Abstract
Pro-apoptotic Bax is a soluble and monomeric protein under normal physiological conditions. Upon its activation substantial structural rearrangements occur: The protein inserts into the mitochondrial outer membrane and forms higher molecular weight oligomers. Subsequently, the cells can undergo apoptosis. In our studies, we focused on the structural rearrangements of Bax during oligomerization and on the protein stability. Both protein conformations exhibit high stability against thermal denaturation, chemically induced unfolding and proteolytic processing. The oligomeric protein is stable up to 90 °C as well as in solutions of 8 M urea or 6 M guanidinium hydrochloride. Helix 9 appears accessible in the monomer but hidden in the oligomer assessed by proteolysis. Tryptophan fluorescence indicates that the environment of the C-terminal protein half becomes more apolar upon oligomerization, whereas the loop region between helices 1 and 2 gets solvent exposed.
Keywords: Bcl-2 proteins, Apoptosis, Conformational changes, Protein structure, Protein stability
Introduction
In order to maintain the survival of complex organisms, cells need a strict regulation of division and cell death programs. Members of the Bcl-2 protein family are essential factors in this regulation (Cory and Adams 2002; Kuwana and Newmeyer 2003). The Bcl-2 proteins can be divided into three subgroups: First the anti-apoptotic Bcl-2 like proteins, second the pro-apoptotic multi-domain proteins, and third the pro-apoptotic BH3 only proteins. Human Bax alpha (abbreviated Bax) is probably the best studied example within this family and belongs to the proapoptotic multi-domain proteins. Moreover, Bax is one of the key factors known to initiate apoptosis in vertebrates. Deletion of Bax and its homolog Bak in mice leads to prenatal death or severe deformation in different tissues (Lindsten et al. 2000).
Monomeric Bax protein is cytosolic and inactive. Upon activation, it can undergo a conformational change to form an oligomeric protein complex that inserts into the MOM (Antonsson et al. 2001; Youle and Karbowski 2005). By this, Bax permeabilizes the membrane for cytochrome c (Kuwana et al. 2002) and downstream apoptotic events can be initiated. The conformational change of Bax is regulated by an ever growing number of proteins [e.g. Bid and Bim (Chipuk et al. 2006; Kuwana et al. 2005; Lovell et al. 2008), Bif1 (Cuddeback et al. 2001; Takahashi et al. 2005), p53 (Chipuk et al. 2004; Mihara et al. 2003), ASC (Ohtsuka et al. 2004), Ku70 (Sawada et al. 2003), 14-3-3 (Nomura et al. 2003) or Humanin (Guo et al. 2003)]. In vitro, activation and oligomerization can be induced by mixing Bax with detergents (Antonsson et al. 2000; Antonsson et al. 2001; Hsu and Youle 1997, 1998; Kuwana et al. 2002; Suzuki et al. 2000). Obviously, in living cells Bax activation has to be strictly regulated since premature activation may leads to sudden cell death while blocked activation favors early cancer development.
The structure of monomeric Bax has been determined by NMR methods (Suzuki et al. 2000) and shows a globular fold composed of nine alpha helices, termed α1 to α9. In the structure, α5 is hidden in the hydrophobic core and α9 is attached to a hydrophobic cleft on the protein surface. In contrast, the structure of oligomeric Bax is unknown, but it was demonstrated by biochemical methods that α5, α6 and α9 may be inserted into the OMM (Annis et al. 2005; Garcia-Saez et al. 2004; Nechushtan et al. 1999; Wolter et al. 1997).
The aim of this work was to study the conformational changes upon Bax oligomerization in order to understand the underlying architecture of the oligomeric complex. In our study, we compared monomeric and oligomeric Bax (in detergents or reconstituted in liposomes) using CD-spectroscopy, tryptophan fluorescence (TF), protease digestion, size exclusion chromatography (SEC) and chemical cross-linking. Our results indicate that Bax is extraordinarly stable in the oligomeric form. During monomer to oligomer transition the protein undergoes the following conformational changes: (a) helix α9 becomes protected against protease attack whereas (b) α1 moves in a protease accessible position. (c) The tryptophans in α5 to α9 experience a more hydrophobic environment. (d) The monomer shows an unusually high energy barrier for unfolding but needs a hydrophobic environment to adopt the active conformation. (e) The oligomerization even further stabilizes the protein against unfolding. (f) In the complex, the protein is tightly packed and protected towards thermal and chemical stress.
Material and methods
ClustalW alignment In order to perform the alignment, amino acid sequences from different vertebrate species annotated as Bax isoform alpha [human (Q07812), rat (Q63690), mouse (Q07813), bovine (O022703)] or complete sequences having an equivalent start codon assignment [cat (Q8SQ43), dog (Q8HYUS), zebra fish (Q919N4)] as well as having only a short extension [xenopus (Q98U13)] were used. Sequences with N-terminal extensions or deletions within the sequence were excluded, e.g. the sequence from Pan troglodytes (chimpanzee), which shows only two differences in the part homologous to the human Bax alpha, but is elongated by 70 amino acids at the N-terminus.
Expression and purification of human Bax alpha We essentially followed the procedure described by Suzuki et al. (2000). The purity of the protein was analyzed by SDS-PAGE and LC-MS (calculated mass: 21,184 Da; determined mass: 21,183 Da).
Bax reconstitution in liposomes Lipid mixtures from E. coli or bovine heart extracts (Avanti polar lipids Inc. Alabaster, AL) were dissolved in chloroform, dried by evaporation to form a thin film, and resuspended in buffer 1 (20 mM Tris-HCl, 100 mM NaCl; pH 7.5) by repeatedly vortexing, freezing and thawing. The final lipid concentration was 20 mg/ml. The solution was passed through a membrane with 200 or 400 nm pores (Avestin Inc. Ottawa, Canada) for at least 25 times. Bax pre-incubated with 0.1% DDM was mixed with the vesicles pre-incubated with 0.1% DDM to a final concentration of 0.25 mg/ml protein and 12 mg/ml lipid. Biobeads (BioRAD, Hercules, CA) were added to remove excess detergent. For protein insertion the solution was incubated for 3 h at 30 °C at 300 rpm. To remove free protein as well as aggregates, a nycodenz gradient was used. The gradient was layered as followed: the liposome/Bax solution in 10% nycodenz (dissolved in buffer 1) was overlaid with 5% nycodenz in buffer 1 and finally pure buffer 1. The gradient was spun at 200,000×g for 30 min at 10 °C. A pellet and a swimming lipid band (fuzzy when bovine heart lipids were used) were separated. The latter was mixed with buffer 1 and centrifuged under identical conditions to remove the nycodenz by sedimentation.
