Summary
During protein synthesis, it is often necessary for the ribosome to form a complex with a membrane-bound channel, the SecY/Sec61 complex, in order to translocate nascent proteins across a cellular membrane. Structural data on the ribosome-channel complex are currently limited to low-resolution cryo-electron microscopy maps, including one showing a bacterial ribosome bound to a monomeric SecY complex. Using that map along with available atomic-level models of the ribosome and SecY, we have determined, through molecular dynamics flexible fitting (MDFF), an atomic-resolution model of the ribosome-channel complex. We characterized computationally the sites of ribosome-SecY interaction within the complex and determined the effect of ribosome binding on the SecY channel. We also constructed a model of a ribosome in complex with a SecY dimer by adding a second copy of SecY to the MDFF-derived model. The study involved 2.7-million-atom simulations over altogether nearly 50 ns.
Introduction
While practically all protein synthesis begins at the ribosome, one of the most complex molecular machines present in all organisms, the direction a newly formed or forming protein takes next varies. A number of proteins are synthesized directly into the cytoplasm where they remain. However, for proteins not destined for the cytoplasm, including, e.g., secretory and membrane proteins, the possession of an N-terminal signal sequence targets them to the membrane-bound protein-conducting channel, the SecY (as it is known in the bacterial and archaeal cytoplasmic membrane) or Sec61 (in the eukaryotic endoplasmic reticulum membrane) translocon (Osborne et al., 2005; Rapoport, 2007; Papanikou et al., 2007; Driessen and Nouwen, 2008; Mandon et al., 2009). When this targeting occurs prior to the end of translation, in a process known as co-translational translocation, the ribosome must dock to the channel and insert the nascent chain while it is still being synthesized (Halic and Beckmann, 2005).
Significant work has been carried out to elucidate the nature of the ribosome-translocon complex, including its arrangement and the oligomeric state of the translocon, although multiple hypotheses remain. Fluorescence-quenching experiments led to the suggestion that the ribosome forms a tight seal with a large channel (40-60 Å wide when open) (Crowley et al., 1993; 1994; Hamman et al., 1997). A number of low-resolution cryo-electron microscopy (cryo-EM) maps, however, revealed a gap between the ribosome and the translocon; from these maps it was also concluded that the eukaryotic translocon is comprised of three to four Sec61s with the pore likely being formed at their interface (Beckmann et al., 1997; Ménétret et al., 2000; Beckmann et al., 2001; Morgan et al., 2002; Ménétret et al., 2005). A higher resolution map of the complex displayed a dimer of SecY bound to a translating ribosome, leading to the hypothesis that two separate channels in the monomers could fuse to form a larger one in the dimer (Mitra et al., 2005). More recent cryo-EM maps have shown both a monomer of SecY (Ménétret et al., 2007) and a monomer of Sec61 (Ménétret et al., 2008) beneath a non-translating ribosome, suggesting that the SecY/Sec61 monomer is the active channel and that dimers or tetramers may form for reasons other than forming a larger channel. The location of SecY/Sec61 in the maps containing a monomer does not match that of either copy of SecY in the map containing a dimer, leaving open the question of the proper arrangement of ribosome and translocon and whether the arrangement may change depending on the functional state of the ribosome.
Despite a wealth of low-resolution structural data on ribosome-translocon complexes, high-resolution data are only available for the components (i.e., ribosome and translocon) separately. The crystal structure of a single SecY (the archaeal SecYEβ) led to the hypothesis that instead of being formed at the interface of monomers, the channel exists within a single monomer (see Figure 1A,B) (van den Berg et al., 2004). The structure of a SecY monomer displays an hourglass-shaped pore with a constriction region in the center formed by a handful of hydrophobic residues, the so-called pore ring. During translocation, the nascent chain is in close contact with the pore ring (Cannon et al., 2005; Bol et al., 2007), which has been shown to be capable of expanding to accommodate the incoming polypeptide in simulations (Gumbart and Schulten, 2006; Tian and Andricioaei, 2006). The periplasmic half-channel is also blocked by a small mobile helical “plug” domain (van den Berg et al., 2004). For the exit of membrane proteins into the lipid bilayer, a lateral gate is seen at the interface between two pseudosymmetric halves of SecY composed of transmembrane segments (TMs) 1-5 and 6-10 (van den Berg et al., 2004; White and von Heijne, 2008). The insertion of a signal sequence at the lateral gate initiates channel opening apparently by destabilizing the interactions holding the plug in place, causing it to move out of the channel (Plath et al., 1998; Smith et al., 2005; Li et al., 2007; Gumbart and Schulten, 2008).
Figure 1.
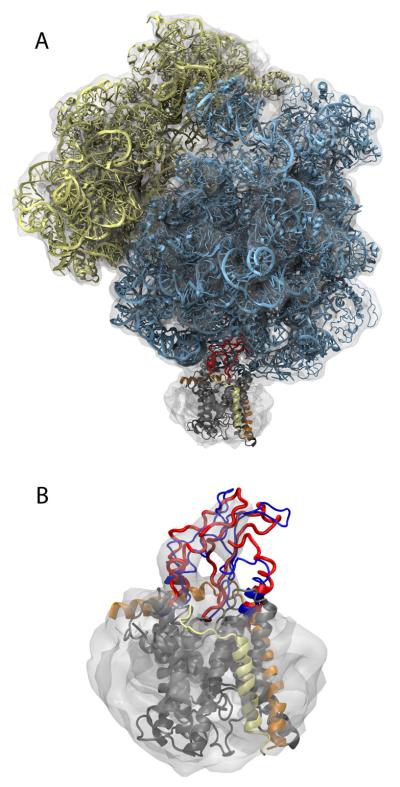
SecYEβ monomer, alone and in complex with the ribosome. (A) SecYEβ viewed from the cytoplasmic side. SecY is in grey with loop 6/7 highlighted in purple, loop 8/9 in red, the plug in blue, and the lateral gate in green. SecE is shown in orange and Secβ in yellow. (B) SecYEβ viewed from the plane of the membrane in the same representation as in (A). (C) Simulation system assumed for the SecYEβ monomer in complex with a ribosome. SecYEβ is shown as in parts A and B. The large subunit of the ribosome is shown in cyan and the small subunit in yellow. The membrane is in yellow with its phosphorus atoms in orange. The surrounding water is indicated as a light blue background. Ribosome, channel, and membrane are also shown in Movie S1 in Supplemental Data.
