Abstract
Selenium (Se) is a trace element with important roles in human health. Several selenoproteins have essential functions in development. However, the cellular and tissue distribution of Se remains largely unknown because of the lack of analytical techniques that image this element with sufficient sensitivity and resolution. Herein, we report that X-ray fluorescence microscopy (XFM) can be used to visualize and quantify the tissue, cellular and subcellular topography of Se. We applied this technique to characterize the role of Se in spermatogenesis and identified a dramatic Se enrichment specifically in late spermatids, a pattern that was not seen in any other elemental maps. This enrichment was due to elevated levels of the mitochondrial form of glutathione peroxidase 4 and was fully dependent on the supplies of Se by Selenoprotein P. High-resolution scans revealed that Se concentrated near the lumen side of elongating spermatids, where structural components of sperm are formed. During spermatogenesis, maximal Se associated with decreased phosphorus, whereas Zn did not change. In sperm, Se was primarily in the midpiece and co-localized with Cu and Fe. XFM allowed quantification of Se in the midpiece (0.8 fg) and head (0.14 fg) of individual sperm cells, revealing the ability of sperm cells to handle the amounts of this element well above its toxic levels. Overall, the use of XFM allowed visualization of tissue and cellular Se and provided important insights in the role of this and other trace elements in spermatogenesis.
Keywords: selenium, X-ray fluorescence microscopy, spermatogenesis, male reproduction, trace elements
Introduction
Selenium (Se) is an essential trace element that occurs in proteins in the form of selenocysteine (Sec), the 21st naturally occurring amino acid in the genetic code.1. Sec is structurally similar to cysteine (Cys) except that the Se atom in Sec is present in place of the sulfur (S) atom in Cys. S and Se belong to the same group in the periodic table (group VIA), but their unique physicochemical properties lead to important differences in their nucleophilicity, redox potential and ionization properties. For example, at physiological, pH free Sec (pKa 5.2) is almost completely ionized while Cys (pKa 8.3) is protonated.2,3
Sec is encoded by an in-frame stop codon, UGA, and its synthesis and insertion require a complex machinery known as the selenosome.3-6 Organisms that co-translationally incorporate Sec are found in all three domains of life: prokaryotes, archaea, and eukaryotes. Mammalian sets of Sec-containing proteins (selenoproteomes) consist of 24-25 selenoproteins.7 Well-characterized selenoproteins include enzymes with antioxidant functions and other oxidoreductases.3,8 Knockout of several selenoprotein genes, such as cytosolic or mitochondrial thioredoxin reductases, lead to embryonic lethality.9-11 However, the functions of the majority of mammalian selenoproteins are unknown.
In mammals, Se is essential for spermatogenesis and therefore for male fertility.12 In livestock and experimental rodents, moderate to severe Se deficiency leads to phenotypes that range from impaired sperm motility and morphological alterations of the mid-piece architecture to breakage of sperm head and tail and infertility.12-15 Tracer studies with 75Se demonstrated that Se is present within the mid-piece of spermatozoa and associated with a Cys-rich structural protein of the mitochondrial sheath, first named “selenoflagellin” and later MCS for “mitochondrial capsule selenoprotein”.16-19 This protein was further renamed “sperm mitochondrion-associated Cys-rich protein” (SMCP) as it turned out to be not a selenoprotein.20 Ursini and collaborators finally identified phospholipid hydroperoxide glutathione peroxidase (GPx4 or PHGPX) as the selenoprotein component of the mid-piece structure.21 GPx4 is a peroxidase that reduces lipid hydroperoxides with reduced glutathione (GSH).22 In developing spermatids, GPx4 is a soluble enzyme, but it is cross-linked with other structural proteins in the mitochondrial sheath in sperm.21 This protein occurs in 3 forms. The cytosolic form, the most abundant form of the enzyme, is present in most cell types and is essential during embryonic development in mice. Mitochondrial (mGPx4) and nuclear (nGPx4) forms are synthesized from the same gene and are predominantly expressed in testes during spermatogenesis.23,24
Germ cell development requires that the chromatin in the sperm nucleus becomes highly condensed. During chromatin condensation, histones are replaced by the more basic protamines.25,26 nGPx4 may act as a protamine thiol peroxidase and form disulfide cross-links among these proteins thus stabilizing and protecting DNA.23,24
Another selenoprotein, Selenoprotein P (SelP), is relevant to the process of spermatogenesis, although it is not expressed in germ cells. Mouse or human SelP contains 10 selenocysteine residues and has a role in Se transport from liver to other organs. Apolipoprotein E receptor 2 (ApoER2) is expressed in Sertoli cells and mediates SelP uptake in mouse testes.27 SelP−/− mice show abnormal spermatozoa and a reduced Se content in testes.41
It was recently suggested that yet another selenoprotein, thioredoxin-glutathione reductase (TGR), promotes isomerization of disulfide bonds formed between GPx4 and target proteins during nonproductive cross-linking.28,29 TGR is also highly expressed in testis after puberty and belongs to the family of animal thioredoxin reductases (TRs).
In addition to Se, Zn plays important roles in spermatogenesis. The main pool of Zn in sperm nuclei is bound to the protamine-chromatin complexes, especially protamine 2.30-32 Zn is also thought to be present in high levels in the outer dense fibers of sperm flagella during development, but is eliminated during maturation in the epididymis in order for sperm to be motile.30 Reports on Fe and Cu in male reproduction are not conclusive and show a more general requirement of Fe and Cu as protein cofactors. Little is known about the roles of other trace elements in sperm development.
The cellular and subcellular distribution of Se within organs and cells and the possible relocalization and trafficking of this trace element under physiological and pathophysiological conditions have not been previously examined. This is because accurate analytical techniques with sufficient sensitivity to quantify and localize trace amounts of Se on the subcellular level in tissues and cells are not available. At the same time, the role of Se in human health and disease has received much attention.33-35 Information on Se distribution between and within tissues and cells has the potential to provide a better understanding of the use, transport and storage of this trace element in mammals and other organisms.