Secondary structure determination by circular-dichroism spectroscopy The concentration of Bax (in buffer 2; 20 mM Tris, pH 8.8) was adjusted to 0.1 mg/ml. The CD-spectra and melting curves were recorded on a Jasco J715 spectropolarimeter (Jasco, Gross Umstadt, Germany) with a Jasco PFD 350S Peltier type FDCD attachment for temperature control using a 0.1 mm quartz cuvette. Two spectra were accumulated per measurement using a data pitch of 0.1 nm, a scan speed of 20 nm s−1 and 1 nm slit width. The content of secondary structure was calculated using the program CDNN (Bohm et al. 1992). Notably, samples of Bax did not show any precipitation during the temperature increase which was tested by UV-spectroscopy. If buffer 1 was used instead of buffer 2, only neglectable differences were visible in the spectra.
Protease digestion Monomeric and oligomeric Bax samples were mixed with subtilisin or proteinase K at a stoichiometry of 1:200 and incubated for one hour on ice. The reaction was stopped either by (a) treatment with the denaturing SDS-gel loading buffer and subsequent boiling for 5 min or (b) freezing in liquid nitrogen and storage at −80 °C before further analysis. Protein samples were subjected to SDS-PAGE and blotted onto a PVDF membrane (transfer buffer: 25 mM Tris-HCl, 192 mM glycine, 20% methanol) for subsequent N-terminal sequencing of individual bands (using a gas-phase sequencer Procise 492cLC, Applied Biosystems, Foster City, CA). Sample mixtures were also analyzed by ESI-MS. After Bax reconstitution in liposomes, the protein concentration was estimated by the band intensity on a SDS-gel. Consequently, the estimation was less accurate than the estimation by UV spectroscopy. Furthermore, since ESI-MS did not work with Bax liposomes only N-terminal sequencing was performed.
Tryptophan fluorescence Fluorescence emission spectra were recorded on a Perkin-Elmer spectrometer (LS50B, Waltham MA). Bax (concentrations of 0.25 to 1 µM), and free acetylated-tryptophan (4 µM) were excited at 280 nm at a slit width of 5 nm to detect the emitted fluorescence in the range between 300 and 400 nm.
Cross-link experiments Protein samples were adjusted to a concentration of about 0.5 mg/ml in the presence or absence of 0.5% DDM. These samples were slowly heated to 50–90 °C and the temperature was kept constant for 600 s. Afterwards, the samples were immediately cooled on ice. 10 µl samples were mixed with 1 µl ammonium peroxydisulfate (APS, 25 mM) and 2 µl ruthenium (II) Tris-bipyridyldication (5 mM Sigma-Aldrich) in the dark and then immediately exposed to illumination with visible light (400 to 700 nm, generated by a Xenon lamp, 100 W Leica, filters: KG4, GG 395 nm). The photo-induced reaction was stopped by the addition of 5 µl SDS-gel loading buffer.
Miscellaneous To induce oligomerization, Bax was mixed with 0.5% DM (Anatrace), 0.5% DDM (Anatrace) or 2% OG (Anatrace) in buffer 1. The samples were incubated under shaking for at least 8 h at 4 °C. The oligomerization was analyzed by size exclusion chromatography on a Superdex 200 column using SMART FPLC (GE Healthcare). ESI-MS was performed on a micrOTOF LC (Bruker Daltonics, Billerica, MA). Absorption spectra were collected on a Shimadzu UV-1700 UV-visible spectrophotometer. Bax structure was illustrated with Pymol using the PDB file 1F16 of monomeric Bax (Suzuki et al. 2000). Data were plotted using origin 6.1.
Results
Primary structure analysis of Bax
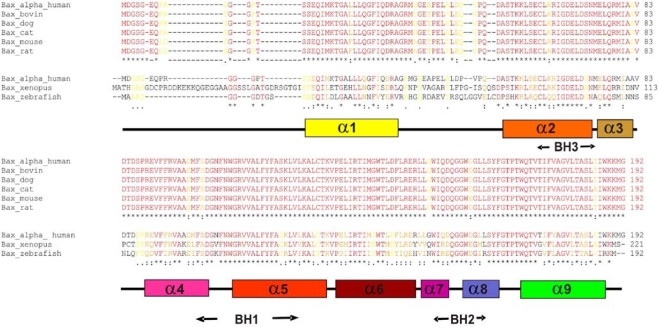
Bax alpha is a 21 kDa splice variant of human Bax that is composed of 192 amino acids. The protein contains three of four known Bcl-2 homology domains, BH1-3 (Fig. 1a), and the NMR structure (Suzuki et al. 2000) revealed nine alpha helices, α1–α9 (Fig. 1a, b). While studies on Bax proteins from other sources are rare, human Bax has been extensively described in the literature. In order to locate conserved residues which may be relevant for the conformational change or oligomer formation, the Bax protein was aligned with Bax orthologs from different mammalian and vertebrate origin (see Fig. 2). The overall similarity of the mammalian orthologs was too high (91% identity; see Fig. 2) to identify conserved residues relevant for protein function. However, after alignment of human Bax alpha with sequences from the vertebrate species Xenopus laevis and Danio rerio (see Fig. 2) less conserved sequence sections became apparent. The overall sequence homology dropped to 76% and only 44% of the amino acid residues were identical. The C-terminal part including the entire structural parts supposedly involved in the activation process starting at the BH1 domain (α5–α9, amino acids 98 to 192 in Bax) is significantly higher conserved (90% homology) than the N-terminal part of the protein (62% homology). The conserved amino acids were highlighted in the NMR-structure model of monomeric human Bax alpha (shown in Fig. 1c). A surface exposed domain and the protein core were shown to be most conserved among Bax from different species. The invariant surface region is formed by amino acids of α2, the loop between α4 and α5, part of α5 as well as the region of α7 to α9. In other words, all three Bcl-2 homology domains are involved (see Fig. 1a–c). In addition, the conserved surface region is surprisingly hydrophobic (as indicated by the green hue in Fig. 1d), which might indicate a specific role in membrane interaction or oligomerization.
Fig. 1.
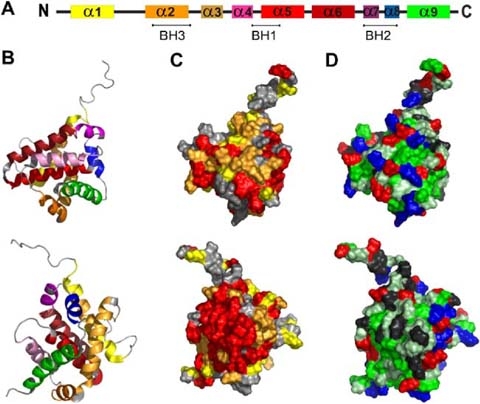
Representation of conserved regions, and hydrophobicity in the Bax structure. The secondary as well as a cartoon of the ternary structure of Bax (PDB:1F16) are shown in a and b, respectively. Helices are colored according to the code: α1 yellow, α2 fawn, α3 orange, α4 pink, α5 light red, α6 dark red, α7 purple, α8 blue, α9 green. c Shows a representation of the sequence conservation of the Bax surface. Conserved amino acids were identified by ClustalW alignment of human Bax alpha and its orthologs of Xenopus laevis and Danio rerio and were highlighted in the published NMR structure of human Bax alpha (Suzuki et al. 2000). Amino acids identical in all sequences are shown in red. Substitutions which were classified as highly conserved residues are marked in orange and semi-conserved ones are displayed in yellow. d Demonstrates the hydrophobicity of the surface exposed amino acids (according to the hydropathy index by Kyte and Doolittle 1982). Acidic [E,D] and basic [K,R] amino acids are shown in red and blue, respectively. Hydrophobic amino acids [I,V,L,A,C,F,M] are shown in green and slightly hydrophobic residues [G,T,S,W] in light green. Others [Q,N,P,H,Y] are marked in grey. b–d Show two views of the molecule tilted by 180°
Fig. 2.
