Abstract
Objective:
To report the experience gained over 4 years in working with the German SARI project (Surveillance of Antimicrobial Use and Antimicrobial Resistance in Intensive Care Units), and to compare SARI with data from the Swedish STRAMA and the US AUR surveillance system.
Methods:
Prospective unit and laboratory based surveillance was carried out in 40 German ICUs from 2001 through 2004. WHO 2004 definitions of defined daily doses (DDD) per 1,000 patient days (pd) were used to express antimicrobial consumption (AD). Apart from the proportion of resistant isolates (RP), the incidence density of resistant isolates (RD) was calculated on the basis of the number of resistant isolates per 1,000 pd. To determine the changes over time, the Wilcoxon signed rank test for paired samples was used.
Results:
From 1/2001 through 12/2004, 40 ICUs provided data on 53,399 isolates, a total of 789,569 DDD and 597,592 pd. Total AD ranged from 427 to 2,798, with the median being 1,351. There was no statistically significant change in total antimicrobial use, but a statistically significant decrease was observed in the use of aminoglycosides. RD was highest for MRSA with 4.4 resistant isolates/1,000 pd followed by imipenem resistant Pseudomonas aeruginosa with 1.7 resistant isolates/1,000 pd. The corresponding RPs were 21.5% and 23.2%. Over the 4-year period (2001–2004), significant increases were seen in the RDs of third generation cephalosporin and ciprofloxacin resistant Escherichia coli. In 2004, the mean RD reached 0.28 and 1.41, respectively. In comparison, the RP of selected pathogens was highest in the US ICUs and lowest in Swedish ICUs, with the exception of imipenem resistant P. aeruginosa.
Conclusion:
Antibiotic consumption remained stable over a period of 4 years, (the mean being 1,321 DDD/1,000 pd). The same applied to the situation regarding resistance in Staphylococcus aureus, enterococci and P. aeruginosa. For most pathogens the RP was higher in SARI ICUs than in Swedish ICUs, but lower than in US ICUs.
Introduction
Antimicrobial use is the key driver of resistance, either because it selects a resistant mutant, or because it allows the emergence of a resistant pathogen in colonisation flora [1, 2]. In many countries, the high frequency of resistant bacterial pathogens presents a major public health problem. Moreover, such resistance is slow to reverse or may even be irreversible, and the therapeutic options are therefore reduced [3].
In order to influence the spread of resistant pathogens and develop sustainable systems, the European Commission and the World Health Assembly (WHA) Resolution therefore encourage the appropriate and cost-effective use of antimicrobials. It is also necessary to detect resistant pathogens and to monitor antimicrobial consumption volumes, their patterns of use and the impact of infection control measures [4, 5].
However, the relationship between antibiotic use and the emergence of resistance is complex. The type of micro-organism, the resistance mechanisms involved and the relative importance of the different epidemiological variables will determine the likelihood of an intervention being successful. Until these interactions are fully explored, it seems reasonable to promote the prudent use of antibiotics as the single most important measure to stop increases in resistance trends. Comparison of data as a benchmarking instrument is an important initial step, and serves to help ICUs recognize problems and improve the way in which antimicrobials are used [6].
This paper presents data from SARI over a period of 4 years and compares them with data from other ICU surveillance systems, i.e. STRAMA and AUR.
Methods
SARI was initiated in February 2000 with the objective of collecting monthly prospective data from SARI-ICUs actively participating in the German hospital infection surveillance system (KISS) [13]. In our analysis, we concentrated on the period 2001 through 2004, because in 2000 the data set only covered 16 ICUs. From 1/2001 through 12/2004, 40 ICUs provided data and were included in the analysis. The 40 ICUs were located in 24 geographically distinct hospitals. Of this number, 15 belonged to university hospitals. Fourteen ICUs were interdisciplinary, 15 were surgical and 11 were medical. ICU bed numbers ranged from 6 to 26, with a median of 12.
A report comparing local monitoring data for each ICU and SARI benchmark data (i.e. aggregate summary data on all SARI ICUs) was initially presented to each ICU every 3 months. Since 2004, this report has been compiled every 6 months. In addition, a 2-day workshop took place once a year. Monthly antimicrobial usage data were obtained from computerised hospital databases. Consumption was expressed as defined daily doses (DDD) and normalised per 1,000 patient-days (AD = antimicrobial usage density), 1 DDD being the standard adult daily dose of an antimicrobial agent for 1 day’s treatment defined by the WHO (data are recalculated by DDD index 2004) [14].