Synchrotron X-ray fluorescence microscopy (XFM) is a recently developed method ideally suited to study the intracellular distribution and speciation of selected trace elements.36 Third generation, high brilliance sources for hard X-rays provide sufficient coherent flux to acquire quantitative elemental maps of trace metals with sub-micron spatial resolution in cell and tissue samples.37,38 XFM allows simultaneous measurement of a large number of different trace elements with excellent sensitivity.37,38 In this work, we found that this imaging technique can be used to visualize and quantify the tissue and subcellular topography of Se and other trace elements during spermatogenesis in mice. We found Se is enriched in spermatids and explained this finding by the SelP-dependent increase in mGPx4 selenoprotein levels.
RESULTS
Increased levels and spatial reorganization of Se in late stages of germ cell development
We found that XFM can be adapted for imaging Se at high quality in biological samples, including tissue slices and dried cells. As Se is known to have a role in male reproduction, we used this method for the analysis of spatial distribution of Se within seminiferous tubuli of mouse testis and in sperm cells. Analysis of seminiferous tubuli from paraffin-embedded testis sections (5 μm) revealed a sharp increase of Se in spermatids compared to earlier stages of spermatogenic cells (Fig. 1 and 2). Simultaneously, we imaged P, S, Ca, Zn, Fe, Cu and Co, but none of these elements paralleled this specific enrichment.
Fig 1.
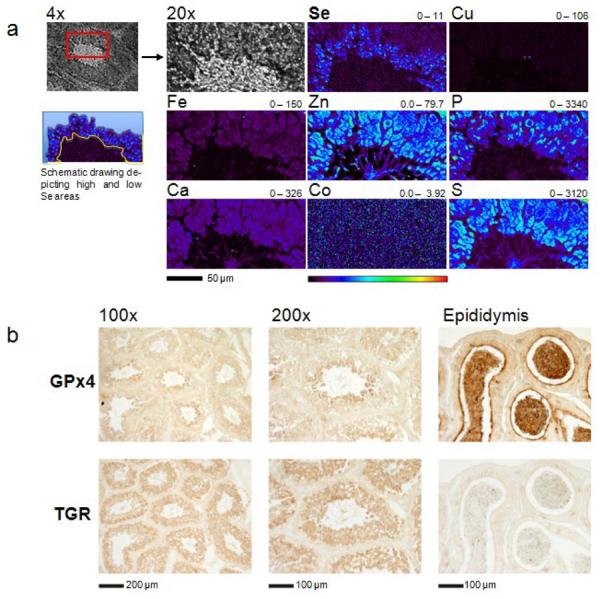
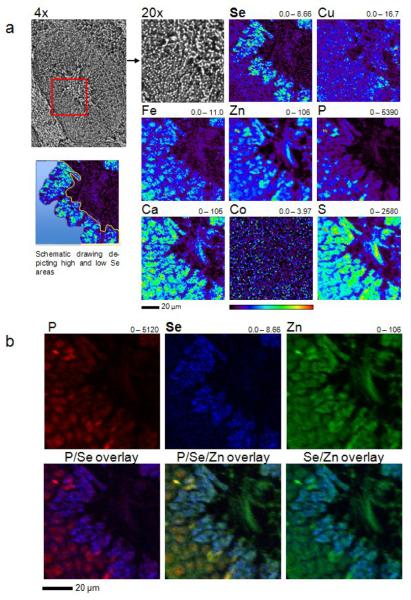
Se enrichment in late spermatids. (a) An XFM scan of a mouse seminiferous tubule. A paraffin embedded testis section (5 μm) of a C57BL/6J wild-type mouse was mounted on a silicon nitride window and both light microscope and XFM images were obtained. The corresponding element and its maximum and minimum threshold value in ng/cm2 are given above each image. The rainbow-colored scale bar relates to the signal intensity measured as ng/cm2 in each spot, with dark pixels representing areas of low concentration and a gradient to bright pixels depicting increasing concentrations. A scale bar (50 μm) is shown below the elemental maps. A schematic drawing visualizes the areas, which are described as “low Se” (area filled with light blue color) and “high Se” (area enclosed by the blue line from one side and the yellow line from the other side) areas. The “low Se” area comprises spermatogonia and spermatocytes, while spermatids constitute the “high Se” area. The scan was obtained by using 12.8-keV incident energy with a dwell time of 1.3 sec per pixel and 1 μm steps through the sample. (b) Immunohistochemical localization of GPx4 and TGR in mouse seminiferous tubuli and epididymis with antibodies specific for these enzymes. Scale bars are shown below the immunostaining images.
Fig 2.
XFM analysis of a mouse seminiferous tubule. (a) A paraffin embedded mouse testis section (5 μm) was analyzed with XFM. The corresponding element and its maximum and minimum threshold values in ng/cm2 are given above each image. The rainbow-colored scale bar relates to the signal intensity measured as ng/cm2 in each spot, with dark pixels representing areas of low concentration and a gradient to bright pixels depicting increasing concentrations. A scale bar (20 μm) is shown below the elemental maps. A schematic drawing visualizes the areas, which are described as “low Se” (area filled with light blue color) and “high Se” (area enclosed by the blue line from one side and the yellow line from the other side) areas. The “low Se” area comprises spermatogonia and spermatocytes, while spermatids constitute the “high Se” area. The scan was obtained by using 12.8-keV incident energy with a dwell time of 1.3 sec per pixel and 1 μm steps through the sample. (b) Overlays of P, Se and Zn elemental maps. A scale bar (20 μm) is shown below the elemental maps.