Amino acid sequence alignment of orthologous Bax alpha representatives from mammals and vertebrates. Amino acids identical in all sequences are colored in red and labeled with asterisks. Substitutions of highly conserved residues are marked in orange with colons and of semi-conserved residues are displayed in yellow indicated with periods. The secondary structure (for color code see Fig. 1a) as well as the position of the BH domains of Bax are also shown
Comparison of the secondary structures of monomeric and oligomeric Bax For the structural comparison of monomeric and oligomeric Bax, the protein was expressed in Escherichia coli and purified in its monomeric form as specified in (Suzuki et al. 2000) (see Fig. 3a). According to previous studies (Antonsson et al. 2000; Antonsson et al. 2001; Kuwana et al. 2002), the purified protein was mixed with detergents such as OG, DM and DDM to induce oligomerization. In order to investigate the influence of the individual detergents more thoroughly, we used detergents of various alkyl chain lengths (8, 10 and 12) as well as different head group moieties.
Fig. 3.
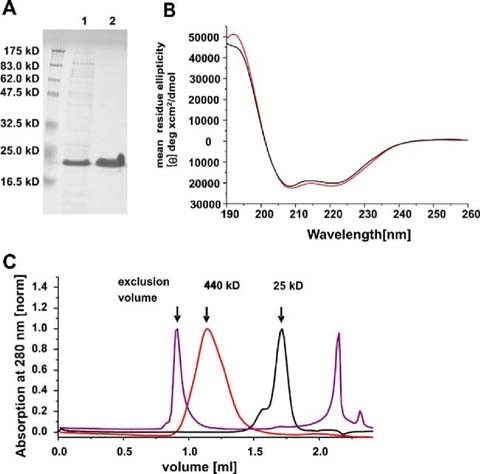
Purification, folding and oligomerization of the Bax protein. The SDS gel shown in a demonstrates the purity of Bax: (1) after the chitin affinity chromatography (first purification step) (2) after anion exchange chromatography (second purification step). Molecular weight standard bands are indicated on the left margin. The calculated molecular weight of Bax is 21 kD. b CD spectra of monomeric (black) and oligomeric Bax in 0.5% DDM (red). c SEC analysis of monomeric and oligomeric Bax as well as monomeric Bax pre-incubated at 90 °C. Monomeric Bax is shown in black and eluted as a monomer with a small amount of dimers. Oligomeric Bax (in 0.5% DDM) is shown in red and monomeric Bax pre-incubated at 90 °C in purple. The arrows mark the exclusion volume as well as the elution maxima of ferritin (440 kD) and chymotrysiongen A (25 kD)
The secondary structures of Bax in the monomeric and oligomeric forms were compared by CD spectroscopy (Fig. 3b). Only minor structure differences were induced upon detergent-induced oligomerization. Monomeric and oligomeric Bax in 0.5% DDM showed the typical CD spectrum of a purely α-helical protein (maximum at 192 nm, minima at 208 nm and 222 nm, shown in Fig. 3b). Oligomerization of Bax in 0.5% DM or 2% OG showed comparable results (data not shown). The α-helical content was determined to be about 60% for the monomer and about 64% for the oligomer. This is in close agreement with previously published CD spectra of monomeric human Bax alpha that displayed approximately 66% α-helix content (Yethon et al. 2003). As well, the NMR structure of monomeric Bax shows that 65% of the amino acids are contained in helices (Suzuki et al. 2000). From these data, we concluded that Bax was properly folded in the monomeric and the oligomeric form and only a small portion of the secondary structure was restructured upon detergent-induced oligomerization. The oligomerization of Bax in detergents was later confirmed by SEC (see Fig. 3c).
Bax reconstituted in liposomes was not analyzed by CD due to insufficiently accurate protein concentration determination (see Material and Methods).
The C-terminal domain of Bax is in close contact to the hydrophobic detergent environment after oligomerization As outlined above, the sequence comparison of Bax orthologs showed a significantly higher conservation of the C-terminal part of the protein (see Fig. 2). All six tryptophans are located in this region (Fig. 4a) and except Trp188 (which is replaced by arginine in the zebra fish sequence), are conserved (see Fig. 2). We recorded TF in order to analyze the influence of detergents or membrane insertion on the local environment of the tryptophan residues during oligomerization.
Fig. 4.
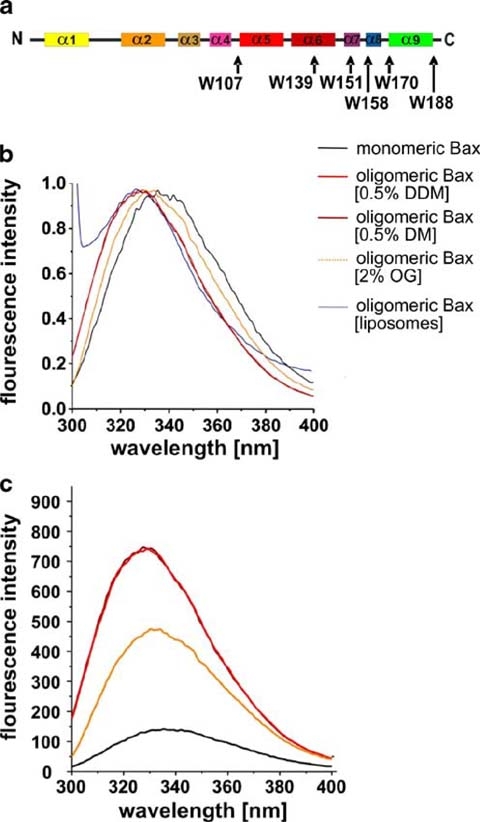
Tryptophan fluorescence of Bax. The positions of the tryptophans in the secondary structure of Bax are highlighted by arrows and corresponding labels in a. The color code is the same as described for Fig. 1a. TF spectra (normalized to 1) of monomeric and oligomeric Bax in different detergents as well as Bax reconstituted in liposomes are shown in b. Not Normalized TF spectra are shown in c with the same color code as indicated in b
The emission maximum of monomeric Bax was determined at 336 nm (Fig. 4b) showing the tryptophans in an at least partially solvent accessible position (Royer 2006). This is in line with the NMR structure (Suzuki et al. 2000) where all tryptophanes are at least partially solvent exposed. The oligomerization of Bax in detergent or liposomes caused a blue shift of the TF emission maximum up to 9 nm (shown in Fig. 4b emission maximum in DDM: 328 nm, in DM: 328 nm, in OG: 331 nm, in liposomes of bovine heart lipid extracts: 327 nm). Additionally, the intensity of the TF increased upon oligomerization (three- to fourfold for DM and DDM, twofold for OG, not measured for Bax reconstituted in liposomes; see Fig. 4c).