In order to address the nature of the ribosome-translocon complex, in particular the role of the Escherichia coli ribosome in channel opening, we have modeled and simulated an atomic-level structure of the ribosome in complex with a SecYEβ monomer. The model of the complex was developed by fitting individual structures of the ribosome and the channel into the cryo-EM map of a ribosome-SecY-monomer complex (Ménétret et al., 2007) using the molecular dynamics flexible fitting (MDFF) method (Trabuco et al. (2008; 2009); see Methods), which was recently successfully applied for resolving the structure of a functional intermediate of the ribosome (Villa et al., 2009). By simulating the resulting complex in its native membrane/water environment, we are able to characterize the atomic-scale interactions that bind the ribosome to the channel. We observe a slight destabilizing effect of the bound ribosome on SecY's plug, the destabilization being enhanced through the inclusion of a second copy of SecYEβ in an additional simulation. The association between the ribosome and channel is only minimally disturbed during simulated translocation of a polypeptide from the exit tunnel into SecY; however, mutating conserved arginines in the SecY binding loops to glutamate is found to decrease the strength of association. In total, we carried out nearly 50 ns of simulations of the 2.7-million-atom system, which resulted in one of the largest molecular dynamics simulations published to date. The simulation period is sufficient to apply the MDFF method (Trabuco et al., 2008; Villa et al., 2009; Hsin et al., 2009) and to relax and equilibrate the ribosome-SecY interface.
Results
We began simulations of the ribosome-channel complex by fitting the crystallographic structures of the E. coli ribosome and the Methanococcus jannaschii SecYEβ, both with modifications (see Methods), into the cryo-EM map of the ribosome-SecY-monomer complex (Ménétret et al., 2007). We then carried out equilibrium simulations of the complex, analyzing both the interactions between the ribosome and SecYEβ as well as the effect of the ribosome on SecY's structure and dynamics. We examined the change in the interactions between SecY and the ribosome upon mutation of conserved residues in SecY's ribosomal binding loops, a mutation which is known to eliminate binding between the ribosome and channel (Ménétret et al., 2007). We also simulated translocation of a partially extended and partially helical polypeptide through the exit tunnel and into the channel. Finally, we tested if the ribosome-SecY model derived permits placement of a second copy of SecY.
Flexible fitting of the atomic structures into the cryo-EM map
We flexibly fitted the initial model of the ribosome-channel complex into the cryo-EM map using MDFF (Trabuco et al., 2008; 2009) in multiple stages over the course of approximately 11 ns (see Methods). This simulation time is comparable with those needed in other applications of MDFF (Trabuco et al., 2008; Villa et al., 2009; Hsin et al., 2009). The resolution of the channel portion of the map is relatively low (17 Å compared to 9.6 Å for the ribosome), possibly due to a natural flexibility of the channel in the prepared samples (Ménétret et al., 2007); alternatively, the presence of bound lipids and detergents may reduce the apparent resolution of the channel, such as in the case of the yeast V-ATPase (Ménétret et al., 2008; Diepholz et al., 2008). The low resolution of the channel in the map provides insufficient detail to guide flexible fitting of the atomic structure of SecYEβ; therefore, we constrained most of SecYEβ during fitting, except for those parts which interact directly with the ribosome, which are well resolved in the map.
Loops 6/7 and 8/9 of SecY, i.e., the loops between helices 6 and 7 and 8 and 9, respectively, which insert into the ribosome's protein exit tunnel, were not constrained during fitting (see Figure 2). Loop 6/7 expands slightly, resulting in a maximum RMSD of 5.4 Å for the backbone compared to the initial structure. Loop 8/9 is more stable under the influence of the ribosome and the map, exhibiting a maximum RMSD of 4.4 Å. The largest change in SecY comes from the C-terminus, which shifts significantly to increase its interaction with ribosomal protein L24. The repositioning of part of L24 also allows it to interact with loop 6/7; the area of interaction between L24 and SecY increases over 50% during fitting, from 305 Å2 to 467 Å2.
Figure 2.
Molecular dynamics flexible fitting of the ribosome-translocon complex. (A) Fitted structure. The ribosome and SecYEβ are colored as in Figure 1 except that both loops 6/7 and 8/9 are shown in red. The cryo-EM map used for fitting is shown in grey, transparent, contoured at 1.67σ above the mean. (B) Fitting of SecYEβ. Only parts of SecY near the ribosome (e.g., loops 6/7, 8/9, and the C-terminus) were free during fitting. Blue represents the starting structure and red the final one. See Figure S1 for a stereo view and Movie S2 for an overview of the fitting procedure, both in Supplemental Data.
After fitting the ribosome and channel structures to the map, we prepared a system containing the complex with membrane, water, and ions, involving altogether 2.7 million atoms (see Methods). Initial equilibration of water and lipids was carried out for 1.5 ns, after which we allowed the entire system to move freely. Secondary structure restraints (as described in Trabuco et al. (2008)) were maintained for the ribosome-channel complex for an additional 2.5 ns. We simulated the complex for a total of 15.5 ns. This relatively short simulation time is sufficient to relax the components of the ribosome and SecY involved in interactions localized at the interface, which are the focus of this study.
