Abstract
Both Ikaros and Notch are essential for normal T cell development. Collaborative mutations causing a reduction in Ikaros activity and an increase in Notch activation promote T cell leukemogenesis. Although the molecular mechanisms of this cooperation have been studied, its consequences in thymocyte development remain unexplored. In this study, we show that Ikaros regulates expression of a subset of Notch target genes, including Hes1, Deltex1, pTa, Gata3, and Runx1, in both Ikaros null T cell leukemia lines and Ikaros null primary thymocytes. In Ikaros null leukemia cells, Notch deregulation occurs at both the level of Notch receptor cleavage and expression of Notch target genes, because re-expression of Ikaros in these cells down-regulates Notch target gene expression without affecting levels of intracellular cleaved Notch. In addition, abnormal expression of Notch target genes is observed in Ikaros null double-positive thymocytes, in the absence of detectable intracellular cleaved Notch. Finally, we show that this role of Ikaros is specific to double-positive and single-positive thymocytes because derepression of Notch target gene expression is not observed in Ikaros null double-negative thymocytes or lineage-depleted bone marrow. Thus, in this study, we provide evidence that Ikaros and Notch play opposing roles in regulation of a subset of Notch target genes and that this role is restricted to developing thymocytes where Ikaros is required to appropriately regulate the Notch program as they progress through T cell development.
The development of T cells from multipotent progenitors into lineage-committed cells is dependent upon instructive signals from the thymic microenvironment and the genetic regulatory networks that transmit these signals. The NF Ikaros is a largely hematopoietic-specific zinc-finger regulatory protein that is essential for normal T cell development (1). Ikaros also functions as a tumor suppressor in the T cell lineage (2–4). It has been reported by many groups that simultaneous deregulation of Ikaros expression and the Notch pathway cooperate in leukemogenesis, in both mice and humans (5–8). Significantly, Notch is also essential for T cell development (9, 10), suggesting that an interaction between Ikaros and the Notch pathway could also be essential in T cell developmental processes.
The Notch receptor is a transmembrane protein that undergoes two proteolytic cleavage events upon recognition of its extracellular ligand (members of the Delta or Serrate/Jagged family) (11). These cleavages free the intracellular domain, which travels to the nucleus. In the nucleus, intracellular cleaved Notch (ICN)3 regulates transcription of Notch target genes through its binding to and activation of the transcriptional repressor CSL (designated RBP-Jκ/CBF-1 in mammals). It has recently been shown that Ikaros and recombination signal binding protein for immunoglobulin κJ (RBP-J) are able to bind to the same DNA sequences in EMSAs (5), suggesting that they may compete for binding. However, we have recently shown that Ikaros and RBP-J bind cooperatively to repress expression of the Notch target gene Hes1 in a leukemia T cell line (12). The role of Ikaros in Notch target gene repression in the developing thymocyte has not yet been explored.
The mammalian Notch family consists of four receptors: Notch1, 2, 3, and 4 (13). Although thymocytes express Notch1, 2, and 3 (14), a nonredundant essential role in T cell development has only been established for Notch1 (10). Notch3, although not essential for T cell development (15), has been shown to play a role in regulation of the pre-TCR checkpoint, β-selection. A role for Notch2 in T cell development has not been shown. Within the lymphocyte lineage, the major defined role for Notch2 is in development of marginal zone B cells (16). The differential roles of these Notch proteins, as largely determined in genetically engineered mice, suggests that each has at least a subset of unique gene targets. However, to date, it is unknown how this specific activation is accomplished, because a paradigm has been established that activation is mediated through the same DNA binding factor, RBP-J. In fact, it has been shown that activated forms of Notch1 and Notch3 are both able to up-regulate the canonical Notch target genes, Hes1, Deltex1, and pTa when they are ectopically over-expressed (17, 18). Whether both are capable of doing so when expressed at endogenous levels in developing thymocytes is unknown.
Thymocyte differentiation can be subdivided into four major stages of development in which progenitors are characterized by their cell surface expression of the CD4 and CD8 coreceptors. The most primitive T cell precursors exhibit a phenotype of CD4−CD8−, and thus are designated double-negative (DN) cells. Upon proper rearrangement of the TCRβ-chain, these cells up-regulate both CD4 and CD8 to become CD4+ CD8+ double-positive (DP) cells. At this stage, DP thymocytes undergo both positive and negative selection and eventually adopt a CD4+ or CD8+ single-positive fate (19). During T cell development, expression of Notch target genes is highly regulated. Notch target gene expression is “on” at the DN thymic progenitor stage, but is turned “off” by the DP stage (14, 20, 21). Deregulated expression can have catastrophic consequences. In transgenic mice, when Notch target gene expression cannot be shut down due to constitutive expression of ICN, leukemia results (22). Regulated expression is so important that two layers of regulation have been identified for Notch target genes. First, the activating stimulus, ICN, is only generated at precise times during T cell development. ICN, RBP-J, and the coactivator Mastermind-like (MAML) form a tertiary complex that is required for Notch target gene activation (23). Secondly, Notch target genes are actively repressed. The repression complex is less well defined and has eluded purification to date. However, it has been reported that histone deacetylase 1, C-terminal binding protein/C-terminal binding protein interacting protein, silencing mediator of retinoid and thyroid hormone receptors/nuclear receptor corepressor, Msx2-interacting nuclear target protein, CBF1-interacting corepressor, and SMRT/HDAC1-associated repressor protein are corepressors that associated with RBP-J for gene repression (24–28).
Despite the importance of Notch in leukemogenesis and T cell development, only a handful of direct Notch target genes have been identified in developing T cells. Even fewer genes have been identified as direct targets for Ikaros regulation in developing T cells. In this report, we use a combination of Ikaros null (Ik−/−) leukemia lines and Ik−/−genetically engineered mice to unravel the interdependent roles of Ikaros and Notch in regulation of Notch target gene expression in developing T cells. More specifically, we demonstrate that Ikaros and Notch antagonistically coregulate at least a subset of Notch target genes. These include genes encoding Hes1, Deltex1, and pre-TCR, all three of which are established Notch target genes (29–32). We show that, even in the absence of ICN, Notch target genes are abnormally expressed in DP Ik−/− thymocytes, providing evidence that Ikaros is an obligate repressor of these genes. Despite the increase in Notch1 target gene expression in Ik−/− thymocytes, we show that interaction of Notch with its ligands is still required for T cell development and proliferation, providing evidence that Ikaros deficiency alone does not result in constitutive activation of the complete program of Notch target genes.
Materials and Methods
Mice
Ik−/− mice (C57BL/6 × SV129) were generated by intercrossing of (Ik+/−) heterozygotes. Genotypes were assessed by PCR analysis of tail DNA as previously described (3). Analyses of thymus populations occurred between 3 and 4 wk of age before the onset of leukemia. All animal procedures were approved by the Northwestern University Animal Care and Use Committee.
Cell lines and cell culture
JE131 and D510 cell lines have been previously described (4, 33). All cell lines were maintained in RPMI 1640 medium (Life Technologies) supplemented with 10% bovine growth serum (BGS; HyClone), 50 μM 2-ME, and 500 U of penicillin-streptomycin (Pen-Strep) per ml (RPMI complete). OP9 monolayers were maintained in OP9 medium (αMEM supplemented with 20% FBS and 500 U Pen-Strep). Cocultures were maintained in Coculture medium (high glucose DMEM supplemented with 10% FBS, 1× (each) Glutamax, HEPES, sodium pyruvate, Pen-Strep, and 55 μM 2-ME.