In order to understand the influence of detergents on the TF emission independent of the influence of the detergents on the protein, the emission spectrum of acetylated tryptophan was recorded. The addition of detergent caused nearly no shift in emission (data not shown), demonstrating that all blue shifts shown were due to conformational changes in the protein backbone and not to polarity shifts of the buffer environment.
Protease treatment of heterologously expressed Bax and Bax in mammalian cell extracts The increased hydrophobicity of the C-terminal part of Bax upon oligomerization implies that parts of the protein may become buried in the core of the oligomer and might, therefore, be inaccessible for proteases. Goping et al. (Goping et al. 1998) performed proteinase K digestion of Bax in mammalian cell extracts of FL5.12 cells before and after induction of apoptosis (by growth factor IL-3 withdrawal). The authors observed that upon initiation of apoptosis and Bax activation the accessibility of Bax to proteinase K changed. Their data indicate, that proteinase K treatment caused an N-terminal truncation of the activated Bax in natural membranes of apoptotic cells whereas the N-terminus remained unchanged in the monomeric protein in untreated cells. In contrast, the monomeric protein was cleaved at the C-terminus.
We repeated the protease treatment with heterologously produced Bax to analyze, first, if the recombinant monomeric Bax and the detergent induced Bax oligomers adopt conformations comparable to those of inactive, monomeric and active, oligomeric Bax in mammalian cells, respectively. Second, we intended to analyze the cleavage sites in both conformations in order to understand the conformational change and further explore the conformation of the oligomeric form.
On SDS gels, the band pattern after proteinase K or subtilisin treatment of the heterologously produced monomeric and oligomeric Bax resembled that of Bax in mammalian cell extracts before and after induction of apoptosis, respectively (Goping et al. 1998) (see Fig. 5a and Western blots in Goping et al. 1998). Moreover, mass analysis of the digested proteins revealed that the detergent induced oligomer was truncated at the N-terminus, whereas the monomeric form was cleaved at the C-terminus and its N-terminus remained intact. Both observations are in line with the data of Goping et al. (see Table 1 and Fig. 5b).
Fig. 5.
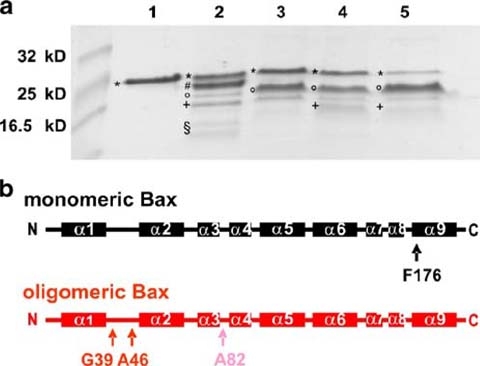
Analysis of subtilisin treated Bax. Fragments of differently treated Bax samples after proteolysis with subtilisin are shown on a coomassie-stained 17% SDS gel in a. Untreated monomeric Bax was loaded onto lane 1, subtilisin treated monomeric Bax is shown in lane 2, oligomeric Bax (in 0.5% DDM) after proteolysis with subtilisin refers to lane 3. Monomeric and oligomeric (in 0.5% DM) Bax both pre-heated to 90 °C are presented in lane 4 and lane 5, respectively. Subsequent analysis of proteolytic products after heating revealed different fragments of Bax, which are labeled with asterisks for full length Bax (aa 1–192 and aa 1–191), number sign for aa 1–176, aa 1–174 and aa 1–173, degree symbol for aa 39–192, aa 47–192, aa 39–191 and aa 47–191, plus symbol aa 82–192 and aa 83–192 as well as section symbol for aa 1–39 and aa1– 46. In b the main cleavage sites in monomeric (black) and oligomeric (red) Bax are highlighted by arrows
Table 1.
Proteinase K cleavage fragments of Bax
| Identified proteolytic fragments | 40°C | 90°C | ||
|---|---|---|---|---|
| Monomer | Oligomer (detergent) | Monomer | Oligomer (detergent) | |
| aa 1–192 | <30% | <40% | – | – |
| aa 1–38 | Traces | – | ||
| aa 1–46 | ||||
| aa 1–176 | >50% | |||
| aa 1–174 | <20% | – | – | – |
| aa 1–172 | ||||
| aa 1–171 | ||||
| aa 39–192 | <10% | >50% | >80% | >80% |
| aa 47–192 | – | |||
| aa 39–176 | Traces | |||
| aa 82–192 | – | <10% | <10% | <10% |
| 82–191 | ||||
| 83–191 | ||||
| 83–192 | ||||
| 85–192 | ||||
| 85–191 | ||||
| aa 53–192, aa | – | – | <10% | <10% |
| 58–192 | ||||
Amounts of fragments identified by mass analysis of monomeric and oligomeric Bax after proteinase K digestion with and without preincubation of Bax at 90 °C; aa amino acid.
Further analysis of subtilisin (Fig. 5a) or proteinase K (Fig. 7a and Table 1) treated monomeric Bax identified a fragment lacking 16 C-terminal residues as the major cleavage product. This was accompanied by fragments of lower abundance lacking 18, 19 and 21 C-terminal residues. Thus, the C-terminal helix α9 was protease digested in the monomeric form suggesting that it is not tightly attached to the protein, whereas the N-terminus was quite stable (see Table 1).
Fig. 7.
