Abstract
Age-related changes in brain function include those affecting learning, memory, and sleep-wakefulness. Sleep-wakefulness is an essential behavior that results from the interaction of multiple brain regions, peptides and neurotransmitters. The biological function(s) of sleep, however, remains unknown, due to a paucity of information available at the cellular level. Aged rats exhibit alterations in the circadian and homeostatic influences associated with sleep-wake regulation. We recently showed that alterations in cortical profiles occur after timed bouts of spontaneous sleep in young rats. Examination of the cellular response to sleep-wake in old rats may thus provide insight(s) into the biological function(s) of sleep. To test this hypothesis, we monitored cortical profiles in the frontal cortex of young and old Sprague-Dawley rats after timed bouts of spontaneous sleep-wake behavior. Proteins were separated by two-dimensional electrophoresis (2-DE), visualized by fluorescent staining, imaged, and analyzed as a function of behavioral state and age. Old rats showed a 6-fold increase in total protein expression, independent of the behavioral state at sacrifice. When analyzed according to age and behavioral state, there was a decrease (~46%) in the number of phospho-spots present during SWS in aged animals. SWS-associated spots present only in old animals were associated with multiple functions including vesicular transport, cell signaling, oxidation state, cytoskeletal support, and energy metabolism. These data suggest that the intracellular response to the signaling associated with spontaneous sleep is affected by age and is consistent with the idea that the ability of sleep to fulfill its’ function(s) may become diminished with age.
Keywords: two-dimensional electrophoresis (2DE), mass spectrometry, sleep-associated proteins, spontaneous sleep bouts, aging
1. Introduction
Aging is associated with a generalized and progressive reduction in the body’s ability to respond to stress that is thought to arise from damage to intracellular organelles and macromolecules (i.e., DNA, proteins, and lipids) as a result of the accumulation of reactive oxygen and nitrogen species, ROS and RNS, respectively; (Balaban et al., 2005; Calabrese et al., 2006; Joseph et al., 2005). The covalent binding of these species affects the activity of a host of proteins, thereby affecting protein function and turnover (Stadtman, 1990; Stadtman, 1992) and has been documented in bacteria (Cabiscol et al., 2000; Nystrom, 2002), fly (Sohal, 2002), and rat (Butterfield et al., 1999; Kanski et al., 2003) models of aging. In the rat, the functional categories of covalently modified proteins are similar in muscle and brain and include proteins involved in biosynthesis, energy production, cytoskeletal dynamics and signal transduction (Butterfield et al., 1999; Kanski et al., 2003; Poon et al., 2004a; Poon et al., 2004b; Poon et al., 2006).
The brain is particularly susceptible to oxidative stress, with high levels of unsaturated fatty acids, iron/ascorbate, and oxygen consumption and relatively low levels of proteins involved in antioxidant defense (Floyd, 1999; Poon et al., 2004a; Poon et al., 2004b; Poon et al., 2006). The severity of symptoms can vary. Mitochondrial dysfunction, for example, has been linked to neurodegenerative disorders like Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease (Genova et al., 2004), while “normal” aging is associated with alterations in a broad spectrum of brain functions that include learning, memory, and sleep (Calabrese et al., 2006).
Sleep is a relatively simple behavior with complex anatomical and physiological underpinnings. The existing data indicate that sleep is an essential behavior, regulated by a combination of circadian and homeostatic influences (Kryger et al., 2000). The biological function(s) remains unknown, though there is general agreement that sleep serves a restorative function(s). Key to the elucidation of the biological function (s) is an understanding of how signals associated with sleep impact the cellular milieu. Early intracellular studies from the 1970’s suggested that sleep may replenish proteins. Studies using rapid eye movement (REM) sleep deprivation to investigate the function of REM sleep showed that protein synthesis occurs during REM sleep, suggesting a restorative function of sleep (Bobillier et al., 1971; Drucker-Colin et al., 1979). A subsequent study that examined 14C-leucine absorption into cerebral protein showed that protein synthesis increased during SWS compared with REM sleep and waking (Ramm and Smith, 1990). These early and seemingly conflicting results may reflect the sensitivity of the methods employed or the relatively large areas tested. Sleep research has greatly benefited from recent technological advances in high throughput analyses of mRNA and protein profiles (Basheer et al., 2005; Cirelli et al., 2004; Terao et al., 2006; Vazquez et al., 2008). The results of these recent studies identified proteins/mRNAs associated with ATP generation/storage, the oxidation reduction state and cytoskeletal metabolism, suggesting roles for sleep in the maintenance of energy metabolism, re-dox state and synaptic plasticity in young rats. There is thus considerable overlap in the functional categories associated with sleep and those affected by aging.
Aging negatively impacts sleep-wake behavior in clinically compromised and otherwise healthy elderly humans (Avidan, 2005; Montgomery and Dennis, 2004; Prinz, 1995). In aged humans, there is a decrease in “deep” (or “restorative”) sleep and an increase in “light” or early stages of sleep that leads to more frequent awakenings and changes in sleep timing (Bliwise, 1993; Prinz, 1995). In addition, the ability to compensate for sleep loss after prolonged waking is diminished (Bonnet and Rosa, 1987; Carskadon and Dement, 1985). Other experimental models including rodents (Shiromani et al., 2000), flies (Cirelli, 2006), and zebrafish (Zhdanova et al., 2008) also exhibit age-related alterations in sleep. In aged rodents (Shiromani et al., 2000), the inability to compensate for sleep loss is accompanied by reductions in protein/mRNA (Cunha et al., 1995; Sperlagh et al., 1997) and/or alterations in protein function (Basheer et al., 2005) and protein profiles (Pawlyk et al., 2007). Taken together, these results provide evidence that the intracellular response to sleep is diminished during aging in brain regions associated with sleep-wakefulness under both normal and stressful conditions.