The ICUs indicated the number of isolates tested per month belonging to the following sentinel bacterial species: Staphylococcus aureus, coagulase negative staphylococci (CNS), Enterococcus faecalis, Enterococcus faecium, Pseudomonas aeruginosa, Enterobacter cloacae, Citrobacter spp., Serratia marcescens, Acinetobater baumannii, Stenotrophomonas maltophilia, Streptococcus pneumoniae, Escherichia coli and Klebsiella pneumoniae; furthermore, on the number of isolates tested against sentinel antibiotics and the number of isolates resistant to these antibiotics. They were specified as resistant by the participating ICUs using the identification method routinely employed in their clinical laboratory. The data were presented using interpretive criteria recommended by the German Industrial standard (DIN) or the National Committee for Clinical Laboratory Standards (NCCLS) [15, 16]. Calculation was done irrespective of the interpretive criteria applied. To exclude copy strains, all the laboratories were obliged to use the same protocol. A copy strain was defined as an isolate of the same species showing the same susceptibility pattern throughout the period of 1 month in the same patient, regardless of the site of isolation.
Data on hospital and patient characteristics were obtained from the KISS system, i.e. type of hospital, number of ICU beds, type of ICU (medical, surgical, interdisciplinary), number of patient-days, number of device days and mean length of stay. The ICUs collected data on nosocomial infections (NI) for each patient according to the definitions given by the Centers for Disease Control and Prevention (CDC) [13].
The proportion of resistant isolates was calculated by dividing the number of resistant isolates by the total number of isolates belonging to the species tested against this antibiotic, multiplied by 100 (RP). The incidence density of resistant isolates was defined as being the number of resistant isolates per 1,000 patient days (pd) (RD).
For analysis of temporal changes in resistance densities and antimicrobial consumption, we included all the SARI-ICUs that had provided data in 2001, 2002, 2003 and 2004. We then compared the data provided by the individual ICUs in the year 2001 with that of the same ICU in 2004 (n = 34). Additionally, in the analysis of temporal changes in RD we included only those ICUs that had provided at least ten isolates per antimicrobial/pathogen per year for testing, because RDs are statistically unsafe if the number of isolates tested is insufficient. To determine the changes between the two terminal years (year 2001 vs year 2004), we chose the Wilcoxon signed rank test for paired samples as described by Fridkin et al. [17]. The statistics are based on the ranking of absolute values of differences. Pairs with large differences have greater weight than pairs with small differences.
To test for differences in RD in selected pathogens according to the type of ICU (medical, surgical, interdisciplinary), hospital size (> 600 beds) and university status, Kruskal-Wallis and Wilcoxon tests for independent samples were employed.
Statistical analyses were performed using SAS version 8.01 and EpiInfo version 6.04.
Results
A total of 789,569 DDD and 597,592 pd were included in the analysis. Table 1 shows pooled antibiotic consumption in accordance with the WHO classification over a period of 4 years for all 40 SARI ICUs by ICU type. The total antimicrobial ADs ranged from 427 to 2,798, with a median of 1,351 and a mean of 1,321. They did not differ significantly by type of ICU, size of hospital (> 600 beds) or university status. Penicillins with β-lactamase inhibitor were the antimicrobial group with the highest AD in all types of ICU. In medical ICUs, these were followed by quinolones, penicillins with extended spectrum and macrolides, in surgical ICUs by quinolones and carbapenems and in interdisciplinary ICUs by second generation cephalosporins and quinolones. There was no statistically significant change in total antimicrobial use from 2001 through 2004, but statistically significant changes were found in the antimicrobial classes used. The median administration of extended spectrum penicillins increased from 75.3 to 102.3 (mainly due to the use of piperacillin); the administration of penicillins with β-lactamase inhibitor combinations decreased from 258.5 to 191.5 (mainly due to a decrease in the use of piperacillin-tazobactam) and the administration of aminoglycosides also dropped from 59.6 to 28.4).
Table 1.