Sequence analyses
Total RNA isolation and cDNA synthesis was performed as described below. Notch1 heterodimerization domain (HD) and Proline, Glutamine, Serine, and Threonine-rich domain (PEST) regions were amplified using previously described primers (34). PCR products were purified using a Promega Wizard SV PCR Clean-up system. Analysis and alignment were performed using DNA Strider. The Notch1 reference sequence was obtained with GenBank accession number NM_008714.
Protein preparation and immunoblotting
Protein extracts were prepared by whole cell lysis with 420 mM NaCl Lysis Buffer (20 mM Tris (pH 7.5), 0.1% BSA, 1 mM EDTA, 1% Nonidet P-40) supplemented with protease inhibitors. Protein extracts were separated by gel electrophoresis on a SDS-polyacrylamide gel and transferred to a PVDF membrane overnight at 4°C. Membranes were blocked for at least 1 h in TBS-5% milk. Abs against Notch1 (Santa Cruz Biotechnology) were diluted 1/500 in TBS-5% milk and incubated with the membrane for 1 h at room temperature. Abs against cleaved Notch1-val1744 (Cell Signaling Technology) were diluted 1/500 in TBS-Tween 5% BSA and incubated with the membrane overnight at 4°C. Blots were washed with TBS and incubated with HRP-conjugated Ab for 1 h at room temperature. Proteins were visualized by incubation with ECL reagent and exposure to film.
Retroviral constructs
The murine stem cell virus (MSCV) IRES GFP, MSCV IRES H-2Kk, and MSCV IRES Ik-1 GFP and MSCV IRES Ik-1 H-2Kk constructs were generated as described previously (4). MSCV DNMAML GFP was a gift of Dr. Warren Pear (University of Pennsylvania, Philadelphia, PA).
Retroviral transduction and cell sorting
MSCV constructs were transfected into the Phoenix ecotropic packaging cells using Lipofectamine reagent (Invitrogen). Viral supernatants were harvested at 48 and 72 h postinfection. Filtered supernatants were used to infect cells using 0.5–1.0 ml of supernatant per 2 × 106 cells supplemented with 8/μg/ml polybrene in a 24-well tissue culture dish. Plates were centrifuged at 500 × g for 2 h at 32°C. Supernatants were removed and cells were cultured with RPMI complete medium. Successfully transduced cells were sorted by H-2Kk expression with the MiniMacs system (Miltenyi Biotec) as previously described (4). Purity was consistently >95%.
Abs and flow cytometry analysis
All Abs were from eBiosciences unless otherwise stated. For flow cytometric analysis, the following Abs were used: anti-CD4 (GK1.5), anti-CD8 (53–6.7), MACSelect control FITC Ab (Miltenyi Biotec), and anti-H-2Kk (H100–27.R55) (Miltenyi Biotec). Abs were allophycocyanin, FITC, or PE conjugates. For staining, cells were plated in microwell staining plates at a density of 5 × 105 to 1 × 106 cells per well. Fluorochrome-conjugated Abs were added to cells and incubated on ice for 15 min. Cells were washed three times, and analyzed by flow cytometry on a FACSCalibur (BD Biosciences) flow cytometer using Flow Jo software. Cell cycle analysis was performed as previously described (4).
Thymic progenitor isolation and in vitro T cell differentiation
Thymuses were harvested from 3- to 4-wk-old Ik−/− mice and wild-type littermates. Single cell suspensions were generated by disruption with frosted slides and pipetting through a 70-μm nylon filter in RPMI complete medium. Cells were washed in PBS-2% BGS-1 mM EDTA. Enrichment of DN cells was performed by density centrifugation over a 1.086M Optiprep gradient (Accurate Chemical and Scientific Corporation). These cells were then incubated with α-CD4 and α-CD8 Abs (eBioiscience) for 30 min, followed by incubation with BioMag Goat Anti-Rat IgG beads (Qiagen) for 30 min. Ab-positive cells were removed through magnetic depletion, and the negative fractions (DNs) were washed and resuspended in PBS-2% BGS-1 mM EDTA. Purity was consistently >95%. DNs were plated onto subconfluent OP9 GFP and OP9 DL1 monolayers in 12-well dishes. Cocultures were maintained in coculture medium supplemented with 1 ng/ml rmIL-7 (Biovision) and 5 ng/ml rhFlt3-L (PeproTech). For inhibitor assays, equal amounts of γ-secretase inhibitor XII [8 mM] (Calbiochem) or DMSO (carrier) were added to cultures. Developmental progression was assessed after 4 days of coculture by flow cytometric analyses of CD4 and CD8 expression.
Bone marrow progenitor isolation
Bone marrow was isolated by flushing femurs and tibias from wild-type or Ik−/− mice with RPMI complete medium using a 25-gauge syringe. Single cell suspensions were generated by pipetting medium through a 70-μm nylon filter. For lineage depletion, cells were incubated with α-B220, α-CD11b, α-Ter119, α-CD4, and α-CD8 Abs (eBioiscience) for 30 min, followed by incubation with BioMag Goat Anti-Rat IgG beads (Qiagen) for 30 min. Ab-positive cells were removed through magnetic depletion, and the negative fractions (Lin-) were washed and used for RNA isolation.
Rt-PCR
RNA was prepared from sorted primary thymocytes or from retrovirally transduced JE131, DO11, and Tu5 cells (sorted for H-2Kk expression) at 24 h postinfection using the SV Total RNA Isolation System (Promega). cDNA was generated with a Superscript III kit (Invitrogen), and used for quantitative real-time RT-PCR (qRT-PCR) analyses (Bio Rad iQ5 Real Time PCR machine). The iQ SYBR Green Supermix (Bio-Rad) was used. Primer sequences were generated by the Beacon Design program and synthesized by IDT DNA Technologies. Sequences are available upon request.
Results
Constitutive Notch target gene expression is observed in some, but not all, Ikaros-deficient T leukemia cell lines
To dissect the role of Ikaros in regulating Notch target gene expression, we used a panel of T cell lines derived from Ikaros-deficient mice with spontaneously arising T cell leukemia. It was previously reported that T leukemia cell lines arising from Ikaros hypomorphic (IkL/L) mice constitutively express ICN, the active form of Notch (5). Therefore, we first surveyed the panel of Ikaros-deficient T cell lines for expression of ICN. Two cell lines, JE131 (4) and DO11, were generated from genetically engineered Ik−/− mice. The Tu5 cell line was generated from a genetically engineered “dominant negative” (DN+/−) Ikaros mouse, in which targeted deletion of exons 3 and 4 of the Ikaros gene results in expression of non-DNA binding isoforms from the targeted allele (2, 35). A dominant negative effect of these isoforms on the ability of DNA-binding competent Ikaros to bind DNA and activate transcription has been demonstrated (36), hence the “DN” designation. Because the leukemia that arises in DN+/− mice displays loss of heterozygosity (LOH), the Tu5 cell line only expresses non-DNA binding Ikaros isoforms (2). The D510 cell line was derived from a transgenic mouse that expressed high-levels of a non-DNA binding Ikaros isoform exclusively in T cells (37). D510 expresses normal levels of DNA binding competent Ikaros from the endogenous Ikaros alleles (37). Jurkat cells are a human T cell leukemia line that constitutively expresses ICN (38) and, therefore, were used as a positive control.