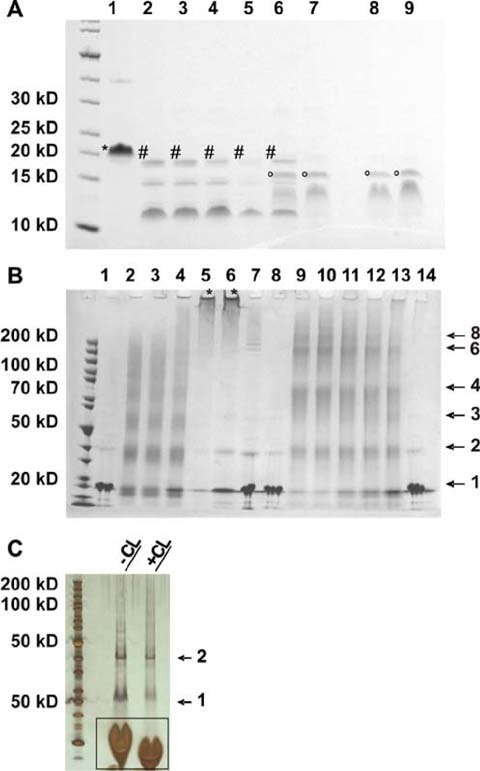
Properties of heat treated Bax. Monomeric and oligomeric Bax were incubated at different temperatures for 10 min, cooled on ice and treated with proteinase K for 1 h on ice (a) or cross linked (b). Cross link oligomers in liposomes are shown in c. Separation was done on 4–12% gradient SDS gels and analyzed. a Untreated monomeric Bax was loaded onto lane 1, proteinase K treated monomeric Bax pre-incubated at different temperatures were loaded onto lanes 2 through 7: lane 2 4 °C, lane 3 50 °C, lane 4 60 °C, lane 5 70 °C, lane 6 80 °C and lane 7 90 °C. In lane 8 and 9 oligomeric Bax (in 0.5% DM) was pre-incubated at different temperatures: lane 8 4 °C, lane 9 90 °C. Mass analysis of the proteolytic products after pre-heating revealed differently sized Bax fragments. The labeling is equal to that shown in Fig. 5a. b Monomeric and oligomeric Bax proteins cross-linked after pre-incubation at different temperatures: Lane 1 untreated monomeric Bax pre-incubated at 4 °C, lane 2 to 6: monomeric Bax after pre-incubation at different temperatures and cross-linking for 5 s: lane 2 4 °C, lane 3 50 °C, lane 4 70 °C, lane 5 80 °C, lane 6 90 °C, lane 7 monomeric Bax pre-incubated at 90 °C without cross-linking, lane 8 untreated oligomeric Bax (0.5% DDM) pre-incubated at 4 °C, lane 9–13 oligomeric Bax (0.5% DDM) pre-incubated at different temperatures and after cross-linking for 5 s, lane 9 4 °C, lane 10 50 °C, lane 11 70 °C, lane 12 80 °C, lane 13 90 °C, lane 14 oligomeric Bax (0.5% DDM) pre-incubated at 90 °C without cross-linking. The main visible band and the number of cross linked monomers per band are indicated by arrows and numbers on the right margin. The asterisk indicates aggregates or big oligomers that did not enter the gel. c Demonstration of a silver stained SDS-gel of reconstituted Bax oligomers (without heat treatment) before and after cross-linking. The arrows indicates monomers and dimers. The bands framed by the black box are not protein but lipid bands
By contrast, the detergent induced oligomers showed no proteolytic cleavage at the C-terminus. However, the N-terminal part of the protein was cleaved after Ser4, Met38, Leu45, and, though less frequent, after Ala81 and Ala82 (see Fig. 5b). The cut at Ser4 demonstrates that a cleavage within the first 12 N-terminal amino acids, that were described to be very flexible (Suzuki et al. 2000), is possible only in oligomeric Bax, but the same domain is inaccessible in the monomeric protein. It also showed that Bax oligomerization provokes remarkable differences in the C- and N-terminus of the protein, which is in agreement with the TF data and previously reported observation (Roucou and Martinou 2001 and references therein). Briefly, we observed that oligomerization led to exposure of N-terminal parts of Bax, especially the loop between α1 and α2 that contains Met38 and Leu45, whereas α9 was protected from protease attack and must be in a shielded environment likely to be located in the core structure of the oligomer.
Since we were unable to do mass spectrometry analysis with Bax inserted in liposomes (see Material and Methods), proteolytic degradation of Bax liposomes was more difficult to follow. However, N-terminal sequencing clearly identified a Bax fragment lacking 38 N-terminal residues after treatment with proteinase K. Since cleavage after Met38 also occurred in detergent induced oligomeric Bax, the Bax conformations in both membrane mimicking environments seems not to vary much.
Monomeric Bax is more temperature sensitive than oligomeric Bax. Yethon et al. (2003) reported differences in the melting behavior of monomeric Bax and Bax inserted in liposomes (DOPC: DOPE: DOPS: PI: CL, 43: 27: 9: 9: 12 molar ratio). They showed, that monomeric Bax is very resistant to temperature depending denaturation and heat stability even increases in the presence of lipids. However, it remains unclear which part of the protein unfolds during heat treatment and if detergent has a similar stabilizing effect as liposomes.
In order to test the heat stability of oligomeric Bax in detergent, we recorded melting curves of the protein. The curves of monomeric Bax looked comparable to those of Yethon et al. (2003). Moreover, the stability of Bax in detergent and in liposomes seems to be similar, as far as both increase stability a lot compared to the monomeric form (see melting curves in Fig. 6 upper and lower panel and Yethon et al. 2003).
Fig. 6.
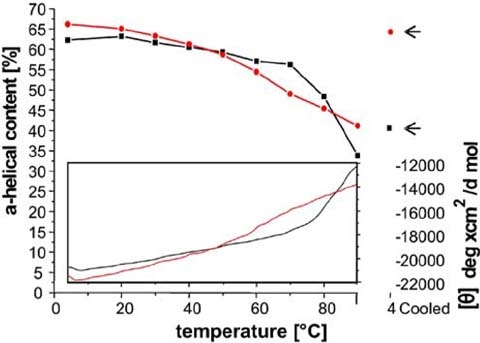
Melting behavior of heat treated monomeric and oligomeric Bax. The thermal stability of monomeric (black) and oligomeric (red) Bax is shown. The upper panel depicts the decrease of the alpha helical content of Bax during the temperature increase, estimated from CD spectra taken at different temperatures. The alpha helical content of Bax heated to 90 °C and recooled to 4 °C is highlighted by arrows. The lower inset panel indicates the decrease of the ellipticity at 222 nm measured during the whole temperature increase
Monomeric Bax showed a sigmoidal melting curve and started to unfold at temperatures higher than 75 °C. But even at 90 °C melting was not completed (shown in Fig. 6 lower panel) and a CD-spectrum at 90 °C revealed still α-helical line shape with a remaining α-helical content of 35% (see Fig. 6 upper panel). Subsequent cooling to 4 °C resulted in a renaturation of 45% α-helices (see Fig. 6 upper panel). Addition of detergent (DDM) at 90 °C or after heating caused a better but still only partly refolding of the helices (up to 50% α -helices, data not shown).
