Abstract
Background
Emerging evidences suggest that enteric glial cells (EGC), a major constituent of the enteric nervous system (ENS), are key regulators of intestinal epithelial barrier (IEB) functions. Indeed EGC inhibit intestinal epithelial cells (IEC) proliferation and increase IEB paracellular permeability. However, the role of EGC on other important barrier functions and the signalling pathways involved in their effects are currently unknown. To achieve this goal, we aimed at identifying the impact of EGC upon IEC transcriptome by performing microarray studies.
Results
EGC induced significant changes in gene expression profiling of proliferating IEC after 24 hours of co-culture. 116 genes were identified as differentially expressed (70 up-regulated and 46 down-regulated) in IEC cultured with EGC compared to IEC cultured alone. By performing functional analysis of the 116 identified genes using Ingenuity Pathway Analysis, we showed that EGC induced a significant regulation of genes favoring both cell-to-cell and cell-to-matrix adhesion as well as cell differentiation. Consistently, functional studies showed that EGC induced a significant increase in cell adhesion. EGC also regulated genes involved in cell motility towards an enhancement of cell motility. In addition, EGC profoundly modulated expression of genes involved in cell proliferation and cell survival, although no clear functional trend could be identified. Finally, important genes involved in lipid and protein metabolism of epithelial cells were shown to be differentially regulated by EGC.
Conclusion
This study reinforces the emerging concept that EGC have major protective effects upon the IEB. EGC have a profound impact upon IEC transcriptome and induce a shift in IEC phenotype towards increased cell adhesion and cell differentiation. This concept needs to be further validated under both physiological and pathophysiological conditions.
Background
The intestinal epithelial barrier (IEB) is the first boundary between the organism and the luminal environment. It plays a dual role by allowing the passage of nutrients and electrolytes but preventing the passage of pathogens. The maintenance of its homeostasis is of utmost importance for the survival of the organism. The IEB is formed by a monolayer of specialized intestinal epithelial cells (IEC) under constant renewal and maintained together via various cell-to-cell and cell-to-matrix interactions. The IEB is part of a complex network of specialized cell types constituting its microenvironment such as immune cells, subepithelial fibroblasts, endothelial cells or luminal bacteria. Emerging evidences suggest that under physiological conditions, the IEB's functions are actively regulated by its cellular microenvironment [1-3]. For instance, myofibroblasts have been shown to enhance epithelial cell proliferation and intestinal epithelial restitution [4]. In addition, microbiota have been shown to control both the maturation and the maintenance of the IEB [5].
The enteric nervous system (ENS) is also a major constituent of the cellular microenvironment of the IEB. Indeed IEB and, in particular, the proliferative compartment of the crypts are densely innervated by nerve fibres originating mainly from the submucosal plexus. Recent data have shown that, besides controlling secretory processes, activation of enteric neurons can reduce IEC proliferation and barrier permeability, in particular via the release of vasoactive intestinal peptide (VIP) [6-8]. Enteric neurons innervating the IEB are also closely associated with enteric glial cells (EGC), the major constituent of the ENS.
For many years, EGC have been considered as mainly passive and structural cells supporting neurons and ganglions. However, this concept has lately been revisited mainly focused on the role played by astrocytes in the central nervous system (CNS) [9-11]. Besides controlling and regulating neuronal functions, increasing evidence suggests that EGC could be major regulators of IEB functions, similar to astrocytes controlling blood brain barrier functions [10]. Supporting this concept, recent data have demonstrated that EGC can profoundly inhibit IEC proliferation, in part via the liberation of TGF-β1 [12]. EGC also decrease IEB paracellular permeability via the release of S-nitrosoglutathione (GSNO) [13]. Furthermore, in vivo lesions of EGC network increase IEB paracellular permeability and IEC proliferation and, at term, lead to major lethal intestinal inflammation [13-15]. However, the role of EGC in the control of other major IEC functions such as cell differentiation, cell-to-cell or cell-to-matrix adhesion, and the associated regulatory pathways remains largely unknown.
Therefore, in our study, we combined transcriptomic studies as well as functional studies to determine the impact of EGC on the regulation of major genes and functions involved in IEB homeostasis. Microarray approach was used to identify EGC-induced modifications in gene expression profiling of proliferating Caco-2. The identified genes and related functional pathways are consistent with the concept that EGC are a major constituent of the IEB microenvironment favoring barrier protection.
Results and Discussion
Enteric glial cells modulate intestinal epithelial cells transcriptome
Microarray experiments
We performed microarray analysis of EGC influence on the transcriptome of Caco-2 cells using oligonucleotide chips (Cancerochips) developed at West Genopole transcriptome core facility of Nantes. These microarrays contain around 6,864 genes and are dedicated to gene expression studies in Caco-2 cell line as well as to gene expression signature studies of multiple tumors. Caco-2 cells were cultured onto Transwell filters in the absence or presence of EGC seeded at the bottom of the wells for 8 or 24 hours. The Transwell filters did not allowed any contact between IEC and EGC, thus implicating only paracrine communication between the two cell types.
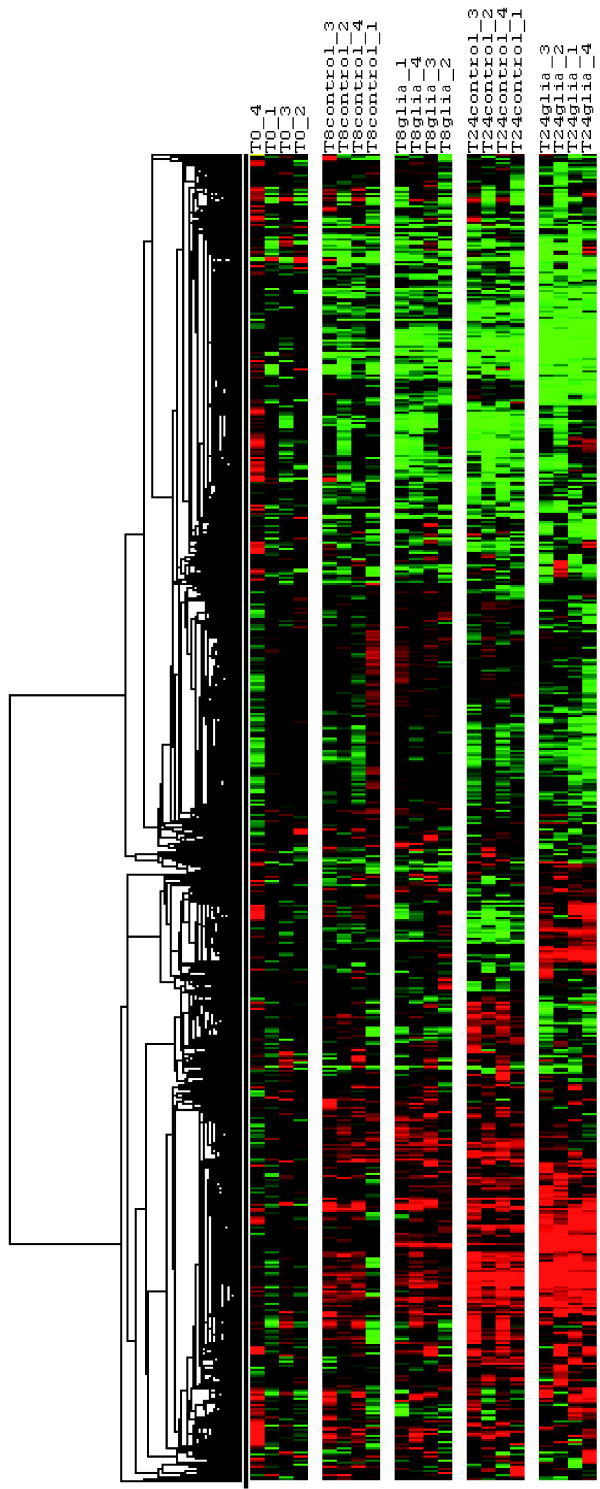
Hierarchical clustering of the whole data showed the impact of the time of culture as well as the impact of the presence of EGC on the transcriptional profiling of IEC, i.e. Caco-2 cells (Figure 1). We observed changes in IEC transcriptome over the 24 hours of culture in control condition. At 8 hours, differences in transcriptome profiling already existed in control condition as compared to t = 0. In general, the observed changes in differentially expressed genes between t = 0 and t = 8 hours in control conditions were increased in the same way of regulation when reaching t = 24 hours (Figure 1). These changes might be due to the growth and differentiation of the proliferating IEC over the 24 hours of culture. We observed no major differences in gene expression profiling between IEC cultured alone and IEC cultured in presence of EGC at 8 hours of culture. In contrast, at 24 hours, EGC presence led to consistent and major changes in IEC gene expression profiling (Figure 1).
Figure 1.
Hierarchical clustering of expression data. Four individual microarrays were used per condition. Hierarchical clustering was performed on genes using Gene Cluster. Each ratio was normalized to the median of the t = 0 hour-condition values of the corresponding gene. Each column represents an individual array (T0: t = 0 hour condition samples; T8control: t = 8 hours of culture without EGC; T8glia: t = 8 hours of culture in presence of EGC; T24control: t = 24 hours of culture without EGC; T24glia: t = 24 hours of culture in presence of EGC). Each line represents one individual gene. The clustering shows the impact of the time of culture on gene expression profiling in Caco-2 cells. The EGC-induced modulation of IEC transcriptome is highly visible at t = 24 hours.
Gene expression modulated by EGC
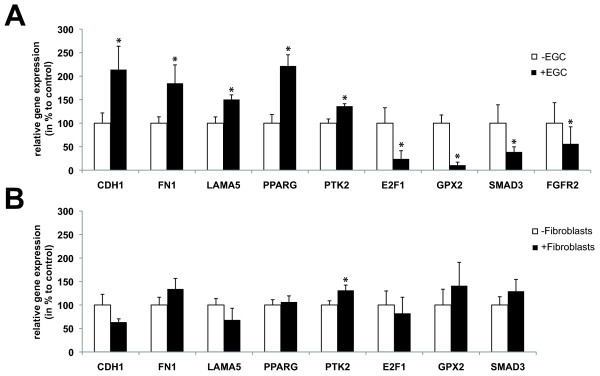
Using Genespring software, we aimed to identify statistically significant differences in gene expression profiling between Caco-2 cells cultured alone or in presence of EGC. After 8 h of culture, no significant difference in gene expression profiling was found between IEC cultured alone (control condition) or in presence of EGC ("glia" condition). However, after 24 hours of culture, we identified 116 genes differentially expressed between control and EGC conditions by using two different strategies. Benjamini and Hochberg False Discovery Rate method was used to determine 98 differentially expressed genes between control and glia conditions at t = 24 hours, and we also selected 27 genes with a two-fold change and Student's t-test p-value less than or equal to 0.05. Among the 116 differentially expressed genes, 46 genes were down-regulated and 70 were up-regulated in IEC cultured with EGC as compared to control (Table 1, 2). Quantitative PCR was also performed on various genes to validate the microarray results. In particular, results showed an EGC-induced increase of CDH1, FN1, LAMA5, PPARG, PTK2 mRNA expression in IEC and a decrease of E2F1, FGFR2, GPX2 and SMAD3 mRNA expression in IEC, similar to the data obtained with microarrays (Figure 2A). We next sought to determine the specificity of EGC effects upon IEC transcriptome by characterizing the impact of fibroblasts on the expression of these genes in IEC. Under identical culture conditions, we showed that fibroblasts increased expression of PTK2 but did not significantly modify gene expression of CDH1, FN1, LAMA5, PPARG, E2F1, GPX2 and SMAD3 in IEC (Figure 2B).
Table 1.