ICN was expressed in the T cell leukemia lines that lack expression of DNA-binding Ikaros isoforms (JE131, DO11, and Tu5). In contrast, ICN was not detected in the D510 T cell leukemia cell line, which expresses DNA-binding competent Ikaros isoforms (Fig. 1A). In two of the three ICN-expressing cell lines (JE131 and Tu5), ICN was smaller than its expected mobility, which suggests that the ICN protein in these leukemia lines is truncated. To further explore the mechanism of aberrant ICN expression in the Ikaros-deficient leukemia cell lines, we performed sequence analysis of Notch1 regions that are commonly mutated in T-acute lymphoblastic leukemia (T-ALL) (39, 34). Mutational analysis of the C-terminal PEST region, which is involved in regulating protein stability, revealed that, in the JE131 and Tu5 cells, the Notch1 sequence contains nucleotide insertions resulting in a frameshift and premature termination of translation (Fig. 1B). In the JE131 cells, the insertion occurs in a mutational hotspot, which was previously identified in murine T-ALLs (34). Additional mutations were identified in the HD domain, which maintains stable association of Notch receptor subunits. Three of the Ikaros-deficient leukemia lines (JE131, Tu5, and D510) contain point mutations in the amino terminal HD-domain (Fig. 1B); however, these mutations do not produce the L to P substitutions that are most commonly found in T-ALLs, and therefore their functional significance is unknown. Interestingly, although the DO11 cell line expresses ICN, we did not identify any mutations in the HD domain or PEST domains. This suggests a different mechanism for ICN stabilization in these cells. Lack of Notch1 mutations have been identified in other ICN-expressing T cell lines, such as Jurkat (39), providing a precedent for this observation.
FIGURE 1.
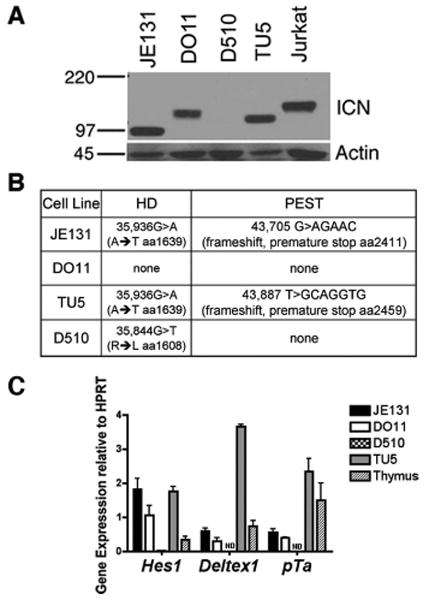
Notch target genes are constitutively expressed in Ikaros-deficient leukemia cell lines that express ICN. A, Western blot analysis was used to assay for the presence of ICN in whole cell lysates prepared from Ikaros deficient T leukemia cell lines. Blots were probed with Abs directed against ICN that has been properly cleaved at val1744 (expected mobility: 110kD). Blots were also probed with anti-actin Abs as a loading control. B, Summary of sequence analysis of Notch1 using cDNA (PEST) or DNA (HD) from Ikaros deficient leukemia cell lines. HD = heterodimerization domain (exon 26 and exon 27). PEST = Proline, Glutamine, Serine, and Threonine-rich domain (exon 34). C, qRT-PCR analysis was performed using cDNA from ICN positive (JE131, DO11, TU5) and ICN negative (D510) Ikaros-deficient leukemia cell lines. ND = Not Detected. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined by the Pfaffl method and are shown as ratios ((Etarget) −CT target:(Ereference) −CT reference). E = efficiency of PCR; target = gene of interest; reference = HPRT. Experiments were performed at least twice in duplicate. Error bars indicate SEM.
These results indicate that Notch pathway activation through production of ICN is a common event in Ikaros-deficient leukemia and might suggest a requirement for Notch deregulation in the leukemogenic process observed in Ikaros-deficient mice. However, the D510 cell line did not express ICN, suggesting that this is not true in all cases. The possibility exists that Notch-dependent gene regulation may be activated in an ICN-independent manner in D510. Therefore, we next assessed the expression of three well-established Notch target genes (Hes1, Deltex1, and pTa) in D510 cells. Quantitative real-time RT-PCR (qRT-PCR) analyses revealed that D510 cells express very low (e.g., Hes1) or undetectable (e.g., Deltex1 and pTa) levels of Notch-induced transcripts. However, high levels of expression are observed in JE131, DO11, and Tu5 cells (Fig. 1C), which all: 1) lack DNA-binding Ikaros and 2) contain ICN.
Reintroduction of Ikaros into an Ik−/− T leukemia cell line down-regulates Notch target gene expression without affecting levels of ICN
We next sought to dissect the contributions of: 1) the lack of Ikaros expression and 2) aberrant ICN generation, to the expression of Notch-induced transcripts observed in the Ikaros-deficient, ICN-expressing T cell leukemia lines. High-level expression of Hes1, Deltex1, and pTa in these cells could be exclusively the result of the presence of ICN. Alternatively, lack of Ikaros could also contribute to this phenotype; in this case, restoration of Ikaros should down-regulate Notch target gene expression. Indeed, restoration of Ikaros expression using retroviral transduction results in down-regulation of Hes1, Deltex1, and pTa expression in all three Ikaros-deficient, ICN-expressing cell lines (Fig. 2A).
FIGURE 2.
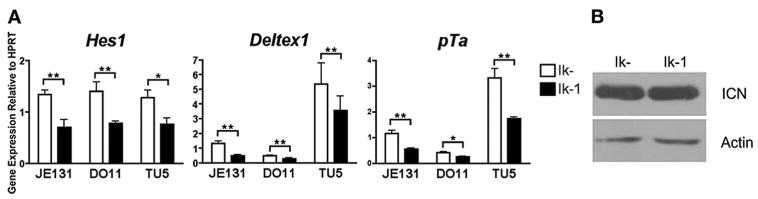
Ikaros can repress Notch target gene expression in the presence of ICN. A, Twenty-four hours postinfection, JE131, DO11, and Tu5 cells successfully infected with MSCV IRES H-2Kk (Ik-) or MSCV IRES Ik-1 H-2Kk (Ik-1) retroviruses were purified using H-2Kk as a marker of successful transduction. cDNA was prepared and analyzed by qRT-PCR for expression of Hes1, Deltex1, and pTa. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined as described in the legend for Fig. 1. Experiments were performed at least three times in duplicate from two independent RNA samples. p values were calculated using paired student T tests (*, p ≤ 0.05; **, p ≤ 0.03). Error bars represent SEM. B, Whole cell extracts from successfully transduced JE131 cell populations prepared as described above were subjected to Western blot analysis using Abs targeted against ICN. Blots were also probed with anti-actin Abs as a loading control.
Ikaros could be initiating down-regulation of Notch target genes through direct repression or through regulation of ICN production. For example, Ikaros could regulate Notch target gene expression by controlling levels of ICN through, for example, regulation of the proteolytic enzymes necessary for ICN generation (e.g., γ-secretase). Therefore, levels of ICN were compared in JE131 cells transduced with Ikaros, in which Notch target gene expression is down-regulated, to levels in JE131 cells infected with a retrovirus that expresses no Ikaros protein, in which Notch target genes are robustly expressed. Levels of ICN remained high in Ikaros-transduced JE131 cells, proving that, at a time point at which Ikaros expression drives down-regulation of Notch target genes, it does not affect levels of ICN (Fig. 2B). These data support a role for Ikaros downstream of ICN in regulating expression of Notch target genes.