Interactions between the ribosome and the channel
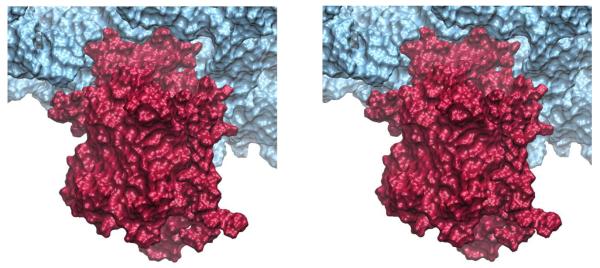
To characterize the connection between the ribosome and channel, shown in Figure 3, we monitored hydrogen bonds and hydrophobic/hydrophilic interactions that formed between them during equilibration of the full system (see Table 1). The first two connections (see Figure 4), involving the 6/7 and 8/9 cytosolic loops of SecY, contribute the majority of interactions; the C-terminus of SecY and part of SecE also contribute to the interactions. As expected, the nonessential Secβ forms no interactions with the ribosome (Kalies et al., 1998). On average there are 5-8 hydrogen bonds formed between the ribosome and each of loops 6/7 and 8/9. Primary contacts between loop 6/7 and the 23S rRNA involve Arg255 and Arg256 in SecY, along with Tyr248; interactions on the RNA side include both backbone and bases. Hydrogen bonds between ribosomal proteins and loop 6/7 of SecY are also observed. In particular, one discerns that Arg243 interacts strongly with residues 34 to 36 in L29, and that Gln261 and Ser262 interact with Ser34 and Gln36 of L29, respectively. Additionally, Tyr258 interacts with L24, and Gly254 with L23. In contrast to loop 6/7, loop 8/9-RNA hydrogen bonds almost exclusively engage the RNA backbone. The highly conserved Arg357 interacts most strongly with the 23S rRNA, while Gly359, Lys364, and Tyr365 interact less strongly.
Figure 3.
Insertion of loops 6/7 and 8/9 of SecY into the ribosome. The ribosome (blue) and SecYEβ (red) are shown as molecular surfaces. Loops 6/7 and 8/9 are near the top of the stereo image.
Table 1.
Prominent interactions between SecYEβ and the ribosome observed during simulation. Residue numbers are taken from E. coli with the exception of the starred residues (Arg369 in connection 2 and His427 and Ile430 in connection 4), which are not absolutely conserved between M. jannaschii and E. coli.
| SecYEβ residue | Ribosome residue | Connection | Interaction Type |
|---|---|---|---|
| Arg243 (SecY) | Ser34-Gly35 (L29) | 1 | H-bond |
| Arg243 (SecY) | Gln36 (L29) | 1 | hydrophilic |
| Val245 (SecY) | Val16 (L23) | 1 | hydrophobic |
| Tyr248 (SecY) | Gua1337 (23S) | 1 | H-bond |
| Ala249 (SecY) | Pro49 (L24) | 1 | hydrophobic |
| Lys250 (SecY) | His70 (L23) | 1 | H-bond |
| Gly254 (SecY) | His70 (L23) | 1 | H-bond |
| Arg255 (SecY) | Ura62 (23S) | 1 | H-bond |
| Arg256 (SecY) | Ura90 (23S) | 1 | H-bond |
| Tyr258 (SecY) | Pro49 (L24) | 1 | H-bond |
| Tyr258 (SecY) | Val48-Pro49 (L24) | 1 | Hydrophobic |
| Gln261 (SecY) | Ser34 (L29) | 1 | H-bond |
| Ser262 (SecY) | Gln36 (L29) | 1 | H-bond |
| His264 (SecY) | Gln38 (L29) | 1 | hydrophilic |
| Arg357 (SecY) | Gua1317-Cyt1319 (23S) | 2 | H-bond |
| Arg357 (SecY) | Gua1334-Cyt1335 (23S) | 2 | H-bond |
| Gly359 (SecY) | Ade1336 (23S) | 2 | H-bond |
| Lys364 (SecY) | Gua1317 (23S) | 2 | H-bond |
| Tyr365 (SecY) | Ade1392 (23S) | 2 | H-bond |
| Arg369* (SecY) | Val16-Ser17 (L23) | 2 | H-bond |
| Arg369* (SecY) | Ser21 (L23) | 2 | H-bond |
| Arg369* (SecY) | Glu25 (L23) | 2 | H-bond |
| Glu74 (SecE) | Lys26 (L23) | 3 | H-bond |
| Lys81 (SecE) | Leu93 (L23) | 3 | H-bond |
| Trp84 (SecE) | Phe26 (L29) | 3 | hydrophobic |
| Pro85 (SecE) | Phe26 (L29) | 3 | hydrophobic |
| His427* (SecY) | Asn52 (L24) | 4 | H-bond |
| Ile430* (SecY) | Ala50 (L24) | 4 | H-bond |
| Ile430* (SecY) | Pro47 (L24) | 4 | hydrophobic |
Figure 4.
Interactions between a SecYEβ monomer and the ribosome during simulation. On the left, all of SecYEβ, colored as in Figure 1, is shown with the relevant interactions numbered. The C-terminus is highlighted in blue. On the right, each site of interaction is shown in more detail. On top, Arg255, 256, and 357 are shown in a blue, space-filling representation. In the middle, residues from L23 (cyan), L29 (red) and SecE (orange) that interact are highlighted in licorice, colored according to their type (blue for basic, red for acidic, green for polar, and white for hydrophobic). On the bottom, hydrogen bonds between the C-terminus of SecY (blue) and L24 (cyan) are shown. Stereo views of all parts are given in Figure S2 in Supplemental Data. Detailed 360° views of the connections are provided in Supplemental Data (Movie S3).
A third ribosome-channel connection was recognized from the EM map in which SecE is seen to contact the ribosome. In simulation, Lys81 of SecE interacts predominantly with the C-terminus of L23, although it also intermittently forms a salt bridge with Glu52. In addition, residues Trp84 and Pro85 are found to interact with Phe26 of L29. The contacts all fall within the conserved cytoplasmic domain of SecE (approximately residues 71-89 in E. coli) recognized previously (Murphy and Beckwith, 1994; Matsuo et al., 2003). The observed interactions suggest that the conserved region of SecE is important for ribosome binding, in agreement with previous simulations which also suggested channel-partner binding as one of SecE's primary roles (Gumbart and Schulten, 2007).
A fourth ribosome-channel connection was observed between the C-terminus of SecY and L24. While present in the original fit of the complex, this connection became stronger during MDFF, as noted above. The interaction between the two proteins resembles the initial stages of a β sheet, with the most prevalent hydrogen bonds involving His437 and Ile440 of SecY and residues 50 to 55 in L24. The structure of SecY in complex with SecA, the bacterial post-translational translocation partner, also displays contacts between the C-terminal region of SecY and the channel partner, and mutation (Tyr429Asp) or deletion of this region is known to inhibit SecA-mediated translocation (Chiba et al., 2002; Mori et al., 2002; Zimmer et al., 2008). Based on the interactions observed here, the C-terminal region of SecY is relevant for ribosome binding as well.
