Abstract
Although the nonamelogenin proteins, ameloblastin and enamelin, are both low-abundance and rapidly degrading components of forming enamel, they seem to serve essential developmental functions, as suggested by findings that an enamel layer fails to appear on teeth of mice genetically engineered to produce either a truncated form of ameloblastin (exons 5 and 6 deleted) or no enamelin at all (null). The purpose of this study was to characterize, by direct micro weighing, changes in enamel mineralization occurring on maxillary and mandibular incisors of mice bred for these alterations in nonamelogenin function (Ambn+/+, +/−5,6, −5,6/−5,6, Enam+/+, +/− ,−/−). The results indicated similar changes to enamel-mineralization patterns within the altered genotypes, including significant decreases by as much as 50% in the mineral content of maturing enamel from heterozygous mice and the formation of a thin, crusty, and disorganized mineralized layer, rather than true enamel, on the labial (occlusal) surfaces of incisors and molars along with ectopic calcifications within enamel organ cells in Ambn−5,6/−5,6 and Enam−/− homozygous mice. These findings confirm that both ameloblastin and enamelin are required by ameloblasts to create an enamel layer by appositional growth as well as to assist in achieving its unique high level of mineralization.
Keywords: ameloblastin, enamel, enamelin, knockout, mineralization
Enamel, like dentin, bone, and cementum, is created by an appositional growth process in which formative cells continuously secrete organic components of an extracellular matrix from a polarized (apical) surface while slowly moving away in the opposite direction until the desired final thickness of the matrix is achieved (1). Formative cells in dentin, bone, and cementum are programmed to utilize fibrillar type 1 collagen as the main support element for mineral (2–4). This protein, constructed from individual tropocollagen subunits, requires time to polymerize and stiffen and become receptive to interactions with other non-collagenous proteins secreted by the cells to promote mineralization and cause the mineral crystallites to deposit in intimate spatial association with the mature collagen fibrils (3). Hence, in collagen-based hard tissues there is always a non-mineralized ‘prematrix’ of variable thickness at its outermost forming surface.
This is not the case for enamel, where epithelial cells responsible for creating the hard outer protective covering for the crowns of teeth (ameloblasts) utilize a different mechanism to allow mineralization to occur within an organic-rich extracellular matrix. To make enamel, ameloblasts produce and secrete large quantities of proline and glutamine-rich matrix proteins (amelogenin, ameloblastin, and enamelin) possessing unusual physical properties (5). Amelogenin has a relatively low molecular weight but is expressed at very high levels, while ameloblastin and enamelin are relatively high-molecular-weight glycoproteins that are expressed at relatively low levels (5, 6). Along with this triad of enamel-specific matrix proteins, ameloblasts also release a matrix metalloproteinase (MMP-20) that rapidly cleaves all newly secreted enamel matrix proteins into various large derivative fragments (7). In addition to this, thin ribbons of lightly carbonated calcium hydroxyapatite, possibly evolving rapidly from amorphous calcium phosphate precursors (8), are seen in close physical proximity to the plasma membranes of ameloblasts at sites where the matrix proteins and MMP-20 are secreted (5, 9). The mineral ribbons are not arranged randomly within the forming enamel layer but are guided into spatially complex three-dimensional patterns that are termed the rod and inter-rod growth sites which are situated at the tips and around the bases of large apical cytoplasmic extensions from ameloblasts known as Tomes’ processes (9, 10). This phase of amelogenesis is called the secretory stage, in which the forming enamel, including its outermost surface into which the Tomes’ processes are inserted, initially contains about 15% mineral by dry weight, which changes to an average of 30% mineral by weight or more, depending upon species, when appositional growth of the enamel layer is terminated (1). The enamel has a soft jelly-like consistency in its native hydrated state in vivo.
Once the full thickness of the enamel layer has been produced and the ameloblasts have retracted their Tomes’ processes back into their cell bodies, a thin coating of inter-rod-type matrix is formed to smooth over any irregularities that exist at the outermost enamel surface (10). The cells then undergo morphological and functional changes during a short transition phase to reduce their secretory activity for matrix proteins and MMP-20 and to increase their absorptive and ion-transport activities (1). It is at this time that ameloblasts start expressing and secreting a serine proteinase (Klk4) that functions to break down any intact protein or fragments present within the enamel layer (7). Ameloblasts also secrete components of a basal lamina that contains a distinct protein called amelotin (11). This lamina covers the enamel surface and provides a means for ameloblasts to adhere to the enamel surface via hemidesmosomes until the tooth erupts (11, 12). These events collectively demarcate the start of the maturation stage of enamel development. It is during this phase that ameloblasts undergo periodic changes in apical morphology (ruffle-end/smooth-ended) and in apical/basal locations of cellular tight junctions through a process called ameloblast modulation (1). The purpose of the maturation stage is to create conditions within the enamel layer that are optimized to promote growth in width and breadth of the thin mineral ribbons formed during the secretory stage. When the maturation stage is complete, ameloblasts will have modulated many times and the mineral crystals will have expanded substantially in volume to fill the internal spaces formerly occupied by protein and water (1). It is by this process that enamel becomes one of the hardest tissues produced biologically, at 95% mineral by weight.
Over the past decade mouse models engineered for loss of function of amelogenin (13), ameloblastin (14, 15), enamelin (16), MMP-20 (17), and Klk4 (18) have been reported, along with transgenic models, for expressing or overexpressing various proteins (e.g. 19–24). These models have provided important information about the expression pattern of individual enamel matrix components and what happens grossly to the enamel layer when any one of its five major protein constituents is missing (or overexpressed). For loss of function, this includes (i) frail, grossly hypomineralized enamel having normal rod/inter-rod organization and thickness (i.e. no Klk4) (18), (ii) thin enamel with disrupted rod patterns and a tendency to break away from the dentin (i.e. no MMP-20) (17), (iii) very thin and rodless enamel (i.e. no amelogenin) (13), and (iv) no enamel at all (i.e. no ameloblastin or enamelin) (14–16). Understanding cause-and-effect relationships in most of these models has been slow and hampered by a lack of comprehensive information about the exact functional role(s) that amelogenin, ameloblastin, and enamelin serve individually and collectively in amelogenesis. It is nevertheless clear from these models that the loss of function of any single one of the four components normally secreted during the active appositional growth phase (MMP-20, amelogenin, ameloblastin, enamelin) reduces enamel thickness marginally (no MMP-20), greatly (no amelogenin) or totally (no ameloblastin, no enamelin). Hence, no compensation of one component for another occurs in this system and any flaws result in the creation of inferior quality mineral. It is also clear from the loss-of-function models that some as-yet-to-be defined relationship exists among secreted amelogenin, its processing by MMP-20, and the formation of enamel rods by ameloblasts (25). In addition, active secretion of both native ameloblastin and enamelin must occur or no enamel layer will form, consistent with their pivotal status in the genetic evolution of tooth enamel (26–28).
Ameloblastin and enamelin have very different primary amino acid sequences and sizes, with ameloblastin almost three times smaller in core molecular weight and less extensively modified post-translationally than enamelin (65 vs. 186 kDa post-translational sizes) (6). Ameloblastin also undergoes a different and less-complex fragmentation sequence than enamelin, and the distribution pattern of its fragments within the forming enamel layer is not the same as enamelin (6, 7). It is therefore of interest that loss of function of either protein seems to lead to the same end result, which is formation of an abnormally thin and disorganized mineralized layer on the crowns of teeth. The objective of this study was to characterize, by sequential strip microdissections (29), similarities and differences in the amount of mineral that develops within the enamel of incisors taken from various genotypes (+/+, +/−, −/−) of mice engineered to express a mutated form of ameloblastin lacking exons 5 and 6 that causes enamel agenesis in the homozygous state (Ambn−5,6/−5,6) (14, 15) compared with the enamelin null mouse model where true enamel formation also fails to occur (16).
Material and methods
Animals
All procedures for housing and breeding of mice and removal of maxillary and mandibular jaw samples for analysis were approved and monitored by Animal Care Committees of the Université de Montré al and the University of Michigan. Ameloblastin loss-of-function jaw samples (Ambn+/−5,6; Ambn−5,6/−5,6) were obtained from a breeding colony maintained at the Université de Montréal that were offspring of the original exon 5 and 6-deleted mouse model described by Fukumoto et al. (14, 15). Enamelin loss-of-function jaw samples (Enam+/−; Enam−/−) were obtained from a breeding colony created and maintained at the University of Michigan (16). The Enam gene in these mice was deleted and replaced with a LacZ nuclear reporter gene (16). Wild-type mice (C57BL/6; Ambn+/+; Enam+/+) were either purchased as needed (Charles River Canada, St-Constant, QC, Canada) or housed and bred on site (University of Michigan). Genotyping in all cases was carried out by polymerase chain reaction (PCR) analyses of genomic DNA obtained from tail biopsies. Some mice from the Ambn group were fixed by perfusion and the incisors processed, after decalcification, for embedding and sectioning in Epon as described by Wazen et al. (15).