Pooled mean and percentiles of the distribution of antimicrobial use density (AD = DDD/1,000 patient days [pd]), 1/2001–12/2004, all SARI ICUs (n = 40), the median AD of surgical, medical and interdisciplinary ICUs and temporal differences between 2001 vs 2004 (data on 34 SARI ICUs providing antimicrobial use data from 2001 to 2004).
| Pooled data of all SARI-ICUs 01/2001–12/2004 | Pooled data by type of ICU 01/2001–12/2004 | Data of 34 SARI-ICUs, 2001–2004 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AD | Median AD | Median AD | No. of ICUs with a change from year 2001 to 2004 | |||||||
| Antimicrobial group (ATC WHO)/Antimicrobials | Pooled mean | 25% percentile | Median | 75% percentile | Medical (n = 11) | Surgical (n = 15) | Interdisc. (n = 14) | Year 2001 | Year 2004 | Increase/decrease/no change |
| Total antimicrobial use | 1,321.3 | 1,038.6 | 1,350.5 | 1,582.8 | 1,468.8 | 1,151.8 | 1,338.3 | 1,247.1 | 1,384.6 | 22/12/0 |
| Penicillins with beta-lactamase inhibitor | 277.2 | 111.9 | 219.1 | 384.9 | 216.8 | 221.4 | 217.5 | 258.5 | 191.5 | 8/26/0* |
| Quinolones | 141.3 | 102.7 | 125.8 | 172.3 | 128.0 | 123.8 | 125.8 | 130.6 | 138.3 | 19/15/0 |
| Second generation cephalosporins | 118.1 | 58.6 | 107.8 | 166.4 | 78.7 | 110.9 | 145.6 | 106.2 | 104.7 | 19/15/0 |
| Third generation cephalosporins | 107.5 | 84.1 | 102.8 | 140.4 | 92.4 | 101.2 | 106.3 | 100.6 | 112.6 | 19/15/0 |
| Penicillins with extended spectrum | 96.0 | 30.1 | 81.8 | 156.5 | 114.7 | 32.8 | 121.2 | 75.3 | 102.3 | 25/9/0* |
| Carbapenems | 81.4 | 41.0 | 70.8 | 122.3 | 68.0 | 120.2 | 59.1 | 66.5 | 65.7 | 19/15/0 |
| Macrolides | 70.2 | 31.6 | 60.1 | 106.5 | 103.5 | 32.0 | 60.1 | 61.7 | 56.2 | 15/18/1 |
| Imidazoles | 52.3 | 0.0 | 52.0 | 101.6 | 42.3 | 34.5 | 89.4 | 53.1 | 50.5 | 11/14/9 |
| First generation cephalosporins | 43.2 | 0.0 | 1.4 | 56.7 | 1.1 | 1.0 | 7.7 | 3.5 | 3.8 | 10/9/15 |
| Aminoglycosides | 41.4 | 19.0 | 40.4 | 64.4 | 40.2 | 37.5 | 47.0 | 59.6 | 28.4 | 8/26/0* |
| Beta-lactamase resistant penicillins | 39.1 | 0.1 | 12.5 | 36.0 | 27.2 | 10.4 | 10.1 | 9.0 | 20.7 | 17/11/6 |
| Glycopeptides | 36.7 | 17.5 | 30.0 | 50.2 | 29.8 | 39.2 | 18.1 | 24.8 | 29.5 | 17/17/0 |
| Sulfonamides and trimethoprim combinations | 29.7 | 0.1 | 21.0 | 32.2 | 28.0 | 22.1 | 14.4 | 16.9 | 9.3 | 11/15/8 |
| Beta-lactamase sensitive penicillins | 18.2 | 0.1 | 15.9 | 29.3 | 26.7 | 7.3 | 16.1 | 5.5 | 16.7 | 20/7/7* |
| Lincosamides | 14.1 | 0.0 | 11.7 | 26.1 | 15.4 | 8.8 | 12.9 | 11.0 | 12.4 | 12/13/9 |
| Tetracyclines | 7.1 | 0.0 | 3.8 | 16.6 | 8.6 | 0.0 | 7.1 | 0.0 | 0.5 | 12/9/13 |
| Fourth generation cephalosporins | 2.0 | 0.0 | 0.0 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5/4/25 |
Other antimicrobials and inhibitors (J01CG) are not listed
The laboratory data covered 53,399 isolates (29,160 gram positives and 24,239 gram negatives). The number of SARI-isolates i.e. the 13 sentinel microorganisms per 1,000 pd ranged from 17.3 to 164.6, with the median lying at 80.0. The number of annual SARI-isolates ranged from 33 to 1199, with the median being 316. RD and RP did not differ significantly in university and non-university ICUs or by type of ICU. However, in ICUs located in hospitals with more than 600 beds the RD and RP of imipenem resistant P. aeruginosa were significantly higher than smaller hospital ICUs (p = 0.046 and 0.043, respectively).