Based on this data, we hypothesized that aging would have pronounced effects on protein profiles across spontaneous sleep-wake bouts. In addition, we hypothesized that the functional categories of proteins affected by aging would be related to those associated with sleep in young animals. To test this hypothesis, we compared protein profiles from the frontal cortex of young and old rats following timed bouts of spontaneous sleep. Proteins separated by two-dimensional electrophoresis were sequentially stained with ProQ Diamond and SYPRO Ruby to visualize phosphorylated and total protein, respectively. Spots showing SWS- and age- related were isolated, eluted, and identified by a combination of mass spectroscopy analyses
2. Results
2.1 Sleep and wake parameters across age
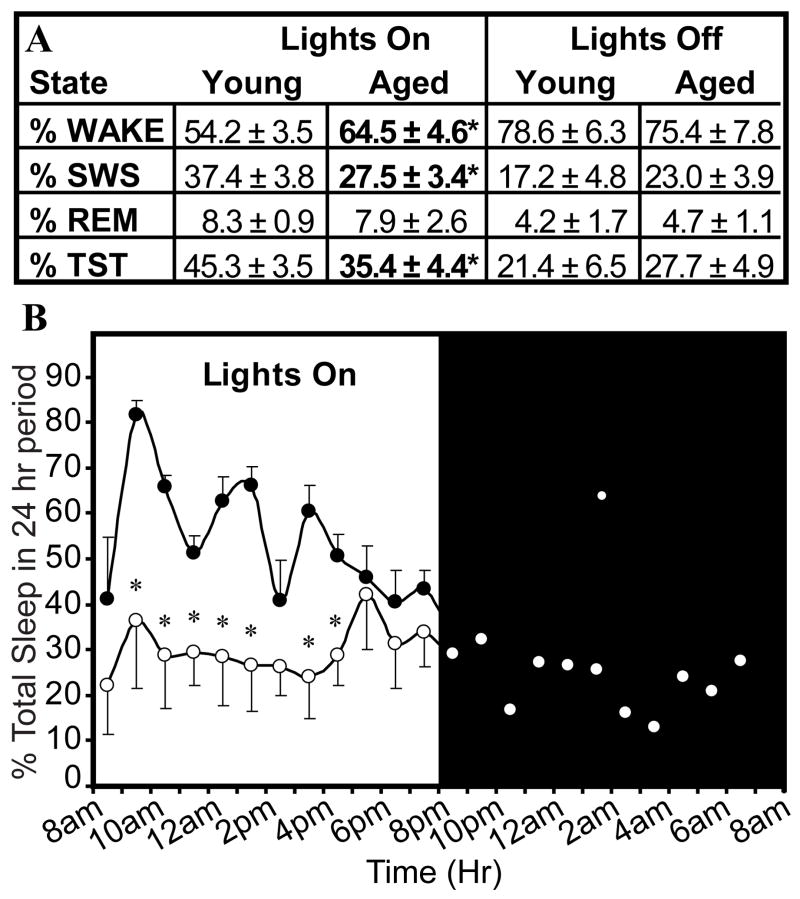
Old rats had decreased percentages of slow wave sleep (SWS; t= 3.87, df= 6, *p=0.004), total sleep time (%TST; t= 3.54, df= 6, *p=0.006) and an increased percentage of waking (W; t= −3.56, df= 6, *p=0.005) during the lights-on period only (Figure 1A, Figure 1B), consistent with previous results (Van Gool and Mirmiran, 1983).
Figure 1.
Analysis of spontaneous sleep-wake behavior in young and old rats over a 24 hour period. The amount of SWS and total sleep time (%TST) were decreased during the lights-on period in aged rats compared young rats (*p<0.05). No significant differences were observed between age groups during the lights-off period. The percentages of Wake, SWS, and REM sleep in young rats over 24 h were comparable to previous reports (Tobler and Borbely, 1986).
2.2 Protein expression is increased in aged rats
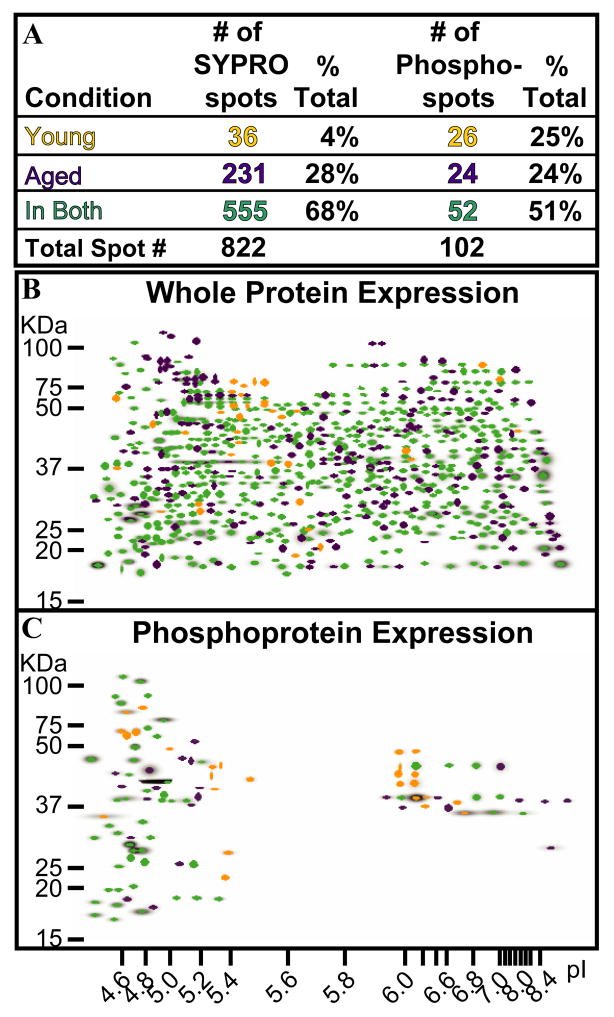
Master gel images from young and old animals were generated and analyzed for SYPRO Ruby (Figure 2B) and Pro-Q Diamond (Figure 2C) stains. A total of 822 protein spots (Figure 2A, left) were distinguished in the SYPRO Ruby Master image, 68% of which were present in both age groups. Only 4% of the spots (n=36) were unique to young animals. The number of SYPRO Ruby spots present only in aged animals, however, accounted for 28% (n=231) of the total number of spots visualized, a 7-fold increase over that of young rats. A total of 102 phosphorylated spots was visualized using Pro-Q Diamond (Figure 2A, right), with approximately 51% of the phosphorylated proteins present in both W and SWS states (n=52). The number of phosphorylated spots, however, did not vary between young (n=26) and old rats (n= 24; Figure 2A).
Figure 2.
Total and phosphoprotein analyses of young and old rats as a function of age. (A). Numerical analyses of SYPRO Ruby and ProQ Diamond profiles. (B). In the SYPRO Ruby Master image, spots are colored-coded to denote age-related protein expression. (C). ProQ Master image shows phosphoprotein spots expressed as a function of age. Green spots were present independent of age; purple spots; found only in old animals; gold spots; present only in young rats.
2.3 Protein expression is altered by behavioral state in young and aged rats
Table 1 (left panel) shows the analysis of SYPRO Ruby spots as a function of age and behavioral state. In young rats, ~3% were present in both W and SWS states (n=18). State-related expression also accounted for nearly 3% of the total visualized in young rats (W, ~1%; SWS, ~2%). In old animals, approximately 18% (n=142) were visualized in both behavioral states, while about 12% of the spots showed state-related expression. Of the state-related spots visualized in aged rats, ~4% spots (n=30) were W-related and ~8% spots (n=142) were unique to SWS. Thus, there was an increase in the number of SYPRO Ruby spots in aged rats that was independent of behavioral state. The Pro-Q Diamond staining showed that there was no change in the number of phosphorylated spots visualized in either age group (Figure 2A). However, a decrease in the number of phosphorylated spots during SWS was evident in old rats (Table 1, right panel). In young rats, phosphorylated spots accounted for ~27% of the total spots visualized, compared to only 12% in aged rats. The marked decrease in the number of phosphorylated proteins in aged rats during sleep (~56%) thus provides evidence that cellular activities associated with SWS are altered as a function of age.
Table 1.