In addition to interactions with the channel, a number of interactions between the ribosome and the membrane are formed. Helices H7, H9, H54, and H59 of the 23S rRNA contact the membrane, along with ribosomal proteins L23, L24, and L29 (see Figure S3 in Supplemental Data). The ribosome-lipid connection allows the ribosome to maintain its angle relative to the membrane plane, estimated to be 20° based on the cryo-EM density and, thus, to maintain also the gap between ribosome and channel (Ménétret et al., 2007). The ribosome-membrane contact is a feature of the overall placement of SecY as seen in the EM map and should not be affected by long-time relaxation; however, the contact may be superseded by interactions with a second copy of SecY, as experiments have indicated additional copies of the channel may be present in functioning ribosome-translocon complexes (Schaletzky and Rapoport, 2006).
Effects of ribosome binding on the channel
To determine the effects of the ribosome, if any, on the channel, we compared the behavior of the channel alone and in complex with the ribosome. It was found that on a time scale of 14 ns, a difference in behavior is small but recognizable. The root mean-square fluctuations (RMSF) for all residues in SecY are presented in Figures 5A and B for the ribosome-bound and unbound channels. The most obvious difference is seen for the ribosome-binding loops. In the ribosome-free SecY, loops 6/7 and 8/9 have significant flexibility, as reflected in the RMSF; this flexibility has been observed also in other simulations of SecY on the same time scale (Haider et al., 2006). In contrast, in the ribosome-bound channel, loops 6/7 and 8/9 are less mobile. Disturbances due to the ribosome are primarily found on the cytoplasmic side of the channel, due to its proximity to the ribosome. The plug, which serves to close the channel on its periplasmic side, exhibits greater fluctuations when the ribosome is bound compared to when it is not. Although the corresponding difference in RMSF is small, it is in line with previous observations for plug-deletion mutants simulated on the same time scale (Gumbart and Schulten, 2008).
Figure 5.
Effects of ribosome binding on SecY. (A) Root mean-square fluctuations (RMSF) for the ribosome-bound SecY (red) and SecY alone in the membrane (black). The RMSF was calculated over the last 12 ns of the simulation. The positions of TMs 1 through 10 are indicated in the plot. (B) SecYEβ, with SecY colored according to the difference of the two RMSF curves in (A). Red represents regions which fluctuate more in the ribosome-bound SecY compared to SecY alone. (C) RMSF for SecY alone with loops 6/7 and 8/9 free (black) and immobilized (red).
The increased fluctuations of the plug are supported by Principal Component Analysis (PCA), a method which finds the dominant correlated motions present in a simulation trajectory that are often indicative of the long-time behavior of biopolymers beyond the sampled time scale. The first mode computed via PCA displays a downward motion of the plug, away from the channel center, in the simulation of the ribosome-SecY complex. However, in the simulation of SecY alone, the first mode given by PCA displays a motion of the plug towards the lateral gate, maintaining the closed state of the channel (see Figure S4 in Supplemental Data).
We also simulated SecY alone, but with its ribosome-binding loops 6/7 and 8/9 held fixed. Fixing the loops mimics the ribosome's restraining effect, allowing one to determine the relative importance of the mechanical and electrostatic effects of ribosome binding. By comparing their RMSFs one finds that the disturbance to the plug is even larger when the loops are held fixed compared to when the ribosome is bound; there is also a small disturbance to one half of the lateral gate (TM7; see Figure 5C). Interactions (primarily hydrophobic) of the plug with the rest of the channel over the course of the simulation were also examined; the frequency of interactions between plug and TM3 decreased significantly with respect to the simulation in which the loops were unrestrained (see Table S1 in Supplemental Data). This decrease is likely due to the increased fluctuations of TM3 and may explain in part the increased fluctuations of the plug (Figure 5C). As in the case of the ribosome-SecY complex, PCA reveals a downward motion of the plug in the first mode, actually a larger one than for the ribosome-SecY complex (see Figure S4C in Supplemental Data). Thus, a mechanical interaction between the channel partner and SecY, in which SecY's binding loops are restrained, is sufficient to increase fluctuations in the plug. However, electrostatic interactions, including additional contacts with SecE or the C-terminus of SecY, may also play a role in destabilizing the closed state of the channel.
Mutations in the ribosome-binding loops of SecY
Loops 6/7 and 8/9 in SecY form the most prominent interactions with the ribosome by inserting into the ribosome's polypeptide exit tunnel. A number of positively charged residues within these loops are known to be required for ribosome binding to the channel in both bacteria and eukaryotes (Raden et al., 2000; Cheng et al., 2005; Ménétret et al., 2007). Specifically, mutating Arg255 and Arg256 in loop 6/7 or the highly conserved Arg357 in loop 8/9 to glutamate abrogates ribosome binding in E. coli (Ménétret et al., 2007); the locations of the three amino acids are indicated in Figure S5A in Supplemental Data. To characterize the effects of these mutations on the ribosome-channel complex, we mutated all three residues, Arg255, Arg256, and Arg357 in SecY, to glutamate and simulated the resulting complex for 14 ns.
The effect of the three mutations is most pronounced in the change of the electrostatic potential of SecY. A significant positive potential near loops 6/7 and 8/9, shown in Supplemental Figure S5B, is eliminated as a result of the mutations, indicating why the negatively charged ribosome does not bind to the SecY mutant. In simulation of the ribosome-SecY-mutant complex, hydrogen bonds between loops 6/7 and 8/9 of SecY and the ribosome were reduced, particularly those involving the mutated residues of SecY. Further details on the impact of mutations on the electrostatic potential of SecY and on the interactions between SecY and the ribosome are provided in Supplemental Data.