List of the genes up-regulated by enteric glial cells in intestinal epithelial cells.
| Gene Symbol | Genbank | Description | % up-regulation (/control) | Fold difference |
|---|---|---|---|---|
| TXNIP | NM_006472 | thioredoxin interacting protein | 217,60 | 3,18 |
| ANKRD1 | NM_014391 | ankyrin repeat domain 1 (cardiac muscle) | 169,22 | 2,69 |
| FN1 | U42593 | fibronectin 1 | 152,16 | 2,52 |
| TUBB3 | NM_006086 | tubulin, beta 3 | 149,94 | 2,50 |
| MGLL | AJ270950 | monoglyceride lipase | 135,97 | 2,36 |
| METTL7A | NM_014033 | methyltransferase like 7A | 132,96 | 2,33 |
| PKN2 | NM_006256 | protein kinase N2 | 128,33 | 2,28 |
| / | XM_166201 | synonyms: KIAA0056, MGC104671; Homo sapiens KIAA0056 protein (hCAP-D3), mRNA. | 115,09 | 2,15 |
| EPB41L2 | NM_001431 | erythrocyte membrane protein band 4.1-like 2 | 110,50 | 2,11 |
| AASS | AJ007714 | aminoadipate-semialdehyde synthase | 110,15 | 2,10 |
| ACTG2 | NM_001615 | actin, gamma 2, smooth muscle, enteric | 102,56 | 2,03 |
| B4GALT5 | NM_004776 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 | 102,38 | 2,02 |
| SNX2 | NM_003100 | sorting nexin 2 | 102,38 | 2,02 |
| VIP | NM_003381 | vasoactive intestinal peptide | 102,33 | 2,02 |
| EIF4A2 | NM_001967 | eukaryotic translation initiation factor 4A, isoform 2 | 102,14 | 2,02 |
| / | NM_019027 | RNA-binding protein | 101,66 | 2,02 |
| POLR3F | NM_006466 | polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa | 94,46 | 1,94 |
| PNRC1 | NM_006813 | proline-rich nuclear receptor coactivator 1 | 93,24 | 1,93 |
| NPPB | NM_002521 | natriuretic peptide precursor B | 83,97 | 1,84 |
| / | BC017857 | Homo sapiens cDNA clone IMAGE:4690793, with apparent retained intron. | 82,20 | 1,82 |
| KRT8 | NM_002273 | keratin 8 | 81,96 | 1,82 |
| SAT1 | NM_002970 | spermidine/spermine N1-acetyltransferase 1 | 80,24 | 1,80 |
| ASS1 | NM_000050 | argininosuccinate synthetase 1 | 76,73 | 1,77 |
| S100A11P | NM_021039 | / | 75,22 | 1,75 |
| SLC7A7 | NM_003982 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 72,31 | 1,72 |
| HAGH | NM_005326 | hydroxyacylglutathione hydrolase | 69,54 | 1,70 |
| BNIP3L | AF536326 | BCL2/adenovirus E1B 19 kDa interacting protein 3-like | 69,19 | 1,69 |
| / | AF195968 | PRR5-ARHGAP8 fusion | 68,01 | 1,68 |
| BNIP3 | NM_004052 | BCL2/adenovirus E1B 19 kDa interacting protein 3 | 67,84 | 1,68 |
| IL18 | NM_001562 | interleukin 18 (interferon-gamma-inducing factor) | 67,14 | 1,67 |
| RDM1 | BC038301 | RAD52 motif 1 | 67,11 | 1,67 |
| FAM107B | NM_031453 | family with sequence similarity 107, member B | 65,70 | 1,66 |
| PLAC8 | NM_016619 | placenta-specific 8 | 63,77 | 1,64 |
| SMARCA1 | NM_139035 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | 63,72 | 1,64 |
| PLOD2 | NM_000935 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 62,87 | 1,63 |
| TMSB4Y | NM_004202 | thymosin, beta 4, Y-linked | 62,77 | 1,63 |
| SCPEP1 | NM_021626 | serine carboxypeptidase 1 | 60,96 | 1,61 |
| LAMA5 | NM_005560 | laminin, alpha 5 | 60,53 | 1,61 |
| LAMC1 | NM_002293 | laminin, gamma 1 (formerly LAMB2) | 59,89 | 1,60 |
| METAP1 | BC030054 | methionyl aminopeptidase 1 | 59,55 | 1,60 |
| IQGAP2 | NM_006633 | IQ motif containing GTPase activating protein 2 | 58,98 | 1,59 |
| C1orf43 | NM_015449 | chromosome 1 open reading frame 43 | 56,86 | 1,57 |
| CASP4 | NM_001225 | caspase 4, apoptosis-related cysteine peptidase | 55,71 | 1,56 |
| BTG1 | NM_001731 | B-cell translocation gene 1, anti-proliferative | 54,63 | 1,55 |
| SLC2A1 | K03195 | solute carrier family 2 (facilitated glucose transporter), member 1 | 54,34 | 1,54 |
| DCTN2 | NM_006400 | dynactin 2 (p50) | 52,68 | 1,53 |
| TOP2A | NM_001067 | topoisomerase (DNA) II alpha 170 kDa | 52,42 | 1,52 |
| KRT18 | NM_000224 | keratin 18 | 51,16 | 1,51 |
| LAMC1 | M55210 | laminin, gamma 1 (formerly LAMB2) | 50,37 | 1,50 |
| PRC1 | BC005140 | protein regulator of cytokinesis 1 | 50,13 | 1,50 |
| IMPDH2 | NM_000884 | IMP (inosine monophosphate) dehydrogenase 2 | 49,46 | 1,49 |
| / | AF202922 | LRP16 protein | 48,20 | 1,48 |
| PLD3 | NM_012268 | phospholipase D family, member 3 | 46,99 | 1,47 |
| RNF4 | NM_002938 | ring finger protein 4 | 44,88 | 1,45 |
| / | AC060225 | Homo sapiens 3 BAC RP11-23J16 complete sequence. | 42,10 | 1,42 |
| SMARCA1 | NM_003069 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | 41,18 | 1,41 |
| DYNLT3 | NM_006520 | dynein, light chain, Tctex-type 3 | 40,06 | 1,40 |
| PPARG | NM_015869 | peroxisome proliferative activated receptor, gamma | 38,92 | 1,39 |
| GLRX | AF069668 | glutaredoxin (thioltransferase) | 37,78 | 1,38 |
| PTK2 | NM_153831;NM_005607 | PTK2 protein tyrosine kinase 2 | 37,72 | 1,38 |
| CDH1 | NM_004360 | cadherin 1, type 1, E-cadherin (epithelial) | 36,87 | 1,37 |
| RNASE4 | NM_002937 | ribonuclease, RNase A family, 4 | 31,40 | 1,31 |
| CTSH | NM_004390 | cathepsin H | 29,45 | 1,29 |
| MKI67 | NM_002417 | antigen identified by monoclonal antibody Ki-67 | 29,28 | 1,29 |
| EIF2A | NM_032025 | eukaryotic translation initiation factor 2A, 65 kDa | 26,54 | 1,27 |
| TGFBI | BC000097 | transforming growth factor, beta-induced, 68 kDa | 25,95 | 1,26 |
| MLLT3 | NM_004529 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | 22,42 | 1,22 |
| APOBEC3B | NM_004900 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B | 22,20 | 1,22 |
| ADD3 | NM_019903 | adducin 3 (gamma) | 20,82 | 1,21 |
| FTH1 | NM_002032 | ferritin, heavy polypeptide 1 | 15,97 | 1,16 |
Table 2.
List of the genes down-regulated by enteric glial cells in intestinal epithelial cells.
| Gene Symbol | Genbank | Description | % down-regulation (/control) | Fold difference |
|---|---|---|---|---|
| CARD12 | AF376061 | caspase recruitment domain family, member 12 | 83,43 | 6,04 |
| KLK14 | NM_022046 | kallikrein 14 | 62,46 | 2,66 |
| FGFR2 | NM_022970 | fibroblast growth factor receptor 2 | 57,12 | 2,33 |
| BDP1 | NM_018429 | B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB | 56,58 | 2,30 |
| SFRP4 | NM_003014 | secreted frizzled-related protein 4 | 55,26 | 2,24 |
| C6 | NM_000065 | complement component 6 | 54,78 | 2,21 |
| PRKCD | NM_006254 | protein kinase C, delta | 54,67 | 2,21 |
| / | XM_066534 | Homo sapiens diacylglycerol kinase, kappa (DGKK), mRNA. | 52,44 | 2,10 |
| C20orf133 | NM_001033086 | chromosome 20 open reading frame 133 | 52,36 | 2,10 |
| PRKCQ | NM_006257 | protein kinase C, theta | 50,50 | 2,02 |
| CDK5R1 | NM_003885 | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 50,42 | 2,02 |
| RPP40 | NM_006638 | ribonuclease P 40 kDa subunit | 47,41 | 1,90 |
| SLC30A1 | AF323590 | solute carrier family 30 (zinc transporter), member 1 | 46,22 | 1,86 |
| TIMM8A | NM_004085 | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 41,07 | 1,70 |
| EBNA1BP2 | NM_006824 | EBNA1 binding protein 2 | 36,39 | 1,57 |
| ITGAE | NM_002208 | integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) | 36,17 | 1,57 |
| NOL1 | NM_006170 | nucleolar protein 1, 120 kDa | 33,86 | 1,51 |
| C6orf66 | NM_014165 | chromosome 6 open reading frame 66 | 33,81 | 1,51 |
| NOL5A | NM_006392 | nucleolar protein 5A (56 kDa with KKE/D repeat) | 33,30 | 1,50 |
| BAG1 | U46917 | BCL2-associated athanogene | 32,19 | 1,47 |
| / | AF123534 | nucleolar protein NOP5/NOP58 | 32,14 | 1,47 |
| ASAH1 | AK025211 | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 29,98 | 1,43 |
| TINAGL1 | AF236150 | tubulointerstitial nephritis antigen-like 1 | 29,62 | 1,42 |
| AADAC | NM_001086 | arylacetamide deacetylase (esterase) | 29,48 | 1,42 |
| HSPA14 | AF112210 | heat shock 70 kDa protein 14 | 29,34 | 1,42 |
| PSMC6 | NM_002806 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 29,31 | 1,41 |
| HNRPDL | D89678 | heterogeneous nuclear ribonucleoprotein D-like | 28,40 | 1,40 |
| SAMHD1 | NM_015474 | SAM domain and HD domain 1 | 28,12 | 1,39 |
| TP53RK | NM_033550 | TP53 regulating kinase | 26,99 | 1,37 |
| MARK2 | NM_004954 | MAP/microtubule affinity-regulating kinase 2 | 26,41 | 1,36 |
| CCR9 | NM_031200 | chemokine (C-C motif) receptor 9 | 24,74 | 1,33 |
| RGL1 | NM_015149 | ral guanine nucleotide dissociation stimulator-like 1 | 24,20 | 1,32 |
| E2F1 | NM_005225 | E2F transcription factor 1 | 23,90 | 1,31 |
| PSMC1 | NM_002802 | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | 23,75 | 1,31 |
| IMP3 | NM_018285 | IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) | 23,48 | 1,31 |
| RNU3IP2 | BC023662 | RNA, U3 small nucleolar interacting protein 2 | 23,41 | 1,31 |
| SMAD3 | NM_005902 | SMAD, mothers against DPP homolog 3 (Drosophila) | 21,49 | 1,27 |
| GPX2 | NM_002083 | glutathione peroxidase 2 (gastrointestinal) | 21,27 | 1,27 |
| LSP1 | NM_002339 | lymphocyte-specific protein 1 | 21,21 | 1,27 |
| FGG | NM_021870 | fibrinogen gamma chain | 18,37 | 1,23 |
| C20orf94 | NM_001009608 | chromosome 20 open reading frame 94 | 16,04 | 1,19 |
| PPIL1 | NM_016059 | peptidylprolyl isomerase (cyclophilin)-like 1 | 14,74 | 1,17 |
| HOXB2 | NM_002145 | homeobox B2 | 13,91 | 1,16 |
| APOH | NM_000042 | apolipoprotein H (beta-2-glycoprotein I) | 13,17 | 1,15 |
| PRSS23 | NM_007173 | protease, serine, 23 | 10,41 | 1,12 |
| IARS2 | NM_018060 | isoleucine-tRNA synthetase 2, mitochondrial | 10,02 | 1,11 |
Figure 2.
Enteric glial cells EGC) and fibroblasts differentially modulated intestinal epithelial cell (IEC) transcriptome. (A). Real-time quantitative PCR studies on CDH1(n = 5), FN1 (n = 7), LAMA5 (n = 6), PPARG (n = 5), PTK2 (n = 5), E2F1 (n = 7), FGFR2 (n = 6), GPX2 (n = 8), SMAD3 (n = 7) gene expression in IEC cultured for 24 hours alone (- EGC) or in presence of EGC(+ EGC) confirmed that EGC significantly modulate the level of expression of genes identified by the microarrays data analysis as differentially expressed in IEC cultured in presence of EGC (*p < 0.05; Mann-Whitney test). (B). In contrast, real-time quantitative PCR studies on CDH1 (n = 5), FN1 (n = 5), LAMA5 (n = 5), PPARG (n = 5), PTK2 (n = 5), E2F1 (n = 5), GPX2 (n = 5), SMAD3 (n = 5) gene expression in IEC cultured for 24 hours alone (- fibroblasts) or in presence of fibroblasts (+fibroblasts) showed a differential regulation of gene expression as compared to EGC effects (*p < 0.05; Mann-Whitney test).