Sample preparations
Seven-week-old (Ambn group) and 8-week-old (Enam group) male and female mice were anaesthetized and killed. Hemi-maxillae and hemi-mandibles were removed and cleaned of skin, hair, and adhering muscular tissues. The hemi-jaws were then either immediately immersed in liquid nitrogen for 5 h (Ambn group) or flash-frozen in prechilled containers immersed in dry ice and shipped on dry ice to Montreal (Enam group). All hemi-jaws were freeze-dried for 48 h at −55°C (Labconco, Kansas City, MO, USA) and stored thereafter at 4°C with Drierite (Fisher Scientific, Whitby, ON, Canada) in sealed containers. The bony caps covering the labial aspects of freeze-dried incisors were removed and either discarded or processed for weighing on a separate tooth-by-tooth basis, as described below for enamel. The underlying enamel organs similarly were either discarded or cut into a series of 1-mm-long strips from the apical to the incisal ends of the tooth. Each strip was lifted off the tooth using a curved dental tool, placed in individual 12-mm-diameter aluminum pans, and processed for weighing. The exposed surfaces underneath the cell layers were wiped gently with dry Kimwipes (Kimberly-Clark, Roswell, GA, USA). Some teeth (19 from the Ambn group and 28 from the Enam group) representing all genotypes were mounted with carbon tape, without any further processing, on metal stubs and examined in a JEOL JMS-6460LV variable-pressure scanning electron microscope using the backscatter mode at 25 kV and 52–60 p.s.i. pressure. On most incisors, the enamel layer or thin calcified material present along the labial surface was transected using a scalpel blade into a series of 1-mm-long strips from the beginning of the secretory stage to an incisal location where the enamel was normally fully mature and so hard that it bent the scalpel blade when attempting to cut away the strip. Each strip was removed and placed in a separate aluminum pan. Samples of enamel-free dentin were also taken from each incisor and placed in their own individual aluminum pans. In addition, the first, second, and third molars with fully intact roots were carefully dissected out of the alveolar bone, cleaned of any adhering periodontal tissues, and placed in separate aluminum pans.
Microweighing, ashing, and data analyses
The aluminum pans containing the bone, enamel organ, enamel, dentin or molar samples were placed in an oven and heated for about 12 h at 45°C. The samples were cooled to room temperature and weighed on an SC2 microbalance (Sartorius, Göttingen, Germany) to obtain their ‘before ashing dry weights’. Each sample was transferred to a separate, small 1.3-ml capacity Coors crucible (Fisher Scientific) and placed inside an Isotemp muffle furnace (Fisher Scientific) where they were burned (ashed) at 575°C for 18 h. The crucibles were cooled and each sample was reweighed to obtain its ‘after ashing mineral weight’. This procedure vaporizes organic material and any bound water, leaving behind the mineral residue contained within the sample (29). The mineral gain for enamel strips was calculated by subtracting the mineral weight of a previous strip from the mineral weight of the current strip, sequentially across all strips in a series. The percentage mineral by weight was calculated using the equation (ashed weight/total dry weight) × 100. Weight data for enamel strips, and for pieces of dentin and bone, were collected from a minimum of 12 maxillary and 12 mandibular incisors per genotype (12 × 2 × 6 = 144 hemi-jaws overall). Weight data for enamel organ cell strips were obtained from a separate set of maxillary and mandibular incisors from four wild-type mice and from six null mice in the Ambn and Enam groups [2 × (4 + 6) × 2 = 40 hemi-jaws]. Weight data for intact maxillary and mandibular molars were obtained from four mice per genotype (2 × 4 × 6 = 48 hemi-jaws × 3 molars per hemi-jaw = 144 molars sampled overall). Means and confidence intervals for weight data and univariate factorial analysis of variance (anova) were obtained using Version 8 of Statistica for Windows (Statsoft, Tulsa, OK, USA). Principal component analyses of covariances were obtained using Version 3.1 of the Multivariate Statistical Package (Kovach Computing Services, Pentraeth, UK). For enamel organ cells, eight variables were defined to represent the raw mineral weight or the percentage mineral by weight detected by stage or stage equivalent (secretion and early, mid, and late maturation) in the two jaws (maxilla, mandible). For hard tissues, seven variables were assigned to represent the normalized mineral weights in nearly mature enamel, bone, and molars, or the percentage mineral by weight in these tissues for the two jaws as well as within dentin samples from mandibular incisors.
Results
Mineral content in enamel organ cell strips
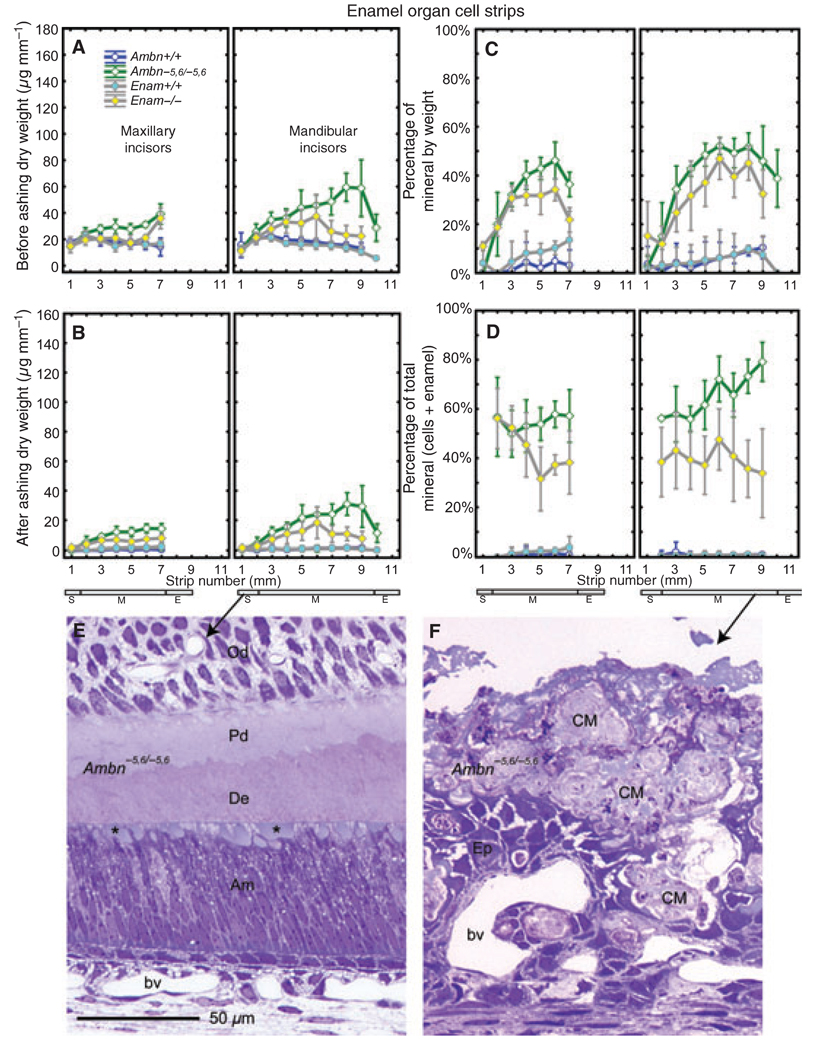
Enamel organ cell strips removed from maxillary and mandibular incisors of wild-type mice (Ambn+/+, Enam+/+) had dry weights of 15–20 µg mm−1 across the length of the tooth (Fig. 1A). They showed, as expected, no significant mineral content (Fig. 1B), although there was sometimes an occasional small flake of bone and/or enamel present along with the freeze-dried cell material in some strips (Fig. 1C, maximum at 10 ± 5% late maturation). Enamel organ cell strips taken from the incisors of loss-of-function mice (Ambn−5,6/−5,6, Enam−/−) consistently showed higher dry weights than their wild-type counterparts, especially across the region of the tooth where the maturation stage was normally located and for cell strips removed from the mandibular incisors of Ambn−5,6/−5,6 mice (Fig. 1A). Sometimes focal areas of calcified material covering these teeth adhered tightly to the cells and were lifted off with them. A mineral residue was detected following ashing in all cell strips removed from Ambn−5,6/−5,6 and Enam−/− incisors (Fig. 1B), which represented as much as 50 ± 5% mineral by weight in the positional equivalent of the late maturation stage (Fig. 1C, strip 6 maxillary; strips 6–9, mandibular). In global terms, the mineral associated with cell strips accounted for between 50 and 73% of the total mineral present labially on the incisors of Ambn−5,6/−5,6 mice (Fig. 1D, sum of cell strips + extracellular calcified material covering dentin; see section on enamel strips), but it represented only 32–56% of the total labial mineral determined for incisors from Enam−/− mice (Fig. 1D). Histological sections from Ambn−5,6/−5,6 mice indicated that small globular patches of material were associated with apical surfaces of ameloblasts in the secretory stage (Fig. 1E), whereas very large and irregular masses of material were found more incisally between and at the surface of disorganized remnants of the enamel organ (Fig. 1F).