Translocation of a polypeptide from ribosome to channel
An obvious question that arises from the modeled structure of the ribosome-channel complex is if protein translocation can proceed without disrupting the connection between ribosome and channel. The potential for disruption is particularly high within the exit tunnel, where loops 6/7 and 8/9 of SecY insert. From the original cryo-EM map, two conformations of the tip of loop 6/7 were found, one with 70% occupancy, which was used in the initial model, and one with 30% occupancy, which was proposed to leave a greater opening in the exit tunnel for polypeptide translocation (Ménétret et al., 2007). In our simulations of the ribosome-channel complex, loop 6/7 maintained its initial conformation. To determine if this conformation would hinder the passage of a polypeptide chain through the exit tunnel, we simulated, using steered MD and a reduced model of the ribosome (see Methods), the translocation of an alanine polypeptide that included six extended residues followed by 20 helical residues (Ala26). Ala26 was positioned initially such that only the N-terminus protruded from the ribosomal exit tunnel (see Figure 6A). Previous studies using normal mode analysis of the ribosome exit tunnel suggested that motions inherent to the exit tunnel facilitate polypeptide translocation (Kurkcuoglu et al., 2008). However, all residues in the reduced ribosome model used here are restrained to their initial configuration (see Methods), preventing any conformational changes in the ribosome that may take place during polypeptide translocation. Thus, only conformational changes in SecY, and in its interactions with the ribosome, are accounted for.
Figure 6.
Translocation of an alanine polypeptide through the ribosome's exit tunnel and into the channel. In all panels, the ribosome is shown as cyan space-filling spheres and SecYEβ is shown as in Figure 1. The translocating polypeptide, Ala26, is shown in green. The system is shown at (A) t=0 ns, with only the tip of Ala26 visible outside the ribosome, (B) t=3 ns, (C) t=6 ns, and (D) t=9 ns. The full translocation process is shown in Movie S5 in Supplemental Data.
Over the course of the 10-ns simulation, Ala26 was translocated 50 Å. The short simulation time, sufficient for the present purposes of exploring an undetermined ribosome-SecY translocation route, is comparable to simulation times used in prior translocation studies of an individual SecY (Gumbart and Schulten, 2006; 2008). As shown in Figure 6, despite the gap between the channel and the exit tunnel of the ribosome, the polypeptide can easily bridge the two separate environments. Additionally, the exit tunnel is large enough to accommodate the two loops of SecY as well as an α-helix. Interactions between the loops and the ribosome are only minimally affected by the passing polypeptide. The average of 5.5 hydrogen bonds between loop 6/7 of SecY and the ribosome is maintained during translocation while hydrogen bonds between loop 8/9 and the ribosome decrease from an average of 7.5 to 5.5. The original conformation of loop 6/7 is maintained during the simulation and does not impede translocation of Ala26.
Binding of the ribosome to a SecY dimer
As noted in the introduction, both monomers and dimers of SecY bound to the ribosome have been seen in cryo-EM maps, but the reasons for the formation of dimers are still unknown (Breyton et al., 2002; Mitra et al., 2005; Ménétret et al., 2007). Based on the map of the SecY dimer in complex with the ribosome, a “front-to-front” orientation of SecY monomers, in which the lateral gates face each other, was proposed (Mitra et al., 2005). In this orientation, both gates could open in order to form a larger channel. Alternatively, a “back-to-back” orientation of SecY monomers in which the two SecE TMs are in contact had also been suggested (Breyton et al., 2002; van den Berg et al., 2004; Gumbart and Schulten, 2006). Based upon our model of the ribosome-SecY-monomer complex, the second copy of SecY in the front-to-front orientation could only interact with helix H59 of 23S due to the gap between the ribosome and the channel on its front side. In the back-to-back orientation, however, the second copy of SecY can form a number of interactions with both ribosomal proteins and RNA (see Figure 7 and Figure S6 in Supplemental Data). Therefore, we chose to simulate the complex formed between the ribosome and a back-to-back SecYEβ dimer for 15.5 ns. We characterized the interactions formed over time between the ribosome and the SecY dimer, finding an increased stability of the ribosome-SecY-dimer complex compared to the ribosome-SecY-monomer complex. However, as the placement of the second SecY is neither supported nor guided by EM density, the model arrived at is hypothetical and serves here mainly to illustrate the possible nature of a ribosome-SecY-dimer complex. The properties of the hypothetical complex are described in detail in Supplemental Data.
Figure 7.
Complex between the ribosome and a SecY dimer. The ribosome and two copies of SecYEβ in a back-to-back conformation are shown.
Discussion
By combining separate atomic-resolution structures with a cryo-EM density map, we have modeled the complex formed by the ribosome and the translocon preceding protein translocation. This complex, at nearly 2.7 million atoms including water and membrane, is among the largest simulated to date. Simulations are reaching the million-atom mark with increasing frequency, however (Klein and Shinoda, 2008); prominent examples include atomic-scale simulations of a virus (1 million atoms) (Freddolino et al., 2006), arrays of light-harvesting proteins (1 million) (Chandler et al., 2008) and BAR domains (2.3 million) (Yin et al., 2009), the flagellum (2.4 million) (Kitao et al., 2006), and the ribosome (1.9-2.64 million) (Sanbonmatsu et al., 2005; Sanbonmatsu and Tung, 2007; Ishida and Hayward, 2008). Our simulations of the ribosome-channel complex began with flexible fitting via MDFF of the crystal structures of the ribosome and channel to a cryo-EM map in order to produce a model in agreement with the physiological state of the complex in the cryo-EM experiments. Because the fitting is also an MD simulation itself, the resulting structure is stereochemically accurate and thus suitable for further simulations. Our subsequent simulations revealed not only the atomic-level details of the interactions between the ribosome and the channel, but also how the ribosome can prepare the channel for translocation. Given the large size of the system investigated, computer power limited overall simulation time to 50 ns. However, the availability of the EM map to construct the complex and the focus on local properties (ribosome-SecY interface) made a study over the limited time scale feasible.
Interactions between the ribosome and channel observed during equilibration of the complex are limited to four primary connections, namely loops 6/7 and 8/9 of SecY with the 23S rRNA as well as ribosomal proteins L23, L24, and L29; the C-terminus of SecY with L24; and SecE with L23 and L29. Three of these four connections have analogues in the recently solved SecA-SecY structure, the one involving SecE being absent (Zimmer et al., 2008); additionally, the C-terminus of SecY is known to be required for SecA-mediated translocation (Chiba et al., 2002; Mori et al., 2002). A cryo-EM map of a eukaryotic 80S ribosome bound to Sec61 displays many features similar to the bacterial system studied here, particularly the orientation of the channel beneath the ribosome and the insertion of loops 6/7 and 8/9 into the ribosome's exit tunnel (Ménétret et al., 2008). Furthermore, none of the four connections involve ribosomal signatures, i. e. proteins or rRNA residues present in one domain of life but not in others (Roberts et al., 2008). Thus, it appears that most of the sites of interaction on the channel are common to all organisms, even for different channel partners.