A comparison of the median length of stay revealed that between 2001 and 2004 the length of stay increased in 15 and decreased in 20 ICUs; however, this was not statistically significant (median length of stay in 34 ICUs — 4.2 in 2001 and 3.8 in 2004). From 2001 through 2004, mean deviceassociated nosocomial infection rates, expressed as NI/1,000 device days were as follows: urinary tract infections 3.1 (range 0.3–8.3), blood stream infections 1.8 (range 0.2–4.6), and pneumonia 6.8 (range 2.0–17.3). The use of invasive devices did not change significantly over time (2001 vs 2004).
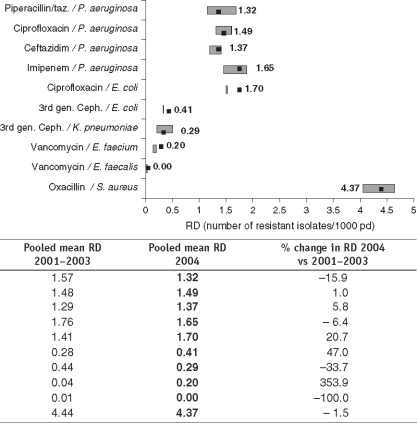
With respect to resistance, pooled data over 4 years showed the highest mean RD of 4.4 resistant isolates per 1,000 pd for MRSA i.e. methicillin resistant S. aureus (mean resistance proportion 21.5%) followed by imipenem resistant P. aeruginosa with an RD of 1.7 and a mean RP of 23.2% (Table 2). Furthermore, data sets on RD from the year 2001 were compared with those of the year 2004. Significant increases were found for the RD of third generation resistant E. coli (over 4 years 14 ICUs showed an increase and only 6 ICUs a decrease in RP), as well as for the RD of ciprofloxacin resistant E. coli (20 ICUs showed an increase and only 5 ICUs a decrease) (Table 2). Figure 1 shows the pooled mean RD for January through December 2004 for selected antimicrobial-resistant pathogens. Next to or overlapping this point is the 95% confidence interval of mean RD over the previous 3 years (2001–2003). The most dramatic increase in mean RD was observed for vancomycin resistant E. faecium (354%), followed by third generation resistant E. coli (47%), ciprofloxacin resistant E. coli (21%) and ceftazidime resistant P. aeruginosa (6%). In contrast, vancomycin resistant E. faecalis decreased by 100% and third generation resistant K. pneumoniae by 34%. At −2%, MRSA remained fairly stable.
Table 2.
Pooled mean and key percentiles of resistance densities (RD = number of resistant isolates/1,000 pd and antimicrobial resistance proportions (RP = number of resistant isolates/100 tested isolates) and) of selected pathogens from SARI ICUs (n = 40), 2001–2004 and temporal differences in RD between 2001 vs 2004 (data of 34 SARI ICUs providing resistance data from 2001 through 2004).
| Pathogen | Resistant to | Rooled RD (RP%) of all SARI-ICUs 01/2001–12/2004 | RD from 34 SARI ICUs providing resistance data from 2001 to 2004 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No tested | Pooled mean | 25% percentile | Median | 75% percentile | No ICUs** in analysis | Year 2001 median RD | Year 2004 median RD | No. of ICUs with an increase/decrease/no change of RD from 2001 to 2004 | ||
| S. aureus | MethiciLLin | 12,238 | 4.4 (21.5%) | 1.92 (12.0%) | 3.34 (21.2%) | 4.8 (32.1%) | 30 | 2.7 | 3.7 | 18/12/0 |
| E. faecalis | Vancomycin | 5,371 | 0.0 (0.1%) | 0.0 (0%) | 0.0 | 0.0 | 27 | 0 | 0 | 0/1/26 |
| E. faecium | Vancomycin | 1,945 | 0.1 (2.6%) | 0.0 (0%) | 0.0 | 0.1 (3.3%) | 11 | 0 | 0 | 2/3/6 |
| K. pneumoniae | Third gen. cephalosporins | 3,205 | 0.4 (7.5%) | 0.0 (0%) | 0.1 (2.0%) | 0.3 (6.5%)) | 20 | 0 | 0.2 | 9/3/8 |
| E. coli | Third gen. cephalosporins | 8,110 | 0.3 (2.3%) | 0.0 (0%) | 0.2 (1.7%) | 0.5 (3.5%) | 27 | 0 | 0.2 | 14/6/7* |
| Ciprofloxacin | 7,152 | 1.5 (12.3%) | 0.5 (8.3%) | 1.3 (12.0%) | 2.0 (16.6%) | 27 | 1.1 | 1.8 | 20/5/2* | |