Fig. 1.
Mineral content in enamel organ cells on incisors. Means plots ± 95% confidence intervals showing initial dry weight (A), the mineral weight after ashing (B), and the percentage mineral by weight (C, representing B/A) for 1-mm-long enamel organ strips microdissected from maxillary (left graph each panel) and mandibular (right graph each panel) incisors of wild-type (Ambn+/+, Enam+/+) and homozygous (Ambn−5,5/−5,6 and Enam−/−) mice. The stages of amelogenesis on these teeth are indicated by the small rectangles at the bottom of each column (S, secretory stage; M, maturation stage; E, erupted portion). Panel D uses data from panel B and from panel B of Fig. 3 to illustrate the quantity of mineral that is associated with enamel organ cells relative to the total quantity of mineral detected in cell layers and underlying enamel layer at each strip position. Mineral is rarely detected within enamel organ cell strips under normal conditions (A–D, circles) but is present in increasingly larger amounts across the length of incisors from homozygous Ambn−5,5/−5,6 and Enam−/− mice (A–D, diamonds). Panels E and F, longitudinal 1-µm-thick Epon sections of incisors from Ambn−5,6/−5,6 mice stained with Toluidine blue, illustrating small globular masses of material (E, asterisk) seen at the apical surfaces of tall columnar secretory-stage ameloblasts (E, Am) before they depolarize and become disorganized, and the much larger and irregular masses of calcified material (F, CM) found later during the maturation stage in association with an epithelium (F, Ep) that is unrecognizable as an enamel organ. Od, odontoblasts; Pd, predentin; bv, blood vessel.
Scanning electron microscopy appearance of exposed enamel surfaces on incisors
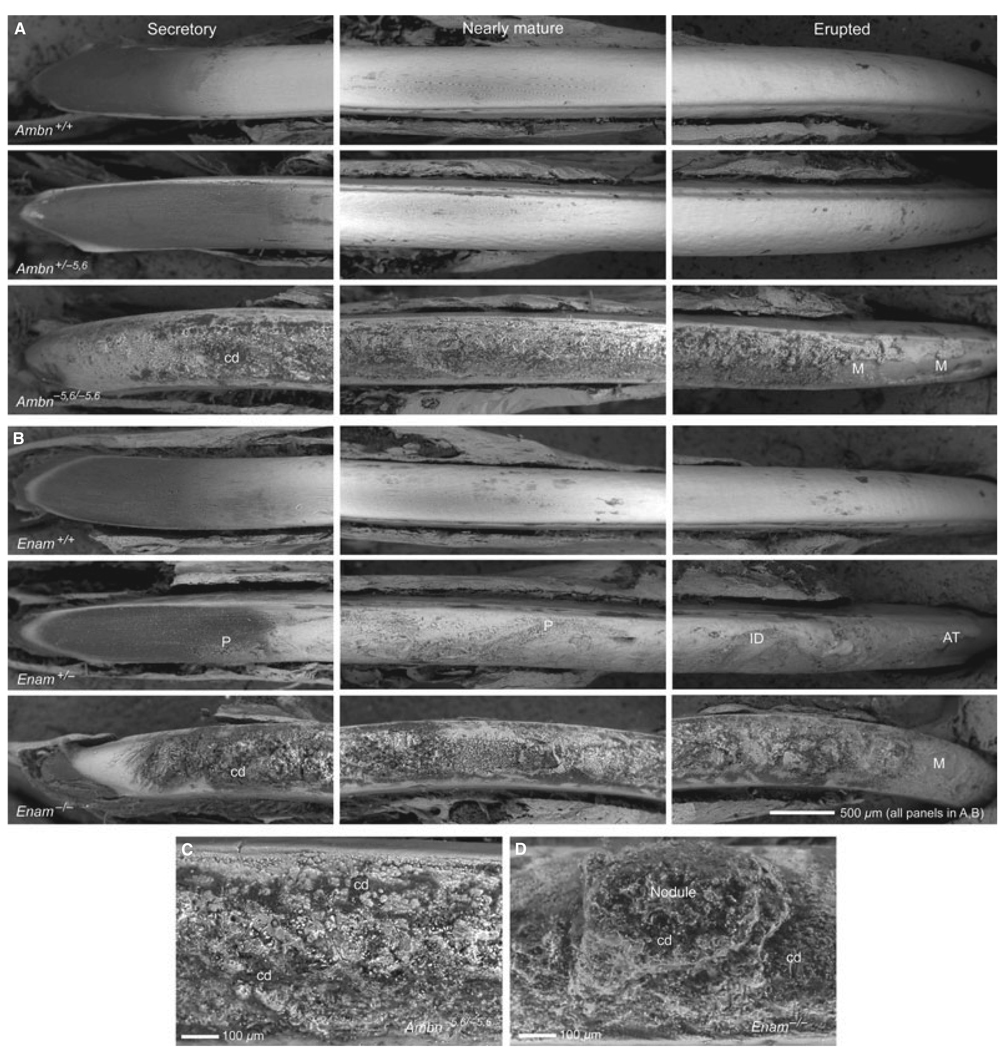
Under low magnification, the enamel surfaces of freeze-dried incisors from wild-type mice that had been wiped with Kimwipes following removal of covering enamel organ cell layers appeared relatively smooth from apical (secretory) to incisal (erupted) ends (Fig. 2A, Ambn+/+; Fig. 2B, Enam+/+). The area of the secretory zone typically appeared dark when anal-ysed using backscatter electron imaging as a result of its high content of organic material, whereas more incisally located maturing and mature areas of the enamel had a white and slightly undulating appearance. The enamel surfaces of incisors from heterozygous Ambn+/−5,6 mice appeared similar to wild-type controls (Fig. 2A) but those from heterozygous Enam+/− mice were rough and mildly deformed and there was evidence of compression and severe labial attrition of the enamel at the erupted incisal ends (Fig. 2B). The surfaces of homozygous Ambn−5,6/−5,6 and Enam−/− incisors did not appear to be covered by enamel but were coated with a thin and crusty calcified material that was very rough in texture and interspersed with nodular masses and cellular debris (Fig. 2A–C). The erupted portions of these teeth also showed areas where the calcified material appeared to have fallen off the dentin, especially in the case of incisors from Ambn−5,6/−5,6 mice (Fig. 2A,B).
Fig. 2.
Scanning electron microscopy of incisors. Backscatter electron imaging of the appearance of developing enamel along the labial surfaces of incisors denuded of overlying enamel organ cell layers from the apical (secretory), middle (nearly mature), and incisal (erupted) ends of teeth from wild-type (A, Ambn+/+; B, Enam+/+), heterozygous (A, Ambn+/−5,6; B, Enam+/−), and homozygous (A,C Ambn−5,6/−5,6; B,C Enam−/−) mice. In this technique, dark areas contain less mineral than white areas. The labial surfaces of incisors from wild-type mice (+/+) and from heterozygous Ambn+/−5,6 mice are relatively smooth, whereas the labial surfaces of incisors from heterozygous Enam+/− mice are rougher with pitted (P) and indented (ID) surfaces in some areas and signs of severe attrition (AT) in other areas near the incisal tips. The labial surfaces of incisors from homozygous Ambn−5,6/−5,6 and Enam−/− mice show no enamel but are covered instead by a thin layer of rough and poorly organized calcified material that often appears missing (M) in focal areas on the erupted portions of the tooth. The calcified material covering Ambn−5,6/−5,6 incisors is crusty with numerous small spherical masses and cellular debris (cd) embedded in it (C), whereas the calcified material covering Enam−/− incisors appears more flaky and often contains very large and irregular nodular masses extending out of the surface crust (D, nodule).
