, Pierce JM, Tian B, Watson MJ, Chari RS, Ballard DW, Brasier AR, Dermody TS. Identification of an NF-kappaB-dependent gene network in cells infected by mammalian reovirus. J Virol. 2006;80:1077–86. doi: 10.1128/JVI.80.3.1077-1086.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–66. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 17.Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, Shahab A, Zhao XD, Hibberd M, Wei CL, Lim B, Ng HH, Ruan Y, Chin KC. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol Cell. 2007;27:622–35. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 19.Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci U S A. 2006;103:5899–904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 21.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–61. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 22.Lipniacki T, Paszek P, Brasier AR, Luxon B, Kimmel M. Mathematical model of NF-kappaB regulatory module. J Theor Biol. 2004;228:195–215. doi: 10.1016/j.jtbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Cheong R, Bergmann A, Werner SL, Regal J, Hoffmann A, Levchenko A. Transient IkappaB kinase activity mediates temporal NF-kappaB dynamics in response to a wide range of tumor necrosis factor-alpha doses. J Biol Chem. 2006;281:2945–50. doi: 10.1074/jbc.M510085200. [DOI] [PubMed] [Google Scholar]
- 24.Cho KH, Shin SY, Lee HW, Wolkenhauer O. Investigations into the analysis and modeling of the TNFalpha-mediated NF-kappaB-signaling pathway. Genome Res. 2003;13:2413–22. doi: 10.1101/gr.1195703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihekwaba AE, Broomhead DS, Grimley RL, Benson N, Kell DB. Sensitivity analysis of parameters controlling oscillatory signalling in the NF-kappaB pathway: the roles of IKK and IkappaBalpha. Syst Biol (Stevenage) 2004;1:93–103. doi: 10.1049/sb:20045009. [DOI] [PubMed] [Google Scholar]
- 26.Joo J, Plimpton S, Martin S, Swiler L, Faulon JL. Sensitivity Analysis of a Computational Model of the IKK NF-{kappa}B I{kappa}B{alpha} A20 Signal Transduction Network. Ann N Y Acad Sci. 2007;1115:221–39. doi: 10.1196/annals.1407.014. [DOI] [PubMed] [Google Scholar]
- 27.O'Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, Levchenko A, Hoffmann A. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–7. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 29.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–5. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 30.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 1993;7:1266–76. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 31.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–5. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 32.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki K, Nogawa H, Nishijima M. Identification of mouse MD-2 residues important for forming the cell surface TLR4-MD-2 complex recognized by anti-TLR4-MD-2 antibodies, and for conferring LPS and taxol responsiveness on mouse TLR4 by alanine-scanning mutagenesis. J Immunol. 2003;170:413–20. doi: 10.4049/jimmunol.170.1.413. [DOI] [PubMed] [Google Scholar]
- 34.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–10. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 36.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–8. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 37.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand DA, See V, White MR. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–6. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson G, Paraoan L, Spiller DG, Wilde GJ, Browne MA, Djali PK, Unitt JF, Sullivan E, Floettmann E, White MR. Multi-parameter analysis of the kinetics of NF-kappaB signalling and transcription in single living cells. J Cell Sci. 2002;115:1137–48. doi: 10.1242/jcs.115.6.1137. [DOI] [PubMed] [Google Scholar]
- 39.Friedrichsen S, Harper CV, Semprini S, Wilding M, Adamson AD, Spiller DG, Nelson G, Mullins JJ, White MR, Davis JR. Tumor necrosis factor-alpha activates the human prolactin gene promoter via nuclear factor-kappaB signaling. Endocrinology. 2006;147:773–81. doi: 10.1210/en.2005-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayot F, Jayaprakash C. NF-kappaB oscillations and cell-to-cell variability. J Theor Biol. 2006;240:583–91. doi: 10.1016/j.jtbi.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 41.Lipniacki T, Kimmel M. Deterministic and stochastic models of NFkappaB pathway. Cardiovasc Toxicol. 2007;7:215–34. doi: 10.1007/s12012-007-9003-x. [DOI] [PubMed] [Google Scholar]
- 42.Lipniacki T, Puszynski K, Paszek P, Brasier AR, Kimmel M. Single TNFalpha trimers mediating NF-kappaB activation: stochastic robustness of NF-kappaB signaling. BMC Bioinformatics. 2007;8:376. doi: 10.1186/1471-2105-8-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 44.Barken D, Wang CJ, Kearns J, Cheong R, Hoffmann A, Levchenko A. Comment on “Oscillations in NF-kappaB signaling control the dynamics of gene expression”. Science. 2005;308:52. doi: 10.1126/science.1107904. author reply 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson DE, Horton CA, See V, Johnson JR, Nelson G, Spiller DG, Kell DB, White MRH. Response to Comment on “Oscillations in NF-{kappa}B Signaling Control the Dynamics of Gene Expression”. Science. 2005;308:52b. [Google Scholar]
- 46.De Lorenzi R, Gareus R, Fengler S, Pasparakis M. GFP-p65 knock-in mice as a tool to study NF-kappaB dynamics in vivo. Genesis. 2009;47:323–9. doi: 10.1002/dvg.20468. [DOI] [PubMed] [Google Scholar]
- 47.Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21:3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- 48.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 49.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–3. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol. 2007;3:1871–78. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–9. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Batsilas L, Berezhkovskii AM, Shvartsman SY. Stochastic model of autocrine and paracrine signals in cell culture assays. Biophys J. 2003;85:3659–65. doi: 10.1016/S0006-3495(03)74783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen H, Nelson G, Nelson DE, Kennedy S, Spiller DG, Griffiths T, Paton N, Oliver SG, White MR, Kell DB. Automated tracking of gene expression in individual cells and cell compartments. J R Soc Interface. 2006;3:787–94. doi: 10.1098/rsif.2006.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihekwaba AE, Broomhead DS, Grimley R, Benson N, White MR, Kell DB. Synergistic control of oscillations in the NF-kappaB signalling pathway. Syst Biol (Stevenage) 2005;152:153–60. doi: 10.1049/ip-syb:20050050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Lentiviral expression construct and activation of NF-κB in cells reconstituted with p65-EGFP.
Fig. S2. Metrics of variability in the TNF-α response.
Fig. S3. Parameter fitting to single-cell data.
Fig. S4. Clonal cells stimulated with LPS.
Fig. S5. Cells unresponsive to LPS.
Fig. S6. Lack of correlation between NF-κB dynamics and spatial location.
Fig. S7. TLRdel MEFs (EGFP-p65) treated with LPS.
Fig. S8. TLRdel MEFs (EGFP-p65) treated with TNF-α.
Fig. S9. TLRdel MEFs (EGFP-p65) cultured with wild-type MEFs and treated with LPS and soluble TNFR (sTNFRII).
Fig. S10. Coculture plating density influences the activation of NF-κB in TLRdel cells.
Fig. S11. Trif-/- MEFs exposed to LPS.
Fig. S12. MyD88-/- MEFs exposed to LPS.
Fig. S13. Total cellular fluorescence during a typical experiment.
Fig. S14. Single-cell fit distances compared to global variation between cells.
Table S1. Parameter distributions from single-cell fitting.
MATLAB files for simulations