Hierarchical clustering of differentially expressed genes
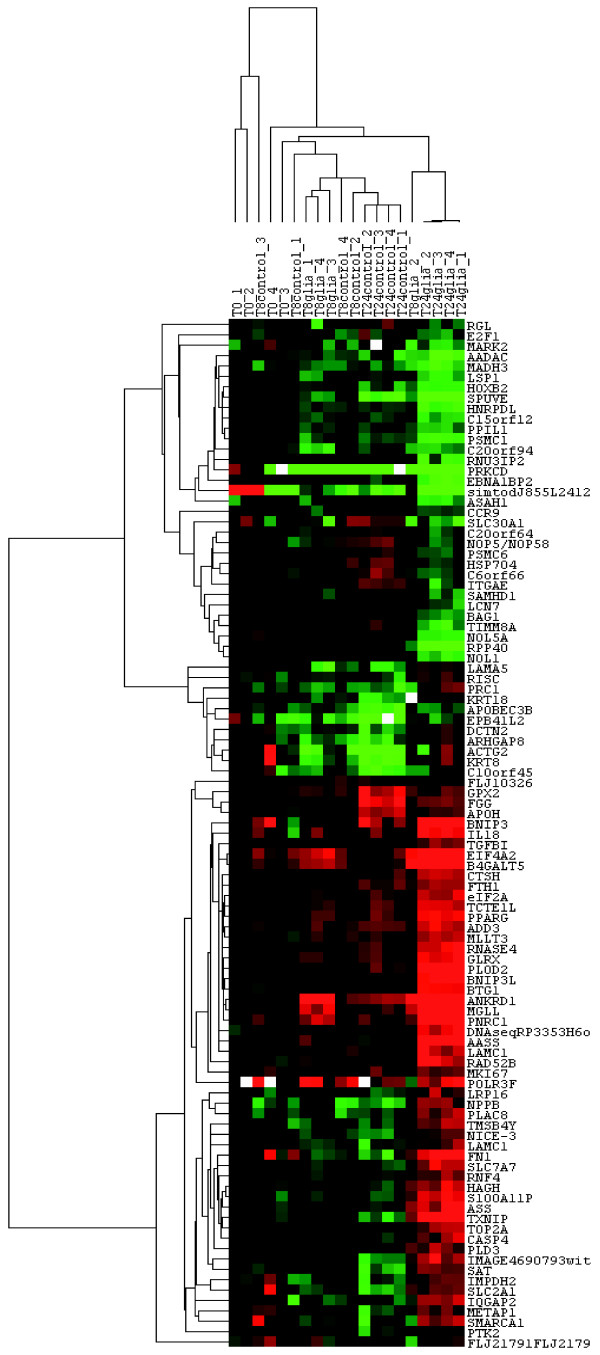
Hierarchical clustering was used to visualize the expression profile of the 116 genes induced or repressed by EGC after 24 hours of culture (Figure 3).
Figure 3.
Hierarchical clustering of the 116 identified genes expression data. Four individual microarrays were used per condition. Hierarchical clustering was performed on conditions and on the 116 genes identified with Genespring. Each ratio was normalized to the median of the t = 0 hour-condition values of the corresponding gene. Each column represents an individual array (T0: t = 0 hour condition samples; T8control: t = 8 hours of culture without EGC; T8glia: t = 8 hours of culture in presence of EGC; T24control: t = 24 hours of culture without EGC; T24glia: t = 24 hours of culture in presence of EGC). Each line represents one individual gene. The clustering reveals clusters of genes with similar pattern of expression among the different conditions. The cluster also shows the distance between the five conditions demonstrating major changes induced by the culture with EGC at t = 24 hours.
All these genes exhibit a differential expression between control and EGC conditions at t = 24 hours. Furthermore, some of them already exhibited a slight difference in expression profile between control and EGC conditions at 8 hours. These results indicate that EGC effects on genes identified as differentially expressed in IEC at 24 hours probably started as early as at 8 hours, even though the modifications were not statistically significant.
Two groups of samples exhibited a very different profile from other samples: EGC condition at t = 24 hours and controls at t = 24 hours (Figure 3). These observations confirm that 1) no major changes existed between control and EGC conditions at t = 8 hours and 2) that the 24 hour-time of culture impacted on gene expression profiling in IEC, likely reflecting differentiation of IEC over the time of culture.
EGC regulate IEC functions
Gene network interactions
Biological interactions among the 116 genes of the gene set provided by Genespring analysis were identified using Ingenuity Pathways Analysis. Among the 116 genes differentially expressed, Ingenuity identified 92 genes contributing to a total of 10 functional networks (Table 3). Each of the 6 first networks contained at least 14 genes that were associated with cell-to-cell signalling and interaction, cellular growth and proliferation, cell morphology, cellular movement, cell death and cell cycle. The 116 genes were also classified into Ingenuity cellular and molecular pathways as well as into Ingenuity signalling pathways (Table 4 and 5). All the functions described above and identified by building functional networks among our gene set were found in the 25 cellular and molecular functions obtained with Ingenuity (Table 4). Moreover, these 6 functions were among the 10 first functions presenting the highest score (Table 4). Finally, the signalling pathways identified by the Ingenuity analysis of our gene set were also relevant to those 6 functions (Table 5). The limit of Ingenuity analysis for our study is that it is not restricted to one specific organ or cell, so that all the results of Ingenuity analysis could not be transposed directly to the regulation of IEC functions by EGC. We therefore performed an "epithelial" specific analysis of the major functions identified with Ingenuity.
Table 3.
Lists of differentially expressed genes involved in functional networks regulated in intestinal epithelial cells by enteric glial cells.
| Ingenuity© top functions | Genes list | Score |
|---|---|---|
| Cell-To-Cell Signaling and Interaction Cell Morphology Tissue Development |
CDH1, FN1, ITGAE, KRT8, KRT18, MGLL, NPPB, PKN2, PPARG, PRKCD, PSMC1, PSMC6, PTK2, SAT, SMAD3, TGFBI, VIP | 26 |
| Cellular Growth and Proliferation Cancer Gene Expression |
B4GALT5, BAG1, BNIP3 (includes EG:664), CCR9, DCTN2, EBNA1BP2, HCAP-D3, LSP1, MLLT3, PNRC1, PPIL1, PRC1, RNF4, SAT, SLC30A1, TP53RK | 24 |
| Cell Morphology Cellular Development Cardiovascular System Development and Function |
AASS, ACTG2, BNIP3L, BTG1, CTSH, DYNLT3, EIF4A2, GLRX, MKI67, PLOD2, RNASE4, SLC2A1, SLC7A7, TXNIP | 20 |
| Cellular Movement Hematological System Development and Function Immune Response |
APOBEC3B, ASAH1, C6, FTH1, IL18, IQGAP2, LAMA5, LAMC1, NOL5A, NOP5/NOP58, PLD3, PRKCQ, PRSS23, SCPEP1 | 20 |
| Cell Death Cancer Gastrointestinal Disease |
APOH, ASS, BDP1, BTG1, CARD12, CASP4, E2F1, FGFR2, FGG, NOL1, RGL1, SMARCA1, SNX2, TOP2A | 20 |
| Cell Cycle Gastrointestinal Disease Developmental Disorder |
ADD3, ANKRD1, CDK5R1, EPB41L2, GPX2, HNRPDL, HOXB2, IMPDH2, MARK2, PKN2, PLAC8, POLR3F, SFRP4, TUBB3 | 20 |
| RNA Post-Transcriptional Modification | IMP3 | 2 |
| RNA Post-Transcriptional Modification | EIF2A | 2 |
| Protein Trafficking Cellular Compromise Auditory and Vestibular System Development and Function |
TIMM8A | 1 |
| RNA Post-Transcriptional Modification RNA Damage and Repair |
RPP40 | 1 |
Table 4.
Lists of differentially expressed genes involved in cellular and molecular functions regulated in intestinal epithelial cells by enteric glial cells.
| Ingenuity© cellular and molecular functions | Genes list | Score |
|---|---|---|
| RNA Post-Transcriptional Modification | NOP5/NOP58, NOL5A, EBNA1BP2, RNU3IP2, IMP3 | 5.17 |
| Cell Death | GPX2, SAT, PRKCD, B4GALT5, TXNIP, TOP2A, CDH1, PKN2, ASAH1, LSP1, BNIP3L, NPPB, CDK5R1, ANKRD1, VIP, CASP4, GLRX, FGFR2, KRT18, BAG1, PRKCQ, BTG1, PPARG, C6ORF66, SLC2A1, CARD12, SMAD3, FTH1, LAMA5, PTK2, IL18, MLLT3, FN1, PLAC8, KRT8, TGFBI, BNIP3, E2F1, SFRP4 | 4.85 |
| Cell-To-Cell Signaling and Interaction | APOH, PRKCD, SMAD3, CDH1, PKN2, LAMA5, PTK2, VIP, IL18, FN1, FGG, KRT8, KRT18, BAG1, ITGAE, LAMC1, PPARG, TGFBI, E2F1 | 3.92 |
| Cellular Development | SMAD3, PRKCD, CDH1, LAMA5, VIP, PTK2, IL18, CDK5R1, FGFR2, FN1, PLAC8, PRKCQ, LAMC1, PPARG, E2F1 | 3.92 |
| Cell Morphology | SMAD3, PRKCD, CDH1, LAMA5, LSP1, DCTN2, VIP, IL18, PTK2, CDK5R1, FN1, KRT8, KRT18, PPARG, TGFBI, E2F1 | 3.88 |
| Cellular Assembly and Organization | APOH, PRKCD, SMAD3, TOP2A, CDH1, DCTN2, EBNA1BP2, NPPB, PTK2, CDK5R1, HCAP-D3, FN1, FGG, KRT8, KRT18, MARK2, LAMC1, PPARG, BNIP3, E2F1 | 3.88 |
| Carbohydrate Metabolism | FN1, B4GALT5, NPPB, PTK2, IL18 | 3.11 |
| Cellular Movement | HOXB2, CCR9, C6, B4GALT5, SMAD3, PRKCD, MGLL, CDH1, LSP1, LAMA5, DCTN2, NPPB, CDK5R1, VIP, PTK2, IL18, FN1, BAG1, ITGAE, PPARG, TGFBI | 3.08 |
| Cellular Growth and Proliferation | SAT, PRKCD, TXNIP, SMAD3, CDH1, FTH1, LAMA5, SLC30A1, EBNA1BP2, VIP, IL18, PTK2, MLLT3, FGFR2, FN1, PLAC8, BAG1, PRKCQ, BTG1, LAMC1, PPARG, BNIP3, E2F1, SFRP4 | 3.05 |
| Cell Cycle | HCAP-D3, FN1, TXNIP, PRKCD, TOP2A, DCTN2, PPARG, EBNA1BP2, E2F1, VIP | 2.57 |
| Molecular Transport | FGFR2, SAT, FN1, PRKCD, MGLL, BAG1, FTH1, PPARG, NPPB, PTK2, VIP, IL18 | 2.45 |
| Nucleic Acid Metabolism | SAT, BAG1, NPPB, VIP | 2.45 |
| Small Molecule Biochemistry | APOH, SAT, PRKCD, B4GALT5, MGLL, FTH1, ASAH1, NPPB, PTK2, IL18, VIP, ASS, GLRX, FGFR2, FN1, BAG1, PPARG | 2.45 |
| Cellular Function and Maintenance | CCR9, SMAD3, CDH1, SLC30A1, PTK2, IL18, CDK5R1, FN1, FGG, KRT18, ITGAE, PPARG, BNIP3 | 2.25 |
| DNA Replication, Recombination, and Repair | HCAP-D3, FN1, SMAD3, PRKCD, TOP2A, FTH1, DCTN2, EBNA1BP2 | 2.17 |
| Gene Expression | APOH, SMAD3, PRKCD, CDH1, PKN2, VIP, IL18, FN1, BAG1, PRKCQ, RNF4, PPARG, E2F1 | 2.13 |
| Cell Signaling | ASS, FN1, PRKCD, PPARG | 2.02 |
| Amino Acid Metabolism | ASS, FTH1 | 1.94 |
| Cellular Compromise | PRKCD, KRT18, TIMM8A, PPARG, E2F1 | 1.94 |
| Drug Metabolism | GLRX, FTH1, NPPB, IL18, VIP | 1.94 |
| Lipid Metabolism | FGFR2, APOH, SAT, FN1, MGLL, ASAH1, PPARG, NPPB, VIP | 1.94 |
| Post-Translational Modification | PRKCD, BAG1, PRKCQ | 1.94 |
| Protein Folding | BAG1 | 1.94 |
| Protein Synthesis | BAG1, IL18 | 1.94 |
| Vitamin and Mineral Metabolism | FGFR2, FTH1, PPARG | 1.94 |
Table 5.