Ectopic expression of dominant negative MAML1 is capable of down-regulating Notch target gene expression in an Ik−/− T leukemia cell line
We next assessed the contribution of ICN to Notch target gene expression in JE131 cells by determining whether interfering with ICN-induced transcriptional activation would be sufficient to produce some of the effects of Ikaros reintroduction. MAML is a coactivator for Notch target gene expression (11, 23, 40, 41). There are three MAML proteins in vertebrates, MAML1, MAML2, and MAML3, all of which are expressed in T cells. A truncated form of MAML1 has been generated that can associate with ICN but lacks the C-terminal transcriptional activation domain. It interferes with the activity of all of the Notch receptors, and therefore of has been designated “dominant negative” (DNMAML1) (42).
Retroviral transduction of DNMAML1 into JE131 cells was able to down-regulate expression of the Notch target genes Hes1, Deltex1, and pTa (Fig. 3A) to a greater extent than reintroduction of Ikaros. Analyses were then extended to include a wider variety of transcriptional targets of Notch signaling, including Gata3, Runx1, CD25, NRARP, Notch1, Notch3, and HuD (43–47). Re-expression of Ikaros resulted in down-regulation of most of the Notch target genes examined, suggesting that Ikaros is involved in the regulation of a wide array of Notch-induced transcripts (Fig. 3B). Expression of DNMAML1 in the JE131 cells also resulted in down-regulation of these additional Notch target genes (Fig. 3, A and B). Note that for a subset of these genes (Deltex1, pTa, Gata3, Runx1, and NRARP), Ikaros was just as efficient in down-regulating their expression as DNMAML1.
FIGURE 3.
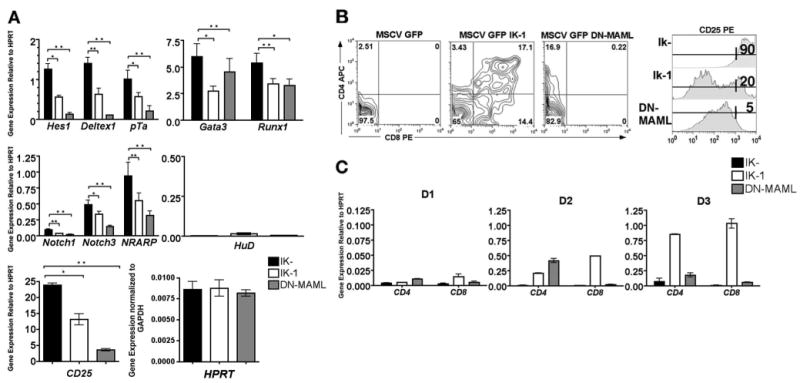
Ectopic expression of DNMAML1 is capable of mediating similar effects as Ikaros re-expression in an Ikaros null T cell line. A, Twenty-four hours postinfection, JE131 cells successfully infected with MSCV IRES GFP (Ik-), MSCV IRES Ik-1 GFP (Ik-1), or MSCV IRES DNMAML-1 GFP (DNMAML) retroviruses were sorted using GFP. cDNA was prepared and analyzed by qRT-PCR. Expression levels of the indicated Notch target genes were assessed. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Equivalent levels of HPRT were observed in each sample as compared with levels of the housekeeping gene, Gapdh. Values were determined as described in the legend for Fig. 1. Experiments were performed at least three times in duplicate from two independent RNA samples. p values were calculated using paired Student t tests (*, p ≤ 0.05; **, p ≤ 0.02). Error bars represent SEM. Paired Student t tests were also performed to compare gene expression levels in Ik-1 vs DNMAML-transduced cells, yielding p values >0.05 for Deltex1, pTa, Gata3, Runx1, and NRARP, and p values <0.05 for Hes1, Notch1, Notch3, and CD25. B, JE131 cells were infected with either MSCV IRES GFP (Ik-), MSCV IRES Ik-1 GFP (Ik-1), or MSCV IRES DNMAML-1 GFP (DNMAML) retroviruses. Four days postinfection, cells were stained with fluorochrome-conjugated Abs against CD4, CD8, and CD25. Plots shown are data obtained for GFP-positive cells. C, Twenty-four (D1), 48 (D2), and 72 h (D3) postinfection, JE131 cells successfully infected with MSCV IRES GFP (Ik-), MSCV IRES Ik-1 GFP (Ik-1), or MSCV IRES DNMAML-1 GFP (DNMAML) retroviruses were sorted using GFP. cDNA was prepared and analyzed by qRT-PCR for CD4 and CD8 expression. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined as described in the legend for Fig. 1 and are shown above each bar. Experiments were performed at least twice in duplicate. Error bars represent SEM.
Interestingly, high-level expression of the RNA-binding protein HuD, which is specifically induced by Notch3, but not Notch1, was not observed in JE131 cells (17). Likewise, reintroduction of Ikaros had no effect on HuD expression. Together, these data suggest that Ikaros' role in Notch target gene regulation may be limited to the regulation of Notch1-induced transcripts (Fig. 3A).
T cell differentiation marker expression is differentially affected by re-expression of Ikaros vs ectopic expression of DNMAML1 in an Ik−/− T leukemia cell line
Reintroduction of Ikaros into the JE131 cell line induces the expression of a variety of T cell differentiation markers (4). JE131 cells display a cell surface phenotype of CD4−CD8−CD25high, which is indicative of thymocytes at the late DN stage of development. Retroviral transduction of Ikaros induces cell surface expression of both CD4 and CD8, as well as down-regulation of CD25 (4) (Fig. 3B), events which occur as thymocytes progress to the DP stage. To determine whether these events could be elicited as a result of Ikaros' role in down-regulation of Notch target gene expression, JE131 cells were transduced with DNMAML1 and the effect on expression of these markers was monitored. Ectopic expression of DNMAML1 was sufficient to induce up-regulation of CD4 and down-regulation of CD25 at both the mRNA and protein levels, but could not induce up-regulation of CD8 (Fig. 3,B and C). Ikaros-mediated induction of CD4 expression increases over time, peaking at 72 h (D3), whereas retroviral expression of DNMAML1 results in CD4 up-regulation that peaks at 48 h (D2) after infection. These data indicate that Ikaros' ability to regulate CD4 and CD25 expression in JE131 cells is likely dependent on its ability to regulate the Notch pathway, whereas this is not the case for CD8.
In Ik−/− thymocytes, Notch target genes can be expressed in the absence of ICN
Levels of ICN are most robust at the DN stage of thymocyte development and virtually nonexistent at the DP stage, due, at least partially, to lack of Notch1 expression in DP thymocytes (48). This parallels levels of Notch target gene expression, which have also been reported, for the most part, to be highest at the DN stage (14, 20, 21). To determine whether Notch target gene expression is deregulated in primary Ik−/− thymocytes before transformation, expression levels of several Notch target genes were compared in Ik−/− and wild-type DN and DP thymocyte subsets. Expression levels were not significantly different in Ik−/− DN thymocytes compared with their wild-type counterparts (Fig. 4A). In contrast, expression levels of the majority of the genes were significantly up-regulated in Ik−/− as compared with wild-type DP thymocytes (Fig. 4A). We also compared expression levels of the Notch3-induced transcript HuD in Ik−/− and wild-type thymocyte subsets. Comparable expression levels of HuD were observed in both DN and DP thymocytes (Fig. 4A). These data, which are consistent with results generated from the Ik−/− cell line, suggest that deregulation of Notch target genes in Ik−/− mice may be specific to Notch1 targets.