Numerical analyses of SYPRO Ruby and ProQ Diamond protein expression as a function age and of behavioral state (s).
| A | SYPRO Ruby | Phosphoprotein | ||||||
|---|---|---|---|---|---|---|---|---|
| YOUNG % | AGED % | YOUNG % | AGED % | |||||
| State | (n) | Total | (n) | Total | (n) | Total | (n) | Total |
| Wake | 6 | 1% | 30 | 4% | 0 | – | 2 | 3% |
| SWS | 12 | 2% | 59 | 8% | 21 | 27% | 9 | 12% |
| In Both | 18 | 3% | 142 | 18% | 5 | 6% | 13 | 17% |
| Total # | 591 | 786 | 78 | 76 | ||||
2.4 Identification of SWS-associated spots in aged animals
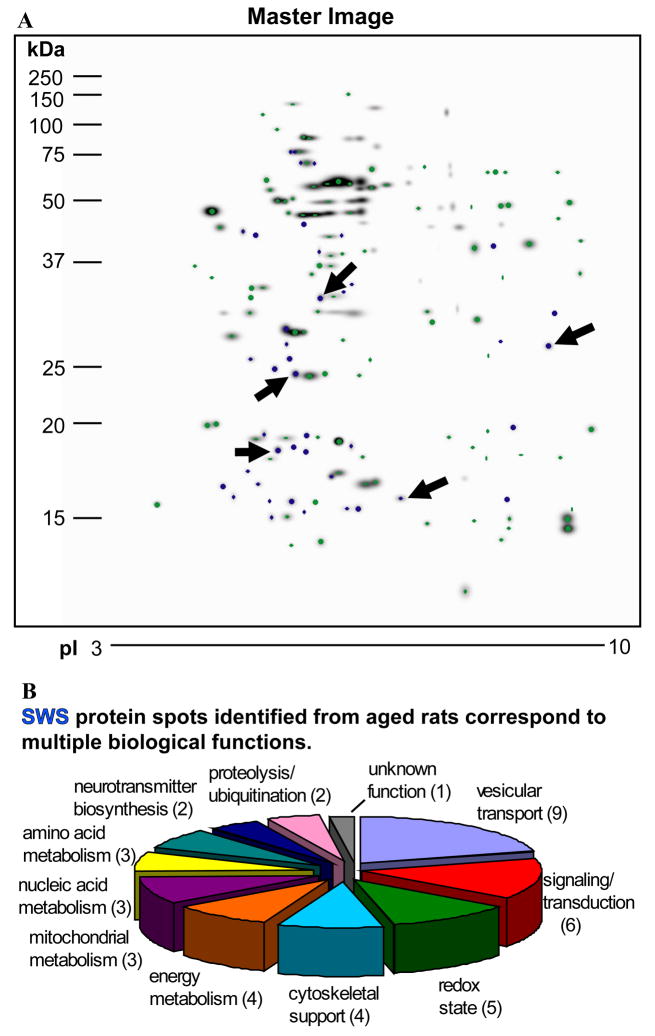
SWS-specific spots (n=5) from aged animals were excised from Gel Code Blue stained gels, eluted and identified by MS/MS analyses (Figure 3A). A total of 42 proteins were identified from these spots with high confidence from old rats sacrificed during SWS (Figure 3A, arrows). A pie chart shows the functional categories associated with the identified proteins (Figure 3B). Table 2 is a complete listing of all proteins identified from the MS analyses. Proteins identified included those associated with vesicular transport (n=9), cell signaling/transduction (n=6), oxidation state (n=5), cytoskeletal support (n=4), energy metabolism (n=4), mitochondrial (n=3), nucleic acid (n=3), and amino acid metabolism (n=3), neurotransmitter biosynthesis (n=2) and proteolysis (n=2). Vesicular transport proteins included ADP-ribosylation factor 1, ADP-ribosylation factor-like protein 8B, Rab11B, Rab 3A, Rab 2A, Rab18, tumor protein D52-like protein, alpha-synuclein, and vesicle-associated membrane protein (Becher et al., 1999; Cao et al., 2006; Haraguchi et al., 2006; Khvotchev et al., 2003; Malagon et al., 2005; Mattson et al., 2002; Sudhof, 2000; Suzuki et al., 2001). Proteins associated with signal transduction included phosphatidylethanolamine-binding protein 1, Rap1B, olfactory marker protein, visinin-like protein 1, translation initiation factor eIF-5A, and dimethylarginine dimethylaminohydrolase 2 (Enwere et al., 2004; George et al., 2006; Hasegawa et al., 2006; Poon et al., 2006; Ribeiro-Neto et al., 2004). Proteins associated with maintenance of the oxidation reduction state included glutathione S-transferase Yb-3, superoxide dismutase, peroxiredoxins 1 and 2, and hemoglobin subunit alpha-1/2. Four of the identified proteins are involved in cytoskeletal support (i.e. profilin-2, platelet-activating factor acetylhydrolase 1b [subunit beta], myelin basic protein and thy-1 membrane glycoprotein; (Mastronardi and Moscarello, 2005; Webster et al., 1999; Witke, 2004; Yan et al., 2003). Proteins associated with the generation of ATP were identified from both cytosolic (i.e. phosphoglycerate mutase 1, 6-phosphogluconolactonase, lactoylglutathione lyase; (Chesler, 2003; Delarue et al., 2007; Ikemoto and Ueda, 2003; Thornalley, 1990) and mitochondrial (i.e. prohibitin, NADH dehydrogenase Fe-S protein 3, S1 chain; (Kushnareva et al., 2002; Mihara and Omura, 1996; Vessal et al., 2006) compartments. Proteins linked to nucleic acid metabolism (i.e. guanylate kinase, histone H2A, histone H4; (Funke et al., 2005; Kim et al., 2001), amino acid metabolism (i.e. dihydropteridine reductase, phosphoserine phosphatase, histidine triad nucleotide-binding protein 1;(de Koning, 2006; Kitzerow and Henrich, 2001; Thony et al., 2000) and neurotransmitter biosynthesis (protein kinase C inhibitor protein 1, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase; (Ichimura et al., 1987; Toker et al., 1992) were also identified. Finally, proteins associated with degradation via proteolysis or ubiquitination mechanisms were also identified (i.e. proteasome 28 subunit, alpha; signalosome subunit 8; (Lykke-Andersen and Wei, 2003; Rechsteiner and Hill, 2005).
Figure 3.
Identification of protein spots in aged rats. (A). Large polyacrylamide gels were matched and analyzed according to age and behavioral state. Green spots indicate proteins common to both age (young and old) and behavioral state (W and SWS). Blue spots represent proteins expressed only in aged rats during SWS. Five of these spots (denoted by arrows) were chosen for MS analyses and identification. (B). SWS-related spots in aged animals were analyzed by mass spectroscopy. Proteins were associated with a variety of biological functions including vesicular transport, neurotransmitter synthesis, cytoskeletal support, energy metabolism, proteolysis, and the redox state of the cell. SWS-specific spots (shown in blue, panel A) identified in aged animals were associated with a variety of biological functions ranging from vesicular transport, neurotransmitter synthesis, cytoskeletal support, energy metabolism, proteolysis, and the redox state of the cell. Of note, when a protein had known multiple classes of functions, it was assigned to the category that was best known based on the literature.
Table 2.
Proteins identified from SWS- and age-unique selected 2DE gel spots.