Mineral content in enamel strips
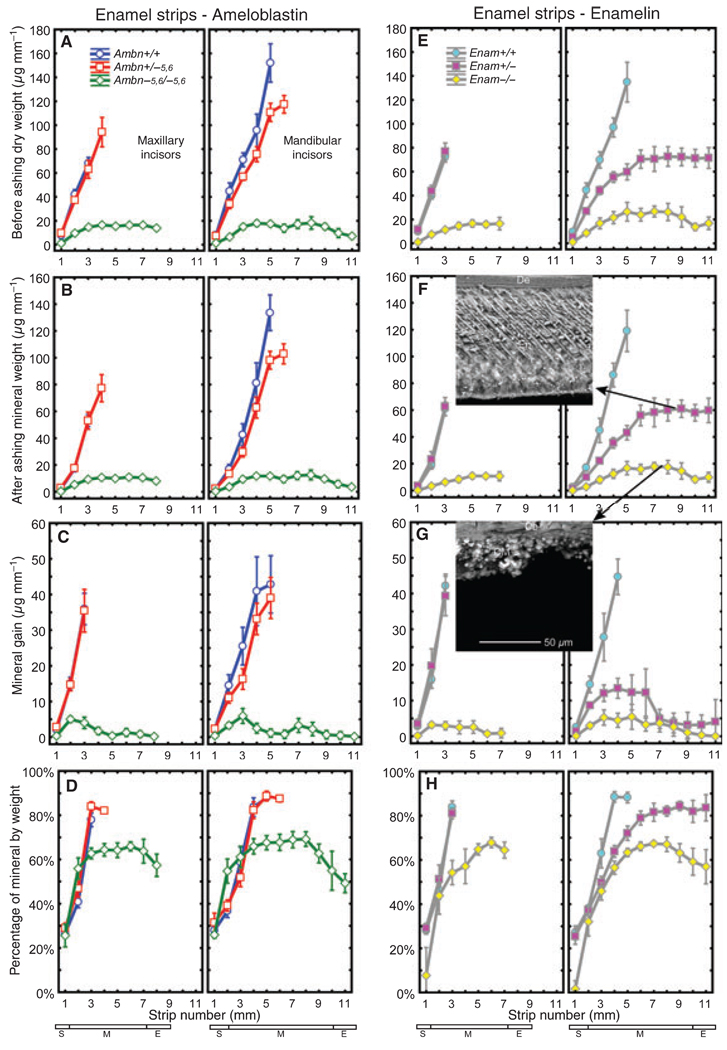
Consistent with results reported previously (29), the dry weight and the mineral weight of enamel strips removed from the maxillary incisors of normal mice (+/+) increased rapidly and progressively over 3 mm in the apical region of the tooth (Fig. 3A,B,E,F). The same result was obtained for enamel strips from mandibular incisors of normal mice (+/+) except that more strips could be prepared before the enamel was too hard to cut with a scalpel blade (5 mm), and the gross dry weight and mineral weight of strips from mandibular incisors was higher by as much as twofold (Fig. 3A,B,E,F). The rate at which mineral accrued (mineral gain) within maturing enamel on maxillary and mandibular incisors of normal mice (+/+) was around 35–40 µg mm−1 (Fig. 3C,G). The mean dry weight, mineral weight, and estimated mineral gain in strips removed from maxillary incisors of heterozygous Ambn+/−5,6 and Enam+/− mice were identical to those of wild-type mice except that it was possible to remove one additional strip (4 mm) from the maxillary incisors of Ambn+/−5,6 mice (Fig. 3A–C, E–G). The mean dry weight, mineral weight, and mineral gain in strips removed from the mandibular incisors of heterozygous Ambn+/−5,6 mice, however, was consistently lower than in wild-type mice at all strip positions, especially within the most mature areas (Fig. 3A–C). Like maxillary incisors, it was also possible to routinely remove one additional strip (6 mm) from these teeth (Fig. 3A–C). Enamel strips could be removed along the entire length of mandibular incisors from heterozygous Enam+/− mice (Fig. 3E–G). All strips showed about half the dry weight and mineral weight and about one-third the mineral gain per strip compared with Enam+/+ control teeth (Fig. 3E–G). Enamel strips could also be removed along the entire length of maxillary and mandibular incisors of homozygous Ambn−5,6/−5,6 and Enam−/− mice (Fig. 3A–C, E–G). These strips were thin and fragile across the whole length of the incisors, and they had only about 15% of the dry weight and mineral weight, and they showed only about 10% as much mineral gain per strip, compared with wild-type controls (Fig. 3A–C, E–G). In relative terms, the percentage mineral by weight in enamel strips from maxillary and mandibular incisors of wild-type mice (Ambn+/+, Enam+/+) and heterozygous mice (Ambn+/−5,6, Enam+/−) showed the expected increases from 30% in forming enamel to 50% as enamel began to mature to 85–90% in nearly mature enamel (Fig. 3D,H). For enamel strips removed from the mandibular incisors of heterozygous Enam+/− mice, the progressive increase in percentage mineral by weight was more gradual along the length of the tooth, and the peak value reached was 84% as opposed to 90% reached more apically in controls (Fig. 3H). Increases in percentage mineral by weight also occurred in the thin calcified material coating the labial surfaces of incisors from homozygous Ambn−5,6/−5,6 and Enam−/− mice (Fig. 3D,H). This material showed peak values of around 68% mineral by weight as well as decreases thereafter, to around 50–60% mineral by weight, in approaching the incisal tips of the incisors.
Fig. 3.
Mineral content in developing enamel on incisors. Mean plots ± 95% confidence intervals showing initial dry weight (A, E), mineral weight after ashing (B, F), mineral gain per strip (C, G) and percentage mineral by weight (D, H representing B/A and F/E) for 1-mm-long enamel strips microdissected from maxillary (left graph each panel) and mandibular (right graph each panel) incisors of wild-type (Ambn+/+, Enam+/+), heterozygous (Ambn+/−5,6, Enam+/−), and homozygous (Ambn−5,6/−5,6, Enam−/−) mice. Stages of amelogenesis on these teeth are indicated by the small rectangles at the bottom of each column (S, secretory stage; M, maturation stage; E, erupted portion). A gene dosage effect for mineral content in enamel is evident on mandibular incisors of heterozygous mice, which is modest in the case of ameloblastin (B, Ambn+/−5,6, 23% reduction) and pronounced in the case of enamelin (F, Enam+/−, 50% reduction). The calcified material covering the labial surfaces of incisors in Ambn−5,6/−5,6 and Enam−/− mice (see Fig. 2) contains only 10% as much mineral as normal (B, F) and is considerably less mineralized compared with nearly mature enamel (D, H; 68% vs. 90% mineral by weight). Insets to F and G, scanning electron microscopy images of longitudinally fractured freeze-dried mandibular incisor enamel from an Enam+/− mouse (F, nearly mature) and disorganized calcified material (CM) from an Enam−/− mouse (G); the magnification bar in panel G applies to both insets. Despite the 50% reduction in mineral content, rod structure and enamel thickness both appear to be relatively normal in Enam+/− mice (inset, F) but not in Enam−/− mice (inset, G). The average enamel thickness measured on mandibular incisors was 100 ± 9, 71 ± 33, and 40 ± 16 µm in Enam+/+, Enam+/−, and Enam−/− mice. De, dentin.