FIGURE 4.
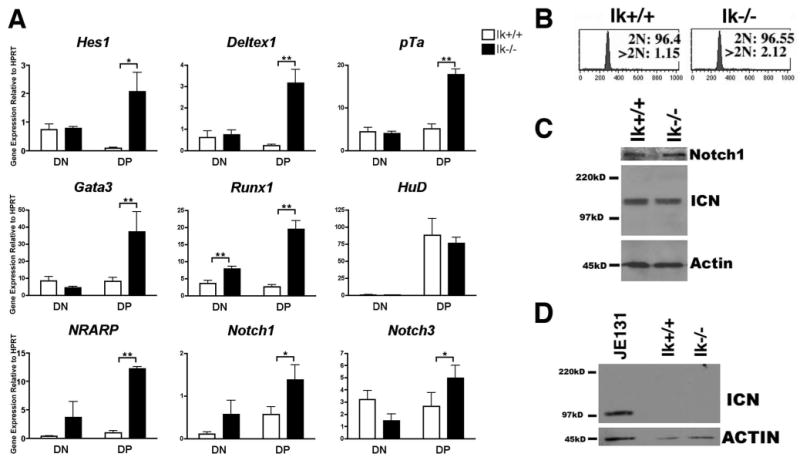
Notch target gene expression is deregulated in primary Ikaros null thymocytes in the absence of deregulated ICN production. A, qRT-PCR analysis was performed using cDNA prepared from sorted thymocyte subsets (double negative, DN; double positive, DP) from Ikaros null (Ik−/−) and wild-type (Ik+/+) mice. Expression levels of the indicated Notch target genes were assessed. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined as described in the legend for Fig. 1. Experiments were performed at least three times in duplicate from three independent RNA samples. p values were calculated using paired Student t tests (*, p ≤ 0.05; **, p ≤ 0.02). Error bars represent SEM. Note that there are no significant differences between expression levels in Ik+/+ and Ik−/− DN thymocytes except in the case of Runx1. B, Flow cytometry histograms depicting propidium iodide staining of permeabilized wild-type (Ik+/+) and Ikaros null (Ik−/−) thymocytes. Cells with 2N DNA were designated as being in the G0 or G1 phases of the cell cycle whereas cells with >2N DNA were designated as being in S, G2, or M. C, Western blot analysis was performed using 30 μg of whole cell extract from Ik+/+ and Ik−/− thymuses and Abs that recognize total Notch1 or ICN that has been properly cleaved at val1744. Blots were also probed with anti-actin Abs as a loading control. ICN blot was exposed to autoradiography film for 5 min. D, Western blot analysis of sorted Ik+/+ and Ik−/− DP thymocytes was used to assess the presence of ICN using Abs that recognize properly cleaved ICN. Blots were also probed with anti-actin Abs as a loading control. Whole cell extracts prepared from the JE131 cell line served as a positive control. ICN blot was exposed to autoradiography film for 15 min.
One explanation for the deregulation of Notch1 target gene expression observed in Ik−/− DPs could be that Ik−/− CD4+CD8+ cells are not true DP thymocytes. For example, because Ikaros has been implicated in regulation of both CD4 and CD8 (49, 50), the possibility exists that these cells are actually DN thymocytes misexpressing these coreceptor molecules. However, analyses of expression levels of cell surface markers revealed similar levels of CD25, CD5, TCRβ, and CD69 on Ik−/− and wild-type DPs (Ref. 51 and data not shown), alleviating this concern. Additionally, cell cycle analysis showed that Ik−/− thymocytes are not abnormally proliferating, as would be the case if they had become transformed (Fig. 4B).
Because deregulated expression of ICN is observed in most Ikaros-deficient leukemia cell lines, we next asked whether lack of Ikaros results in an increase in levels of ICN in thymocytes before transformation. Western blot analyses revealed that levels of ICN are comparable in protein extracts prepared from Ik−/− and wild-type thymuses (Fig. 4C). However, the high-level expression of several Notch-induced transcripts in Ik−/− DP thymocytes suggested that ICN might be aberrantly present within this subpopulation. To investigate this possibility, protein extract was prepared from sorted DP Ik−/− and wild-type thymocytes. Western blot analyses demonstrated that ICN was not generated in Ik−/− DP thymocytes (Fig. 4D). Sequence analyses of Notch1 expressed in Ik−/− thymocytes also demonstrated that PEST and HD regions were identical in sequence to that of Notch1 in wild-type thymocytes (data not shown). Taken together, these data provide evidence that, in the absence of Ikaros, expression of Notch target genes can occur independently of ICN in DP thymocytes.
The Notch/DL1 signaling pathway is not constitutively active in nontransformed Ik−/− primary thymocytes
Signals delivered through Notch are required for T cell development (10). The aberrantly high level of Notch target gene expression in Ik−/− DP thymocytes led us to hypothesize that, in the absence of Ikaros, thymic progenitors may not be dependent on active Notch signaling for developmental progression. To test this, the OP9 DL1 ex vivo T cell development system was used. In this system, progenitors are cocultured with an OP9 bone marrow stromal cell line stably expressing the Notch ligand Delta-like 1 (OP9 DL1) (52). Using this assay, DN progenitors are able to differentiate to the DP stage. Differentiation is dependent on Notch ligand expression, because it is not observed when progenitors are cocultured with the OP9 GFP stromal cell line, which lacks expression of DL1 (52, 53).
Ik−/− progenitors, like their wild-type counterparts, were unable to differentiate beyond the DN stage when cocultured with OP9 GFP cells, suggesting that activation of Notch signaling via DL1 is required for their differentiation (Fig. 5A). To further test this, a pharmacological γ-secretase inhibitor (GSI) was used to block Notch signaling in progenitors cocultured with the OP9 DL1 cells. GSI blocks cleavage of Notch, preventing formation of ICN. Ik−/− DN thymocytes, like their wild-type counterparts, could not differentiate to the DP stage in the presence of GSI (Fig. 5B). Taken together, these data show that Ik−/− progenitors require the presence of exogenous (DL1) Notch ligand to differentiate beyond the DN stage, providing evidence that lack of Ikaros does not result in overall activation of Notch signaling.
FIGURE 5.
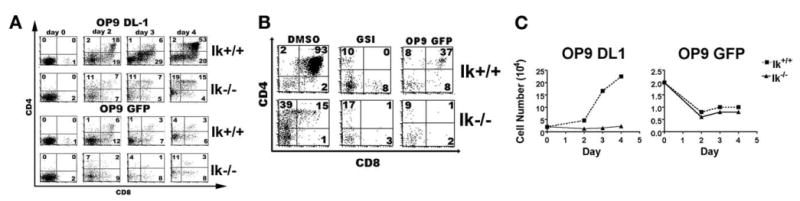
Ikaros null thymic progenitors require interaction with Notch ligand to differentiate and proliferate. A, Equal numbers of DN thymic progenitors from wild-type (Ik+/+) and Ikaros null (Ik−/−) mice were cocultured with the OP9 DL1 (62) or OP9 GFP (bottom) stromal cell line. Flow cytometric plots of cells stained with fluorochrome-conjugated Abs against CD4 and CD8 indicate the differentiation status of cells in each culture over time (days 0 to 4). Experiment was performed four times. Representative plots are displayed. B, DN thymic progenitors from Ik+/+ and Ik−/− mice were cultured on OP9 DL1 stroma in the presence or absence of γ-secretase inhibitor XII (GSI) over a 4-day period. OP9 GFP cultures serve as a control for differentiation in the absence of exogenous Notch signals. Experiment was performed twice. Representative plots are displayed. C, Proliferation curves are from the cultures whose differentiation status is depicted in A. Experiment was performed three times. Representative data are shown.