Mass spectroscopy dentification of proteins from SWS- and age-unique selected 2DE gel spots.
| ID # | Accession|Entry name | Protein name | Theor MW (Da) | Theor pI |
|---|---|---|---|---|
| 1 | P84079|ARF1_RAT | ADP-ribosylation factor 1 | 20697 | 6.31 |
| 2 | Q66HA6|ARL8B_RAT | ADP-ribosylation factor-like protein 8B | 21539 | 8.67 |
| 3 | P37377|SYUA_RAT | Alpha-synuclein | 14515 | 4.74 |
| 4 | P27139|CAH2_RAT | Carbonic anhydrase 2 (EC 4.2.1.1) | 28983 | 6.88 |
| 5 | Q6P4Z9|CSN8_RAT | COP9 signalosome complex subunit 8 | 23236 | 5.09 |
| 6 | P11348|DHPR_RAT | Dihydropteridine reductase (EC 1.5.1.34) | 25552 | 7.67 |
| 7 | Q6MG60|Q6MG60_RAT | Dimethylarginine dimethylaminohydrolase 2 | 29688 | 5.66 |
| 8 | Q3T1J1|IF5A1_RAT | Eukaryotic translation initiation factor 5A-1 | 16832 | 5.07 |
| 9 | P08009|GSTM4_RAT | Glutathione S-transferase Yb-3 (EC 2.5.1.18) | 25681 | 6.84 |
| 10 | Q71RR7|Q71RR7_RAT | Guanylate kinase (EC 2.7.4.8) | 21910 | 5.44 |
| 11 | P01946|HBA_RAT | Hemoglobin subunit alpha-1/2 | 15329 | 7.81 |
| 12 | P62959|HINT1_RAT | Histidine triad nucleotide-binding protein 1 | 13779 | 6.64 |
| 13 | P02262|H2A1_RAT | Histone H2A | 14077 | 10.90 |
| 14 | P62804|H4_RAT | Histone H4 | 11367 | 11.36 |
| 15 | Q6P7Q4|LGUL_RAT | Lactoylglutathione lyase (EC 4.4.1.5) | 20820 | 5.12 |
| 16 | P02688|MBP_RAT | Myelin basic protein S | 21502 | 11.24 |
| 17 | Q9DCT2|NDUS3_MOUSE1 | NADH dehydrogenase iron-sulfur protein 3, mitochondrial (EC 1.6.5.3) | 30149 | 6.67 |
| 18 | P08523|OMP_RAT | Olfactory marker protein | 18853 | 5.12 |
| 19 | Q63716|PRDX1_RAT | Peroxiredoxin-1 (EC 1.11.1.15) | 22109 | 8.27 |
| 20 | P35704|PRDX2_RAT | Peroxiredoxin-2 (EC 1.11.1.15) | 21784 | 5.34 |
| 21 | P31044|PEBP1_RAT | Phosphatidylethanolamine-binding protein 1 | 20801 | 5.47 |
| 22 | P25113|PGAM1_RAT | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) | 28832 | 6.67 |
| 23 | Q5M819|SERB_RAT | Phosphoserine phosphatase (EC 3.1.3.3) | 24967 | 5.49 |
| 24 | O35264|PA1B2_RAT | Platelet-activating factor acetylhydrolase 1B, subunit beta (EC 3.1.1.47) | 25581 | 5.57 |
| 25 | Q9EPC6|PROF2_RAT | Profilin-2 | 15002 | 6.55 |
| 26 | P67779|PHB_RAT | Prohibitin | 29820 | 5.57 |
| 27 | Q6P9V7|Q6P9V7_RAT | Proteasome (Prosome, macropain) 28 subunit, alpha | 28635 | 5.63 |
| 28 | O35509|RB11B_RAT | Ras-related protein Rab-11B | 24488 | 5.64 |
| 29 | Q5EB77|RAB18_RAT | Ras-related protein Rab-18 | 22976 | 5.11 |
| 30 | P05712|RAB2A_RAT | Ras-related protein Rab-2A | 23535 | 6.08 |
| 31 | P63012|RAB3A_RAT | Ras-related protein Rab-3A | 24970 | 4.85 |
| 32 | Q62636|RAB1B_RAT | Ras-related protein Rap-1B [precursor] | 20798 | 5.65 |
| 33 | P07632|SODC_RAT | Superoxide dismutase [Mn] (EC 1.15.1.1) | 15912 | 5.88 |
| 34 | P01830|THY1_RAT | Thy-1 membrane glycoprotein [Precursor] | 18172 | 9.42 |
| 35 | Q6PCT3|TPD54_RAT | Tumor protein D54 | 23992 | 5.80 |
| 36 | Q498R7|CA123_RAT | Uncharacterized protein C1orf123 homolog | 18095 | 5.06 |
| 37 | P63045|VAMP2_RAT | Vesicle-associated membrane protein 2 | 12691 | 7.84 |
| 38 | P62762|VISL1_RAT | Visinin-like protein 1 | 22142 | 5.01 |
| 39 | P35213|1433B_RAT | 14-3-3 protein beta/alpha | 28054 | 4.81 |
| 40 | P68255|1433T_RAT | 14-3-3 protein theta | 27778 | 4.69 |
| 41 | P63102|1433Z_RAT | 14-3-3 protein zeta/delta | 27771 | 4.73 |
| 42 | Q9CQ60|6PGL_MOUSE1 | 6-phosphogluconolactonase (EC 3.1.1.31) | 27254 | 5.55 |
Legend
Search engine reported mouse species.
Blast analysis (see methods) was performed on confidently identified peptide sequences to identify equivalent rodent species.
3. Discussion
Aging is associated with sleep disruptions that diminish the quality of life in the elderly (Phillips and Ancoli-Israel, 2001). Decreased frontal cortex activity during sleep has been correlated with age-associated brain atrophy, synaptic degeneration, neurochemical alterations, and reduced blood flow (Cabeza et al., 2002). We examined putative links between aging and sleep at the protein level. While changes in regional protein expression occurred within minutes of spontaneous sleep-wake bouts in both young and aged animals, our data shows that the cellular response to sleep is dramatically altered by age. The marked increase in the number of SYPRO Ruby spots in old compared to young animals, independent of sleep-wake behavior is consistent with the progressive accumulation of oxidized proteins that accompany aging in healthy as well as those with neurodegenerative diseases (Kanski et al., 2003; Poon et al., 2006). The age-related decrease in phosphoprotein expression during SWS is consistent with a decrease in cellular activities and the dysregulation of cellular activity that occurs following sleep deprivation (Basheer et al., 2005). Further, the SWS-related proteins identified in old rats (Table 2) are associated with the same functional categories as those identified in young animals across spontaneous sleep-wake by both mRNA (Cirelli et al., 2004) and protein analyses (Vazquez et al., 2008). One such protein, superoxide dismutase, is critical in protecting against ROS (Calabrese et al., 2006) and has also been implicated in the disruption of sleep regulation in young animals (Ramanathan et al., 2002) and humans (Christou et al., 2003). In addition, members of the Rab family, profilin, and ADP-ribosylation factors are essential for the maintenance of synaptic plasticity (Witke, 2004). Further, alpha-synuclein, is also critical for synaptic integrity (Hashimoto et al., 2004). Alpha-synuclein abnormalities results in Lewy body inclusions observed in Parkinson’s disease (PD), and have also been observed in patients with REM sleep behavior disorder (RBD; (Turner et al., 2000). Our results are thus consistent with an age-related dysfunctions in antioxidant potential (Sivonova et al., 2007), in macromolecular assemblies required for nucleic acid and protein metabolism (Piwien-Pilipuk et al., 2002), neurotransmitter synthesis (Cruz-Muros et al., 2007) and protein degradation (Dahlmann, 2007). While further studies are needed to provide more detailed insights concerning the role of aging on sleep, our results reveal important clues to understanding both age-associated alterations in sleep architecture and the cellular underpinnings of sleep.