We further analyzed seminiferous tubuli in 4 representative images (Fig. 1, 2) and defined two regions: (i) an outer tubule region spanning the area from the basement membrane to the cells with high Se content, and (ii) the germ cells with high Se content (excluding the tubule lumen). Se content of both regions was quantified by integration of the calibrated X-ray fluorescence signal with MAPS software (Table 1).39 The mean Se amounts in regions (i) and (ii) were 0.78 and 2.28 ng/cm2 respectively, indicating an ∼3-fold increase in Se levels in spermatids (Table 1). To prove that the detected fluorescence corresponds to Se in the areas with medium to high Se levels, we recorded 180 s dwell time point spectra and analyzed the spectra for peaks in the Kα-line (λ = 0.1105 nm) of Se. Spectra with evident peaks in the Kα-line of Se were found to be specific.
Table 1.
Quantification of Selenium, Phosphorus and Zinc in mouse testis. Se, P and Zn were quantified in high and low Se-containing areas of 4 representative XFM scans of seminiferous tubuli from mouse testis. Given are the mean value in ng/cm2 and the maximal amount in ng/cm2 measured in a specific point of this area. MV = mean value.
| High Se Area (spermatids) | Low Se Area (spermatocytes) | |||
|---|---|---|---|---|
| Selenium | Mean [ng/cm2] | Max [ng/cm2] | Mean [ng/cm2] | Max [ng/cm2] |
| Fig. 1 | 1.88 | 11.04 | 0.72 | 4.41 |
| Fig. 2 | 2.65 | 8.66 | 0.94 | 3.07 |
| Fig. S1 | 2.1 | 6.84 | 0.67 | 3.44 |
| Fig. S2 | 2.49 | 7.62 | 0.78 | 4.12 |
| MV | 2.28 | 8.54 | 0.78 | 3.76 |
| Phosphorus | ||||
| Fig. 1 | 59.07 | 2771.78 | 707.96 | 3113.44 |
| Fig. 2 | 64.01 | 3353.93 | 940.21 | 5389.13 |
| Fig. S1 | 56.01 | 3649.28 | 815.44 | 4111.27 |
| Fig. S2 | 55.67 | 2481.2 | 739.12 | 3597.43 |
| MV | 58.69 | 3064.05 | 800.68 | 4052.82 |
| Zinc | ||||
| Fig. 1 | 23.47 | 59.39 | 20.76 | 51.32 |
| Fig. 2 | 35.2 | 80.72 | 31.76 | 130.79 |
| Fig. S1 | 29.98 | 79.24 | 22.97 | 86.59 |
| Fig. S2 | 30.66 | 74.24 | 22.05 | 84.4 |
| MV | 29.83 | 73.4 | 24.39 | 88.28 |
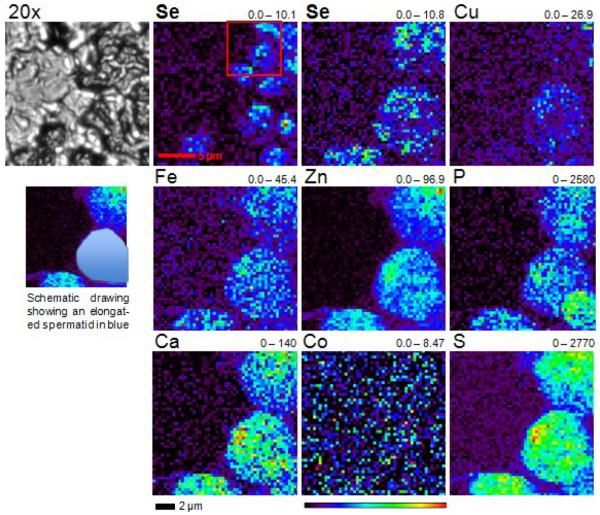
In one seminiferous tubule section, we detected early, slightly elongated spermatids with defined Se content through a fine scan with 0.4 μm resolution (Fig. 3). Interestingly, Se concentrated in these cells at the luminal side, with a mean concentration of ∼3.3 ng/cm2 compared to ∼2.2 ng/cm2 in the remaining part of the cell (Fig. 3 and Table 2). The luminal side develops into the sperm tail. On the other hand, phosphorus concentrated at the opposite apical side of the cell, which develops into the sperm head (Fig. 3 and Table 2). The mean molar Se concentration in whole elongated spermatids was calculated to be ∼24.56 μM. Even though strong Se signals were detected in single spermatids from preparations of seminiferous tubuli, we were unable to detect these signals in the lumen of the tubuli; however, Zn, Ca and S signals were clearly visible (Fig. 1).
Fig 3.
XFM scan of a mouse seminiferous tubule. A paraffin embedded testis section (5 μm) of a C57BL/6J wild-type mouse was mounted on a silicon nitride window and both light microscope and XFM images were obtained. The corresponding element and its maximum and minimum threshold values in ng/cm2 are given above each image. The rainbow-colored scale bar relates to the signal intensity measured as ng/cm2 in each spot, with dark pixels representing areas of low concentration and a gradient to bright pixels depicting increasing concentrations. A scale bar (2 μm) is shown below the elemental maps. A schematic drawing visualizes an early, slightly elongated spermatid in blue. The scan was obtained by using 12-keV incident energy with a dwell time of 4 sec per pixel and 0.4 μm steps through the sample.
Table 2.