Lists of differentially expressed genes involved in signalling pathways regulated in intestinal epithelial cells by enteric glial cells.
| Ingenuity© Signalling Pathway | Genes | Ratio |
|---|---|---|
| Circadian Rhythm Signaling | VIP | 0.046 |
| Cell Cycle: G1/S Checkpoint Regulation | SMAD3, E2F1 | 0.041 |
| Integrin Signaling | ACTG2, FN1, LAMA5, LAMC1, PTK2 | 0.03 |
| Actin Cytoskeleton Signaling | TMSB4Y, FGFR2, FN1, ITGAE, IQGAP2, PTK2 | 0.029 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | TOP2A | 0.029 |
| VEGF Signaling | ACTG2, PTK2 | 0.029 |
| Complement and Coagulation Cascades | FGG, C6 | 0.028 |
| Amyloid Processing | CDK5R1 | 0.028 |
| ERK/MAPK Signaling | PRKCD, PPARG, PTK2 | 0.024 |
| Wnt/β-catenin Signaling | CDH1, MARK2, SFRP4 | 0.022 |
| FGF Signaling | FGFR2 | 0.018 |
| Chemokine Signaling | PTK2 | 0.018 |
| TGF-β Signaling | SMAD3 | 0.016 |
| Protein Ubiquitination Pathway | PSMC6, PSMC1, BAG1 | 0.016 |
| PPAR Signaling | PPARG | 0.015 |
| IGF-1 Signaling | PTK2 | 0.015 |
| Apoptosis Signaling | PRKCQ | 0.015 |
| Neuregulin Signaling | CDK5R1 | 0.015 |
| PTEN Signaling | PTK2 | 0.014 |
| Fc Epsilon RI Signaling | PRKCD | 0.014 |
| T Cell Receptor Signaling | PRKCQ | 0.014 |
| Xenobiotic Metabolism Signaling | PRKCD, PRKCQ | 0.010 |
| NF-κB Signaling | PRKCQ | 0.009 |
| B Cell Receptor Signaling | PRKCQ | 0.009 |
| Ephrin Receptor Signaling | PTK2 | 0.009 |
| Leukocyte Extravasation Signaling | PTK2 | 0.008 |
| Huntington's Disease Signaling | CDK5R1 | 0.007 |
| Axonal Guidance Signaling | PTK2 | 0.004 |
Cell-to cell and cell-to-matrix interaction
EGC regulated the expression of numerous genes involved in the control of IEC adhesive processes. In particular, EGC induced an up-regulation of the expression of all 7 genes with pro-adhesive functions and a down-regulation of the 2 genes with anti adhesive properties, among the gene set found to be differentially expressed in IEC cultured in presence of EGC (Table 6). These genes are crucially involved in the control of cell-to-cell and cell-to-matrix adhesion.
Table 6.
Genes controlling intestinal epithelial cells adhesion and modulated by enteric glial cells.
| Pro-adhesive | Anti-adhesive | ||
|---|---|---|---|
| Gene Symbol | Regulation of gene expression by EGC | Gene Symbol | Regulation of gene expression by EGC |
| CDH1 | up-regulated | CDK5R1 | down-regulated |
| IQGAP2 | up-regulated | KLK14 | down-regulated |
| LAMA5 | up-regulated | ||
| LAMC1 | up-regulated | ||
| FN1 | up-regulated | ||
| PTK2 | up-regulated | ||
| KRT8 | up-regulated | ||
First, EGC concomitantly increased the expression of CDH-1, which encodes E-cadherin, and decreased the expression of CDK5R1. E-Cadherin is the major component of the adherent junction complexes and the level of E-Cadherin in IEC is to be correlated to adhesion complexes formation between IEC [16,17]. Further evidences confirming a pro-adhesive influence of EGC on IEC is the EGC-induced down-regulation of CDK5R1 expression. Indeed, CDK5R1 encodes p35, a regulator of CDK-5 (cyclin-dependent kinase), which induces the degradation of E-Cadherin precursor [18]. In addition, EGC also up-regulated IQGAP2 expression in IEC. This gene encodes for a protein member of IQGAP family that interacts with several molecules controlling cytoskeleton organization, cell adhesion and cell motility such as CDC42 and Rac [19]. Interestingly, IQGAP2 has been shown to mediate E-Cadherin-based cell-to-cell adhesion during development [20]. All these results suggest that EGC enhance cell-to-cell adhesion in IEC.
Our data also demonstrate that EGC modulate the expression of genes that are involved in cell-to-matrix interactions. First, EGC increased expression of several genes encoding proteins of the extracellular matrix such as LAMA5, LAMC1 and FN1. LAMA5 and LAMC1 encode respectively for laminin α5 and γ1 chains which, together with laminin β1 chain, compose laminin-10 [21]. Laminin-10 has been shown to be the most adhesive substratum of laminin isoforms when studying abilities of laminin-2,-5 and -10 in modulating Caco-2 cell adhesion [22]. Furthermore, EGC up-regulated FN1 expression, encoding the fibronectin protein. Interestingly, fibronectin has recently been shown to enhance Caco-2 cell attachment and wound healing [23]. EGC down-regulated KLK14 expression, which encodes KLK (kallikrein) 14, an extracellular serine protease which has been shown to cleave and digest various extracellular matrix proteins such as collagen IV, laminin and fibronectin [24]. In addition, EGC up-regulated PTK2 expression in IEC which may result in increased expression of FAK (Focal Adhesion Kinase) protein, a major regulator of focal adhesions turnover and maturation [25]. Finally, EGC induced an up-regulation of KRT8 expression whose increased expression has recently been shown to cause enhanced adhesion of human breast tumor cells to their extracellular matrix [26].
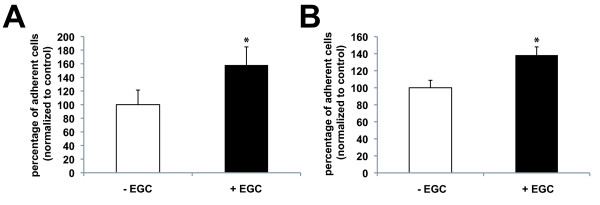
In conclusion, our data suggest that EGC regulation of IEC transcriptome leads to an increase in cell adhesion. In order to functionally validate this hypothesis, we performed in vitro experiments using established adhesion assays. Under these conditions, we first showed that IEC global adhesion was increased after 24 hours of culture with EGC as compared to control (Figure 4A). We next confirmed whether these effects were in part associated with an increase in cell-to-matrix adhesion as the majority of IEC genes regulated by EGC presence appeared to favor cell-to-matrix adhesion. Indeed, cell-to-matrix adhesion assays revealed that EGC significantly increased IEC adhesion to the filter as compared to control (Figure 4B).
Figure 4.
Enteric glial cells (EGC) induced an increase in intestinal epithelial cells(IEC) adhesion. (A):EGC induced a significant increase in IEC total adhesion (i.e. without discriminating cell-to-matrix and cell-to-cell adhesion) after 24 hours of co-culture in presence of EGC (+ EGC) as compared to control (- EGC) (n = 5; p = 0.008; Mann-Whitney test). (B): IEC were significantly more attached to their matrix when they were cultured for 24 hours with EGC (+ EGC) as compared to control (- EGC) (n = 13; p < 0.001; Mann-Whitney test).
Cell differentiation
EGC also regulated the expression of numerous genes involved in IEC differentiation. In particular, EGC up-regulated the expression of 6 genes enhancing differentiation and down-regulated 3 genes known to inhibit IEC differentiation (Table 7).
Table 7.
Genes controlling intestinal epithelial cells differentiation and modulated by enteric glial cells.
| Pro-differentiative | Anti-differentiative | ||
|---|---|---|---|
| Gene Symbol | Regulation of gene expression by EGC | Gene Symbol | Regulation of gene expression by EGC |
| CDH1 | up-regulated | E2F1 | down-regulated |
| PPARG | up-regulated | BAG1 | down-regulated |
| LAMA5 | up-regulated | CDK5R1 | down-regulated |
| PTK2 | up-regulated | FN1 | up-regulated |
| DCTN2 | up-regulated | ||
| DYNLT3 | up-regulated | ||
EGC induced an up-regulation of the expression of pro-differentiative genes or genes associated with enhanced differentiation of IEC such as PPARG, LAMA5, PTK2, CDH-1, DCTN2 and DYNLT3. Indeed, PPARγ, encoding the well-described nuclear receptor superfamily member peroxisome proliferator-activated receptor gamma (PPAR-γ) has been shown to regulate IEC differentiation and its expression has been positively correlated with level of differentiation of Caco-2 and HT29 cells [27,28]. Moreover, a diminution of laminin-a5 in a murine model resulted in a transformation from a small intestinal to a colonic mucosal architecture, suggesting that laminin-α5 has a crucial role in establishing and maintaining the architecture of the small intestine [29]. In addition, it has already been shown that the differentiation of Caco-2 cells was accompanied by an increase in FAK expression [30]. E-Cadherin, whose corresponding gene CDH-1 is up-regulated by EGC, has been largely demonstrated to be involved in the establishment of a differentiated phenotype for IEC. Notably, E-Cadherin has been described to be less expressed at the bottom of the crypts where IEC are undifferentiated [16,31,32]. The down-regulation by EGC of CDK5R1 expression, leading to enhanced levels of E-Cadherin (see previous paragraph), might also further enhance EGC-induced cell differentiation. EGC also increased DCTN2 and DYNLT3 expression, two genes encoding a subunit of dynactin (p50 or dynamitin) and dynein light chain rp3, respectively. Both are involved in post-Golgi movement of vesicles towards apical surface of differentiated enterocytes [33-35], and could therefore reflect increased differentiation of IEC induced by EGC. Intriguingly, although differentiation of the Caco-2 cell line has been shown to be correlated with a down-regulation in fibronectin expression [36], EGC induced an up-regulation in FN1 expression in IEC in our study.
EGC decreased the expression of genes that encode proteins implicated in anti-differentiative pathways such as E2F1, BAG1 and CDK5R1 (discussed above). E2F1 is a gene encoding a protein member of the E2F family of transcription factors and has been shown to be down-regulated in confluent human IEC and differentiated enterocytes [37]. BAG1, encoding a Bcl-2 non-homologous associated molecule, has also been shown to present a decreasing gradient of expression from the base of the crypts to the apex of the villi, suggesting that the down-regulation of BAG1 might reflect a differentiation state of IEC [38].
In conclusion, based on our analysis, EGC-mediated regulation of IEC transcriptome appears to strongly favor IEC differentiation.
Cell motility
EGC regulated in IEC the expression of genes encoding proteins that are known to play a role in IEC motility (Table 8). In particular, EGC induced an increase in FN1 expression in IEC. FN1 has been demonstrated as a major factor in promoting cell migration of IEC and subepithelial fibroblasts, thus favoring epithelial wound healing [39,40]. Interestingly, EGC induced a down-regulation in LSP1 expression in IEC. LSP1 gene encodes for LSP1, a cytoplasmic actin-binding protein, whose overexpression in melanoma cells has been described to inhibit cell motility [41]. EGC-induced up-regulation of PTK2 expression also supports a role of EGC in promoting IEC motility. Indeed, increased FAK protein level promoted epithelial restitution via an increased IEC migration [42,43]. Similarly, the increased PPARγ expression could enhance cell motility as inhibitors of PPARγ inhibit epithelial cell migration [44-46].
Table 8.
Genes controlling intestinal epithelial cells motility and modulated by enteric glial cells.
| Pro-motility | Anti-motility | ||
|---|---|---|---|
| Gene Symbol | Regulation of gene expression by EGC | Gene Symbol | Regulation of gene expression by EGC |
| PPARG | up-regulated | LSP1 | down-regulated |
| FN1 | up-regulated | ||
| PTK2 | up-regulated | ||
Cell proliferation
Expression of genes involved in cell proliferation was differentially regulated in IEC cultured in presence of EGC as compared to control (Table 9). In fact, EGC appeared to modulate the expression of anti-proliferative and pro-proliferative genes toward a dominant anti-proliferative effect (Table 9).