Interactions between the ribosome and the channel might be enhanced through the presence of a second copy of SecYEβ. Modeled in a hypothetical back-to-back configuration, the second copy forms a number of additional contacts to the ribosome that stabilize the complex (see Supplemental Data). The placement of the second copy and the contacts formed agree with a previous cryo-EM map of a Sec61-TRAP-ribosome complex (Ménétret et al., 2005), although the number of Sec61 copies present in that map has recently been questioned (Ménétret et al., 2008). However, experiments have demonstrated that only one in four Sec61s is protected from proteases by ribosome binding, even at high ribosome concentrations, suggesting that the ribosome may bind four copies of Sec61 but interacts asymmetrically with them (Kalies et al., 2008). In our simulation of the SecY dimer, loops 6/7 and 8/9 of the second copy rest on the surface of the ribosome, possibly exposing it enough to be accessible to proteases, whereas the copy with the loops in the ribosome's exit tunnel would remain protected. Crosslinking experiments have demonstrated that SecA also interacts with a SecY dimer asymmetrically, binding to one copy of SecY but inserting the nascent chain into the other (Osborne and Rapoport, 2007).
It was also found that binding of the ribosome has a small but distinct effect on the channel's fluctuations. In particular, fluctuations of the plug are larger when the ribosome is bound than when it is not. This effect is noticeably increased by the presence of a SecYEβ dimer beneath the ribosome, as expected from previous simulations (Gumbart and Schulten, 2006). By simulating SecY alone with loops 6/7 and 8/9 immobilized, it was demonstrated that the increased fluctuations in the plug are primarily a result of restraining these loops, as opposed to more specific interactions with the ribosome. Two recent structures of SecY in complex with the channel partner SecA (Zimmer et al., 2008) as well as with a Fab fragment (Tsukazaki et al., 2008) also illustrate the effects of restraining loops 6/7 and 8/9 of SecY. In both structures the lateral gate is slightly opened, while in the former the plug's position is also shifted. If specific interactions with a channel partner were required to “pre-activate” the channel, one would not expect a Fab fragment to have a similar effect. However, specific interactions between the ribosome and the channel could serve to establish the proper orientation of the channel preceding translocation.
Altogether, our results support the idea that the monomeric SecY is the functional channel. The channel is well positioned below the ribosome to receive the exiting polypeptide chain as shown by the simulated translocation of an alanine polypeptide. Although loops 6/7 and 8/9 of SecY insert into the exit tunnel of the ribosome, they do not interfere with the translocation of the polypeptide. Additionally, interactions between the ribosome and channel involve conserved regions in SecY and SecE, suggesting that the interactions are representative of those in vivo. The gap between the ribosome and the channel persists throughout the simulation, in agreement with the assertion that the membrane seal is formed within a single channel as opposed to at the ribosome-channel interface, as suggested by other models (Hamman et al., 1997). Nonetheless, experiments have demonstrated that additional copies of Sec61 can increase their affinity for ribosomes (Schaletzky and Rapoport, 2006), in agreement with our results for the SecY dimer. However, as neither nascent chain nor other channel partners were present in the cryo-EM map used, the possibility that their presence alters the connection between the ribosome and channel cannot yet be ruled out.
Experimental Procedures
Model of the E. coli ribosome-channel complex
Before simulations of the ribosome-channel complex could be carried out, an atomic-scale model of the complex had to be built. While a model of the E. coli ribosome has been developed in Ménétret et al. (2007), we chose as a starting point a model developed in our lab. This model is more complete than that in Ménétret et al. (2007) and has already been used for simulations (Trabuco et al., 2008; 2009). Briefly, our model of the ribosome is based on the 3.22-Å crystal structure from Berk et al. (2006) (PDB 2I2V/2I2U) with the L1 protuberance and the A-site finger of the 23S rRNA modeled and inserted into the structure. The model is described in detail in the Supplemental Data of Trabuco et al. (2008).
The model of the channel, taken from Ménétret et al. (2007), is based on the crystal structure of the archaeal SecYEβ (PDB 1RHZ) with two modifications. First, loops 6/7 and 8/9 of SecY, which insert into the ribosome's exit tunnel and are longer in the E. coli SecY than the archaeal one, are extended with insertions from the bacterial SecY sequence. Second, SecE's N-terminal amphipathic helix is mutated to match the E. coli sequence (residues 2 to 26 in M. jannaschii corresponding to residues 63 to 87 in E. coli). As SecY and SecE are homologous between archaea and bacteria, residue numbers from E. coli are used in the text where possible; sequence alignments for the channel components can be found in the Supplementary Materials of van den Berg et al. (2004). Secβ and SecG are not homologous (SecG in particular has two TM helices whereas Secβ has only one), although they exist at the same location in their respective complexes. To produce the initial model of the complex, our ribosome model was fitted as a rigid body to that of Ménétret et al. (2007) and then combined with these authors' model of SecYEβ.
Molecular dynamics flexible fitting
In the next step, our model of the ribosome-channel complex was fitted into the cryo-EM map of the ribosome-SecY-monomer complex (Ménétret et al., 2007) using the molecular dynamics flexible fitting (MDFF) method (Trabuco et al., 2008). The MDFF method involves a molecular dynamics (MD) simulation in which external forces proportional to the cryo-EM density gradient are applied, driving atoms into high-density regions of the EM map. Furthermore, secondary structure restraints are applied to protein and RNA molecules in order to prevent structural distortions (Trabuco et al., 2008; 2009). The ribosome structure was fitted in multiple stages using the approach previously employed (Trabuco et al., 2008; Villa et al., 2009); the channel was constrained during these stages. Subsequently, residues at the ribosome-channel interface were fitted to the cryo-EM density while the rest of the structure was constrained (see Figure 2). Simulation times needed for MDFF are typically ~10 ns (Trabuco et al., 2008; Villa et al., 2009; Hsin et al., 2009).