Quantification of trace elements in elongating spermatids. Trace elements were quantified in three different regions (whole cell, Se rich region and P rich region) of an elongating spermatid. Concentrations are given in ng/cm2.
| Mean Value [ng/cm2] | |||
|---|---|---|---|
| whole cell |
Se rich region |
P rich region |
|
| Se | 2.41 | 3.35 | 2.2 |
| Fe | 12.69 | 15.45 | 11.56 |
| Zn | 32.75 | 38.93 | 33.84 |
| Cu | 4.22 | 5.08 | 3.73 |
| P | 875.2 | 860.46 | 1347.08 |
To test that the specific enrichment of Se in late spermatids correlated with an increased expression of selenoproteins, we performed immunostaining of mouse testis sections with antibodies against two most abundant selenoproteins involved in sperm maturation, GPx4 and TGR (Fig. 1). A sharp increase in GPx4 expression in late spermatids was observed, whereas TGR was more uniformly expressed throughout spermatids at different stages (Fig. 1). Thus, the late spermatid and sperm midpiece enrichment of Se are consistent with the elevated GPx4 expression, while TGR may only partially contribute to the Se enrichment in spermatids. Our analyses also showed that GPx4 was present at high levels in epididymal sperm and that TGR was absent in these cells (Fig. 1).
Se enrichment in spermatids can be explained by indirect SelP-dependent delivery of Se for mGPx4
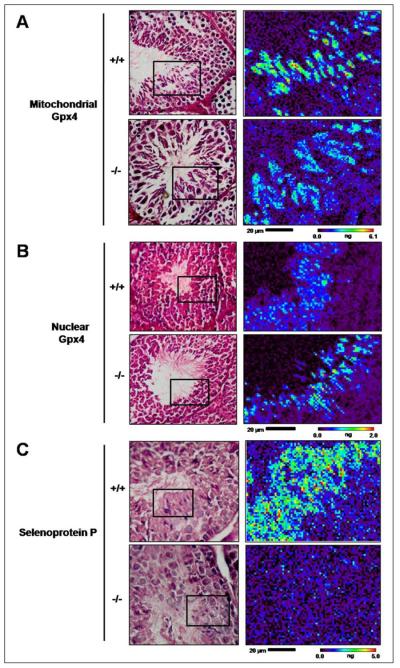
To further examine the basis for the specific enrichment of Se in spermatids, we imaged testis sections from various knockout mouse models characterized by deficiency in individual selenoproteins. Comparison of nGPx4 knockout and isogenic wild-type mice revealed that deficiency in nGPx4 led to minor changes in Se content in testes and did not affect the enrichment of Se in spermatids (Fig. 4A, Table 3). However, mGPx4 knockout led to ∼60% decrease in Se in testes and largely abrogated the enrichment of this element in spermatids (Fig. 4B). We further found that Se content in late spermatids from SelP knockout mice was reduced by ∼77% (Fig. 4C, Table 3) and the remaining Se was uniformly distributed in various cells in the seminiferous tubules. Thus, our XFM analyses visualized both the role of SelP in Se delivery to testes and the specific targeting (indirectly through Sertoli cells) of this Se for the synthesis of the mitochondrial form of GPx4 in spermatids. We also observed that the effect of SelP knockout was more significant than the knockout of mGPx4. Thus, it is likely that SelP delivers Se for other selenoproteins as well, perhaps for TGR, which is another selenoprotein enriched in spermatids.
Fig 4.
Roles of GPx4 and SelP in spermatogenesis in mice. Seminiferous tubules from knockout mice deficient in mitochondrial GPx4 (a), nuclear GPx4 (b) and SelP (c) and appropriate wild type controls were imaged by XFM. Left panels show light microscope images and right panels show Se maps of XFM scans.
Table 3.
Quantification of Se in testis of GPx4 and SelP knockout mice. Se was quantified in high and low Se-containing areas of XFM scans of seminiferous tubuli from mouse testis. Mean value (MV) and the maximal content were measured in regions of interest.
| High Se Area | Low Se Area | |||
|---|---|---|---|---|
| Mouse genotype | Mean [ng/cm2] | Max [ng/cm2] | Mean [ng/cm2] | Max [ng/cm2] |
| WT control for nuclear GPx4 |
4.46 | 13.26 | 1.45 | 4.06 |
| Nuclear GPx4 −/− | 4.86 | 22.53 | 1.45 | 3.39 |
| WT control for mitochondrial GPx4 |
2.86 | 6.43 | 0.68 | 1.83 |
| Mitochondrial GPx4 −/− | 1.21 | 4.34 | 0.56 | 1.83 |
| WT control for SelP | 2.34 | 6.60 | 0.76 | 2.95 |
| SelP −/− | 0.55 | 2.02 | 0.47 | 2.11 |
Phosphorus and zinc imaging during spermatogenesis
Similar to what was shown in a previous study for human microvascular endothelial cells, XFM analysis of seminiferous tubule sections demonstrated elemental distribution of P (DNA) and Zn (transcription factors) typical of eukaryotic cells, with both elements most prevalent in the nucleus (Fig. 1).38 An interesting observation in seminiferous tubuli was that the concentration of P slightly declined during germ cell development to ∼73% of its mean concentration at the beginning of the development (Fig. 1, 2). An overlay of P with Se shows that the P was at its lowest point when Se was maximal (Fig. 2). Zn on the other hand slightly increased (∼22%) in late spermatids, especially in the nuclei and sperm tails (Fig. 2). Fe and Cu, two other essential metal ions, were uniformly distributed with slight variations according to section and cell thickness (Fig. 1 and 2). In the lumen of the tubules, tails of late spermatids were observed that contained Zn, Ca, and S, but lacked Se, Fe and P.