4. Experimental Procedure
4.1. Experimental design and analysis of behavioral state
All animal procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, 7th ed (Council, 1996). Young (~3 mo.; n=9) and aged (~22 mo.; n=8) male Sprague-Dawley rats (250–500g) were implanted with electroencephalographic (EEG) and electomyographic (EMG) to monitor brain and muscle activities, respectively. A catheter was also inserted into the femoral vein to permit sacrifice without contacting the animals. All rats were individually housed (12:12; lights-on, 8 am) and habituated to the recording apparatus with free access to food and water. Spontaneous sleep-wake activity was recorded for 48 h prior to beginning the experimental protocol. Wake (W), slow wave sleep (SWS), and rapid eye movement (REM) sleep states were captured digitally, stored to disk and scored manually. W was defined by the presence of low amplitude, high frequency EEG waves with high EMG activity. SWS was identified by high amplitude slow waves in conjunction with a lower level of EMG activity compared to waking. REM sleep was characterized by the presences of rhythmic theta EEG waves and little or no EMG activity. Sleep-wake states were scored manually in 10 sec epochs. The percents of W, SWS, REM and total sleep time (%TST; the sum of SWS plus REM sleep percentages) were determined. Rats were sacrificed following timed bouts of spontaneous SWS and W as previously described (Greco et al., 1999; Vazquez et al., 2008). Briefly, rats were sacrificed after 10 minutes of continuous waking or following 10 minutes of continuous SWS between 3–4 pm. This time frame is during the lights-on period, the rats’ normal sleep time and corresponds to the latter third of the lights-on phase. All rats were euthanized with an overdose of pentobarbital (200 mg/kg, i.v.) injected into a femoral vein catheter.
4.2. Protein sample lysis, separation and staining
Bilateral frontal cortex tissue (5.2 mm - 4.2 mm anterior to Bregma) (Paxinos and Watson, 1982) was homogenized in buffer containing 7M urea, 2M thiourea, 2% CHAPS, 2% ASB-14, 20 mM Tris, 1% DTT (wt/vol), 0.2% 3/10 ampholytes (vol/vol), and 1% protease inhibitors as described (Vazquez et al., 2008). Briefly, protein lysates (100ug/gel) were first separated according to net charge on IPG strips (pH 3–10 NL; 11 cm). In the second dimension, proteins were separated by molecular mass using Tris-HCL gels (11cm) with a polyacrylamide gradient of 4–15%. Each gel was sequentially stained, first with Pro-Q Diamond (Invitrogen, Carlsbad, CA), followed by SYPRO Ruby (Invitrogen) according to the respective manufacturer’s instructions. Pro-Q Diamond, a fluorescent stain that binds to phosphorylated proteins, was used as a marker of regional cellular activity (Graves and Haystead, 2002; Hunter, 1995; Krebs, 1994; Sun and Tonks, 1994). SYPRO Ruby is a fluorescent stain used to visualize total protein expression patterns. Each lysate was separated and analyzed in duplicate. The use of commercially available pI strips, polyacrylamide gels, high through-put power supplies/equipment that permit the simultaneous separation of up to 12 samples, and sensitive fluorescent stains results in improved between-gel reproducibility for spot detection and resolution (Boonjakuakul et al., 2007; Zhan and Desiderio, 2003a; Zhan and Desiderio, 2003b). To obtain enough protein to ensure accurate MS identification of spots and to verify the results from the combined analyses of individual samples (above), a second series of 2DE separations were performed on lysates (1 mg) that were pooled according to behavioral state prior to separation (1st dimension, pH 3–10 NL strips, 17 cm; 2cd dimension, 10% polyacrylamide Tris-HCl gels). These gels were stained with GelCode Blue® according to the manufacturer’s instructions (Pierce Biotechnology, Rockford, IL). All stained gels were digitally captured using a high resolution scanner (Molecular Imager FX, BioRad). Thus, for these analyses, two complimentary approaches were used to determine age- and state- related expression. In the first analyses, state-related expression from individual profiles were separated on “midi” analytical gels and visualized with a combination of fluorescent stains. In the second set of analyses, individual samples were pooled, separated by 2DE using a giant gel format (17 × 17 cm), and stained with GelCode Blue. Spots that were unique-SWS in old rats were identified by subtractive analysis of the Master gel image generated from the comparison of these gels. Agreement between images generated from individual and pooled sample sets (see below) with regard to isoelectric point, mass, and state-related expression was thus a prerequisite for spot picking and subsequent MS analyses.
4.3. Image analyses
Protein spots were analyzed using PDQuest 2-D Image Analysis Software (BioRad) and have been described in detail (Vazquez et al., 2008). Duplicate samples from each animal (i.e., a total of 34 gels from 17 rats) were analyzed to reduce spot detection errors. Spots were identified as the sum of the intensities of the image pixels within a boundary, where spot height or peak value on an x and y axis were measured (ODs or counts/image units2) and fitted to the scanned spots using Gaussian curves. Gaussian modeling facilitates the identification of overlapping spots, spots in gel streaks, and multiple spots in dense clusters. Errors detected by the automated matching of spots were manually inspected. Spot intensities of equal to or less than 4-fold were considered artifacts and/or background staining. Data was analyzed first as a function of age, independent of behavioral state. The second analysis examined protein expression as a function of age and behavioral state at sacrifice. In each analysis, samples were grouped according to age and/or behavioral state, spot matched, and normalized to one another to create a gel composite. Gel composites were used to generate a Master image for each stain and experimental condition. Averaging parameters (criteria to assist with group comparisons) across experimental groups were set to equal to or greater than 70% consistent with instrument specifications (Zhan and Desiderio, 2003a; Zhan and Desiderio, 2003b). Master gel images for each stain were subsequently generated by combining all spots identified in all gels across all experimental groups. Spots from each behavioral state were compared and matched to the appropriate Master image (i.e., either ProQ Diamond or SYPRO Ruby). Qualitative analysis sets were generated by subtractive (Boolean) analyses to identify spots: 1) common to all conditions as a function of age and behavioral state; 2) present only as a function of age; and 3) unique to either waking or sleep state as a function of age.
4.4. Mass spectrometry and protein identification
Spots selected for protein identification were excised from GelCode Blue® stained gels followed by in-gel tryptic digestion and peptide extraction as previously published (Boonjakuakul et al., 2007). Protein identifications were established by HPLC/ESI/MS/MS analysis of the resulting peptide extracts. Liquid chromatographic separations were performed on an Ultimate Capillary HPLC System (Dionex/LC Packings, Sunnyvale, CA) equipped with a PepMap (Dionex/LC Packings) trap column and a reversed phase C18 nano-column (75 m i.d. × 150 mm packed in-house with Jupiter Proteo C12 end-capped resin, 90 Å pore size, 4 m particle size) and a Famos Micro autosampler. An aliquot of peptide extract (3–4 μL) was loaded onto the trap column with loading solvent (0.1% formic acid) at a flow rate of 20 μL/min. The trap column was washed with the loading solvent for 3 minutes prior to switching it in line with the reversed phase nano-column. The nano-column and elution buffers were maintained at ambient temperature and the mobile phase flow rate was 325 nL/min. The nano-column was equilibrated with 2% B for 20 min prior to sample injection (Solvent A: 2% acetonitrile/0.1% formic acid; Solvent B: 80% acetonitrile/0.08% formic acid). Peptide separation was accomplished using a binary gradient which consisted of a 5 min isocratic wash at 2% B followed by a linear gradient of 2%–50% B over 45 min, and concluded with a column cleanup step of 95% B for 7 min. The column effluent flowed directly into a nanoelectrospray ion source (Protana) on a QSTAR XL quadrupole/quadrupole/time-of-flight (QqTOF) mass spectrometer (Applied Biosystems/MDS SCIEX). Protein identification was accomplished by isolating sequentially eluting peptide populations with a single mass-to-charge ratio (m/z), within the mass spectrometer, fragmenting this population, and measuring the masses of the peptide fragment ions.