Mineral content in incisor dentin, bone, and intact whole molars
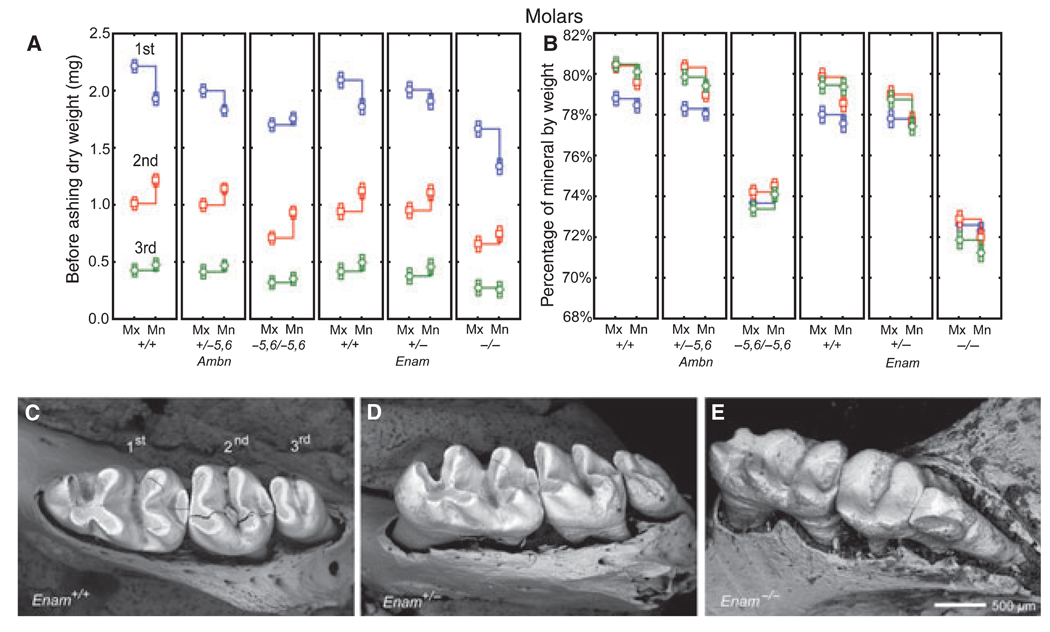
No differences in mineral content by genotype were detected in either dentin or bone (Table 1). Intact whole molars, because of their coronal covering of enamel, showed differences in weights and mineral content related to molar type (first, second, third) as well as to jaw (Mx, Mn) and genotype (Ambn, Enam) (Fig. 4), as seen with incisors (Fig. 3 A,D,E,H). Most notable was a heterozygous effect of a general lowering in total dry weights of maxillary and mandibular first molars from Ambn+/−5,6 mice (Fig. 4A, panels 1 and 2) which was only weakly apparent for maxillary first molars in Enam+/− mice (Fig. 4A, panels 4 and 5). There was also a more dramatic lowering in dry weights of first, second, and third mandibular molars in Enam−/− mice compared with the changes measured for the same molars in Ambn−5,6/−5,6 mice (Fig. 4A, panels 3 and 6), presumably caused, in part, by the severe occlusal wear evident on molars from Enam−/− mice (Fig. 4C,D). The percentage mineral by weight was 78–80% in all molars from wild-type mice (+/+) and heterozygous mice (+/−), about 74% in molars from homozygous Ambn−5,6/−5,6 mice, and around 72% in molars from homozygous Enam−/− mice (Fig. 4B).
Table 1.
Percentage mineral by weight for dentin and bone
| Genotype | Dentin Incisor |
Bone Maxilla |
Bone Mandible |
|---|---|---|---|
| Ambn+/+ | 73.5 ± 0.7 | 66.9 ± 0.9 | 69.7 ± 0.5 |
| Ambn+/−5,6 | 74.4 ± 0.8 | 66.7 ± 0.7 | 69.2 ± 0.8 |
| Ambn−5,6/−5,6 | 74.9 ± 0.5 | 67.3 ± 1.0 | 69.9 ± 0.7 |
| Enam+/+ | 72.9 ± 1.6 | 66.8 ± 0.9 | 68.2 ± 1.8 |
| Enam+/− | 73.4 ± 1.3 | 66.7 ± 1.0 | 67.9 ± 1.5 |
| Enam−/− | 74.4 ± 0.9 | 65.4 ± 1.6 | 68.6 ± 1.0 |
n = 12 incisors or hemi-jaws per genotype.
Fig. 4.
Mineral content in intact whole erupted molars. Mean plots ± 95% confidence intervals showing initial dry weight (A) and percentage mineral by weight (B) for the first, second, and third molars removed from wild-type (A, B; Ambn+/+, Enam+/+), heterozygous (A, B; Ambn+/−5,6, Enam+/−), and homozygous (A, B; Ambn−5,6/−5,6, Enam−/−) mice. Panels C–E: scanning electron microscopy images of the first, second, and third mandibular molar crowns from wild-type (C, Enam+/+), heterozygous (D, Enam+/−), and homozygous (E, Enam−/−) mice; cracks within some molar crowns (C, D) are artifacts from freeze drying. The enamel covering the crowns of molars contributes partly to the overall weight of these teeth but is subject to change as molar crowns wear down by occlusal attrition (C,D). Despite this, weight measurements indicate that the enamel covering the molars are affected similarly as incisors, although a gene dosage effect in the molars of heterozygous mice (A, B; Ambn+/−5,6, Enam+/−) is less pronounced than seen on incisors (Fig. 3A,B,E,H). The molars in Enam+/− mice appear to be normal except for some enhanced attrition evident at the distal sides of the second and third molars (D, asterisk). The molars in Enam−/− mice (E) are severely worn down and there is evidence of bone loss around their roots (see Hu et al. (16) for details). Mn, mandibular; Mx, maxillary.
Summary of main effects: principal component analyses
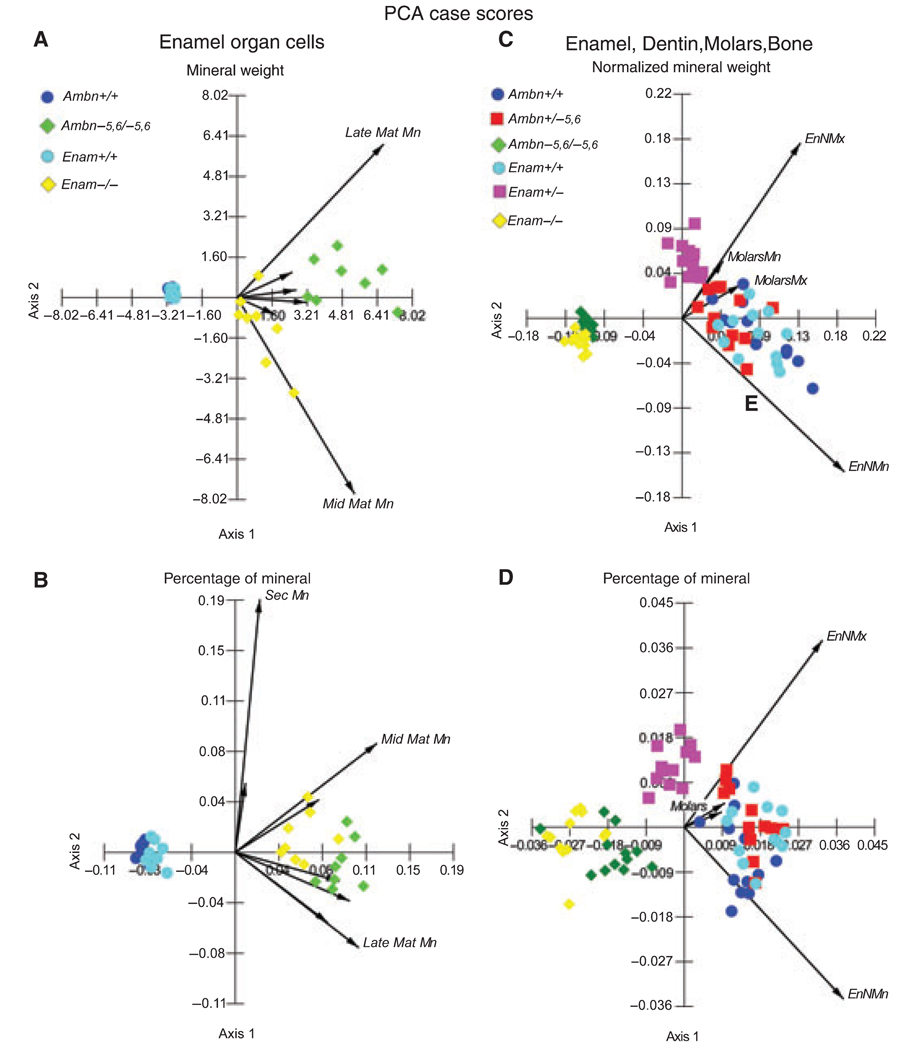
Principal component analyses of mineral weight data showed that the main effects associated with loss of function of ameloblastin or enamelin included increases in mineral content within the enamel organ cell layer (Fig. 5A,B; diamonds on positive x-axes) and decreases in the mineral content within enamel covering the incisors and molars (Fig. 5C,D; diamonds on negative x-axes). Enamel organ cells showed the greatest accumulation of mineral content relative to the positional equivalent of the mid-maturation stage, and especially of the late-maturation stage, on mandibular incisors of Ambn−5,6/−5,6 mice (Fig. 5A,B; long vectors above and below positive x-axis; green diamonds more positive than yellow diamonds). In enamel, the loss of mineral content was slightly more severe for Enam−/− mice than for Ambn−5,6/−5,6 mice (Fig. 5C,D; yellow diamonds consistently more negative than green diamonds on x-axes), and it was more dramatic in the heterozygous state on mandibular incisors for Enam+/− mice compared with Ambn+/−5,6 mice (Fig. 5C,D; magenta squares displaced more negative than on x-axes and positioned 180° opposite the vector representing the mandibular incisor). Factorial anova and Tukey’s honestly significant difference post-hoc comparison tests for equal and unequal n values indicated that there were no differences in mineral weights or percentage mineral by weight between right and left incisors for any given genotype. There were also no differences in the results for enamel organ cells or for enamel at any given stage between Ambn+/+ and Enam+/+ mice, and no significant differences in mineral weights or percentage mineral by weight for calcified material covering the maxillary and mandibular incisors of Ambn−5,6/−5,6 and Enam−/− mice. Enamel organ cells in the positional equivalent of the late-maturation stage, however, contained significantly more mineral in Ambn−5,6/−5,6 mice than in Enam−/− mice. Lastly, mineral weights and percentage mineral by weight for enamel were significantly lower in Enam+/− mice throughout maturation, whereas only nearly mature enamel in Ambn+/−5,6 mice contained significantly less mineral compared with wild-type controls.