Notch signaling is also required for the proliferative expansion that occurs as thymocytes transit from the DN to DP stages (48). This requirement can be easily seen by comparing numbers of thymocytes over time during coculture with OP9 GFP vs OP9 DL1 cells. Wild-type DN thymocytes cocultured with OP9 DL1, but not OP9 GFP, cells expand dramatically (11.2-fold) (Fig. 5C). In contrast, Ik−/− DN thymocytes do not expand significantly when cocultured with OP9 DL1 cells, although they survive minimally better in OP9 DL1 as compared with the OP9 GFP cocultures (Fig. 5C). Therefore, lack of Ikaros cannot compensate for lack of DL1 ligand for inducing thymocyte proliferation.
Deregulation of Notch target gene expression is observed in Ik− single positive thymocytes
When DP thymocytes transition to the SP stages of development, some Notch target genes are induced while others remain in a repressed state (54, 55). To see whether deregulated expression of Notch target genes in Ik−/− mice extends beyond the DP stage of development into more mature thymic populations, cDNA was prepared from sorted CD8+ SP and CD4+ SP thymocyte subsets from young Ik−/− and wild-type mice, and Notch target expression was assessed. Most of the Notch targets analyzed, such as Hes1, pTa, Gata3, and Runx1, exhibited increased expression levels in both CD4+ and CD8+ SP subsets from Ik−/− mice as compared with their wild-type counterparts, similar to what was observed in DP thymocytes (Fig. 6A). However, expression of the Notch target gene, Deltex1 was differentially affected by lack of Ikaros in CD4+ vs CD8+ SP thymocytes (Fig. 6B); Deltex1 expression was up-regulated in CD4+ SP thymocytes, but not CD8+ SP thymocytes in Ik−/− mice.
FIGURE 6.
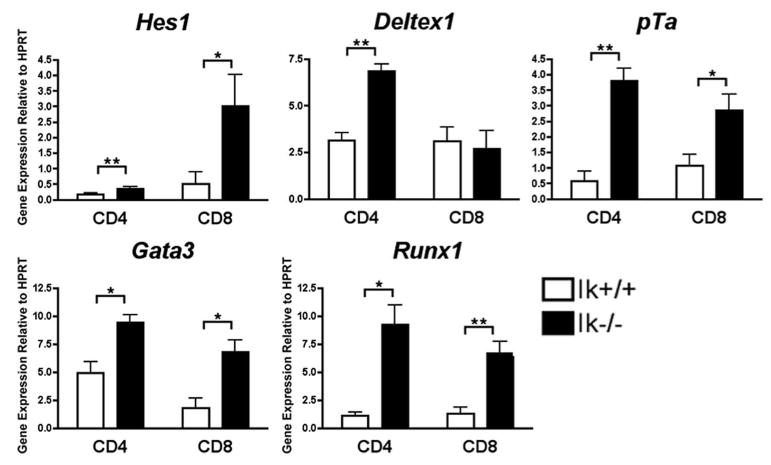
Deregulation of Notch target genes is observed in Ikaros null single positive thymocytes. qRT-PCR analysis was performed using cDNA prepared from sorted CD4+ and CD8+ single positive (19) thymocyte subsets from Ikaros null (Ik−/−) and wild-type (Ik+/+) mice. Expression levels of the indicated Notch target genes were assessed. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined as described in the legend for Fig. 1. Experiments were performed at least three times in duplicate from two independent RNA samples. p values were calculated using paired Student t tests (*, p ≤ 0.03; **, p ≤ 0.01). Error bars represent SEM.
Derepression of Notch target genes is not observed in Ik−/− bone marrow
We next wanted to determine whether Ikaros' role in regulating Notch target genes was limited to populations within the thymus. Because B cell development requires the repression of Notch signaling (9, 10), it is interesting to speculate that derepression of Notch target genes in Ik−/− bone marrow progenitors may contribute to the lack of B cell development observed in these mice. To determine whether Ikaros controls expression of Notch target genes in these early progenitors, cDNA was prepared from lineage depleted bone marrow cells from Ik−/− and wild-type mice, and Notch target gene expression was assessed. Expression levels of these genes were uniformly not increased in Ik−/− compared with wild-type cells (Fig. 7A). Interestingly, the only gene that showed a significant difference in expression levels was Notch1, which was actually more highly expressed in wild-type than in Ik−/− cells. This difference may reflect a positive role for Ikaros in regulating Notch1 expression in bone marrow (BM) progenitors. Alternatively, because alterations in Ik−/− as compared with wild-type BM progenitor populations have been previously documented (56, 57), this disparity might reflect differing levels of Notch1 expression in the different classes of progenitor subpopulations present.
FIGURE 7.
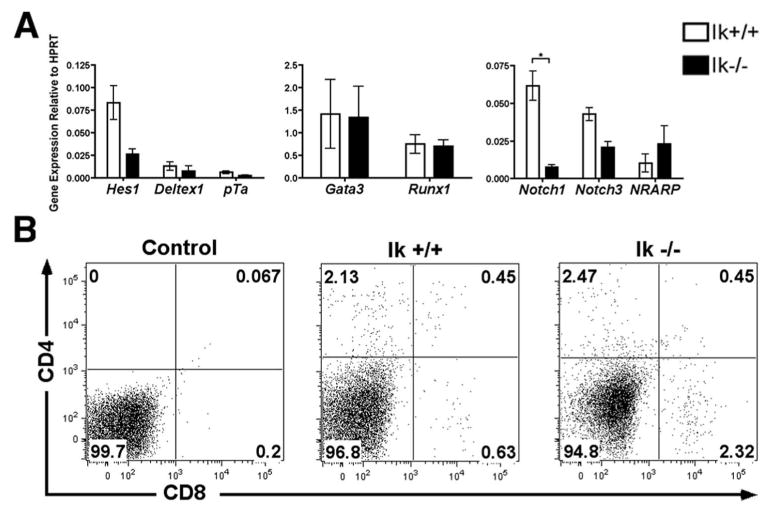
Ikaros null bone marrow progenitors do not display derepressed Notch target gene expression. A, qRT-PCR was performed using cDNA prepared from lineage depleted bone marrow from Ikaros null (Ik−/−) and wild-type (Ik+/+) mice. Expression levels of the indicated Notch target genes were assessed. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined as described in the legend for Fig. 1. Experiments were performed at least three times in duplicate from two independent RNA samples. p values were calculated using paired Student t tests (*, p ≤ 0.02). Error bars represent SEM. B, Whole bone marrow was isolated from Ikaros null (Ik−/−) and wild-type (Ik+/+) mice. Flow cytometric plots of cells stained with fluorochrome-conjugated Abs against CD4 and CD8 are shown. Experiment was performed twice. Representative plots are shown.