Mascot™ (version 2.2.04, Matrix Sciences) software was used for peak detection, creating mass peak lists and performing database searches. The experimentally determined peptide fragment ion masses were used to search a theoretical fragment ion mass database generated by in silico digestion and fragmentation of all Rodentia proteins in the SwissProt database (created 04-23-2009, 462764 sequences, 163773385 residues). Search parameters were employed with trypsin as the enzyme specificity (maximum number of missed cleavages set at 2), carbamidomethylation as a fixed modification of cysteine residues and variable modifications of deamidated Asn and Gln; pyro-Glu; and oxidized Met. The peptide and fragment ion mass tolerances were ± 150 ppm and ± 0.15 Da, respectively. Mascot peptide acceptance criteria required individual ion scores to be > 33 which indicated protein identity or extensive homology at a p value < 0.05 (95% confidence level). The MS/MS spectra for all protein identifications based on ≤ 4 peptide identifications were manually verified. The Mascot search engine automatically performed a decoy database search to determine the false discovery rate (FDR).
Supplementary Material
Acknowledgments
We thank J. Joseph, P. Chang, and K. Shew for expert technical assistance. This study was made possible by support from HL069706, NS045791, The Center for Research on Independent Aging (CRIA), the Center for Advanced Drug Research (CADRE), NIH/NCI P30 CA82103, the Sandler Family Foundation, and the Gordon and Betty Moore Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avidan AY. Sleep disorders in the older patient. Prim Care. 2005;32:563–86. doi: 10.1016/j.pop.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Basheer R, Brown R, Ramesh V, Begum S, McCarley RW. Sleep deprivation-induced protein changes in basal forebrain: Implications for synaptic plasticity. J Neurosci Res. 2005;82:650–8. doi: 10.1002/jnr.20675. [DOI] [PubMed] [Google Scholar]
- Becher A, Drenckhahn A, Pahner I, Margittai M, Jahn R, Ahnert-Hilger G. The synaptophysin-synaptobrevin complex: a hallmark of synaptic vesicle maturation. J Neurosci. 1999;19:1922–31. doi: 10.1523/JNEUROSCI.19-06-01922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliwise DL. Sleep in normal aging and dementia. Sleep. 1993;16:40–81. doi: 10.1093/sleep/16.1.40. [DOI] [PubMed] [Google Scholar]
- Bobillier P, Sakai F, Seguin S, Jouvet M. Deprivation of paradoxical sleep and in vitro cerebral protein synthesis in the rat. Life Sci. 1971;10:1349–1357. doi: 10.1016/0024-3205(71)90186-x. [DOI] [PubMed] [Google Scholar]
- Bonnet MH, Rosa RR. Sleep and performance in young adults and older normals and insomniacs during acute sleep loss and recovery. Biol Psychol. 1987;25:153–72. doi: 10.1016/0301-0511(87)90035-4. [DOI] [PubMed] [Google Scholar]
- Boonjakuakul JK, Gerns HL, Chen YT, Hicks LD, Minnick MF, Dixon SE, Hall SC, Koehler JE. Proteomic and Immunoblot Analyses of Bartonella quintana Total Membrane Proteins Identify Antigens Recognized by Sera from Infected Patients. Infect Immun. 2007;75:2548–61. doi: 10.1128/IAI.01974-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Howard B, Yatin S, Koppal T, Drake J, Hensley K, Aksenov M, Aksenova M, Subramaniam R, Varadarajan S, Harris-White ME, Pedigo NW, Jr, Carney JM. Elevated oxidative stress in models of normal brain aging and Alzheimer’s disease. Life Sci. 1999;65:1883–92. doi: 10.1016/s0024-3205(99)00442-7. [DOI] [PubMed] [Google Scholar]
- Cabeza R, Anderson ND, Locantore JK, McIntosh AR. Aging gracefully: compensatory brain activity in high-performing older adults. Neuroimage. 2002;17:1394–402. doi: 10.1006/nimg.2002.1280. [DOI] [PubMed] [Google Scholar]
- Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- Calabrese V, Giuffrida Stella AM, Calvani M, Butterfield DA. Acetylcarnitine and cellular stress response: roles in nutritional redox homeostasis and regulation of longevity genes. J Nutr Biochem. 2006;17:73–88. doi: 10.1016/j.jnutbio.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Cao Q, Chen J, Zhu L, Liu Y, Zhou Z, Sha J, Wang S, Li J. A testis-specific and testis developmentally regulated tumor protein D52 (TPD52)-like protein TPD52L3/hD55 interacts with TPD52 family proteins. Biochem Biophys Res Commun. 2006;344:798–806. doi: 10.1016/j.bbrc.2006.03.208. [DOI] [PubMed] [Google Scholar]
- Carskadon MA, Dement WC. Sleep loss in elderly volunteers. Sleep. 1985;8:207–221. doi: 10.1093/sleep/8.3.207. [DOI] [PubMed] [Google Scholar]
- Chesler M. Regulation and modulation of pH in the brain. Physiol Rev. 2003;83:1183–221. doi: 10.1152/physrev.00010.2003. [DOI] [PubMed] [Google Scholar]
- Christou K, Markoulis N, Moulas AN, Pastaka C, Gourgoulianis KI. Reactive oxygen metabolites (ROMs) as an index of oxidative stress in obstructive sleep apnea patients. Sleep Breath. 2003;7:105–10. doi: 10.1007/s11325-003-0105-9. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C. Sleep disruption, oxidative stress, and aging: new insights from fruit flies. Proc Natl Acad Sci U S A. 2006;103:13901–2. doi: 10.1073/pnas.0606652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council NR. Guide for the Care and Use of Laboratory Animals. National Academy Press; Washington, D.C.: 1996. [Google Scholar]
- Cruz-Muros I, Afonso-Oramas D, Abreu P, Perez-Delgado MM, Rodriguez M, Gonzalez-Hernandez T. Aging effects on the dopamine transporter expression and compensatory mechanisms. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Cunha RA, Constantino MC, Sebastiao AM, Ribeiro JA. Modification of A1 and A2a adenosine receptor binding in aged striatum, hippocampus and cortex of the rat. Neuroreport. 1995;6:1583–8. doi: 10.1097/00001756-199507310-00029. [DOI] [PubMed] [Google Scholar]
- Dahlmann B. Role of proteasomes in disease. BMC Biochem. 2007;8 Suppl 1:S3. doi: 10.1186/1471-2091-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning TJ. Treatment with amino acids in serine deficiency disorders. J Inherit Metab Dis. 2006;29:347–51. doi: 10.1007/s10545-006-0269-0. [DOI] [PubMed] [Google Scholar]
- Delarue M, Duclert-Savatier N, Miclet E, Haouz A, Giganti D, Ouazzani J, Lopez P, Nilges M, Stoven V. Three dimensional structure and implications for the catalytic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei. J Mol Biol. 2007;366:868–81. doi: 10.1016/j.jmb.2006.11.063. [DOI] [PubMed] [Google Scholar]
- Drucker-Colin R, Zamora J, Bernal-Pedraza J, Sosa B. Modification of REM sleep and associated phasic activities by protein synthesis inhibitors. Exp Neurol. 1979;63:458–67. doi: 10.1016/0014-4886(79)90164-x. [DOI] [PubMed] [Google Scholar]