In order to identify partially unfolded sub-structures of monomeric Bax at increased temperatures, the protein was slowly warmed to 50 °C, 60 °C, 70 °C, 80 °C or 90 °C, followed by a rapid cooling to 4 °C and digestion using proteinase K (proteolytic fragments shown in Fig. 7a). Up to 70 °C the degradation pattern did not change. After heating to 80 °C, a mixture of N-terminal (cleaved after Ala46) and C-terminal truncated Bax (mainly cleaved after Phe176) was identified. Pre-incubation at 90 °C led to a completely inaccessible C-terminus, but cleavage of the 45 N-terminal amino acids (see Fig. 7a and Table 1). In summary the experiment showed that the N-terminus of monomeric Bax became accessible and therefore is likely to be unfolded after heat treatment, whereas the rest of the molecule seems to be stable. Additionally, the cleavage pattern of monomeric heat treated Bax is similar to the one monitored with untreated oligomeric Bax (see Fig. 7a).
Detailed analysis of Bax incubated at 90 °C prior to proteolysis showed that the protein is not monomeric anymore, but forms aggregates or very big oligomers (furthermore called “megaoligomers”) that are larger than normal detergent or liposome-induced oligomers. Megaoligomers were identified by SEC (Fig. 3c) and cross-link analysis (Fig. 7b). However, they are too small to scatter light at 215 nm arguing against protein aggregation.
After cross-linking of oligomeric Bax (in DDM) and subsequent analysis on SDS-gels big oligomers appeared (up to octamers, as shown in Fig. 7b; varying the conditions pointed to even bigger oligomers). Noticeable most oligomers were even numbered (see Fig. 7b). Surprisingly, proteinase K digestion, gel electrophoresis and cross-linking of oligomeric Bax that was pre-incubated at 90 °C showed no differences as to when kept at 4 °C (shown in Fig. 7a, b). From these observations we concluded that Bax oligomers resist temperatures up to 90 °C.
Comparison of both Bax conformations in chaotrophic reagents After recognition of the unusual Bax stability even at elevated temperatures, we were curious to see whether the protein was also resistant towards chemical denaturation. Therefore, the protein was mixed with increasing concentrations of chaotropic reagents. We recorded TF emission spectra to detect the reagent concentration at which denaturation of the protein takes place. Surprisingly, no shift in the emission maxima of either monomeric or oligomeric Bax could be observed in 8 M urea (illustrated in Fig. 8a). At a concentration of 4.5 M guanidinium hydrochloride denaturation of monomeric Bax was induced and at 6 M guanidinium hydrochloride the emission maximum was shifted to 346 nm (see Fig. 8b) indicating protein unfolding. Thus, monomeric Bax showed a very high stability towards chemical denaturation. This behavior was even more pronounced in oligomeric Bax. At 6 M guanidinium hydrochloride the emission maximum of oligomeric Bax in 0.5% DDM was only slightly shifted (4 nm) to 332 nm (see Fig. 8b), indicating a folded protein.
Fig. 8.
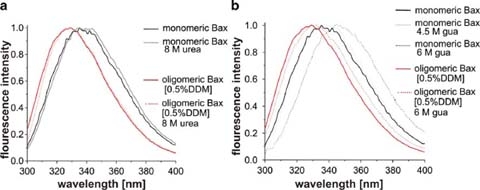
TF of monomeric and oligomeric Bax after treatment with denaturing agents. Treatment with urea is shown in a and treatment with guanidinium hydrochloride in b. The data were normalized and maximal TF emission was set to 1
Discussion
Bax is involved in the intrinsic apoptotic pathway and known to exist in two distinct structural conformations: inactive Bax is monomeric, whereas the active protein forms an oligomer. In vivo it is activated by apoptotic stimuli and, thus, decides on the death or survival of a cell (Kuwana and Newmeyer 2003). During its activation Bax inserts into the MOM and forms a pore by oligomerization (Antonsson et al. 2001; Kuwana et al. 2002; Nechushtan et al. 2001). Through this pore cytochrome c is released promoting further apoptotic events (reviewed in Antignani and Youle 2006; Kuwana and Newmeyer 2003; Reed 2006 and references therein). Activation of Bax is known to involve conformational rearrangements such as the insertion of the helices α5, α6 and α9 into the MOM (Annis et al. 2005).
We address two main questions in this work: First, we intended to identify which part(s) of Bax are involved in structural rearrangements during oligomerization. Furthermore, it was important to assess whether the recombinantly produced monomeric Bax and the detergent induced Bax oligomer folded in the same way as does Bax in non-apoptotic and apoptotic mammalian cells, respectively. Since structural data on Bax conformation in living cells are difficult to obtain and, therefore, are limited we decided to focus on in vitro studies. Detergent induced oligomerization of Bax has previously been used to study conformational rearrangements of Bax (Antonsson et al. 2000; Hsu and Youle 1997). However, these studies did not convincingly reveal that the fold of detergent oligomerized Bax corresponds to that in apoptotic cells. In order to better mimic the natural environment of Bax we also reconstituted the protein into liposomes.
SEC and secondary structure analysis showed that the recombinantly produced Bax was monomeric and folded as previously reported in the literature (Suzuki et al. 2000; Yethon et al. 2003). Detergent induced Bax oligomerization was confirmed by SEC and chemical cross-linking (refer to Fig. 3c and Fig. 7b). The conformation of liposome reconstituted Bax was not accessible by SEC, however, chemical cross-linking yielded mainly dimers (shown in Fig. 7c). Our inability to detect higher number Bax oligomers in liposomes may be explained by the chemical cross-linker’s charge which renders it unlikely to move freely in the hydrophobic lipid environment. It appears that possible other oligomerization sites were buried within the lipid matrix and were, thus, inaccessible to the cross-linker.
The secondary structures of monomeric and detergent solubilized Bax were similar and showed an alpha helical content of about 60–65% (CD-spectroscopy data shown in Fig. 3b). Previously reported secondary structures of monomeric and liposome reconstituted Bax also contained approximately 65% alpha helix (Suzuki et al. 2000; Yethon et al. 2003).
In order to identify structurally important regions in Bax we initiated our studies with an in silico analysis. The alignment of Bax amino acid sequences from different vertebrate species revealed a highly conserved C-terminal half of the proteins whereas the N-terminal halfwith exception of helix α2 is less conserved (see Fig. 2). This implies that the C-terminus of the Bax proteins is more likely involved in Bax function than the N-terminal half (except α2). By projecting the conserved amino acids onto the NMR structure of Bax (Suzuki et al. 2000) a hydrophobic region on the protein surface which is composed of α2 (BH3), the loop between α4 and α5, a part of α5 (BH1) as well as the region of α7 to α9 (containing BH2) became evident (see Fig. 1). The high conservation and the accumulation of BH domains within this region implicate an important role in Bax function. For Bax to act as a cytochrome c pore the protein must run through a series of distinct events: (1) activation by an appropriate trigger, followed by (2) its membrane insertion and (3) either subsequent or simultaneous oligomerization. It appears conceivable that the highly conserved surface region is relevant for at least one of the above outlined events.