A functional consequence of aberrant Notch activation in BM progenitors is the inappropriate appearance of CD4+CD8+ DP pre-T cells in the bone marrow (9, 58). Therefore, as a second test to examine whether lack of Ikaros results in derepression of Notch target genes in BM progenitors, whole bone marrow isolated from Ik−/− and wild-type mice was monitored for cell surface expression of CD4 and CD8 using flow cytometry. No evidence of CD4+CD8+ T cells was observed in Ik−/− BM (Fig. 7B).
Taken together, these data provide strong evidence that the role of Ikaros as a repressor of Notch target genes does not extend to progenitor cells in the BM.
Discussion
Proper regulation of the Notch pathway depends on several layers of regulation in developing T cells, which include precise control of Notch signaling and active repression of Notch-induced transcription. Concomitant deregulation of Ikaros and Notch has been observed in T cell leukemia, suggesting an interaction between the two molecules in the transformation of thymocytes. In this study, we sought to explore a potential interaction between Ikaros and Notch during thymic T cell development.
Analyses of a panel of Ikaros-deficient leukemia cell lines revealed deregulation of the Notch pathway at the level of constitutive ICN expression as well as at the level of Notch target gene expression. We hypothesized several ways in which lack of Ikaros would yield this phenotype. For example, Ikaros could be involved in the regulation of Notch target genes through direct repression or through regulation of Notch signaling components. Re-expression of Ikaros in JE131 cells resulted in down-regulation of Notch target genes without reducing levels of ICN, suggesting that Ikaros regulates Notch target genes through a mechanism downstream of ICN production. In support of a model of direct repression, we have recently demonstrated direct binding of Ikaros to both Hes1 and Deltex1 regulatory regions in JE131 cells (12).
Retroviral transduction of DNMAML1 identified a broad array of genes as targets of ICN-induced transcription in JE131 cells. By comparing the effects of DNMAML1 vs Ikaros transduction on expression of these genes, we demonstrated that they are almost uniformly antagonistically coregulated by Ikaros and Notch. These include Notch target genes, such as Hes1 and Notch3, whose expression were almost completely extinguished upon DNMAML expression but down-regulated to a lesser extent by Ikaros, as well as Deltex1, pTa, Gata3, and Runx1 whose expression were reduced equivalently upon Ikaros and DNMAML1 expression. One explanation for these differing results could be that there is a difference in the relative contribution of the RBP-J activating complex, consisting of RBP-J, MAML, and ICN, to high-level expression of these genes in JE131 cells. It is important to note that, in this system, transduction of Ikaros, as well as DNMAML1, also resulted in down-regulation of Notch1 expression itself, suggesting that this might be another mechanism by which Ikaros can down-regulate Notch target gene expression in vivo. However, no decrease in levels of ICN are seen in Ikaros transduced JE131 cells at 24 h, when decreases in Notch target gene expression are readily observed, supporting another more direct mechanism for Ikaros in repression of Notch target genes. The lack of ICN down-regulation in the face of decreased Notch1 expression at this early time point may be due to the increased stability of ICN that has been reported to occur in leukemia cells (5, 59). In a similar vein, the presence of high levels of ICN in Ikaros-transduced JE131 cells may contribute to the reduced ability of Ikaros to down-regulate some of the Notch1 targets (i.e., Notch1, Hes1) in comparison to DNMAML1, because it would be opposing repression of these genes.
The extensive ability of Ikaros to down-regulate genes that are also down-regulated by DNMAML in JE131 cells provides evidence for a global role for Ikaros in the regulation of RBP-J/ICN-induced transcripts in thymocytes. However, the lack of high-level expression of the Notch3-specific target gene HuD (17) in JE131 as well as the normal HuD expression observed in Ik−/− thymocytes suggests that Ikaros-mediated regulation of the Notch pathway may be specifically restricted to Notch1 target genes. At this time, this is a difficult hypothesis to test. Unfortunately, few target genes have been defined that are specific for a given Notch receptor and little has been reported concerning mechanisms of specificity. More importantly, for our studies, little is known about the composition of the RBP-J-containing repressor complex. Many corepressors have been identified, but whether different complexes function downstream of the different Notch receptors is a possibility that has not been addressed.
Ikaros' role in mediating repression of the Notch pathway is supported by analyses of Notch target gene expression in Ik−/− thymocytes in which deregulated expression of these genes is almost exclusively limited to populations, such as the DP subset, in which their expression is actively repressed. Therefore, we hypothesize that Ikaros may be essential for the rapid and tight shut-down of Notch target gene expression that must occur as cells transit from the DN to the DP stage. However, it is important to note that over-expression of Notch target genes was also observed in Ik−/− SP thymocytes. This observation is particularly significant because it highlights the fact that Notch target gene deregulation is a function of Ikaros' role in gene regulation rather than some artifact of a transformed population within the Ik−/− DP thymocyte subset.
Genetic studies in mice have provided a useful tool to study the various functions of regulatory proteins in both Notch pathway activation and repression (Fig. 8). Conditional inactivation of Notch1 is sufficient to produce defects in T cell lineage commitment, but does not necessarily result in a failure to express Notch target genes, such as pTa (60), possibly due to redundancy among the Notch receptors. Inducible knockouts of RBP-J exhibit normal expression patterns of the Notch targets Hes1 and Hes5 (61). Because RBP-J mediates both transcriptional repression and activation of Notch target genes, this phenotype might reflect the contradictory effects of both impaired repression and impaired activation. When Notch target gene repression is specifically targeted, however, a different picture emerges. The genetic ablation of Mint, which has been shown to function as a RBP-J corepressor through its interaction with CtIP/CtBP, results in the derepression of the Notch targets Hes1, Deltex1, and NRARP in DN progenitors (28). Phenotypic analyses of thymocytes derived from Mint−/− fetal liver cells in adoptive transfer experiments, as well as Mint−/− fetal thymocytes, revealed that Mint functions primarily within the DN population (28), in contrast to Ikaros whose function in Notch target gene repression appears to be most apparent at the DP and SP stages of thymocyte development (Fig. 8). Notch target gene derepression can be observed in another mouse model, in which the gene encoding the DNA-binding protein Zbtb7a (LRF) has been conditionally inactivated. The phenotype observed in Ikaros-deficient mice is in contrast to that observed in LRF-deficient mice, in which aberrant Notch target gene expression is dependent upon ICN expression (58). Therefore, LRF likely inhibits the Notch pathway by influencing upstream signaling rather than repressing downstream Notch target genes (58). In this study, we provide several lines of evidence that suggests that Ikaros functions downstream of ICN in its regulation of at least a subset of Notch target genes. First, we have shown that Ikaros down-regulates expression of Hes1, Deltex1, and pTa without altering ICN expression, and secondly, we have demonstrated that DP thymocytes require Ikaros expression to properly repress Notch target genes even in the absence of detectable ICN. In addition, LRF and Ikaros appear to function in different cellular subsets. Whereas the phenotype of the LRF-deficient mice suggests that LRF functions primarily in hematopoietic stem cells and common lymphoid progenitors, we have demonstrated that Ikaros' role is restricted to committed T cell populations in the thymus.
FIGURE 8.
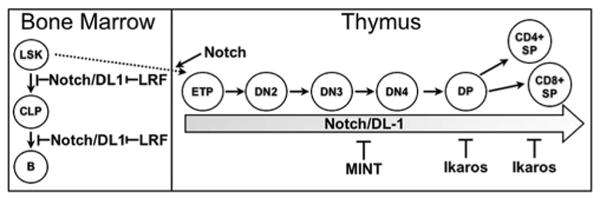
Model depicting corepressors involved in Notch target gene expression throughout hemato lymphoid development. Diagram depicts the different transcriptional regulators involved in negative regulation of Notch target gene expression throughout various stages of hemato lymphoid development. LRF opposes Notch in BM progenitors. MINT represses Notch target gene expression in DN thymocytes; whereas, Ikaros is required for repression of Notch target genes in DP and SP thymocytes. LRF controls Notch target gene expression by modulating Notch signaling, whereas, both MINT and Ikaros regulate Notch target gene expression downstream of ICN.