- Enwere E, Shingo T, Gregg C, Fujikawa H, Ohta S, Weiss S. Aging results in reduced epidermal growth factor receptor signaling, diminished olfactory neurogenesis, and deficits in fine olfactory discrimination. J Neurosci. 2004;24:8354–65. doi: 10.1523/JNEUROSCI.2751-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd RA. Antioxidants, oxidative stress, and degenerative neurological disorders. Proc Soc Exp Biol Med. 1999;222:236–45. doi: 10.1046/j.1525-1373.1999.d01-140.x. [DOI] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–45. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- Genova ML, Pich MM, Bernacchia A, Bianchi C, Biondi A, Bovina C, Falasca AI, Formiggini G, Castelli GP, Lenaz G. The mitochondrial production of reactive oxygen species in relation to aging and pathology. Ann N Y Acad Sci. 2004;1011:86–100. doi: 10.1007/978-3-662-41088-2_10. [DOI] [PubMed] [Google Scholar]
- George AJ, Holsinger RM, McLean CA, Tan SS, Scott HS, Cardamone T, Cappai R, Masters CL, Li QX. Decreased phosphatidylethanolamine binding protein expression correlates with Abeta accumulation in the Tg2576 mouse model of Alzheimer’s disease. Neurobiol Aging. 2006;27:614–23. doi: 10.1016/j.neurobiolaging.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Graves PR, Haystead TA. Molecular biologist’s guide to proteomics. Microbiol Mol Biol Rev. 2002;66:39–63. doi: 10.1128/MMBR.66.1.39-63.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco MA, McCarley RW, Shiromani PJ. Choline acetyltransferase expression during periods of behavioral activity and across natural sleep-wake states in the basal forebrain. Neuroscience. 1999;93:1369–74. doi: 10.1016/s0306-4522(99)00201-8. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Yanaka N, Nogusa Y, Sumiyoshi N, Eguchi Y, Kato N. Expression of ADP-ribosylation factor-like protein 8B mRNA in the brain is down-regulated in mice fed a high-fat diet. Biosci Biotechnol Biochem. 2006;70:1798–802. doi: 10.1271/bbb.60168. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Wakino S, Tanaka T, Kimoto M, Tatematsu S, Kanda T, Yoshioka K, Homma K, Sugano N, Kurabayashi M, Saruta T, Hayashi K. Dimethylarginine dimethylaminohydrolase 2 increases vascular endothelial growth factor expression through Sp1 transcription factor in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:1488–94. doi: 10.1161/01.ATV.0000219615.88323.b4. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Kawahara K, Bar-On P, Rockenstein E, Crews L, Masliah E. The Role of alpha-synuclein assembly and metabolism in the pathogenesis of Lewy body disease. J Mol Neurosci. 2004;24:343–52. doi: 10.1385/JMN:24:3:343. [DOI] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–36. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Ichimura T, Isobe T, Okuyama T, Yamauchi T, Fujisawa H. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. [DOI] [PubMed] [Google Scholar]
- Ikemoto A, Ueda T. Identification of a nerve ending-enriched 29-kDa protein, labeled with [3-32P]1,3-bisphosphoglycerate, as monophosphoglycerate mutase: inhibition by fructose-2,6-bisphosphate via enhancement of dephosphorylation. J Neurochem. 2003;85:1382–93. doi: 10.1046/j.1471-4159.2003.01777.x. [DOI] [PubMed] [Google Scholar]
- Joseph JA, Shukitt-Hale B, Casadesus G, Fisher D. Oxidative stress and inflammation in brain aging: nutritional considerations. Neurochem Res. 2005;30:927–35. doi: 10.1007/s11064-005-6967-4. [DOI] [PubMed] [Google Scholar]
- Kanski J, Alterman MA, Schoneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic Biol Med. 2003;35:1229–39. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- Khvotchev MV, Ren M, Takamori S, Jahn R, Sudhof TC. Divergent functions of neuronal Rab11b in Ca2+-regulated versus constitutive exocytosis. J Neurosci. 2003;23:10531–9. doi: 10.1523/JNEUROSCI.23-33-10531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choy HE, Nam KH, Park SC. Transglutaminase-mediated crosslinking of specific core histone subunits and cellular senescence. Ann N Y Acad Sci. 2001;928:65–70. doi: 10.1111/j.1749-6632.2001.tb05636.x. [DOI] [PubMed] [Google Scholar]
- Kitzerow A, Henrich B. The cytosolic HinT protein of Mycoplasma hominis interacts with two membrane proteins. Mol Microbiol. 2001;41:279–87. doi: 10.1046/j.1365-2958.2001.02524.x. [DOI] [PubMed] [Google Scholar]
- Krebs EG. The growth of research on protein phosphorylation. Trends Biochem Sci. 1994;19:439. doi: 10.1016/0968-0004(94)90125-2. [DOI] [PubMed] [Google Scholar]
- Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. Saunders; Philadelphia: 2000. [Google Scholar]
- Kushnareva Y, Murphy AN, Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem J. 2002;368:545–53. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Wei N. Gene structure and embryonic expression of mouse COP9 signalosome subunit 8 (Csn8) Gene. 2003;321:65–72. doi: 10.1016/s0378-1119(03)00836-9. [DOI] [PubMed] [Google Scholar]
- Malagon MM, Cruz D, Vazquez-Martinez R, Peinado JR, Anouar Y, Tonon MC, Vaudry H, Gracia-Navarro F, Castano JP. Analysis of Rab18 and a new golgin in the secretory pathway. Ann N Y Acad Sci. 2005;1040:137–9. doi: 10.1196/annals.1327.017. [DOI] [PubMed] [Google Scholar]
- Mastronardi FG, Moscarello MA. Molecules affecting myelin stability: a novel hypothesis regarding the pathogenesis of multiple sclerosis. J Neurosci Res. 2005;80:301–8. doi: 10.1002/jnr.20420. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Duan W, Chan SL, Cheng A, Haughey N, Gary DS, Guo Z, Lee J, Furukawa K. Neuroprotective and neurorestorative signal transduction mechanisms in brain aging: modification by genes, diet and behavior. Neurobiol Aging. 2002;23:695–705. doi: 10.1016/s0197-4580(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Mihara K, Omura T. Cytoplasmic chaperones in precursor targeting to mitochondria: the role of MSF and hsp 70. Trends Cell Biol. 1996;6:104–8. doi: 10.1016/0962-8924(96)81000-2. [DOI] [PubMed] [Google Scholar]
- Montgomery P, Dennis J. A systematic review of non-pharmacological therapies for sleep problems in later life. Sleep Med Rev. 2004;8:47–62. doi: 10.1016/S1087-0792(03)00026-1. [DOI] [PubMed] [Google Scholar]
- Nystrom T. Aging in bacteria. Curr Opin Microbiol. 2002;5:596–601. doi: 10.1016/s1369-5274(02)00367-3. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–13. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Sterotaxic Coordinates. Academic Press; Sydney, Australia: 1982. [Google Scholar]
- Phillips B, Ancoli-Israel S. Sleep disorders in the elderly. Sleep Med. 2001;2:99–114. doi: 10.1016/s1389-9457(00)00083-6. [DOI] [PubMed] [Google Scholar]