Selenium in the midpiece and head of sperm cells
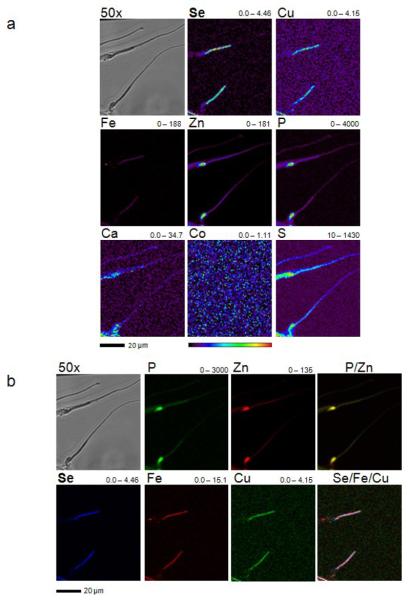
Unlike late spermatids seen in seminiferous tubuli of whole testis sections, Se showed additional spatial compartmentalization in single, separated sperm cells extracted from mouse testes (Fig. 5). Se accumulated in the midpiece structure of sperm, but was also detected in sperm heads (Fig. 5). Se content of the mid-piece was 0.79 fg (≈ 10.13 × 10−18 mol), whereas the head had 0.14 fg (≈ 1.773 × 10−18 mol) Se, which is an ∼ 5.6-times lower level than that in the midpiece (Table 4). To calculate the molar concentration of Se in the head and the midpiece structure of mouse sperm, we took the average width and length of these structures from the literature and calculated their average volume by assuming a spherical form for the head (86.64 × 10−15 l) and a cylindrical form for the midpiece (26.9 × 10−15 l).39 Then we divided the mole by the calculated average volume. The molar concentration of Se in the midpiece was calculated to be ∼377 μM, 18 times higher than in the head (∼20 μM). In both structures, the amount of Se was well above toxic levels of this element, indicating the ability of sperm cells to handle toxic Se amounts. Overlay images of Se with Cu and Fe clearly show an overlap between Se and these metals in the midpiece structure. The total amount of Cu (0.59 fg) was slightly lower than that of Se (0.79 fg), whereas the total amount of Fe was ∼ 3-times higher (Table 4).
Fig 5.
XFM analysis of mouse sperm cells. (a) Sperm was extracted from testis of a C57BL/6J mouse, transferred on a silicon nitride window and air dried. Light microscope and XFM images were obtained. The corresponding element and its maximum and minimum threshold values in ng/cm2 are given above each image. The rainbow-colored scale bar relates to the signal intensity measured as ng/cm2 in each spot, with dark pixels representing areas of low concentration and a gradient to bright pixels depicting increasing concentrations. A scale bar (20 μm) is shown below the elemental maps. The scan was obtained by using 12-keV incident energy with a dwell time of 1.7 sec per pixel and 0.5 μm steps through the sample. (b) The elements P, and Zn and Se, Fe, and Cu determined in (a) were used to make overlays to determine co-localization. A scale bar (20 μm) is shown below the elemental maps.
Table 4.
Quantification of the total amount of indicated trace elements in the defined regions of sperm cells. The total amount of certain trace elements, given in fg, was determined in two representative XFM scans of mouse sperm cells. MP, midpiece.
| Total Amount [fg] | |||
|---|---|---|---|
| MP of Sperm 1 |
MP of Sperm 2 |
Mean Value |
|
| Se | 0.74 | 0.84 | 0.79 |
| Fe | 2.44 | 2.54 | 2.49 |
| Cu | 0.53 | 0.65 | 0.59 |
| Zn | 5.3 | 8.53 | 6.91 |
| P | 115.35 | 186.4 | 150.87 |
| Total Amount [fg] | |||
|
Sperm-Head 1 |
Sperm-Head 2 |
Mean Value |
|
| Se | 0.11 | 0.17 | 0.14 |
| Fe | 0.26 | 0.46 | 0.36 |
| Cu | 0.1 | 0.18 | 0.14 |
| Zn | 16.1 | 18.53 | 17.32 |
| P | 383.34 | 416.3 | 399.82 |
Phosphorus and zinc mainly localize in sperm heads
As observed for seminiferous tubule samples, P and Zn also concentrated in the nuclei of sperm (Fig. 5). Quantifying the concentration of both elements in the sperm head revealed that P was ∼ 23-times higher than Zn (Table 4). P could also be detected at a low level in the midpiece structure (∼ 150 fg), whereas Zn was present throughout the tail in decreasing amounts from the head to the tail (Fig. 5, Table 2).
DISCUSSION
Our study is the first where Se was directly imaged with high quality and spatial resolution both during spermatogenesis and generally in biological systems. We found that Se could be visualized at high resolution in paraffin-embedded tissue sections and dried cells using the recently developed, powerful technique of XFM.
Previous studies have demonstrated the essential role of Se in spermatogenesis and male fertility in mammals.12 With XFM, we show that Se is specifically concentrated in spermatids (male germ cells) after the 2nd meiotic cell division. Moreover, in intact sperm cells, we demonstrate the specific enrichment of Se in the midpiece and to a lower extent in the sperm head. We found that the detected trace elemental distribution is not altered during chemical fixation, and we could detect the same specific distribution in preparations of sperm samples from seminiferous tubuli without fixation and paraffin embedding (Fig. 5, Table 4). Our data support and greatly extend the previous findings discussed in the introduction with other methods.12-15,21,23,24
A major plasma selenoprotein, SelP, may supply Se to testis due to presence of a Se-rich C-terminal segment in SelP.40,41 It was reported recently that ApoER2 mediates the uptake of plasma SelP through Sertoli cells.27 SelP was not detected in spermatocytes, suggesting that this protein may be processed by Sertoli cells and that Se is provided in another form to germ cells.43 Our data show SelP delivers the bulk of Se to testes to satisfy the increased needs for this element during spermatid development. However, since this protein is localized to the basal membranes in semiferious tubule, SelP is not the protein enriched in spermatids, but it supplies Se specifically for this stage of spermiogenesis. 43
To determine which Se species were visualized by XFM in spermatids, we analyzed testis slices from knockout mouse models deficient in the most relevant selenoprotein in testes, GPx4. This protein is present in high levels in both late spermatids and spermatozoa. We found that the observed Se enrichment in spermatids could be largely accounted for by the mitochondrial form of GPx4. On the other hand, knockout of the nuclear GPx4 form did not change the Se map. Altogether, our data show that the elevation in Se levels in spermatids is due to the high expression of mGPx4 and that the source of Se for this protein is SelP.