Fig. 5.
Principal component analysis (PCA) graphs. Principal component analysis case scores (symbols) and main eigenvectors (arrows) are plotted for mineral weight and percentage mineral by weight in enamel organ cells (A, B) and hard tissues (enamel, dentin, molars, bone) (C, D), as determined for samples taken from wild-type (Ambn+/+, Enam+/+), heterozygous (Ambn+/−5,6, Enam+/−), and homozygous (Ambn−5,6/−5,6, Enam−/−) mice. The x- and y-axes of these graphs represent the magnitude of component scores computed across all cases for each variable from the covariance matrix of variables. In this type of analysis, stages of development, tooth type, and hard tissue type are variables and genotype is represented as subcategories within each variable (see the Materials and methods for details). These graphs show clearly that loss of function of either ameloblastin (green diamond) or enamelin (yellow diamond) is associated with a trend for increasing amounts of mineral to accumulate within cells (A, B), especially relative to ameloblastin. There is also a trend for decreasing amounts of mineral to be present in forming enamel (C, D) with intermediate effects in heterozygous mice (squares), especially in the case of enamelin (magenta squares). Mn, mandibular; Mx, maxillary; Sec, secretory stage; Mat, maturation stage; En, enamel; EnN, nearly mature enamel on incisors.
Discussion
Tissue microdissection, weighing, and hot ashing (29, 30) are unquestionably very tedious and time-consuming ways to obtain information about genetically induced changes in the mineral content of developing enamel, but it is an approach that has certain advantages when invasiveness of tissue sampling is not a concern (31), as in the present investigation. The main advantage of direct weighing is that it gives a number that represents the actual mass of material that is present within a sample. It is not a calculated ‘best guess’, but the sample’s real weight. We have found, by experience, that there can be as much as 30% variation between animals within a given group and we prefer to make conclusions about weights based on grand means established from as large a pool of animals as is practical, which is generally greater than is possible using X-ray-based techniques. Considerable insights into the effects of an experimental condition on enamel development can also be obtained simply by observing enamel surfaces during dissection (Fig. 2) or by determining whether fewer or a greater number of strips than normal can be cut and removed from the incisors (Fig. 3). Although beyond the scope of the present investigation, microdissected samples of forming and maturing enamel removed from teeth at known developmental times/stages (e.g. as defined by cumulative strip number along the length of incisors) can also be useful for other purposes such as characterization of protein, proteinase, and/or mineral (15, 29).
The results from this study agree fully with the general idea that ameloblastin and enamelin serve essential tooth enamel-specific functions and if either one of them is missing or dysfunctional at the beginning of the secretory stage, then no enamel will form on the crowns of teeth (27, 28). What these functions might be are currently unclear, but of the many possibilities that exist the one we favor the most is that ameloblastin and enamelin could represent two separate or one collective critical factor(s) required for the formation and maintenance of the mineralization front in close physical proximity to the apical membranes of ameloblasts as the enamel layer grows in thickness by appositional growth during the secretory stage (5, 9, 32–35). This does not mean that one or both of these proteins (or their fragments) are themselves inducers of mineralization, only that both proteins must be present within the extracellular environment near ameloblasts for the mineral-induction event to occur. If this is true, then considerable caution will need to be exercised in interpreting results from in vitro studies with these proteins because the behavior of each protein individually in a test tube will not necessarily reflect accurately their co-dependence or the exact nature of the functions they serve in vivo.
Ambn−5,6/−5,6 and Enam−/− mice also appear to be incapable of upregulating the production and secretion of large amounts of amelogenin, which forms the bulk mass of organic material within normal, developing enamel (15, 16). Hence, in these nonamelogenin loss-of-function models there are apparently few or no proteins initially released at the dentin surface, unlike the amelogenin null model where secretion of normal amounts of ameloblastin and enamelin leads to the creation of a mineralization front, but ameloblasts then are unable to generate much appositional growth within the enamel layer as a result of their inability to produce and secrete amelogenin (13, 25). Ameloblasts in Ambn−5,6/−5,6 and Enam−/− mice in the absence of an enamel-based mineralization front seem to resort to activating alternative mineralization pathways in an attempt to cover the coronal dentin surface with some form of protective coating (Fig 2 and Fig 3). Coronal enamel organ cells do not normally create this type of extracellular mineralizing matrix and, although cause-and-effect relationships are unclear at this time, it appears that the intracellular controls which operate under these altered conditions are inadequate or unable to prevent development of epithelial pathologies and ectopic calcifications within the enamel organ cell layer, which, in the case of Ambn−5,6/−5,6 mice becomes more and more pronounced over time (Fig. 1B,F) (15). The mechanisms and pathways by which ameloblasts in Ambn−5,6/−5,6 and Enam−/− mice detect and respond to problems with the mineralization front are currently unknown.
The findings in heterozygous Ambn+/−5,6 mice, and especially in Enam+/− mice, of a gene dosage effect relative to the total quantity of mineral that forms within developing enamel (Fig. 3B,F), provides additional evidence supporting the idea of an involvement of ameloblastin and enamelin (or their fragments) in mineralization events (6, 16, 34). If these proteins truly are involved in advancing the mineralization front as ameloblasts move away from the dentin, then the gene dosage effect probably results from a decrease in the total number of mineral crystallites that develop per unit volume of enamel in the heterozygous mice (crystallite packing density). This would explain how the enamel layer on mandibular incisors of Enam+/− mice, which contains 50% less total mineral, is not 50% thinner (Fig. 3F) and appears softer and less resistant to abrasion (more porous) despite the fact that the mineral component undergoes characteristic changes from 40 to about 85% mineral by weight as it matures (Fig 2B and Fig 3H, strips 2–9). However, the data in panels G and H of Fig. 3 indicate that there is also a problem with delayed maturation in these incisors, as suggested by the much slower rate at which mineral is gained during early and mid maturation stages (Fig. 3G, strips 3–6) and the incisal shift in the location at which enamel shows its highest percentage of mineral by weight (Fig. 3H, strip 5 in wild-type mice vs. strip 9 in Enam+/− mice). These are indicative of problems with removing organic material from the enamel layer (1, 18), which may, in part, be accentuated by increases in eruption rates (36), as reported for mandibular incisors in Enam+/− mice (16). It is unclear why similar gene dosage effects are not detected in developing enamel covering maxillary incisors (Fig. 3E–F) or to any great extent in developing enamel covering the maxillary or the mandibular molars of heterozygous Enam+/− mice (Fig. 4). This could relate to differences in their embryonic developmental origins or possibly to some environmental factor operating in mandibular incisors that is not influencing mineral formation to the same extent in maxillary incisors and molars. Molars can also be difficult to work with because occlusal wear reduces their absolute weights and alters their enamel-to-dentin ratios.