- Piwien-Pilipuk G, Ayala A, Machado A, Galigniana MD. Impairment of mineralocorticoid receptor (MR)-dependent biological response by oxidative stress and aging: correlation with post-translational modification of MR and decreased ADP-ribosylatable level of elongating factor 2 in kidney cells. J Biol Chem. 2002;277:11896–903. doi: 10.1074/jbc.M109530200. [DOI] [PubMed] [Google Scholar]
- Poon HF, Calabrese V, Scapagnini G, Butterfield DA. Free radicals and brain aging. Clin Geriatr Med. 2004a;20:329–59. doi: 10.1016/j.cger.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Poon HF, Castegna A, Farr SA, Thongboonkerd V, Lynn BC, Banks WA, Morley JE, Klein JB, Butterfield DA. Quantitative proteomics analysis of specific protein expression and oxidative modification in aged senescence-accelerated-prone 8 mice brain. Neuroscience. 2004b;126:915–26. doi: 10.1016/j.neuroscience.2004.04.046. [DOI] [PubMed] [Google Scholar]
- Poon HF, Vaishnav RA, Getchell TV, Getchell ML, Butterfield DA. Quantitative proteomics analysis of differential protein expression and oxidative modification of specific proteins in the brains of old mice. Neurobiol Aging. 2006;27:1010–1019. doi: 10.1016/j.neurobiolaging.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Prinz PN. Sleep and sleep disorders in older adults. J Clin Neurophysiol. 1995;12:139–46. doi: 10.1097/00004691-199503000-00004. [DOI] [PubMed] [Google Scholar]
- Ramanathan L, Gulyani S, Nienhuis R, Siegel JM. Sleep deprivation decreases superoxide dismutase activity in rat hippocampus and brainstem. Neuroreport. 2002;13:1387–90. doi: 10.1097/00001756-200208070-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48:749–53. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Ribeiro-Neto F, Leon A, Urbani-Brocard J, Lou L, Nyska A, Altschuler DL. cAMP-dependent oncogenic action of Rap1b in the thyroid gland. J Biol Chem. 2004;279:46868–75. doi: 10.1074/jbc.M406858200. [DOI] [PubMed] [Google Scholar]
- Shiromani PJ, Lu J, Wagner D, Thakkar J, Greco MA, Basheer R, Thakkar M. Compensatory sleep response to 12 h wakefulness in young and old rats. Am J Physiol. 2000;278:R125–33. doi: 10.1152/ajpregu.2000.278.1.R125. [DOI] [PubMed] [Google Scholar]
- Sivonova M, Tatarkova Z, Durackova Z, Dobrota D, Lehotsky J, Matakova T, Kaplan P. Relationship between antioxidant potential and oxidative damage to lipids, proteins and DNA in aged rats. Physiol Res. 2007;56:757–64. doi: 10.33549/physiolres.931094. [DOI] [PubMed] [Google Scholar]
- Sohal RS. Role of oxidative stress and protein oxidation in the aging process. Free Radic Biol Med. 2002;33:37–44. doi: 10.1016/s0891-5849(02)00856-0. [DOI] [PubMed] [Google Scholar]
- Sperlagh B, Zsilla G, Baranyi M, Kekes-Szabo A, Vizi ES. Age-dependent changes of presynaptic neuromodulation via A1-adenosine receptors in rat hippocampal slices. Int J Dev Neurosci. 1997;15:739–47. doi: 10.1016/s0736-5748(97)00028-2. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences. Free Radic Biol Med. 1990;9:315–25. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–4. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle revisited. Neuron. 2000;28:317–20. doi: 10.1016/s0896-6273(00)00109-4. [DOI] [PubMed] [Google Scholar]
- Sun H, Tonks NK. The coordinated action of protein tyrosine phosphatases and kinases in cell signaling. Trends Biochem Sci. 1994;19:480–5. doi: 10.1016/0968-0004(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Suzuki I, Owada Y, Suzuki R, Yoshimoto T, Kondo H. Localization of mRNAs for six ARFs (ADP-ribosylation factors) in the brain of developing and adult rats and changes in the expression in the hypoglossal nucleus after its axotomy. Brain Res Mol Brain Res. 2001;88:124–34. doi: 10.1016/s0169-328x(01)00036-5. [DOI] [PubMed] [Google Scholar]
- Terao A, Wisor JP, Peyron C, Apte-Deshpande A, Wurts SW, Edgar DM, Kilduff TS. Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip(R) study. Neuroscience. 2006;137:593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thony B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J. 2000;347(Pt 1):1–16. [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I, Borbely AA. Sleep EEG in the rat as a function of prior waking. Electroencephalogr Clin Neurophysiol. 1986;64:74–6. doi: 10.1016/0013-4694(86)90044-1. [DOI] [PubMed] [Google Scholar]
- Toker A, Sellers LA, Amess B, Patel Y, Harris A, Aitken A. Multiple isoforms of a protein kinase C inhibitor (KCIP-1/14-3-3) from sheep brain. Amino acid sequence of phosphorylated forms. Eur J Biochem. 1992;206:453–61. doi: 10.1111/j.1432-1033.1992.tb16946.x. [DOI] [PubMed] [Google Scholar]
- Turner RS, D’Amato CJ, Chervin RD, Blaivas M. The pathology of REM sleep behavior disorder with comorbid Lewy body dementia. Neurology. 2000;55:1730–2. doi: 10.1212/wnl.55.11.1730. [DOI] [PubMed] [Google Scholar]
- Van Gool WA, Mirmiran M. Age-related changes in the sleep pattern of male adult rats. Brain Res. 1983;279:394–8. doi: 10.1016/0006-8993(83)90217-2. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Hall SC, Witkowska HE, Greco MA. Rapid alterations in cortical protein profiles underlie spontaneous sleep and wake bouts. Journal of Cellular Biochemistry. 2008;105:1472–1484. doi: 10.1002/jcb.21970. [DOI] [PubMed] [Google Scholar]
- Vessal M, Mishra S, Moulik S, Murphy LJ. Prohibitin attenuates insulin-stimulated glucose and fatty acid oxidation in adipose tissue by inhibition of pyruvate carboxylase. Febs J. 2006;273:568–76. doi: 10.1111/j.1742-4658.2005.05090.x. [DOI] [PubMed] [Google Scholar]
- Webster MJ, Vawter MP, Freed WJ. Immunohistochemical localization of the cell adhesion molecules Thy-1 and L1 in the human prefrontal cortex patients with schizophrenia, bipolar disorder, and depression. Mol Psychiatry. 1999;4:46–52. doi: 10.1038/sj.mp.4000450. [DOI] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–9. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Yan W, Assadi AH, Wynshaw-Boris A, Eichele G, Matzuk MM, Clark GD. Previously uncharacterized roles of platelet-activating factor acetylhydrolase 1b complex in mouse spermatogenesis. Proc Natl Acad Sci U S A. 2003;100:7189–94. doi: 10.1073/pnas.1236145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Desiderio DM. Differences in the spatial and quantitative reproducibility between two second-dimensional gel electrophoresis systems. Electrophoresis. 2003a;24:1834–46. doi: 10.1002/elps.200305389. [DOI] [PubMed] [Google Scholar]
- Zhan X, Desiderio DM. Spot volume vs. amount of protein loaded onto a gel: a detailed, statistical comparison of two gel electrophoresis systems. Electrophoresis. 2003b;24:1818–33. doi: 10.1002/elps.200305375. [DOI] [PubMed] [Google Scholar]
- Zhdanova IV, Yu L, Lopez-Patino M, Shang E, Kishi S, Guelin E. Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res Bull. 2008;75:433–41. doi: 10.1016/j.brainresbull.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.