Building the simulation system
The full simulation system was built in several stages. First, crystallographic ions associated with the ribosome, 172 Mg2+ and one Zn2+, were placed into the new, fitted structure. The ions' new positions were determined by fitting the coordinating phosphate atoms from the crystal structure to the new model. An additional 1,826 Mg2+ ions were placed in and around the ribosome by the GPU-accelerated code cIonize, which places each ion at a minimum in the electrostatic potential, recalculated after each placement (Stone et al., 2007).
Solvation of the ribosome took place in three steps. First, the primary coordination shell of each Mg2+ ion was completed by placing up to eight water molecules in vacant coordination sites (Eargle et al., 2008). Next, 24,210 water molecules were placed into internal cavities within the ribosome with DOWSER (Zhang and Hermans, 1996), using the Dowser plugin of VMD (Humphrey et al., 1996), which extends DOWSER to support systems containing RNA. Finally, the system was solvated using the program Solvate, which placed an additional 330,163 water molecules in and around the ribosome-channel complex (Grubmüller et al., 1996).
In the final steps, we built a POPC membrane of size 290 Å × 280 Å and placed the ribosome-channel complex such that the hydrophobic belt of SecYEβ corresponds to the membrane interior. Water that overlapped with the membrane was removed. The system was solvated with an additional 330,359 water molecules, this time using the Solvate plugin of VMD (Humphrey et al., 1996). Finally, K+ and Cl− ions were added to establish a concentration of 100 mM. The final system size was 300 × 285 × 335 Å3 and contained 2,679,727 atoms.
Simulation protocols
All molecular dynamics simulations were performed using NAMD 2.7b1, which includes options for grid-steered molecular dynamics as well as internal coordinate restraints (Phillips et al., 2005; Wells et al., 2007). The CHARMM27 force field with the CMAP correction terms was used for all simulations (MacKerell et al., 1998; Foloppe and MacKerell Jr., 2000; MacKerell et al., 2004). A multiple time-stepping protocol was employed for evaluating the potential, with bonded interactions calculated every 1 fs, van der Waals and short-range electrostatic interactions every 2 fs, and long-range electrostatic interactions every 4 fs. For long-range interactions, the particle mesh Ewald (PME) method was used. The PME grid density was never less than 1/Å3. Periodic boundary conditions were assumed for all simulations.
Equilibration of each system was carried out in multiple stages. First, all atoms were constrained except those in the lipid tails, which were allowed to relax for 0.5 ns. In the next stage, the protein and RNA backbones were constrained while the membrane, water, and ions were equilibrated for 1 ns in the NpT ensemble (T=310 K, p=1 atm). Next, all atoms were freed, but secondary structure restraints maintained, for an additional 2.5 ns (see Trabuco et al. (2008) for definition of the restraints). Finally, all external forces were removed and the simulation was continued in the NVT ensemble. For the present study the main simulations involved 2.7 millions atoms and covered overall nearly 50 ns.
Simulated translocation
For the simulated translocation of a polypeptide from the ribosomal exit tunnel into the channel, a reduced system containing 208,000 atoms was used. Starting with the ribosome-SecY-monomer complex resulting from the end of the equilibration, residues of the ribosome within 20 Å of any part of SecYEβ were retained while those farther away were removed. Harmonic restraints with a force constant of k=5 kcal/mol·Å2 were applied to the backbone of all remaining ribosomal residues in order to maintain their structure. The 26-residue alanine polypeptide (Ala26) was then placed in the exit tunnel such that only the N-terminus was exposed at the tunnel's mouth. The simulations followed a protocol used previously for an individual SecY (Gumbart and Schulten, 2008), covering overall translocation times similar to those in the prior study.
After equilibration for 0.5 ns, the Cα-atom of the N-terminal residue of Ala26 was pulled at a velocity of 5 Å/ns toward the center of SecY using steered MD (Izrailev et al., 1997; Sotomayor and Schulten, 2007). The velocity was maintained by attaching the relevant atom to an imaginary point moving at constant velocity via a spring with force constant k=350 pN/Å2. The force required for translocation was typically 200-300 pN with a maximum of 800 pN. As the goal of the simulation was to determine any potential steric barriers presented by SecY to translocation of polypeptides of varying size, the secondary structure of the helix in Ala26 was maintained through weak (k=50 kcal/mol·rad2) dihedral restraints, which permitted intermittent distortions of the helix (Trabuco et al., 2008).
Analysis
VMD was used for analysis and figures (Humphrey et al., 1996). Hydrogen bonds were counted if the donor-acceptor distance was less than 3.5 Å and the angle formed by donor, hydrogen, and acceptor was greater than 145°. Electrostatic potential maps were calculated by solving the Poisson-Boltzmann equation using APBS (Baker et al., 2001) with a grid volume of less than 1 Å3 per point, mobile ions present at a concentration of 150 mM, and protein and solvent dielectric constants of 1.0 and 78.54, respectively.
Supplementary Material
Acknowledgements
The authors thank Chris Akey, J.F. Ménétret, and Tom Rapoport for providing the cryo-EM map and initial model of the complex and for helpful comments on the manuscript. This work was supported by the National Institutes of Health (R01-GM067887 and P41-RR05969) and the National Science Foundation (PHY0822613). The authors acknowledge computer time provided by the Texas Advanced Computing Center and the National Center for Supercomputing Applications through the National Resources Allocation Committee (MCA93S028).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The modeled ribosome-SecYEβ complex has been deposited in the Protein Data Bank (ID code XXXX).