The interpretation of gene dosage effects in heterozygous Ambn+/−5,6 mice (Fig. 3A–D) is much less straightforward. For example, the enamelin loss-of-function model just discussed is a true null condition where a nuclear targeted protein (β-galactosidase), rather than a secreted matrix protein, is transcribed from the autosomal Enam gene (16). The effects observed in Enam+/− mice therefore occur because only half of the normal amount of enamelin is produced in these animals as in wild-type controls. Enamelin is not a very abundant protein to begin with, especially in rodents (6), and the 50% loss in production of this relatively rare protein is probably the reason why the gene dosage effect appears so dramatically in these animals (Fig. 3E–H). Ameloblastin, by contrast, is produced in relatively large amounts and to such an extent that a 50% loss in production of ameloblastin, as occurs in Ambn+/−5,6 mice, may not be enough to bring it to the same critical level as occurs with enamelin. However, there is an additional complicating factor because Ambn+/−5,6 (and Ambn−5,6/−5,6) mice also transcribe and secrete a protein that is identical to normal ameloblastin, except for a sequence of 117 amino acids between positions 67 and 183 on the N-terminal side of the full-length transcript (the numbering includes signal peptide; http://www.ensembl.org/Mus_musculus, transcript ENSMUST00000031226) that were deleted en bloc by genetic engineering (14, 15). It is possible in Ambn+/−5,6 mice that this truncated protein or some of its fragments are partially compensating for one or more functions in mineralization that ameloblastin normally serves, which is independent of its conjoint role with enamelin at the mineralization front where the truncated protein in Ambn−5,6/−5,6 mice is inactive. Hence, it remains possible, until a real Ambn−/− null mice model becomes available, that, similarly to Enam+/− mice, true heterozygous Ambn+/− mice may show a 50% reduction in mineral content in enamel. The effects of the truncated protein in ‘double dose’ conditions, as occurs in Ambn−5,6/−5,6 mice, also needs to be qualified as it may be responsible in whole or in part for causing the higher level of calcification seen in these animals (Fig 1 and Fig 3).
The 117 amino acids of exons 5 and 6 that are missing from the truncated protein produced in Ambn+/−5,6 and Ambn−5,6/−5,6 mice include those which form 90% of the proline-rich region of ameloblastin (http://us.expasy.org/prosite) and the sequence PREHETQQYEYSLPVHPPPLPSQPSLQPHQP that is highly conserved within mammals (http://blast.ncbi.nlm.nih.gov/Blast.cgi). This conserved sequence from exon 6 contains the alternative splicing site (YEYSLPVHPPPLPSQ) within which is located one of the two O-linked glycosylation sites known to be present in ameloblastin (PSQ) (37). Consistent with its high proline content, it also contains several potential Src homology 3 group (SH3) adhesion/binding motifs (six hits spanning the sequence YSLPVHPPPLPSQPSLQPHQP; http://elm.eu.org) (38–41).
Amino acid sequences in ameloblastin for the most part show very poor identity or similarity to sequences within enamelin (6). It is therefore curious that the PPPLPS sequence from the highly conserved region of the deleted segment in ameloblastin shows 100% similarity to a PPPVPN sequence located between residues 101 and 106 in the full-length enamelin mouse transcript (NCBI reference sequence NP_059496.1). It is further noteworthy that the PPPVPN sequence is itself within the proline-rich region of enamelin (http://us.expasy.org/prosite) and it is part of a sequence associated with potential SH3 adhesion/binding motifs (YQMPMWPPPVP; three hits) (38). There are also several additional potential SH3 adhesion/binding sites immediately adjacent within one amino acid residue on the N-terminal (NGMPMPPHMPPQYP; three hits) and C-terminal (WQQPPMPNFPSK; three hits) sides of this site (38). This N-terminally positioned region of enamelin is close to sites that have already been identified as responsible, via point mutations in exon 3 and especially via a frameshift mutation within intron 4 (42) and a single point mutation at position 176 in exon 6 (43), for producing heterozygous and homozygous enamel phenotypes on mandibular mouse incisors identical to those described for the Enam knockout/lacZ knockin mouse model used in this study (16). It is for these reasons that we suspect that part of the mineral-associated activities co-dependent on the presence of ameloblastin and enamelin may involve transient interactions between the apical plasma membranes of ameloblasts and N-terminally located sequences in the vicinity of the PPPLPS sequence in ameloblastin and the PPPVPN sequence in enamelin which the cells somehow may be able to distinguish as distinct from each other (5, 44). It is conceivable that these membrane–protein interactions have to be constantly ‘updated’ through constitutive secretion of the proteins in order to maintain a state of appositional growth, especially considering the speed at which ameloblastin and enamelin are cleaved by MMP-20 following their secretion (7, 45).
The thin calcified material covering the coronal dentin of incisors and molars in Ambn−5,6/−5,6 and Enam−/− mice on morphological grounds alone appears unstructured, non-specific, and pathologic in nature (Fig. 2) (15). It is as if ameloblasts in these animals activated pathways/processes involved in mineralization and then in the absence of the expected appearance of the mineralization front were unable to shut off or control the formation of other types of mineral deposits within the local environment. Although very similar to compact bone in terms of mineral composition (68% mineral by weight), the crusty material on these teeth is neither bone nor does it contain type 1 collagen or cells trapped in structured extracellular lacuna. According to Wazen et al. (15), the organic components of the calcified material in Ambn−5,6/−5,6 mice include small amounts of amelogenin, the engineered truncated form of ameloblastin, and other proteins suggestive of blood contamination, including albumin, bone sialoprotein, and osteopontin. The composition of organic components within the calcified material from Enam−/− mice is not as well characterized but it does contain small amounts of amelogenin in addition to several other high-molecular-weight and low-molecular-weight proteins (16). While the results from this study are consistent with the idea that part of the problem in these animals involves loss of contact and/or focal adhesion of cells to the extracellular matrix (14, 39, 46), there are other loss-of-function models, such as those involving laminin 5 (47) and connexin 43 (48), which show almost the same end result – that is, shut-down of enamel formation and production of a thin, disorganized crusty calcified material on the coronal surfaces of the teeth. It is unlikely therefore that the changes seen in homozygous Ambn−5,6/−5,6 and Enam−/− mice are related to the simple absence of a single specific protein but are probably the end result of many cellular processes that get out of control in these animals unable to form a true enamel layer. Loss of function of ameloblastin seems to lead to greater consequences in development of enamel organ epithelial cell pathologies than loss of function of enamelin, including the formation of labial tumours (46), which are not seen in Enam−/− mice (16). It remains to be established if continuous production and secretion of the truncated ameloblastin protein, as occurs in Ambn−5,6/−5,6 mice throughout the aborted secretory and maturation stages (15), causes these effects or if it is something related to the inherent properties of ameloblastin.
Acknowledgements
Parts of this research were supported by the Canadian Institutes of Health Research (CIHR; ameloblastin) and by the National Institutes of Health (NIDCR/NIH Grant DE11301; enamelin).