Table 9.
Genes controlling intestinal epithelial cells proliferation and modulated by enteric glial cells.
| Pro-proliferative | Anti-proliferative | ||
|---|---|---|---|
| Gene Symbol | Regulation of gene expression by EGC | Gene Symbol | Regulation of gene expression by EGC |
| E2F1 | down-regulated | TXNIP | up-regulated |
| FGFR2 | down-regulated | BTG1 | up-regulated |
| PPIL1 | down-regulated | TP53RK | down-regulated |
| MKI67 | up-regulated | SFRP4 | down-regulated |
The expression of major anti-proliferative and pro-proliferative genes was found to be up-regulated and down-regulated, respectively, by EGC. In particular, PPARG, TXNIP and BTG1 expressions in IEC were up-regulated by EGC. PPARγ activation has been described both in vivo and in vitro to inhibit intestinal epithelial cell proliferation [47,48] and to induce a G1 phase cell cycle arrest [27]. Furthermore, TXNIP encodes the thioredoxin-interacting protein, a negative regulator of thioredoxin. Thioredoxin is an important growth-promoting factor of IEC [49]. Moreover, TXNIP has also recently been suggested to be a tumor suppressor gene in hepatocellular carcinoma [50] and interestingly, TXNIP expression is decreased in colorectal cancer and ulcerative colitis [51]. Similarly, BTG1 has been shown to negatively regulate cell proliferation and to present a maximal expression during G0/G1 phases of the cell cycle in fibroblasts [52]. Further reinforcing the anti-proliferative effects of EGC on IEC is the EGC-induced down-regulation of the expression of pro-proliferative genes such as E2F1, FGFR2 and PPIL1. E2F1 is a gene encoding a protein member of the E2F family of transcription factors that regulate cell cycle progression by modulating expression of proteins required for the G1/S transition. It has been well described that growth stimulatory signals lead to active E2F1 accumulation and S-phase entry [53,54]. FGFR2 encodes a member of the FGF (Fibroblast Growth Factor) receptor family with high affinity for KGF (Keratinocyte Growth Factor) which is a major actor in the mesenchymal stimulation of epithelial cell proliferation [55,56]. Finally, PPIL1, which encodes a cyclophilin-related protein, PPIL1 (peptidyl prolyl isomerase-like protein), implicated in spliceosome activation, has recently been described to be overexpressed in colon tumors and PPIL1 silencing led to an inhibition of colon cancer cell growth [57,58].
These global anti-proliferative effects of EGC upon IEC have to be associated with the EGC-induced modulation of genes that would tend to be pro-proliferative, although these are clearly in reduced numbers. For instance, EGC increase MKI67 expression, which encodes the proliferation marker Ki-67. Indeed, Ki-67 is increasingly expressed during the cell cycle phases [59], excepted in G0 or in cells just escaping from G0 [60]. Its function is still unclear but knock-down for Ki-67 in cancerous cells leads to an inhibition of proliferation mainly via an induction of apoptosis [61,62]. Interestingly, EGC reduced the expression of TP53RK and SFRP4 in IEC that encode proteins involved in anti-proliferative pathways. TP53RK encodes PRPK which is a short kinase that phosphorylates p53, enhancing its transcriptional activity [63] and suppressing cell cycle transition G1/S [64]. SFRP4 encodes the protein sFRP4 (secreted frizzled-related protein), which is an inhibitor of the Wnt-signaling cascade through binding and sequestering Wnt ligand and, thus, has been shown to decrease cell proliferation in many cell lines [65-67].
Taken together, these data suggest that EGC tend to shift IEC transcriptome toward an anti-proliferative phenotype. These results could lead to the identification of specific targets responsible for the anti proliferative effects of EGC previously reported [12]. In addition, this global effect is supported further by the observation that EGC inhibit cell proliferation in part by inducing a cell cycle arrest in G0/G1 phase [11].
Cell survival
EGC differentially regulated in IEC the expression of genes involved in cell death. EGC appeared to modulate the expression of anti-apoptotic and pro-apoptotic genes toward a dominant pro-apoptotic effect (Table 10).
Table 10.
Genes controlling intestinal epithelial cells survival and modulated by enteric glial cells.
| Pro-apoptotic | Anti-apoptotic | ||
|---|---|---|---|
| Gene Symbol | Regulation of gene expression by EGC | Gene Symbol | Regulation of gene expression by EGC |
| BNIP3 | up-regulated | BAG1 | down-regulated |
| CASP4 | up-regulated | ASAH1 | down-regulated |
| CARD12 | down-regulated | GPX2 | down-regulated |
| TUBB3 | up-regulated | ||
Indeed, expressions of pro-apoptotic and anti-apoptotic genes were found to be up-regulated and down-regulated, respectively, by EGC. In particular, BNIP3 and CASP4 expression in IEC were up-regulated by EGC. CASP4, coding for the caspase-4 pro-apoptotic protein has been shown to induce cell death [68,69], like BNIP3 which encodes a pro-apoptotic protein member of Bcl-2 family [70,71]. Conversely, ASAH-1, GPX2 and BAG-1 were down-regulated by EGC. BAG-1 encodes a known anti-apoptotic protein implicated in Bcl-2 signalling pathway [72,73]. ASAH-1 encodes acid ceramidase, an enzyme that catabolizes ceramide into sphingosine by deacylation. Overexpression of acid ceramidase in cells confers on them an increased resistance to cell death induced by various factors such as TNF (tumor necrosis factor) or anti-cancerous drugs [74,75]. Finally, GPX2 encodes a member of the glutathione peroxidase (GPX) family and is a selenoprotein and a glutathione peroxidase. GPX2 is expressed in IEC [76] and inhibits oxidative stress-induced apoptosis in the human breast adenocarcinoma cell line MCF-7 [77].
These global pro-apoptotic effects of EGC upon IEC have to be considered also in view of the EGC-mediated regulation of genes which would tend to be anti-apoptotic, although these are in reduced number. In particular, EGC up-regulated the expression of TUBB3, a gene encoding the class III isotype of β-tubulin. Silencing of class III β-tubulin by siRNA reverted anti-cancer agent-resistant cells to a sensitive phenotype and promoted apoptosis [78,79]. Conversely, EGC inhibited the expression of CARD12 which encodes the CARD12 protein, a member of the CED4/Apaf-1 family and known to induce apoptosis when expressed in cells [80,81].
EGC-induced regulation of genes involved in cell death has probably no clear consequences on IEC survival. This is consistent with a previous study showing that EGC did not modify IEC survival [12].
Conclusion
The present study described the impact of EGC upon the transcriptome of proliferating Caco-2 cells in a validated non-contact co-culture model of EGC and IEC [12,13]. The results obtained confirmed the known role of EGC in the control of some IEB functions and, more interestingly, extended their role in the control of novel major IEB and IEC functions. This study further reinforced the emerging concept that EGC are an important component of the IEB environment with major protective effects. Indeed, the major pathways regulated by EGC in IEC identified with microarrays lead to enhanced cell adhesion, differentiation, and motility, which could favor repair, and reduced cell proliferation.
An important result of this study is the putative identification of genes involved in the anti-proliferative effects of EGC. Indeed, EGC have been shown to have potent anti-proliferative effects upon IEC [11,12]. Interestingly, these effects were associated with an induction of a cell cycle blockade in the G0/G1 phase [11] but were not associated with significant cell death [12]. These results are globally confirmed, as there was no clear trend in the EGC-induced modulation of genes controlling cell survival in IEC but a trend toward an up-regulation of the expression of genes involved in anti-proliferative pathways.
A major finding of our study is that EGC regulated the expression of genes involved in cell adhesion and differentiation toward a global increase of IEC adhesive properties. These results can be analyzed in view of the known effects of EGC upon IEB. Indeed, in vitro studies have shown that EGC increase IEB resistance and decrease IEB paracellular permeability [13]. In the present study, we also demonstrated that EGC could increase global IEB adhesion, in part by increasing cell-to-matrix adhesion. These results are in agreement with in vivo data showing that selective lesions of EGC lead to increased paracellular permeability and major IEB breakdown associated with the development of intestinal inflammation. However, the role of the molecular actors involved in these processes such as fibronectin, laminin or cytokeratin remains to be investigated. EGC might also favor barrier integrity by increasing its resistance to inflammatory stress either by its ability to down-regulate inflammatory genes such as CARD12 or by increasing IEC production of anti-inflammatory mediators such as VIP [82,83].
Another important finding of this study is the observation that EGC might regulate IEC metabolism. In particular, EGC up-regulated genes involved in lipid metabolism such as AADAC, MGLL or APOH, encoding respectively the arylacetamide deacetylase, monoglyceride lipase (MGL) and Apolipoprotein H [84-86]. Interestingly, inhibitors of MGL which is a serine hydrolase that converts 2-arachidonoylglycerol, a ligand of canabinoid receptors, to fatty acids and glycerol, increased gut transit time [87] but its impact on IEB functions remain unknown. EGC also modulated the expression of genes involved in protein metabolism such as CTSH that encodes cathepsin H, a lysosomal cysteine proteinase [88]. In addition, EGC increased the expression of genes involved in arginine metabolic pathway that are SLC7A7 and ASS, which encode respectively for the cationic amino acid transporter y(+)LAT1 and the argininosuccinate synthetase, enzyme catalyzing the penultimate step of the arginine biosynthetic pathways. The functional impact of EGC upon IEC metabolism needs to be investigated in future studies.
Regulation of IEB functions by EGC occurs mainly via paracrine pathways. The majority of EGC effects upon IEB functions are reproduced by glial-derived conditioned medium. In addition, various mediators have been identified as being involved in the control of cell proliferation or paracellular permeability. Our study supports the role of mediators such as TGF-β1 as a regulator of gene pathways modulated by EGC in IEC. In fact, TGF-β1 has been shown to increase the expression of FAK [43], TGFBI [89] or VIP [90]. EGC have also been shown to produce IL-6 [91]. IL-6 has recently been identified as a key molecule involved in IEB barrier protection via increasing both cytokeratin 8 and cytokeratin 18 proteins expression [92], whose mRNA expression were induced by EGC in IEC in our study. In this context, knowledge of genes modulated by EGC could direct future efforts aimed at identifying novel glial mediators involved in EGC control of IEB functions. Our data also further suggest that EGC differentially regulate some IEB functions as compared to fibroblasts, although comparison has only been performed on a limited set of genes and one cannot fully rule out that species differences could also be involved (fibroblasts of human origin vs. enteric glia of rat origin). However, these differences are consistent with the observation that while EGC have anti-proliferative effects on both human and rat IEC [12], fibroblasts increase IEC proliferation [93].
Collectively, our data support the concept that EGC play a key protective role upon IEB homeostasis by reinforcing global barrier functions. Additionally, our study reinforces data suggesting that enteric glia lesions and/or functional defects could be involved in the development of pathologies with altered barrier (such as inflammatory bowel diseases or colorectal cancer) and also be associated with increased barrier susceptibility to pathogen aggression.
Methods
Cell culture
CRL2690 (ATCC), a transformed EGC line isolated from adult rat myenteric plexus, was cultured in DMEM (4.5 g/L glucose; Gibco) supplemented with 10% heat-inactivated FBS, 2 mM glutamine (Gibco), 50 IU/mL penicillin and 50 μg/mL streptomycin. EGC were seeded at a concentration of 30,000 cells/mL in 6- and 12-well plates (Corning, Avon, France). EGC were cultured for an additional 24 hours after having reached confluence prior co-culture with IEC. CCD-18Co, a human colonic fibroblast cell line, was cultured in MEM Alpha Medium (Gibco) supplemented with 10% heat-inactivated FBS, 2 mM glutamine (Gibco), 0.1 mM MEM NEAA (Gibco), 50 IU/mL penicillin and 50 μg/mL streptomycin. Fibroblasts were seeded at a concentration of 130,000 cells/mL in 12-well plates (Corning). Fibroblats were cultured in EGC medium, as described above, for an additional 24 hours after having reached confluence prior co-culture with IEC. Caco-2 cells (ATCC), isolated from a human colonic adenocarcinoma, were cultured in DMEM (4.5 g/L glucose; Gibco) supplemented with 10% heat-inactivated FBS, 2 mM glutamine (Gibco), 50 IU/mL penicillin and 50 μg/mL streptomycin and were seeded at a concentration of 140,000 cells/mL onto porous Transwell filters (6-well and 12-well Transwell clear, 0.40 μm porosity, Corning). Caco-2 cells were processed for experiment 1 day after their seeding. To characterize EGC impact onto IEC transcriptome and functions, IEC seeded onto filters were cultured in presence of EGC seeded at the bottom of the 6-well or 12-well plates.