Taken together, we have demonstrated a role for Ikaros in the tight repression of Notch target genes during thymocyte development. This role manifests itself most dramatically within the DP thymocyte subpopulation in which Notch target gene expression must be tightly repressed. In the absence of Ikaros, Notch target gene expression can be observed in this subset in the absence of detectable ICN. It is interesting to speculate that the role of Ikaros in Notch target gene repression in DP thymocytes could be the first link in the chain that leads to the transformation that occurs within this population with 100% penetrance in Ikaros-deficient mice (2, 3).
Acknowledgments
We thank Dr. Melissa Brown for use of the iCycler as well as valuable advice and support, Dr. Juan Carlos Zuniga-Pflucker for the OP9-DL1 cells, Dr. Warren Pear for the DNMAML1 retroviral construct, and Jennifer Lehman for excellent animal care and genotyping.
Footnotes
This work was supported by the National Institutes of Health (R01 CA104962-01A1) awarded to S. Winandy. S. Chari was supported by The Cellular and Molecular Basis of Disease Training Grant, funded by the National Institutes of Health (T32 NIH T32 GM08061).
Abbreviations used in this paper: ICN, intracellular cleaved Notch; DN, double-negative; DP, double-positive; MAML, Mastermind-like; BGS, bovine growth serum; Pen-Strep, penicillin-streptomycin; HD, heterodimerization domain; PEST, Proline, Glutamine, Serine, and Threonine-rich domain; qRT-PCR, quantitative real-time RT-PCR; T-ALL, T-acute lymphoblastic leukemia; GSI, γ-secretase inhibitor; BM, bone marrow; RBP-J, recombination signal binding protein for immunoglobulin κJ; MSCV, murine stem cell virus.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 2.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 3.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 4.Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S. Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol Cell Biol. 2005;25:1645–1654. doi: 10.1128/MCB.25.5.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR, Thibault C, Barths J, Ghysdael J, et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006;26:209–220. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 7.Beverly LJ, Capobianco AJ. Targeting promiscuous signaling pathways in cancer: another Notch in the bedpost. Trends Mol Med. 2004;10:591–598. doi: 10.1016/j.molmed.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Nieva P, Santos J, Fernandez-Piqueras J. Defective expression of Notch1 and Notch2 in connection to alterations of c-Myc and Ikaros in γ-radiation-induced mouse thymic lymphomas. Carcinogenesis. 2004;25:1299–1304. doi: 10.1093/carcin/bgh124. [DOI] [PubMed] [Google Scholar]
- 9.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 10.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 12.Kathrein KL, Chari S, Winandy S. Ikaros directly represses the Notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem. 2008;283:10476–10484. doi: 10.1074/jbc.M709643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron M. An overview of the Notch signalling pathway. Semin Cell Dev Biol. 2003;14:113–119. doi: 10.1016/s1084-9521(02)00179-9. [DOI] [PubMed] [Google Scholar]
- 14.Felli MP, Maroder M, Mitsiadis TA, Campese AF, Bellavia D, Vacca A, Mann RS, Frati L, Lendahl U, Gulino A, Screpanti I. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int Immunol. 1999;11:1017–1025. doi: 10.1093/intimm/11.7.1017. [DOI] [PubMed] [Google Scholar]
- 15.Kitamoto T, Takahashi K, Takimoto H, Tomizuka K, Hayasaka M, Tabira T, Hanaoka K. Functional redundancy of the Notch gene family during mouse embryogenesis: analysis of Notch gene expression in Notch3-deficient mice. Biochem Biophys Res Commun. 2005;331:1154–1162. doi: 10.1016/j.bbrc.2005.03.241. [DOI] [PubMed] [Google Scholar]
- 16.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 17.Bellavia D, Mecarozzi M, Campese AF, Grazioli P, Talora C, Frati L, Gulino A, Screpanti I. Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 2007;26:1670–1680. doi: 10.1038/sj.emboj.7601626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deftos ML, He YW, Ojala EW, Bevan MJ. Correlating notch signaling with thymocyte maturation. Immunity. 1998;9:777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molnar A, Wu P, Largespada DA, Vortkamp A, Scherer S, Copeland NG, Jenkins NA, Bruns G, Georgopoulos K. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J Immunol. 1996;156:585–592. [PubMed] [Google Scholar]
- 20.Choi JW, Pampeno C, Vukmanovic S, Meruelo D. Characterization of the transcriptional expression of Notch-1 signaling pathway members, Deltex and HES-1, in developing mouse thymocytes. Dev Comp Immunol. 2002;26:575–588. doi: 10.1016/s0145-305x(01)00095-7. [DOI] [PubMed] [Google Scholar]
- 21.Hasserjian RP, Aster JC, Davi F, Weinberg DS, Sklar J. Modulated expression of notch1 during thymocyte development. Blood. 1996;88:970–976. [PubMed] [Google Scholar]
- 22.Aster JC, Pear WS. Notch signaling in leukemia. Curr Opin Hematol. 2001;8:237–244. doi: 10.1097/00062752-200107000-00010. [DOI] [PubMed] [Google Scholar]
- 23.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jκ/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci USA. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, Nakafuku M. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- 30.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Israel A. [Delta]-1 activation of Notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang YH, Li D, Winoto A, Robey EA. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc Natl Acad Sci USA. 2004;101:4936–4941. doi: 10.1073/pnas.0401133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 34.Lin YW, Nichols RA, Letterio JJ, Aplan PD. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 38.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J Biol Chem. 2002;277:50612–50620. doi: 10.1074/jbc.M209529200. [DOI] [PubMed] [Google Scholar]
- 42.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 43.Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, Wolfe MS, Turka LA, Pear WS. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- 44.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krebs LT, Deftos ML, Bevan MJ, Gridley T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev Biol. 2001;238:110–119. doi: 10.1006/dbio.2001.0408. [DOI] [PubMed] [Google Scholar]
- 48.Huang EY, Gallegos AM, Richards SM, Lehar SM, Bevan MJ. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J Immunol. 2003;171:2296–2304. doi: 10.4049/jimmunol.171.5.2296. [DOI] [PubMed] [Google Scholar]
- 49.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. The CD8α gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 50.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2β determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Urban JA, Winandy S. Ikaros null mice display defects in T cell selection and CD4 versus CD8 lineage decisions. J Immunol. 2004;173:4470–4478. doi: 10.4049/jimmunol.173.7.4470. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by δ-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 53.Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A, Maryanski JL, Zuniga-Pflucker JC. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol. 2004;172:5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 54.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 56.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 57.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190:1201–1214. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pear WS, Aster JC. T cell acute lymphoblastic leukemia/lymphoma: a human cancer commonly associated with aberrant NOTCH1 signaling. Curr Opin Hematol. 2004;11:426–433. doi: 10.1097/01.moh.0000143965.90813.70. [DOI] [PubMed] [Google Scholar]
- 60.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early α β lineage thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 61.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 62.Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, et al. Constitutive activation of NF-κB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]