References
- 1.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg M, Septier D, Lécolle S, Chardin H, Quintana MA, Acevedo AC, Gafni G, Dillouya D, Vermelin L, Thonemann B, Schmalz G, Bissila-Mapahou P, Carreau JP. Dentin mineralization. Int J Dev Biol. 1995;39:93–110. [PubMed] [Google Scholar]
- 3.Wiesmann HP, Meyer U, Plate U, Höhling HJ. Aspects of collagen mineralization in hard tissue formation. Int Rev Cytol. 2004;242:121–156. doi: 10.1016/S0074-7696(04)42003-8. [DOI] [PubMed] [Google Scholar]
- 4.Diekwisch TGH. The developmental biology of cementum. Int J Dev Biol. 2001;45:695–706. [PubMed] [Google Scholar]
- 5.Bartlett JD, Ganss B, Goldberg M, Moradian-Oldak J, Paine ML, Snead ML, Wen X, White SN, Zhou YL. Protein–protein interactions of the developing enamel matrix. Curr Top Dev Biol. 2006;74:57–115. doi: 10.1016/S0070-2153(06)74003-0. [DOI] [PubMed] [Google Scholar]
- 6.Hu JC-C, Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14:387–398. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Papagerakis P, Yamakoshi Y, Hu JCC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beniash E, Metzler RA, Lam RSK, Gilbert PUPA. Transient amorphous calcium phosphate in forming enamel. J Struct Biol. 2009;166:133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanci A, Warshawsky H. Characterization of putative secretory sites on ameloblasts of the rat incisor. Am J Anat. 1984;171:163–189. doi: 10.1002/aja.1001710204. [DOI] [PubMed] [Google Scholar]
- 10.Warshawsky H, Josephsen K, Thylstrup A, Fejerskov O. The development of enamel structure in rat incisors as compared to the teeth of monkey and man. Anat Rec. 1981;200:371–399. doi: 10.1002/ar.1092000402. [DOI] [PubMed] [Google Scholar]
- 11.Moffatt P, Smith CE, St-Arnaud R, Simmons D, Wright JT, Nanci A. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochem J. 2006;399:37–46. doi: 10.1042/BJ20060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nanci A, Zalzal S, Kogaya Y. Cytochemical characterization of basement membranes in the enamel organ of the rat incisor. Histochemistry. 1993;99:321–331. doi: 10.1007/BF00269105. [DOI] [PubMed] [Google Scholar]
- 13.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen EH, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 14.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wazen R, Moffatt P, Zalzal SF, Yamada Y, Nanci A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009;28:292–303. doi: 10.1016/j.matbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu JCC, Hu Y, Smith CE, Mckee MD, Wright JT, Yamakoshi Y, Papagerakis P, Hunter GK, Feng JQ, Yamakoshi F, Simmer JP. Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caterina JJ, Skobe Z, Shi J, Ding YL, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 18.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC-C. Hypomaturation enamel defects in Klk4 knockout/lacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong H, White SN, Paine ML, Luo W, Snead ML, Sarikaya M. Enamel structure properties controlled by engineered proteins in transgenic mice. J Bone Miner Res. 2003;18:2052–2059. doi: 10.1359/jbmr.2003.18.11.2052. [DOI] [PubMed] [Google Scholar]
- 20.Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem. 2003;278:19447–19452. doi: 10.1074/jbc.M300445200. [DOI] [PubMed] [Google Scholar]
- 21.Miskin R, Masos T, Shoham Z, Williams-Simons L. Urokinase-type plasminogen activator mRNA is expressed in normal developing teeth and leads to abnormal incisor enamel in alpha MUPA transgenic mice. Transgenic Res. 2006;15:241–254. doi: 10.1007/s11248-006-0006-3. [DOI] [PubMed] [Google Scholar]
- 22.Klopcic B, Maass T, Meyer E, Lehr HA, Metzger D, Chambon P, Mann A, Blessing M. TGF-beta superfamily signaling is essential for tooth and hair morphogenesis and differentiation. Eur J Cell Biol. 2007;86:781–799. doi: 10.1016/j.ejcb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Wen X, Zou Y, Goldberg M, Moats R, Conti PS, Snead ML, Paine ML. Biglycan overexpression on tooth enamel formation in transgenic mice. Anat Rec (Hoboken) 2008;291:1246–1253. doi: 10.1002/ar.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson CW, Li Y, Daly B, Suggs C, Yuan ZA, Fong H, Simmons D, Aragon M, Kulkarni AB, Wright JT. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs. 2009;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett JD, Skobe Z, Lee DH, Wright JT, Li Y, Kulkarni AB, Gibson CW. A developmental comparison of matrix metalloproteinase-20 and amelogenin null mouse enamel. Eur J Oral Sci. 2006;114 Suppl 1:18–23. doi: 10.1111/j.1600-0722.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- 26.Sire JY, Davit-Béal T, Delgado S, Gu X. The origin and evolution of enamel mineralization genes. Cells Tissues Organs. 2007;186:25–48. doi: 10.1159/000102679. [DOI] [PubMed] [Google Scholar]
- 27.Sire JY, Delgado S, Girondot M. Hen’s teeth with enamel cap: from dream to impossibility. BMC Evol Biol. 2008;8:246. doi: 10.1186/1471-2148-8-246. on line. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deméré TA, Mcgowen MR, Berta A, Gatesy J. Morphological and molecular evidence for a stepwise evolutionary transition from teeth to baleen in mysticete whales. Syst Biol. 2008;57:15–37. doi: 10.1080/10635150701884632. [DOI] [PubMed] [Google Scholar]
- 29.Smith CE, Chong DL, Bartlett JD, Margolis HC. Mineral acquisition rates in developing enamel on maxillary and mandibular incisors of rats and mice: implications to extracellular acid loading as apatite crystals mature. J Bone Miner Res. 2005;20:240–249. doi: 10.1359/JBMR.041002. [DOI] [PubMed] [Google Scholar]
- 30.Hiller CR, Robinson C, Weatherell JA. Variations in the composition of developing rat incisor enamel. Calcif Tissue Res. 1975;18:1–12. doi: 10.1007/BF02546222. [DOI] [PubMed] [Google Scholar]
- 31.Neues F, Epple M. X-ray microcomputer tomography for the study of biomineralized endo- and exoskeletons of animals. Chem Rev. 2008;108:4734–4741. doi: 10.1021/cr078250m. [DOI] [PubMed] [Google Scholar]
- 32.Diekwisch TGH, Berman BJ, Gentner S, Slavkin HC. Initial enamel crystals are not spatially associated with mineralized dentine. Cell Tissue Res. 1995;279:149–167. doi: 10.1007/BF00300701. [DOI] [PubMed] [Google Scholar]
- 33.Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen WY, Krebsbach PH, Yamada Y, Hammarström L, Simmer JP, Fincham AG, Snead ML, Smith CE. Comparative immuno-chemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem. 1998;46:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 34.Bouropoulos N, Moradian-Oldak J. Induction of apatite by the cooperative effect of amelogenin and the 32-kDa enamelin. J Dent Res. 2004;83:278–282. doi: 10.1177/154405910408300402. [DOI] [PubMed] [Google Scholar]
- 35.Müller W, Boreiko A, Wang X, Krasko A, Geurtsen W, Custódio MR, Winkler T, Lukic-Bilela L, Link T, Schrö-der H. Morphogenetic activity of silica and bio-silica on the expression of genes controlling biomineralization using SaOS-2 cells. Calcif Tissue Int. 2007;81:382–393. doi: 10.1007/s00223-007-9075-4. [DOI] [PubMed] [Google Scholar]
- 36.Risnes S, Moinichen CB, Septier D, Goldberg M. Effects of accelerated eruption on the enamel of the rat lower incisor. Adv Dent Res. 1996;10:261–269. doi: 10.1177/08959374960100022401. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi K, Yamakoshi Y, Hu JCC, Gomi K, Arai T, Fukae M, Krebsbach PH, Simmer JP. Splicing determines the glycosylation state of ameloblastin. J Dent Res. 2007;86:962–967. doi: 10.1177/154405910708601009. [DOI] [PubMed] [Google Scholar]
- 38.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, Mcguigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SK, Kim SM, Lee YJ, Yamada KM, Yamada Y, Chi JC. The structure of the rat ameloblastin gene and its expression in amelogenesis. Mol Cells. 2003;15:216–225. [PubMed] [Google Scholar]
- 40.Li SSC. Specificity and versatility of SH3 and other praline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Travé G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 42.Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, Shimizu A, Fujimoto N, Kawai A, Miura I, Kaneda H, Kobayashi K, Ishijima J, Maeda T, Gondo Y, Noda T, Wakana S, Shiroishi T. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI) Hum Mol Genet. 2005;14:575–583. doi: 10.1093/hmg/ddi054. [DOI] [PubMed] [Google Scholar]
- 43.Seedorf H, Klaften M, Eke F, Fuchs H, Seedorf U, Hrabe DE, Angelis M. A mutation in the enamelin gene in a mouse model. J Dent Res. 2007;86:764–768. doi: 10.1177/154405910708600815. [DOI] [PubMed] [Google Scholar]
- 44.Wang HJ, Tannukit S, Zhu DH, Snead ML, Paine ML. Enamel matrix protein interactions. J Bone Miner Res. 2005;20:1032–1040. doi: 10.1359/JBMR.050111. [DOI] [PubMed] [Google Scholar]
- 45.Smith CE, Nanci A. Protein dynamics of amelogenesis. Anat Rec. 1996;245:186–207. doi: 10.1002/(SICI)1097-0185(199606)245:2<186::AID-AR7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 46.Fukumoto S, Yamada A, Nonaka K, Yamada Y. Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation. Cells Tissues Organs. 2005;181:189–195. doi: 10.1159/000091380. [DOI] [PubMed] [Google Scholar]
- 47.Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–1323. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flenniken AM, Osborne LR, Anderson N, Ciliberti N, Fleming C, Gittens JEI, Gong XQ, Kelsey LB, Lounsbury C, Moreno L, Nieman BJ, Peterson K, Qu DW, Roscoe W, Shao Q, Tong D, Veitch GIL, Voronina I, Vukobradovic I, Wood GA, Zhu YH, Zirngibl RA, Aubin JE, Bai DL, Bruneau BG, Grynpas M, Henderson JE, Henkelman RM, Mckerlie C, Sled JG, Stanford WL, Laird DW, Kidder GM, Adamson SL, Rossant J. A Gja1 missense mutation in a mouse model of oculodentodigital dysplasia. Development. 2005;132:4375–4386. doi: 10.1242/dev.02011. [DOI] [PubMed] [Google Scholar]