Microarray experiments
Transcriptomic analysis was performed with a microarray of 6,864 genes called "Cancerochip" and available from the West Genopole transcriptome core facility of Nantes. These Cancerochips contained 6,864 probes (50-mers oligonucleotides), each specific of a single gene. These genes were selected to be preferentially and/or differentially expressed in Caco-2 cells and in various tumours. Three replicates of each probe were spotted onto Cancerochips. This allowed the measurement of the probes reproducibility within the array.
Total RNA was extracted from Caco-2 cells cultured on 6-well filters alone or in presence of EGC at t = 0, 8 and 24 hours. Each condition was performed in 4 replicates allowing the measurement of the reproducibility of the cell culture experiments. RNA extraction was performed with RNeasy mini kit (Qiagen) according to the manufacturer's recommendations.
The protocols used for subsequent amplification and labelling were described in the DNA chips platform protocols. Each individual sample was compared to a reference pool consisting of Caco-2 cells transcripts of the four replicates extracted at t = 0 hour. Total RNA (0.5 μg) was amplified and labelled using the Amino Allyl MessageAmp II aRNA Amplification kit (Ambion) and CyDye Post Labelling Reactive Dyes (Amersham). After reverse transcription to synthesize first strand cDNA, second strand cDNA was subsequently synthesized following the manufacturer's protocol. In vitro transcription was then achieved in order to amplify the initial transcripts quantity, concomitantly with aminoallyl-dUTP incorporation to perform labelling with cyanins (Cy5 for the reference and Cy3 for samples). The hybridization of the chips was performed following the protocol of the West Genopole transcriptome core facility of Nantes. After washing, the chips were scanned (Scanexpress- Perkin Elmer).
Data analysis
After acquisition, the scanned images were analyzed using GenePix Pro v5.1 software (Axon). Raw signals were processed using the MADSCAN package http://cardioserve.nantes.inserm.fr/mad/madscan/. Spots with weak, saturated signal or badly shaped were considered as missing values. Print-tip lowess was applied to raw signals to normalize both channels (Cy3 and Cy5) of a same array. Fitting coefficients were calculated on rank invariant spots, assuming that they correspond to ubiquitous genes (genes that did not vary between samples). Sample to reference ratios (Cy3/Cy5) were further calculated, and Log transformed (Expression Logratios). Probes with more than 25% of missing values were rejected.
In order to identify differentially expressed genes, Expression Logratios were analyzed using Genespring v7.0 software (Agilent Technologies). Genes differentially expressed between Caco-2 cells cultured alone or in presence of EGC were searched with Benjamini and Hochberg False Discovery Rate method with a significance threshold of 0.05. This method includes a correction for multi-testing and has been widely used for microarray data [94]. This analysis led to the identification of 98 genes differentially expressed in IEC cultured in presence of EGC as compared to control, i.e. IEC cultured alone at t = 24 hours and none at t = 8 hours. Analysis of variance (ANOVA) using time of culture and presence/absence of ECG as parameters gave very similar results. Data visualization using hierarchical clustering and Volcano-Plot suggested that this strategy might have missed some differentially expressed genes at t = 24 hours; we thus selected an additional set of genes based on expression fold-changes. Twenty seven genes with a fold-change threshold of 2 and a t-test p-value < 0.05 without multi-testing correction were found. Altogether 116 unique genes were found differentially expressed in IEC at t = 24 hours of culture in presence of EGC as compared to control.
Hierarchical clustering was performed after normalization on medians of the ratio values of the t = 0 hour-condition samples. Hierarchical clustering was performed using the Cluster software. It was applied to order either genes and samples or genes only. It creates a visualization of the grouping of genes and samples based on profile similarity, even if it does not provide robustness assessment of the classification.
Among the 116 genes identified with Genespring analysis, 17 of them did not present reliable values at t = 0 hour. Thus, these 17 genes were excluded from hierarchical clustering analyses. As a consequence, clustering analyses only involved 99 genes.
Microarray data were uploaded to GEO database http://www.ncbi.nlm.nih.gov/geo/ and are available under the access number GSE17027.
RT-quantitative PCR
Extraction of total RNA from Caco-2 cells cultured alone, in presence of EGC or fibroblasts for 24 hours was performed with RNeasy Mini kit (Qiagen) according to the manufacturer's protocol. For reverse transcription, 1 μg of purified total RNA was denatured and subsequently processed for reverse transcription using SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer's recommendations. PCR amplifications were performed using the Absolute Blue SYBR green fluorescein kit (ABgene) according to the manufacturer's protocol and run on MyiQ thermocycler (Biorad). The expression of the gene S6 was analyzed in parallel as an internal control.
CDH1 [GenBank: NM_004360]
Forward primer:
5'-GACCAGGACTATGACTACTTGAACG-3'
Reverse primer:
5'-ATCTGCAAGGTGCTGGGTGAACCTT-3'
E2F1 [GenBank: NM 005225]
Forward primer:
5'-CCGCTCGAGGAGAAGTCACGCTATGA-3'
Reverse primer:
5'-CCCAAGCTTTTGGTGATGTCATAGATGC-3'
FN1 [GenBank: NM_054034]
Forward primer:
5'-GCAGGCTCAGCAAATGGTTCAG-3'
Reverse primer:
5'-AGGTAGGTCCGCTCCCACTG-3'
FGFR2 [GenBank: NM_022970]
Forward primer:
5'-GTCCTGCCAAAACAGCAAG-3'
Reverse primer:
5'-CCCCTATGCAGTAAATGGCTA-3'
GPX2 [GenBank: NM_002083]
Forward primer:
5'-gtccttggcttcccttgc-3'
Reverse primer:
5'-tgttcaggatctcctcattctg-3'
LAMA5 [GenBank: NM_005560]
Forward primer:
5'-CCCACCGAGGACCTTTACTGC-3'
Reverse primer:
5'-GGTGTGCCTTGTTGCTGTTGG-3'
PPARG [GenBank: NM_138712/NM_005037/NM_138711/NM_015869]
Forward primer:
5'-ttgctgtcattattctcagtgga-3'
Reverse primer:
5'-gaggactcagggtggttcag-3'
PTK2 [GenBank: NM_153831/NM_005607]
Forward primer:
5'- GAGATCCTGTCTCCAGTCTAC-3'
Reverse primer:
5'- TGCACCTGCTATTTTTAGTTG-3'
SMAD3 [GenBank: NM 005902]
Forward primer:
5'-CCAAGCTTAGAACGGGCAGGAGGAG-3'
Reverse primer:
5'-CACTCGAGTGGTGGCTGTGCAGGTC-3'
S6 [GenBank: NM_001010]
Forward primer:
5'-TGGCAAGATGATCCCAATGA-3'
Reverse primer:
5'-AGCTTCTTTGGGACACCTGCT-3'
Adhesion experiments
Global adhesion assay
IEC adhesion was estimated by performing a "global adhesion assay" that evaluated total IEC adhesion to their environment, i.e. adhesion to neighboring IEC and adhesion to matrix. IEC were cultivated on filters (12-well Transwell clear, 0.40 μm porosity, Corning) alone or in the presence of EGC for 24 hours. IEC were then trypsinized with 0.01% trypsin-EDTA free (Sigma) allowing gentle trypsinization for 30 minutes at 37°C. Non-adherent IEC were harvested and counted in a blind fashion using Malassez slides (VWR international). IEC remaining adhered on filters were trypsinized with 2.5% trypsin with EDTA (Gibco), harvested and counted. Results are expressed in percentage of remaining adherent IEC normalized to the total number of counted IEC (i.e., adherent IEC and non-adherent IEC). Only those series in which the percentage of IEC total adhesion in control condition was comprised between 20 and 70% were analyzed.
Cell-to-matrix adhesion assay
IEC were cultivated on filters (12-well Transwell clear, 0.40 μm porosity, Corning) alone or in presence of EGC for 24 hours. IEC were then trypsinized for 10 minutes with a 2.5% trypsin-EDTA (Gibco). Trypsin was neutralized with IEC culture medium (see above). IEC were subsequently reseeded on filters and incubated for 3 hours at 37°C. Time of incubation has been defined to allow 50% of seeded IEC to adhere to filters in control condition. Following incubation, unseeded cells were harvested and counted in a blind fashion using Malassez slides (VWR international). IEC that had adhered on filters were trypsinized and counted. Results are expressed in percentage of adherent IEC normalized to the total number of counted IEC (i.e., adherent IEC and non-adherent IEC). Only those series in which the percentage of IEC total adhesion in control condition was comprised between 20 and 70% were analyzed.
List of abbreviations
CNS: Central nervous system; EGC: Enteric glial cells; ENS: Enteric nervous system; FAK: Focal adhesion kinase; IEB: Intestinal epithelial barrier; IEC: Intestinal epithelial cells; PPARγ: Peroxisome proliferator-activated receptor gamma; TGF-β1: Transforming growth factor beta-1; VIP: Vasoactive intestinal peptide.
Authors' contributions
LVL designed microarrays studies and functional experiments and carried out microarrays studies. She analyzed the microarrays data and wrote the manuscript. MMM designed and carried out functional experiments and performed RT-qPCR studies. JL supervised microarray studies. IG participated in microarrays hybridization. RT assisted with the bioinformatics analysis. RH contributed to the bioinformatics analysis and wrote the manuscript. MN supervised the project and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Laurianne Van Landeghem, Email: laurianne.van-landeghem@univ-nantes.fr.
Maxime M Mahé, Email: maxime.mahe@univ-nantes.fr.
Raluca Teusan, Email: raluca.teusan@nantes.inserm.fr.
Jean Léger, Email: jean.leger@nantes.inserm.fr.
Isabelle Guisle, Email: isabelle.guisle@nantes.inserm.fr.
Rémi Houlgatte, Email: remi.houlgatte@nantes.inserm.fr.
Michel Neunlist, Email: michel.neunlist@univ-nantes.fr.
Acknowledgements
The authors are grateful to Dr. Damien MASSON for kindly giving PPARγ primers. They also thank the West Genopole transcriptome core facility for providing expertise and technical help. They thank Philippe Aubert for his technical assistance. The authors are thankful to Anne Tomasevich for her help in editing the paper. LVL was supported by a PhD grant of the MNRT and MMM by a PhD grant from Nantes Métropole. This work was supported by a grant from INCa Appel d'Offre Libre 2007 (MOPRESTAGLIA).