In spermatids, chromatin becomes condensed and tightly packed through crosslinks among protamines, and at the same time structural components of the midpiece and tail are formed. 21,23,24 GPx4 contributes to both processes, with its nuclear form serving a role in the sperm head and the mitochondrial GPx4 in the midpiece.21,23,24 It also appears that TGR may partially contribute to the Se increase in spermatids. Immunochemical analyses of mouse seminiferous tubuli showed a dramatic increase in GPx4 in late spermatids and mature sperm (Fig. 1). TGR was more uniformly expressed in spermatids and was absent in the epididymal sperm.
We also observed that higher concentrations of Se were detected on the luminal side of the cells where the tail develops. It is known, but for the first time directly visualized in this study, that Se concentrates in the midpiece of the sperm tail.16,17 The specific enrichment of Se at the luminal site of elongated spermatids should be of importance for the correct development of the tail. Phosphorus, with its highest levels in DNA and therefore in the nucleus, concentrated on the apical side, where the head develops (Fig. 2, Table 2).
In addition to P, Zn was abundant in the nuclei in the testes samples. This element is an important cofactor for numerous transcription factors and other proteins. Some cells might have been even captured in a meiotic state as evidenced by their ring-like P distribution (Fig. 1, upper right corner). During germ cell development, P concentration declined to around ∼73% of its original levels (Fig. 3, Table 1), likely due to reduction in DNA during meiosis, when the daughter chromatids are divided. An overlay of P with Se shows that P is at its lowest point when Se is highest suggesting that all meiotic steps are completed and spermatid development begins (Fig. 3). Zn, on the other hand, is still high in late germ cell development and is seen in high levels in the nucleus of spermatids and both the nucleus and the tail of early sperm cells (Fig. 1, 3, Table 1). This is in agreement with earlier studies, describing the need for Zn in the nuclei of spermatids for chromatin condensation and high amounts of Zn in the tail of early spermatids.25,26,31 In single sperm cells, P and Zn are prevalent in the nucleus, with P being 20 times higher than Zn, consistent with completion of DNA packing. P in the tail is most likely from ATP, the energy source for tail motion. Zn concentration in the tail is lower, consistent with the finding that Zn in the tails of mature sperm has to decrease for full motility.31 The overlap of Cu and Fe with Se in the midpiece structure is reasonable, because both elements are important co-factors in the electron-transport chain of mitochondria, which are tightly packed in the midpiece.
In summary, this is the first study where Se was directly imaged in its native environment with high spatial resolution. We show a high and specific accumulation of Se during spermatid development due to increased synthesis of mGPx4 that is dependent on the supply of Se by a plasma selenoprotein, SelP. Our study is also the first where Se was visualized, localized and quantified in sperm. We suggest that the technique of XFM could provide considerable insights into Se trafficking and distribution in mammals. Advances in XRF will further help increase sensitivity of this method and make it amenable for detection of lower amounts of Se.
Materials and Methods
Tissue and sperm samples
Testes and sperm samples were taken from 3-4 month old C57BL/6J mice fed with a standard rodent chow diet (Harlan Teklad, Madison, WI). Tissues were fixed in 10 % neutral buffered formalin and washed with PBS for 12 h. PBS-washed tissues were embedded in paraffin at the Veterinary Diagnostic Center, University of Nebraska–Lincoln. 5 μm sections of the formalin-fixed, paraffin embedded tissues were cut on a standard microtome. Sections were transferred into a 40 °C water bath and mounted on silicon nitride windows (2 × 2 mm; 200 nm thickness, Silson, Blisworth, UK) for XFM imaging. Sperm was extracted from testes and epididymus, immediately transferred onto silicon nitride windows and air dried. Light microscope images were taken with a Leica DMXR high resolution light microscope (Leica Microsystems, Bannockburn, IL). Following XFM analysis, samples were deparafinized in xylene and stained in hematoxylin (Invitrogen, Carlsbad, CA) and Eosin (Sigma, St. Louis, MO) to reveal fine tissue structure. Images were taken using light Olympus AX70 microscope at UNL Microscopy Core Facility. Tissues from GPx4 knockout24 and SelP knockout mice41 were provided by Drs. Marcus Conrad and Raymond Burk, respectively.
X-ray fluorescence microscopy
Samples were analyzed by XFM at the hard X-ray microprobe (7-17-keV) beamline 2-ID-E of the Advanced Photon Source at the Argonne National Laboratory. Prior to data collection, samples were placed on specimen holders developed to fit the fine tuned motorized stages of both the available light microscope and the X-ray microprobe. The samples were first mounted on a Leica DMXRE microscope (Leica Microsystems, Wetzlar, Germany). The x-, y-, and z-, coordinates of selected target areas in the samples were precisely located on the windows, relative to the four window edges, which served as reference points. In addition, light microscope images of the target areas were taken. The samples were then mounted on the microprobe, and the target areas were relocalized using the coordinates from the light microscope together with the x-, y-, and z-, coordinates of the window edges determined from the microprobe. Undulator-generated incident X-rays of 12.8-keV were monochromatized with a sidebounce Si <220> monochromator. X-rays were focused down to a measured spot size of 0.4 × 0.4 μm with Fresnel zones plate optics (X-radia, Concord, CA). The specimens were raster scanned with 1 μm or 0.3 μm steps, and entire X-ray fluorescence spectra were recorded at each pixel for 1.2-1.7 s or 4 s dwell time, respectively, by using a single element UltraLE Ge energy dispersive detector (Canberra, Meridien, CT). Point spectra of the fluorescence were taken at specific points with a dwell time of 180 s. X-ray fluorescence spectra of thin-film standards NBS-1832 and NBS-1833 (National Bureau of Standards, Gaithersburg, MD) were collected prior to the sample analysis and used for standardization of the data by fitting the sample spectra against the signal derived from the standards.38 Quantification and image processing were performed with MAPS software.38,39,42 Each image represents a two-dimensional projection of the volumetric distribution for each element, which means the thickness of the sample influences the signal.