Interestingly, five of six tryptophans in the Bax sequence are present in the highly conserved surface region (all except Trp139). TF spectroscopy revealed alterations in the environment of the tryptophans upon Bax oligomerization in both, detergent and liposomes. Dependent on the detergent or the lipids, the hydrophobicity of the tryptophan environment increased as indicated by an up to 9 nm blue shift of the TF emission maximum (see Fig. 4b). Moreover, the concomitant increase in fluorescence intensity suggests a diminished accessibility of the tryptophan residues to oxygen (Fig. 4c). Consequently, at least some of the tryptophans are shielded from oxygen which quenches TF. Oxygen concentrations are usually higher in lipid bilayers and in aqueous solution than in the protein core (Altenbach et al. 1994; Dzikovski et al. 2003; Marsh et al. 2006)). Thus, our observations strongly support the notion that at least some of the tryptophan residues take part in the formation of the core oligomeric structure. This idea was also confirmed by the results of the proteinase K digestions discussed below.
The changes in TF emission wavelength and intensity upon Bax oligomerization varied between different detergents and liposomes (see Fig. 4b, c). Whereas the TF of oligomeric Bax in DDM, DM and liposomes was comparable, Bax oligomerization in OG showed a smaller blue shift and a lower increase in TF intensity. This might be due to the smaller alkyl chain and possibly the differ in micellar shape of OG in contrast to the other detergents and the lipids, causing a less effective protection of the hydrophobic parts of the protein (Lipfert et al. 2007).
As mentioned before protease digestion experiments indicated changes in the conserved surface region upon oligomerization. NMR data suggest that in monomeric Bax, α9 is buried in a hydrophobic cleft on the protein surface (Suzuki et al. 2000). In this study, we found that α9 is subject to proteinase K and subtilisin degradation (see Fig. 5) and, thus, must be at least temporarily solvent exposed. In contrast, α9 was not degraded in the oligomeric protein (see Fig. 5). α9 is a very hydrophobic helix and its movement away from the protein surface could enable its insertion into the lipid membrane. Thus, helix α9 may function as a membrane sensor and, subsequently, membrane anchor that promotes Bax activation (also proposed by Nechushtan et al. 1999).
In summary, our data obtained by TF and protease digestion suggest that at least a part of the conserved Bax surface region changes its conformation upon oligomerization and is likely to form the core of the oligomeric protein. This is in line with the data of others who showed that α5 and α9 (as well as α6, which was not included in the region investigated) become membrane inserted upon Bax activation (Annis et al. 2005; Garcia-Saez et al. 2005; Garcia-Saez et al. 2006).
In order to figure out whether our proteinase K cleavage patterns of Bax mirror the situation in vivo or are merely in vitro artifacts, we compared them to proteinase K digestion patterns Goping et al. obtained of Bax in mammalian cell extracts before and after induction of apoptosis (Goping et al. 1998). We were able to show that recombinantly produced monomeric and detergent induced oligomeric Bax had similar cleavage patterns as Bax in cells before and after apoptosis induction, respectively (see Fig. 5 and Goping et al. 1998). This finding indicates a comparable fold of the recombinantly produced Bax and Bax in mammalian cells.
The less conserved N-proximal part of Bax before α2 (see Fig. 2) most noticeably changed its conformation and became solvent exposed upon oligomerization. This region was cleaved by proteinase K and subtilisin from oligomeric Bax in detergent and liposomes but only to a minor extent from the monomeric form (see Fig. 5). The same region was unfolded during heat treatment, which will be discussed later.
Cartron et al. (2003) showed that α1 is important in membrane binding but not membrane insertion (Cartron et al. 2003). α1 deletion (Bax Δ1–37) inhibits membrane insertion of Bax (Cartron et al. 2003) and the mutant protein is unable to fold into the active conformation. Additionally, it is unclear whether it can adopt the normal monomeric conformation. C-terminal to α1 a long (15 amino acids), flexible (shown by NMR (Suzuki et al. 2000)) loop is found in all solved animal Bcl-2 structures. The loop seems to have a regulatory function (reviewed in (Petros et al. 2004) and references therein). In our studies, proteinase K and subtilisin cleaved oligomeric Bax mainly in this loop (see Fig. 5b and Table 1). In vivo Bax is cleaved in this loop by calpain, which leads to increased apoptosis (Altznauer et al. 2004; Gao and Dou 2000). Taking all findings together, α1 seems crucial for Bax to adopt its correct monomeric conformation, but as soon as folding is completed α1 can be removed, thereby enhancing Bax activation.
From the literature it is known that monomeric and oligomeric Bax are very heat stable (Yethon et al. 2003), and that heating to 43 °C activates Bax in mammalian cell extracts (Pagliari et al. 2005). The latter indicates that the conformational change of Bax is facilitated by heat in the presence of membranes. The melting curves we obtained for monomeric and detergent induced oligomeric Bax were very similar to those recorded by Yethon et al. on monomeric Bax and Bax inserted in liposomes (see Fig. 6 and (Yethon et al. 2003)). Bax resisted thermal denaturation in both conformations: The monomer contained 35% residual α-helices at 90 °C (sigmoidal curve shape; see Fig. 6) while the oligomer was even less affected by heat (residual α-helical content >40%, non-sigmoidal curve shape; Fig. 6). This observation raised a couple of new questions: Which parts of the monomeric protein were the most stable ones? Can we identify differences in the protein structure before and after heat treatment? Can the protein also resist chemical unfolding? And why is a protein of a mesophilic, endothermic organism so stable against heat?
Detergent oligomerized Bax showed a melting curve comparable to that observed by Yethon et al. (2003) with Bax reconstituted in lipids. The refolding efficiency surpassed theirs (Yethon et al. 2003) and complete refolding occurred at lower temperatures. Similar to Yethon et al. (2003), we found that monomeric Bax irreversibly unfolded upon heating. After heating to at least 70 °C and subsequent cooling to 4 °C (heat treatment), monomeric Bax showed an aberrant fold, distinct from the normal monomeric or oligomeric Bax conformations. Instead, “monomeric” Bax formed a “megaoligomer” which was never obtained after heat treatment of the oligomeric form (shown in Fig. 7b). In the Bax megaoligomer, the N-terminus preceding α2 became accessible to proteases; whereas α9 at the C-terminus became inaccessible. This resembles the protease accessibility of the oligomeric Bax (see Table 1). Normal Bax oligomers are only formed in the presence of detergents or liposomes. We assume that in the absence of solubilizing molecules, hydrophobic patches emerging at high temperatures due to partial denaturation of the protein cannot be shielded. Hence, upon cooling the hydrophobic parts of the Bax molecules avoid exposure to the hydrophilic environment by assembly into megaoligomers. The hydrophobic α9 (see Fig. 1d) may form part of these patches.