Supplement
Supplemental data include 1 table, 10 figures, 5 movies and a detailed description of the interactions between the ribosome and the mutant SecY as well as between the ribosome and the SecY dimer.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Berk V, Zhang W, Pai RD, Cate JHD. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol R, de Wit JG, Driessen AJM. The active protein-conducting channel of Escherichia coli contains an apolar patch. J. Biol. Chem. 2007;282:29785–29793. doi: 10.1074/jbc.M702140200. [DOI] [PubMed] [Google Scholar]
- Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM, Jr., Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler D, Hsin J, Harrison CB, Gumbart J, Schulten K. Intrinsic curvature properties of photosynthetic proteins in chromatophores. Biophys. J. 2008;95:2822–2836. doi: 10.1529/biophysj.108.132852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon E, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Mori H, Ito K. Roles of the C-terminal end of SecY in protein translocation and viability of Escherichia coli. J. Bacteriol. 2002;184:2243–2250. doi: 10.1128/JB.184.8.2243-2250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Diepholz M, Venzke D, Prinz S, Batisse C, Flörchinger B, Rössle M, Svergun DI, Böttcher B, Féthière J. A different conformation for EGC stator subcomplex in solution and in the assembled yeast V-ATPase: Possible implications for regulatory disassembly. Structure. 2008;16:1789–1798. doi: 10.1016/j.str.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Eargle J, Black AA, Sethi A, Trabuco LG, Luthey-Schulten Z. Dynamics of recognition between tRNA and elongation factor Tu. J. Mol. Biol. 2008;377:1382–1405. doi: 10.1016/j.jmb.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foloppe N, MacKerell AD., Jr. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comp. Chem. 2000;21:86–104. [Google Scholar]
- Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Molecular dynamics simulations of the complete satellite tobacco mosaic virus. Structure. 2006;14:437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Grubmüller H, Heymann B, Tavan P. Ligand binding and molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- Gumbart J, Schulten K. Molecular dynamics studies of the archaeal translocon. Biophys. J. 2006;90:2356–2367. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbart J, Schulten K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- Gumbart J, Schulten K. The roles of pore ring and plug in the SecY protein-conducting channel. J. Gen. Physiol. 2008;132:709–719. doi: 10.1085/jgp.200810062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Hall BA, Sansom MSP. Simulations of a protein translocation pore: SecY. Biochemistry. 2006;45:13018–13024. doi: 10.1021/bi061013d. [DOI] [PubMed] [Google Scholar]
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40-60 Å during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- Hsin J, Gumbart J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Protein-induced membrane curvature investigated through molecular dynamics flexible fitting. Biophys. J. 2009;97:321–329. doi: 10.1016/j.bpj.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ishida H, Hayward S. Path of nascent polypeptide in exit tunnel revealed by molecular dynamics simulation of ribosome. Biophys. J. 2008;95:5962–5973. doi: 10.1529/biophysj.108.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies KU, Rapoport TA, Hartmann E. The beta subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol. 1998;141:887–894. doi: 10.1083/jcb.141.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies KU, Stokes V, Hartmann E. A single Sec61-complex functions as a protein-conducting channel. Biochim. Biophys. Acta. 2008;1783:2375–2383. doi: 10.1016/j.bbamcr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Kitao A, Yonekura K, Maki-Yonekura S, Samatey FA, Imada K, Namba K, Go N. Switch interactions control energy frustration and multiple flagellar filament structures. Proc. Natl. Acad. Sci. USA. 2006;103:4894–4899. doi: 10.1073/pnas.0510285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ML, Shinoda W. Large-scale molecular dynamics simulations of self-assembling systems. Science. 2008;321:798–800. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- Kurkcuoglu O, Kurkcuoglu Z, Doruker P, Jernigan RL. Collective dynamics of the ribosomal tunnel revealed by elastic network modeling. Proteins: Struct., Func., Bioinf. 2008;75:837–845. doi: 10.1002/prot.22292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol. Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jr., Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comp. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Mandon EC, Trueman SF, Gilmore R. Translocation of proteins through the Sec61 and SecYEG channels. Curr. Opin. Cell Biol. 2009;21:501–507. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo E, Mori H, Ito K. Interfering mutations provide in vivo evidence that Escherichia coli SecE functions in multimeric states. Mol. Gen. Genomics. 2003;268:808–815. doi: 10.1007/s00438-003-0803-9. [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Hegde RS, Agular M, Gygi SP, Park E, Rapoport TA, Akey CW. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétret JF, Hegde RS, Heinrich SU, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Schaletzky J, Clemons WM, Jr., Osborne AR, Skånland SS, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DG, Ménétret JF, Neuhof A, Rapoport TA, Akey CW. Structure of the mammalian ribosome-channel complex at 17 Å resolution. J. Mol. Biol. 2002;324:871–886. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- Mori H, Shimizu Y, Ito K. Superactive SecY variants that fulfill the essential translocation function with a reduced cellular quantity. J. Biol. Chem. 2002;277:48550–48557. doi: 10.1074/jbc.M204436200. [DOI] [PubMed] [Google Scholar]
- Murphy CK, Beckwith J. Residues essential for the function of SecE, a membrane component of the Escherichia coli secretion apparatus, are located in a conserved cytoplasmic region. Proc. Natl. Acad. Sci. USA. 1994;91:2557–2561. doi: 10.1073/pnas.91.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell. Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Raden D, Song W, Gilmore R. Role of the cytoplasmic segments of Sec61α in the ribosome-binding and translocation-promoting activities of the Sec61 complex. J. Cell Biol. 2000;150:53–64. doi: 10.1083/jcb.150.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. USA. 2008;105:13953–13958. doi: 10.1073/pnas.0804861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Tung CS. High performance computing in biology: Multimillion atom simulations of nanoscale systems. J. Struct. Biol. 2007;157:470–480. doi: 10.1016/j.jsb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaletzky J, Rapoport TA. Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol. Biol. Cell. 2006;17:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Clemons WM, Jr., DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J. Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K. Accelerating molecular modeling applications with graphics processors. J. Comp. Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- Tian P, Andricioaei I. Size, motion, and function of the SecY translocon revealed by molecular dynamics simulations with virtual probes. Biophys. J. 2006;90:2718–2730. doi: 10.1529/biophysj.105.073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular Dynamics Flexible Fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. 2009 doi: 10.1016/j.ymeth.2009.04.005. Methods In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D, Abramkina V, Aksimentiev A. Exploring transmembrane transport through alpha-hemolysin with grid-steered molecular dynamics. J. Chem. Phys. 2007;127:125101–125110. doi: 10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, von Heijne G. How translocons select transmembrane helices. Annu. Rev. Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- Yin Y, Arkhipov A, Schulten K. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins: Struct., Func., Gen. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.