Immunohistochemistry
Immunohistochemistry was performed with Histostain-Plus (DAB, rabbit) kit (Zymed) according to manufacturer's instructions. Sections were deparaffinized with xylene and rehydrated in a graded series of ethanol. 10% non-immune goat serum was used to block nonspecific binding. The slides were incubated with GPx4 (1:100 dilution) or TGR (1:200 dilution) primary antibodies for 1 h, and washed three times with PBS containing 0.05% Tween 20 (PBST). The biotinylated secondary antibodies were applied to the sections for 10 min. The slides were then washed three times with PBST, and incubated with horseradish peroxidase conjugated streptavidin for 10 min followed by wash with PBST (three times). The staining was performed with DAB chromogen.
Acknowledgements
We thank Wayne Vogl for helpful discussion. We also thank Marcus Conrad for providing tissues from nGPx4 and mGPx4 knockout mice and Raymond Burk and Kristina Hill for tissues from SelP knockout mice. This study was supported by NIH GM065204 (to VNG) and the Intramural Research Program of the Center for Cancer Research, NCI, NIH (to DLH). Use of the Advanced Photon Source was supported by the DOE, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The abbreviations used are
- GPx4 or PHGPX
phospholipid hydroperoxide glutathione peroxidase
- S
Sulfur
- Se
selenium
- Sec
selenocysteine
- SelP
selenoprotein P
- SMCP
sperm mitochondrion-associated Cys-rich protein
- snGPx4
testis nucleus-specific GPx4
- TGR
thioredoxin-glutathione reductase
- TR
thioredoxin reductase
- XFM
X-ray fluorescence microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hatfield DL, Gladyshev VN. How Selenium Has Altered Our Understanding of the Genetic Code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johansson L, Gafvelin G, Arnér ES. Selenocysteine in proteins – properties and biotechnological use. Bioch. Biophys. Acta. 2005;1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine Incorporation Machinery and the Role of Selenoproteins in Development and Health. Prog. Nucl. Acid. Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 4.Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zinoni R, Birkmann A, Stadtman TC, Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1986;83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:1–6. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN. Characterization of Mammalian Selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 8.Gromer S, Eubel JK, Lee L, Jacob J. Human selenoproteins at a glance. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad M, Jakupoglu C, Moreno SG, Lipp S, Banjac A, Schneider M, et al. Essential Role for Mitochondrial Thioredoxin Reductase in Hematopoiesis, Heart Development and Heart Function. Mol. Cell. Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakupoglu C, Przemeck GKH, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, et al. Cytoplasmatic Thioredoxin Reductase Is Essential for Embryogenesis but Dispensable for Cardiac Development. Mol. Cell. Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bondareva AA, Capecci MR, Iverson SV, Li Y, Lopez NI, Lucas OL, Merrill GF, Prigge JR, Siders AM, Wakamiya M, Wallin SL, Schmidt EE. Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome. Free Radic. Biol. Med. 2007;43:911–923. doi: 10.1016/j.freeradbiomed.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiorino M, Roveri A, Ursini F, Brigelius-Flohé R, Flohé L. Selenium and male reproduction. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. 2nd Springer Science+Business Media, LLC; 2006. pp. 323–332. Chap 25. [Google Scholar]
- 13.Wu SH, Oldfield JE, Whanger PD, Weswig PH. Effect of Selenium, Vitamine E, and Antioxidants on Testicular Function in Rats. Biol. Reprod. 1973;8:625–629. doi: 10.1093/biolreprod/8.5.625. [DOI] [PubMed] [Google Scholar]
- 14.Wallace E, Cooper GW, Calvin HI. Effects of Selenium Deficiency on the Shape and Arrangement of Rodent Sperm Mitochondria. Gamete Res. 1983;4:389–399. [Google Scholar]
- 15.Shalini S, Bansal MP. Dietary selenium deficiency as well as excess supplementation induces multiple defects in mouse epididymal spermatozoa: understanding the role of selenium in male fertility. Int. J. Androl. 2008;31:438–449. doi: 10.1111/j.1365-2605.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown DG, Burk RF. Selenium retention in tissues and sperm of rats fed a Torula yeast diet. J. Nutr. 1973;103:102–108. doi: 10.1093/jn/103.1.102. [DOI] [PubMed] [Google Scholar]
- 17.Calvin IH. Selective incorporation of selenium-75 into a polypeptide of the rat sperm tail (1) J. Exp. Zool. 1978;204:445–452. doi: 10.1002/jez.1402040315. [DOI] [PubMed] [Google Scholar]
- 18.Calvin IH, Cooper GW, Wallace E. Evidence That Selenium in Rat Sperm Is Associated With a Cysteine-Rich Structural Protein of the Mitochondrial Capsules. Gamete Res. 1981;4:139–149. [Google Scholar]
- 19.Kleene KC, Smith J, Bozorgzadeh A, Harris M, Hahn L, Karimpour I, Gerstel J. Sequence and developmental expression of the mRNA encoding the seleno-protein of the sperm mitochondrial capsule in the mouse. Dev. Biol. 1990;137:395–402. doi: 10.1016/0012-1606(90)90263-i. [DOI] [PubMed] [Google Scholar]