References
- Powell DW, Adegboyega PA, Di Mari JF, Mifflin RC. Epithelial cells and their neighbors I. Role of intestinal myofibroblasts in development, repair, and cancer. Am J Physiol Gastrointest Liver Physiol. 2005;289(1):G2–7. doi: 10.1152/ajpgi.00075.2005. [DOI] [PubMed] [Google Scholar]
- Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol. 1999;277(2 Pt 1):C183–201. doi: 10.1152/ajpcell.1999.277.2.C183. [DOI] [PubMed] [Google Scholar]
- Blikslager AT, Moeser AJ, Gookin JL, Jones SL, Odle J. Restoration of barrier function in injured intestinal mucosa. Physiol Rev. 2007;87(2):545–564. doi: 10.1152/physrev.00012.2006. [DOI] [PubMed] [Google Scholar]
- Shao J, Sheng GG, Mifflin RC, Powell DW, Sheng H. Roles of myofibroblasts in prostaglandin E2-stimulated intestinal epithelial proliferation and angiogenesis. Cancer Res. 2006;66(2):846–855. doi: 10.1158/0008-5472.CAN-05-2606. [DOI] [PubMed] [Google Scholar]
- Penna FJ, Peret LA, Vieira LQ, Nicoli JR. Probiotics and mucosal barrier in children. Curr Opin Clin Nutr Metab Care. 2008;11(5):640–644. doi: 10.1097/MCO.0b013e32830a70ab. [DOI] [PubMed] [Google Scholar]
- Toumi F, Neunlist M, Cassagnau E, Parois S, Laboisse CL, Galmiche JP, Jarry A. Human submucosal neurones regulate intestinal epithelial cell proliferation: evidence from a novel co-culture model. Neurogastroenterol Motil. 2003;15(3):239–242. doi: 10.1046/j.1365-2982.2003.00409.x. [DOI] [PubMed] [Google Scholar]
- Xue J, Askwith C, Javed NH, Cooke HJ. Autonomic nervous system and secretion across the intestinal mucosal surface. Auton Neurosci. 2007;133(1):55–63. doi: 10.1016/j.autneu.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunlist M, Toumi F, Oreschkova T, Denis M, Leborgne J, Laboisse CL, Galmiche JP, Jarry A. Human ENS regulates the intestinal epithelial barrier permeability and a tight junction-associated protein ZO-1 via VIPergic pathways. Am J Physiol Gastrointest Liver Physiol. 2003;285(5):G1028–1036. doi: 10.1152/ajpgi.00066.2003. [DOI] [PubMed] [Google Scholar]
- Ruhl A. Glial cells in the gut. Neurogastroenterol Motil. 2005;17(6):777–790. doi: 10.1111/j.1365-2982.2005.00687.x. [DOI] [PubMed] [Google Scholar]
- Savidge TC, Sofroniew MV, Neunlist M. Starring roles for astroglia in barrier pathologies of gut and brain. Lab Invest. 2007;87(8):731–736. doi: 10.1038/labinvest.3700600. [DOI] [PubMed] [Google Scholar]
- Neunlist M, Van Landeghem L, Bourreille A, Savidge T. Neuro-glial crosstalk in inflammatory bowel disease. J Intern Med. 2008;263(6):577–583. doi: 10.1111/j.1365-2796.2008.01963.x. [DOI] [PubMed] [Google Scholar]
- Neunlist M, Aubert P, Bonnaud S, Van Landeghem L, Coron E, Wedel T, Naveilhan P, Ruhl A, Lardeux B, Savidge T. Enteric glia inhibit intestinal epithelial cell proliferation partly through a TGF-beta1-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2007;292(1):G231–241. doi: 10.1152/ajpgi.00276.2005. [DOI] [PubMed] [Google Scholar]
- Savidge TC, Newman P, Pothoulakis C, Ruhl A, Neunlist M, Bourreille A, Hurst R, Sofroniew MV. Enteric glia regulate intestinal barrier function and inflammation via release of S-nitrosoglutathione. Gastroenterology. 2007;132(4):1344–1358. doi: 10.1053/j.gastro.2007.01.051. [DOI] [PubMed] [Google Scholar]
- Bush TG, Savidge TC, Freeman TC, Cox HJ, Campbell EA, Mucke L, Johnson MH, Sofroniew MV. Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell. 1998;93(2):189–201. doi: 10.1016/S0092-8674(00)81571-8. [DOI] [PubMed] [Google Scholar]
- Aube AC, Cabarrocas J, Bauer J, Philippe D, Aubert P, Doulay F, Liblau R, Galmiche JP, Neunlist M. Changes in enteric neurone phenotype and intestinal functions in a transgenic mouse model of enteric glia disruption. Gut. 2006;55(5):630–637. doi: 10.1136/gut.2005.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffit F, Perreault N, Jean D, Francoeur C, Herring E, Rancourt C, Rivard N, Vachon PH, Pare F, Boucher MP. Repressed E-cadherin expression in the lower crypt of human small intestine: a cell marker of functional relevance. Exp Cell Res. 2005;302(2):206–220. doi: 10.1016/j.yexcr.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Pecina-Slaus N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int. 2003;3(1):17. doi: 10.1186/1475-2867-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Wang J, Ye Z, Ip NY, Lin SC. CDK5 activator p35 downregulates E-cadherin precursor independently of CDK5. FEBS Lett. 2008;582(8):1197–1202. doi: 10.1016/j.febslet.2008.02.053. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4(6):571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale DR, Watson AJ. Rac-1 and IQGAP are potential regulators of E-cadherin-catenin interactions during murine preimplantation development. Gene Expr Patterns. 2002;2(1-2):17–22. doi: 10.1016/S0925-4773(02)00350-7. [DOI] [PubMed] [Google Scholar]
- Tzu J, Marinkovich MP. Bridging structure with function: structural, regulatory, and developmental role of laminins. Int J Biochem Cell Biol. 2008;40(2):199–214. doi: 10.1016/j.biocel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck N, Gross I, Gendry P, Stutzmann J, Freund JN, Kedinger M, Simon-Assmann P, Launay JF. Laminin isoforms: biological roles and effects on the intracellular distribution of nuclear proteins in intestinal epithelial cells. Exp Cell Res. 2005;303(2):494–503. doi: 10.1016/j.yexcr.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Kolachala VL, Bajaj R, Wang L, Yan Y, Ritzenthaler JD, Gewirtz AT, Roman J, Merlin D, Sitaraman SV. Epithelial-derived fibronectin expression, signaling, and function in intestinal inflammation. J Biol Chem. 2007;282(45):32965–32973. doi: 10.1074/jbc.M704388200. [DOI] [PubMed] [Google Scholar]
- Borgono CA, Michael IP, Shaw JL, Luo LY, Ghosh MC, Soosaipillai A, Grass L, Katsaros D, Diamandis EP. Expression and functional characterization of the cancer-related serine protease, human tissue kallikrein 14. J Biol Chem. 2007;282(4):2405–2422. doi: 10.1074/jbc.M608348200. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6(1):56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Liu F, Chen Z, Wang J, Shao X, Cui Z, Yang C, Zhu Z, Xiong D. Overexpression of cell surface cytokeratin 8 in multidrug-resistant MCF-7/MX cells enhances cell adhesion to the extracellular matrix. Neoplasia. 2008;10(11):1275–1284. doi: 10.1593/neo.08810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bush CR, Necela BM, Su W, Yanagisawa M, Anastasiadis PZ, Fields AP, Thompson EA. RS5444, a novel PPARgamma agonist, regulates aspects of the differentiated phenotype in nontransformed intestinal epithelial cells. Mol Cell Endocrinol. 2006;251(1-2):17–32. doi: 10.1016/j.mce.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Lefebvre M, Paulweber B, Fajas L, Woods J, McCrary C, Colombel JF, Najib J, Fruchart JC, Datz C, Vidal H. Peroxisome proliferator-activated receptor gamma is induced during differentiation of colon epithelium cells. J Endocrinol. 1999;162(3):331–340. doi: 10.1677/joe.0.1620331. [DOI] [PubMed] [Google Scholar]
- Mahoney ZX, Stappenbeck TS, Miner JH. Laminin alpha 5 influences the architecture of the mouse small intestine mucosa. J Cell Sci. 2008;121(Pt 15):2493–2502. doi: 10.1242/jcs.025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy P, Robin H, Kornprobst M, Capeau J, Cherqui G. Enterocytic differentiation of the human Caco-2 cell line correlates with alterations in integrin signaling. J Cell Physiol. 1998;177(4):618–627. doi: 10.1002/(SICI)1097-4652(199812)177:4<618::AID-JCP12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251(5000):1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Laprise P, Chailler P, Houde M, Beaulieu JF, Boucher MJ, Rivard N. Phosphatidylinositol 3-kinase controls human intestinal epithelial cell differentiation by promoting adherens junction assembly and p38 MAPK activation. J Biol Chem. 2002;277(10):8226–8234. doi: 10.1074/jbc.M110235200. [DOI] [PubMed] [Google Scholar]
- Ku NO, Zhou X, Toivola DM, Omary MB. The cytoskeleton of digestive epithelia in health and disease. Am J Physiol. 1999;277(6 Pt 1):G1108–1137. doi: 10.1152/ajpgi.1999.277.6.G1108. [DOI] [PubMed] [Google Scholar]
- Fath KR, Trimbur GM, Burgess DR. Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol. 1994;126(3):661–675. doi: 10.1083/jcb.126.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Sung CH. Cytoplasmic dynein regulation by subunit heterogeneity and its role in apical transport. J Cell Biol. 2001;153(7):1499–1509. doi: 10.1083/jcb.153.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon PH, Simoneau A, Herring-Gillam FE, Beaulieu JF. Cellular fibronectin expression is down-regulated at the mRNA level in differentiating human intestinal epithelial cells. Exp Cell Res. 1995;216(1):30–34. doi: 10.1006/excr.1995.1004. [DOI] [PubMed] [Google Scholar]
- Deschenes C, Alvarez L, Lizotte ME, Vezina A, Rivard N. The nucleocytoplasmic shuttling of E2F4 is involved in the regulation of human intestinal epithelial cell proliferation and differentiation. J Cell Physiol. 2004;199(2):262–273. doi: 10.1002/jcp.10455. [DOI] [PubMed] [Google Scholar]
- Vachon PH, Cardin E, Harnois C, Reed JC, Vezina A. Early establishment of epithelial apoptosis in the developing human small intestine. Int J Dev Biol. 2000;44(8):891–898. [PubMed] [Google Scholar]
- Leeb SN, Vogl D, Grossmann J, Falk W, Scholmerich J, Rogler G, Gelbmann CM. Autocrine fibronectin-induced migration of human colonic fibroblasts. Am J Gastroenterol. 2004;99(2):335–340. doi: 10.1111/j.1572-0241.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Goke M, Zuk A, Podolsky DK. Regulation and function of extracellular matrix intestinal epithelial restitution in vitro. Am J Physiol. 1996;271(5 Pt 1):G729–740. doi: 10.1152/ajpgi.1996.271.5.G729. [DOI] [PubMed] [Google Scholar]
- Howard TH, Hartwig J, Cunningham C. Lymphocyte-specific protein 1 expression in eukaryotic cells reproduces the morphologic and motile abnormality of NAD 47/89 neutrophils. Blood. 1998;91(12):4786–4795. [PubMed] [Google Scholar]
- Hines OJ, Ryder N, Chu J, McFadden D. Lysophosphatidic acid stimulates intestinal restitution via cytoskeletal activation and remodeling. J Surg Res. 2000;92(1):23–28. doi: 10.1006/jsre.2000.5941. [DOI] [PubMed] [Google Scholar]
- Walsh MF, Ampasala DR, Hatfield J, Heide R Vander, Suer S, Rishi AK, Basson MD. Transforming growth factor-beta stimulates intestinal epithelial focal adhesion kinase synthesis via Smad- and p38-dependent mechanisms. Am J Pathol. 2008;173(2):385–399. doi: 10.2353/ajpath.2008.070729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima A, Tomimoto A, Fujita K, Sugiyama M, Takahashi H, Ikeda I, Hosono K, Endo H, Yoneda K, Iida H. Inhibition of peroxisome proliferator-activated receptor gamma activity suppresses pancreatic cancer cell motility. Cancer Sci. 2008;99(10):1892–1900. doi: 10.1111/j.1349-7006.2008.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KR, Choi HN, Lee HJ, Baek HA, Park HS, Jang KY, Chung MJ, Moon WS. A peroxisome proliferator-activated receptor gamma antagonist induces vimentin cleavage and inhibits invasion in high-grade hepatocellular carcinoma. Oncol Rep. 2007;18(4):825–832. [PubMed] [Google Scholar]
- Schaefer KL, Takahashi H, Morales VM, Harris G, Barton S, Osawa E, Nakajima A, Saubermann LJ. PPARgamma inhibitors reduce tubulin protein levels by a PPARgamma, PPARdelta and proteasome-independent mechanism, resulting in cell cycle arrest, apoptosis and reduced metastasis of colorectal carcinoma cells. Int J Cancer. 2007;120(3):702–713. doi: 10.1002/ijc.22361. [DOI] [PubMed] [Google Scholar]