On the contrary, the better secondary structure recovery of Bax in detergents as compared to Bax in liposomes after heat treatment might be due to the lower size and the higher flexibility of the detergent micelles. Although CD spectroscopy does not provide direct evidence, liposomes are expected to be destroyed at elevated temperatures whereas the detergent molecules maintain their solubilizing function. The shape of the melting curve indicated partial unfolding of the detergent induced Bax oligomers. Nevertheless, a part of the oligomer (maybe to hydrophobic core) seems to remain stable allowing complete refolding of the oligomer during cooling with the detergent molecules shielding the hydrophobic patches. Pagliari et al. (2005) also showed by cross linking oligomer assembly after heating Bax monomers lacking α9 to 43 °C. This shows that without α9 Bax resistance against heat seemed to be weakened.
Bax is not only very heat stable but revealed a high resistance against chemical denaturation as well. Again, the monomeric form was affected (see Fig. 8) whereas the oligomer was almost completely stable under all tested conditions. These findings raised the question why a human protein would exhibit such a high stability rarely known from other human proteins. Cancer or virus infected cells seek to suppress apoptosis. Bax represents an ideal target for apoptosis suppression, but its high stability might put up the necessary resistance.
In summary, we showed that Bax is a very indestructible protein, although substantial conformational changes occur during its activation. On the one hand, we detected significant changes in the region preceding α2. Here, α1 seems to have an important function in adopting the monomeric conformation, but loses importance as soon as the folding is complete. In living cells it is cut by calpain leading to Bax activation. On the other hand, a highly conserved, hydrophobic surface region composed of α2, the loop between α4 and α5, parts of α5, and α7 to α9 and, therefore, significant parts of all three BH domains of Bax seem to be important for and involved in the conformational change. Upon oligomerization this region is exposed to a more hydrophobic environment and α9 becomes inaccessible for proteases. In order to understand the oligomeric structure of Bax in more detail, we are currently performing further analyses on the structural properties of the protein in the monomeric as well as oligomeric form.
Acknowledgment
This work was financially supported by DFG research grant ZE522-4/1.We are very grateful to Elisabeth Weyher-Stingl for excellent technical assistance with ESI-MS and CD spectroscopy as well as Reinhardt Mentele for N-terminal sequencing. We thank Professor Dieter Oesterhelt for continuous support and Dr. Birgit Wiltschi for reading the manuscript and giving useful suggestions. We also thank Dr. Tjandra for providing us the pTYB1-Bax plasmid.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. Proc Natl Acad Sci USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altznauer F, Conus S, Cavalli A, Folkers G, Simon H-U. J Biol Chem. 2004;279:5947–5957. doi: 10.1074/jbc.M308576200. [DOI] [PubMed] [Google Scholar]
- Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, Andrews DW. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignani A, Youle RJ. Curr Opin Cell Biol. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Antonsson B, Montessuit S, Lauper S, Eskes R, Martinou JC. Biochem J. 2000;345:271–278. doi: 10.1042/0264-6021:3450271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson B, Montessuit S, Sanchez B, Martinou J-C. J Biol Chem. 2001;276:11615–11623. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- Bohm G, Muhr R, Jaenicke R. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- Cartron P-F, Priault M, Oliver L, Meflah K, Manon S, Vallette FM. J Biol Chem. 2003;278:11633–11641. doi: 10.1074/jbc.M208955200. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Green DR. Cell Death Differ. 2006;13:1396–402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Cuddeback SM, Yamaguchi H, Komatsu K, Miyashita T, Yamada M, Wu C, Singh S, Wang H-G. J Biol Chem. 2001;276:20559–20565. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- Dzikovski BG, Livshits VA, Marsh D. Biophys J. 2003;85:1005–1012. doi: 10.1016/S0006-3495(03)74539-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Dou QP. J Cell Biochem. 2000;80:53–72. doi: 10.1002/1097-4644(20010101)80:1<53::AID-JCB60>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez AJ, Mingarro I, Perez-Paya E, Salgado J. Biochemistry. 2004;43:10930–10943. doi: 10.1021/bi036044c. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez AJ, Coraiola M, Dalla Serra M, Mingarro I, Menestrina G, Salgado J. Biophys J. 2005;88:3976–3990. doi: 10.1529/biophysj.104.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Saez AJ, Coraiola M, Serra MD, Mingarro I, Muller P, Salgado J. FEBS J. 2006;273:971–981. doi: 10.1111/j.1742-4658.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Goping IS, Gross A, Lavoie JN, Nguyen M, Jemmerson R, Roth K, Korsmeyer SJ, Shore GC. J Cell Biol. 1998;143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- Hsu Y-T, Youle RJ. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- Hsu Y-T, Youle RJ. J Biol Chem. 1998;273:10777–10783. doi: 10.1074/jbc.273.17.10777. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Newmeyer DD. Curr Opin Cell Biol. 2003;15:691–699. doi: 10.1016/j.ceb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Cell. 2002;111:331–342. doi: 10.1016/S0092-8674(02)01036-X. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K. Mol Cell. 2000;6:1389–1399. doi: 10.1016/S1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Columbus L, Chu V, Lesley S, Doniach S. J Phys Chem B. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Marsh D, Dzikovski BG, Livshits VA. Biophys J. 2006;90:L49–L51. doi: 10.1529/biophysj.106.081042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. Mol Cell. 2003;11:577–590. doi: 10.1016/S1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Nechushtan A, Smith CL, Hsu YT, Youle RJ. EMBO J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan A, Smith CL, Lamensdorf I, Yoon S-H, Youle RJ. J Cell Biol. 2001;153:1265–1276. doi: 10.1083/jcb.153.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Shimizu S, Sugiyama T, Narita M, Ito T, Matsuda H, Tsujimoto Y. J Biol Chem. 2003;278:2058–2065. doi: 10.1074/jbc.M207880200. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, Aaronson SA, Lee SW. T Cell Biol. 2004;6:121–128. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- Pagliari LJ, Kuwana T, Bonzon C, Newmeyer DD, Tu S, Beere HM, Green DR. PNAS. 2005;102:17975–17980. doi: 10.1073/pnas.0506712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros AM, Olejniczak ET, Fesik SW. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Reed JC. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- Roucou X, Martinou JC. Cell Death Differ. 2001;8:875–877. doi: 10.1038/sj.cdd.4400910. [DOI] [PubMed] [Google Scholar]
- Royer C. Chem Rev. 2006;106:1769–1784. doi: 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- Sawada M, Sun W, Hayes P, Leskov K, Boothman DA, Matsuyama S. T Cell Biol. 2003;5:320–329. doi: 10.1038/ncb950. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Cell. 2000;103:645–654. doi: 10.1016/S0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Karbowski M, Yamaguchi H, Kazi A, Wu J, Sebti SM, Youle RJ, Wang H-G. Mol Cell Biol. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter KG, Hsu Y-T, Smith CL, Nechushtan A, Xi X-G, Youle RJ. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]