- 20.Cataldo L, Baig K, Oko R, Mastrangelo MA, Kleene KC. Developmental expression, intracellular localization, and selenium content of the cysteine-rich protein associated with the mitochondrial capsules of mouse sperm. Mol. Reprod. Dev. 1996;45:320–31. doi: 10.1002/(SICI)1098-2795(199611)45:3<320::AID-MRD9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohé L. Dual Function of the Selenoprotein PHGPx During Sperm Maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 22.Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 23.Pfeifer H, Conrad M, Roethlein D, Kyriakopoulos A, Brielmeier M, Bornkamm GW, Behne D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15:1236–1238. [PubMed] [Google Scholar]
- 24.Conrad M, Moreno SG, Sinowatz F, Ursini F, Kölle S, Roveri A, Brielmeier M, Wurst W, Maiorino M, Bornkamm GW. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol. Cell. Biol. 2005;25:7637–7644. doi: 10.1128/MCB.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog. Nucl. Acid. Res. Mol. Biol. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- 26.Bench G, Corzett MH, Kramer CE, Grant PG, Balhorn R. Zinc is sufficiently abundant within mammalian sperm nuclei to bind stoichiometrically with protamine 2. Mol. Reprod. Dev. 2000;56:512–519. doi: 10.1002/1098-2795(200008)56:4<512::AID-MRD9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 27.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J. Biol. Chem. 2007;282:12290–12297. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun QA, Su D, Novoselov SV, Carlson BA, Hatfield DL, Gladyshev VN. Reaction Mechanism and Regulation of Mammalian Thioredoxin/Glutathione Reductase. Biochemistry. 2005;44:14528–14537. doi: 10.1021/bi051321w. [DOI] [PubMed] [Google Scholar]
- 29.Su D, Novoselov SV, Sun QA, Moustafa ME, Zhou Y, Oko R, Hatfield DL, Gladyshev VN. Mammalian Selenoprotein Thioredoxin-glutathione Reductase. J. Biol. Chem. 2005;280:26491–26498. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- 30.Bertelsmann H, Sieme H, Behne D, Kyriakopoulos A. Is the distribution of selenium and zinc in the sublocations of spermatozoa regulated? Ann. N. Y. Acad. Sci. 2007;1095:204–208. doi: 10.1196/annals.1397.024. [DOI] [PubMed] [Google Scholar]
- 31.Henkel R, Bittner J, Weber R, Hüther F, Miska W. Relevance of zinc in human sperm flagella and its relation to motility. Fertil. Steril. 1999;71:1138–1143. doi: 10.1016/s0015-0282(99)00141-7. [DOI] [PubMed] [Google Scholar]
- 32.Zbuzkova V, Kincl FA. The effect of copper and other metals on spermatogenesis in rats. Acta Endocrinol. 1971;66:379–384. doi: 10.1530/acta.0.0660379. [DOI] [PubMed] [Google Scholar]
- 33.Duffield-Lillico AJ, Dalkin BL, Reid ME, Turnbull BW, Slate EH, Jacobs ET, Marshall JR, Clark LC. Selenium supplementation, baseline plasma selenium status and incidence of prostate cancer: an analysis of the complete treatment period of the Nutritional Prevention of Cancer Trial. B. J. U. Int. 2003;91:608–612. doi: 10.1046/j.1464-410x.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- 34.Stranges S, Marshall JR, Trevisan M, Natarajan R, Donahue RP, Combs GF, Farinaro E, Clark LC, Reid ME. Effects of selenium supplementation on cardiovascular disease incidence and mortality: secondary analyses in a randomized clinical trial. Am. J. Epidemiol. 2006;163:694–699. doi: 10.1093/aje/kwj097. [DOI] [PubMed] [Google Scholar]
- 35.Alanne M, Kristiansson K, Auro K, Silander K, Kuulasmaa K, Peltonen L, Salomaa V, Perola M. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum. Genet. 2007;122:355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 36.Paunesku T, Vogt S, Maser J, Lai B, Woloschak G. X-ray fluorescence microprobe imaging in biology and medicine. J. Cell. Biochem. 2006;99:1489–1502. doi: 10.1002/jcb.21047. [DOI] [PubMed] [Google Scholar]
- 37.Kemner KM, Kelly SD, Lai B, Maser J, O'loughlin EJ, Sholto-Douglas D, Cai Z, Schneegurt MA, Kulpa CF, Jr., Nealson KH. Elemental and redox analysis of single bacterial cells by x-ray microbeam analysis. Science. 2004;306:686–687. doi: 10.1126/science.1103524. [DOI] [PubMed] [Google Scholar]
- 38.Finney L, Mandava S, Ursos L, Zhang W, Rodi D, Vogt S, Legnini D, Maser J, Ikpatt F, Olopade OI, Glesne D. X-ray fluorescence microscopy reveals large-scale relocalization and extracellular translocation of cellular copper during angiogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:2247–2252. doi: 10.1073/pnas.0607238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummins JM, Woodall PF. On mammalian sperm dimensions. J. Reprod. Fert. 1985;75:153–175. doi: 10.1530/jrf.0.0750153. [DOI] [PubMed] [Google Scholar]
- 40.Vogt S. MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J. Phys. IV France. 2003;104:635–638. [Google Scholar]
- 41.Hill KE, Zhou J, McMahan W, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of Selenoprotein P Alters Distribution of Selenium in the Mouse. J. Biol. Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 42.Hill KE, Zhou J, Austin LM, Motley AK, Ham AJL, Olson GE, Atkins JF, Gesteland RF, Burk RF. The Selenium-rich C-terminal Domain of Mouse Selenoprotein P Is Necessary for the Supply of Selenium to Brain and Testis but Not for the Maintenance of Whole Body Selenium. J. Biol. Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 43.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282:12290–12297. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 44.Vogt S, Maser J, Jacobsen C. Data analysis for X-ray fluorescence imaging. J. Phys. IV France. 2003;104:617–622. [Google Scholar]