- Fujisawa T, Nakajima A, Fujisawa N, Takahashi H, Ikeda I, Tomimoto A, Yonemitsu K, Nakajima N, Kudo C, Wada K. Peroxisome proliferator-activated receptor gamma (PPARgamma) suppresses colonic epithelial cell turnover and colon carcinogenesis through inhibition of the beta-catenin/T cell factor (TCF) pathway. J Pharmacol Sci. 2008;106(4):627–638. doi: 10.1254/jphs.FP0071766. [DOI] [PubMed] [Google Scholar]
- Matthiessen MW, Pedersen G, Albrektsen T, Adamsen S, Fleckner J, Brynskov J. Peroxisome proliferator-activated receptor expression and activation in normal human colonic epithelial cells and tubular adenomas. Scand J Gastroenterol. 2005;40(2):198–205. doi: 10.1080/00365520410009573. [DOI] [PubMed] [Google Scholar]
- Takaishi S, Sawada M, Seno H, Kayahara T, Morita-Fujisawa Y, Fukuzawa H, Chiba T. Growth promoting effect of thioredoxin on intestinal epithelial cells. Dig Dis Sci. 2003;48(2):379–385. doi: 10.1023/A:1021952132241. [DOI] [PubMed] [Google Scholar]
- Sheth SS, Bodnar JS, Ghazalpour A, Thipphavong CK, Tsutsumi S, Tward AD, Demant P, Kodama T, Aburatani H, Lusis AJ. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene. 2006;25(25):3528–3536. doi: 10.1038/sj.onc.1209394. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Masuda H, Ishii Y, Nishida Y, Kobayashi M, Asai S. Decreased expression of thioredoxin interacting protein mRNA in inflamed colonic mucosa in patients with ulcerative colitis. Oncol Rep. 2007;18(3):531–535. [PubMed] [Google Scholar]
- Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP. BTG1, a member of a new family of antiproliferative genes. Embo J. 1992;11(4):1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365(6444):349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Mann DJ, Jones NC. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr Biol. 1996;6(4):474–483. doi: 10.1016/S0960-9822(02)00515-8. [DOI] [PubMed] [Google Scholar]
- Rubin JS, Osada H, Finch PW, Taylor WG, Rudikoff S, Aaronson SA. Purification and characterization of a newly identified growth factor specific for epithelial cells. Proc Natl Acad Sci USA. 1989;86(3):802–806. doi: 10.1073/pnas.86.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch PW, Rubin JS, Miki T, Ron D, Aaronson SA. Human KGF is FGF-related with properties of a paracrine effector of epithelial cell growth. Science. 1989;245(4919):752–755. doi: 10.1126/science.2475908. [DOI] [PubMed] [Google Scholar]
- Xu C, Zhang J, Huang X, Sun J, Xu Y, Tang Y, Wu J, Shi Y, Huang Q, Zhang Q. Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with SKIP. J Biol Chem. 2006;281(23):15900–15908. doi: 10.1074/jbc.M511155200. [DOI] [PubMed] [Google Scholar]
- Obama K, Kato T, Hasegawa S, Satoh S, Nakamura Y, Furukawa Y. Overexpression of peptidyl-prolyl isomerase-like 1 is associated with the growth of colon cancer cells. Clin Cancer Res. 2006;12(1):70–76. doi: 10.1158/1078-0432.CCR-05-0588. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Broll R, Bruch HP, Duchrow M. Proliferation marker pKi-67 affects the cell cycle in a self-regulated manner. J Cell Biochem. 2002;87(3):334–341. doi: 10.1002/jcb.10302. [DOI] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710–1715. [PubMed] [Google Scholar]
- Zheng JN, Ma TX, Cao JY, Sun XQ, Chen JC, Li W, Wen RM, Sun YF, Pei DS. Knockdown of Ki-67 by small interfering RNA leads to inhibition of proliferation and induction of apoptosis in human renal carcinoma cells. Life Sci. 2006;78(7):724–729. doi: 10.1016/j.lfs.2005.05.064. [DOI] [PubMed] [Google Scholar]
- Wang R, Luo D, Ma X, Yang W, Chen R, Liu Y, Meng L, Zhou J, Xu G, Lu YP. Antisense Ki-67 cDNA transfection reverses the tumorigenicity and induces apoptosis in human breast cancer cells. Cancer Invest. 2008;26(8):830–835. doi: 10.1080/07357900801941878. [DOI] [PubMed] [Google Scholar]
- Abe Y, Matsumoto S, Wei S, Nezu K, Miyoshi A, Kito K, Ueda N, Shigemoto K, Hitsumoto Y, Nikawa J. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J Biol Chem. 2001;276(47):44003–44011. doi: 10.1074/jbc.M105669200. [DOI] [PubMed] [Google Scholar]
- Abe Y, Takeuchi T, Imai Y, Murase R, Kamei Y, Fujibuchi T, Matsumoto S, Ueda N, Ogasawara M, Shigemoto K. A Small Ras-like protein Ray/Rab1c modulates the p53-regulating activity of PRPK. Biochem Biophys Res Commun. 2006;344(1):377–385. doi: 10.1016/j.bbrc.2006.03.071. [DOI] [PubMed] [Google Scholar]
- Horvath LG, Henshall SM, Kench JG, Saunders DN, Lee CS, Golovsky D, Brenner PC, O'Neill GF, Kooner R, Stricker PD. Membranous expression of secreted frizzled-related protein 4 predicts for good prognosis in localized prostate cancer and inhibits PC3 cellular proliferation in vitro. Clin Cancer Res. 2004;10(2):615–625. doi: 10.1158/1078-0432.CCR-0707-03. [DOI] [PubMed] [Google Scholar]
- Horvath LG, Lelliott JE, Kench JG, Lee CS, Williams ED, Saunders DN, Grygiel JJ, Sutherland RL, Henshall SM. Secreted frizzled-related protein 4 inhibits proliferation and metastatic potential in prostate cancer. Prostate. 2007;67(10):1081–1090. doi: 10.1002/pros.20607. [DOI] [PubMed] [Google Scholar]
- Carmon KS, Loose DS. Secreted frizzled-related protein 4 regulates two Wnt7a signaling pathways and inhibits proliferation in endometrial cancer cells. Mol Cancer Res. 2008;6(6):1017–1028. doi: 10.1158/1541-7786.MCR-08-0039. [DOI] [PubMed] [Google Scholar]
- Lin R, Sun Y, Li C, Xie C, Wang S. Identification of differentially expressed genes in human lymphoblastoid cells exposed to irradiation and suppression of radiation-induced apoptosis with antisense oligonucleotides against caspase-4. Oligonucleotides. 2007;17(3):314–326. doi: 10.1089/oli.2007.0064. [DOI] [PubMed] [Google Scholar]
- Martin CA, Panja A. Cytokine regulation of human intestinal primary epithelial cell susceptibility to Fas-mediated apoptosis. Am J Physiol Gastrointest Liver Physiol. 2002;282(1):G92–G104. doi: 10.1152/ajpgi.2002.282.1.G92. [DOI] [PubMed] [Google Scholar]
- Ray R, Chen G, Velde C Vande, Cizeau J, Park JH, Reed JC, Gietz RD, Greenberg AH. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J Biol Chem. 2000;275(2):1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- Crowley-Weber CL, Payne CM, Gleason-Guzman M, Watts GS, Futscher B, Waltmire CN, Crowley C, Dvorakova K, Bernstein C, Craven M. Development and molecular characterization of HCT-116 cell lines resistant to the tumor promoter and multiple stress-inducer, deoxycholate. Carcinogenesis. 2002;23(12):2063–2080. doi: 10.1093/carcin/23.12.2063. [DOI] [PubMed] [Google Scholar]
- Tang SC. BAG-1, an anti-apoptotic tumour marker. IUBMB Life. 2002;53(2):99–105. doi: 10.1080/15216540211473. [DOI] [PubMed] [Google Scholar]
- Clemo NK, Arhel NJ, Barnes JD, Baker J, Moorghen M, Packham GK, Paraskeva C, Williams AC. The role of the retinoblastoma protein (Rb) in the nuclear localization of BAG-1: implications for colorectal tumour cell survival. Biochem Soc Trans. 2005;33(Pt 4):676–678. doi: 10.1042/BST0330676. [DOI] [PubMed] [Google Scholar]
- Saad AF, Meacham WD, Bai A, Anelli V, Elojeimy S, Mahdy AE, Turner LS, Cheng J, Bielawska A, Bielawski J. The functional effects of acid ceramidase overexpression in prostate cancer progression and resistance to chemotherapy. Cancer Biol Ther. 2007;6(9):1455–1460. doi: 10.4161/cbt.6.9.4623. [DOI] [PubMed] [Google Scholar]
- Strelow A, Bernardo K, Adam-Klages S, Linke T, Sandhoff K, Kronke M, Adam D. Overexpression of acid ceramidase protects from tumor necrosis factor-induced cell death. J Exp Med. 2000;192(5):601–612. doi: 10.1084/jem.192.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S, Wingler K, Schmehl K, Jacobasch G, Kreuzer OJ, Meyerhof W, Brigelius-Flohe R. Cellular and subcellular localization of gastrointestinal glutathione peroxidase in normal and malignant human intestinal tissue. Free Radic Res. 2001;35(6):655–663. doi: 10.1080/10715760100301181. [DOI] [PubMed] [Google Scholar]
- Yan W, Chen X. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J Biol Chem. 2006;281(12):7856–7862. doi: 10.1074/jbc.M512655200. [DOI] [PubMed] [Google Scholar]
- Mhaidat NM, Thorne RF, de Bock CE, Zhang XD, Hersey P. Melanoma cell sensitivity to Docetaxel-induced apoptosis is determined by class III beta-tubulin levels. FEBS Lett. 2008;582(2):267–272. doi: 10.1016/j.febslet.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Kavallaris M, Burkhart CA, Horwitz SB. Antisense oligonucleotides to class III beta-tubulin sensitize drug-resistant cells to Taxol. Br J Cancer. 1999;80(7):1020–1025. doi: 10.1038/sj.bjc.6690507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes BJ, Wang L, Huang WJ, Lavellee M, Manji GA, Brown M, Jurman M, Cao J, Morgenstern J, Merriam S. Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem Biophys Res Commun. 2001;284(1):77–82. doi: 10.1006/bbrc.2001.4928. [DOI] [PubMed] [Google Scholar]
- Lu C, Wang A, Wang L, Dorsch M, Ocain TD, Xu Y. Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem Biophys Res Commun. 2005;331(4):1114–1119. doi: 10.1016/j.bbrc.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rey E, Delgado M. Role of vasoactive intestinal peptide in inflammation and autoimmunity. Curr Opin Investig Drugs. 2005;6(11):1116–1123. [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Trickett JI, Patel DD, Knight BL, Saggerson ED, Gibbons GF, Pease RJ. Characterization of the rodent genes for arylacetamide deacetylase, a putative microsomal lipase, and evidence for transcriptional regulation. J Biol Chem. 2001;276(43):39522–39532. doi: 10.1074/jbc.M101764200. [DOI] [PubMed] [Google Scholar]
- Nakaya Y, Schaefer EJ, Brewer HB Jr. Activation of human post heparin lipoprotein lipase by apolipoprotein H (beta 2-glycoprotein I) Biochem Biophys Res Commun. 1980;95(3):1168–1172. doi: 10.1016/0006-291X(80)91595-8. [DOI] [PubMed] [Google Scholar]
- Ho SY, Delgado L, Storch J. Monoacylglycerol metabolism in human intestinal Caco-2 cells: evidence for metabolic compartmentation and hydrolysis. J Biol Chem. 2002;277(3):1816–1823. doi: 10.1074/jbc.M108027200. [DOI] [PubMed] [Google Scholar]
- Duncan M, Thomas AD, Cluny NL, Patel A, Patel KD, Lutz B, Piomelli D, Alexander SP, Sharkey KA. Distribution and function of monoacylglycerol lipase in the gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2008;295(6):G1255–1265. doi: 10.1152/ajpgi.90500.2008. [DOI] [PubMed] [Google Scholar]
- Bohley P, Seglen PO. Proteases and proteolysis in the lysosome. Experientia. 1992;48(2):151–157. doi: 10.1007/BF01923508. [DOI] [PubMed] [Google Scholar]
- Thapa N, Lee BH, Kim IS. TGFBIp/betaig-h3 protein: a versatile matrix molecule induced by TGF-beta. Int J Biochem Cell Biol. 2007;39(12):2183–2194. doi: 10.1016/j.biocel.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Pitts RL, Wang S, Jones EA, Symes AJ. Transforming growth factor-beta and ciliary neurotrophic factor synergistically induce vasoactive intestinal peptide gene expression through the cooperation of Smad, STAT, and AP-1 sites. J Biol Chem. 2001;276(23):19966–19973. doi: 10.1074/jbc.M011759200. [DOI] [PubMed] [Google Scholar]
- Ruhl A, Franzke S, Collins SM, Stremmel W. Interleukin-6 expression and regulation in rat enteric glial cells. Am J Physiol Gastrointest Liver Physiol. 2001;280(6):G1163–1171. doi: 10.1152/ajpgi.2001.280.6.G1163. [DOI] [PubMed] [Google Scholar]
- Wang L, Srinivasan S, Theiss AL, Merlin D, Sitaraman SV. Interleukin-6 induces keratin expression in intestinal epithelial cells: potential role of keratin-8 in interleukin-6-induced barrier function alterations. J Biol Chem. 2007;282(11):8219–8227. doi: 10.1074/jbc.M604068200. [DOI] [PubMed] [Google Scholar]
- Goke M, Kanai M, Podolsky DK. Intestinal fibroblasts regulate intestinal epithelial cell proliferation via hepatocyte growth factor. Am J Physiol. 1998;274(5 Pt 1):G809–818. doi: 10.1152/ajpgi.1998.274.5.G809. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]