| P. aeruginosa | Imipenem | 4,442 | 1.7 (23.2%) | 0.4 (8.4%) | 1.1 (18.6%) | 2.4 (26.6%) | 21 | 2.0 | 1.1 | 5/16/0 |
| Ceftazidime | 5,005 | 1.3 (15.6%) | 0.4 (6.5%) | 1.0 (11.1%) | 2.0 (20.5%) | 26 | 0.6 | 1.1 | 14/12/0 | |
| Ciprofloxacin | 5,009 | 1.5 (17.6%) | 0.5 (8.6%) | 1.2 (15.4%) | 2.2 (26.2%) | 26 | 1.2 | 1.0 | 10/14/2 | |
| Piperacillin-tazobactam | 4,058 | 1.5 (22.0%) | 0.3 (8.7%) | 0.0 (15.7%) | 3.5 (36.7%) | 21 | 1.6 | 1.4 | 9/12/0 | |
“Resistant to third generation cephalosporins” i.e. cefotaxim or ceftiaxon or ceftazidime; *if p-value < 0.05 by Wilcoxon signed-rank test for paired samples 2001 vs 2004; ** ICUs with on at least ten isolates tested per antimicrobial/pathogen per year
Figure 1.
Comparison of pooled mean resistance densities (RD) of all SARI ICUs for selected antimicrobial-resistance pathogens from 1/2004 to 12/2004 with 1/2001–12/2003;▪=2004 pooled mean RD;{ie307-1} = 2001–2003 95% confidence interval of mean RD.
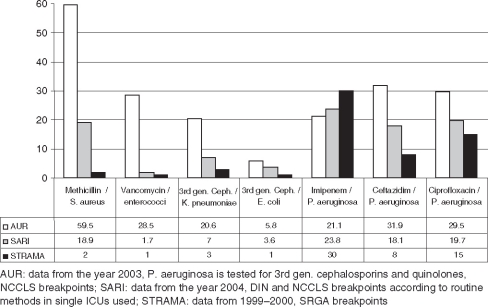
For comparison with the US-American and the Swedish surveillance systems, RP were compared with the latest available data, showing that with the exception of imipenem resistant P. aeruginosa the RP were highest in US-ICUs (Figure 2).
Figure 2.
Comparison of mean resistance proportions (RP=number of resistant isolates/100 tested isolates) of three surveillance systems: the US AUR, the German SARI and the Swedish STRAMA.
Discussion
This study reports the data gained over 4 years of working with SARI, an ongoing surveillance system for antibiotic use and resistance. We discuss the principal findings: (1) In SARI ICUs the mean antibiotic consumption was 1,321 DDD/1,000 pd, and the preferred antibiotic groups were penicillins with beta-lactamase inhibitors and quinolones. (2) Total antibiotic consumption remained stable over 4 years. (3) Resistance densities for third generation cephalosporin and ciprofloxacin resistant E. coli rose significantly over 4 years. Otherwise, the resistance situation remained stable. (4) The RP of selected pathogens was highest in the US ICUs and lowest in Swedish ICUs, with the exception of imipenem resistant P. aeruginosa.
In the hospitals, antimicrobial use ranged from 427 to 2,798 DDD/1,000 pd. This heterogeneity was also seen in 29 Swedish ICUs, where antibiotic consumption ranged from 605 to 2,143 DDD/ 1,000 pd, with a median of 1,147 between 1999 and 2000 [7]. It is a well-known fact that antibiotic consumption in ICUs is generally higher than hospital-wide consumption [18]. The preferred antibiotic groups in all SARI ICUs were penicillins with beta-lactamaseinhibitors (AD = 277) quinolones (AD = 141) and second generation cephalosporins (AD 118). In Swedish ICUs, cephalosporins (AD = 287) and isoxazolyl penicillin (AD = 156) outranged other classes of antimicrobials [7, 9].
Over the 4-year period, we saw a significant decrease in the use of aminoglycosides (26 out of 34 ICUs decreased use) and of penicillins with beta-lactamase inhibitors. However, the latter was due to the replacement of the fixed combination piperacillin/tazobactam by the individual compounds piperacillin and sulbactam, which are less expensive in Germany. There was no change over time in the total use of antibiotics within individual ICUs.
Summarized data are presented both as RD as an indirect parameter of the burden of resistance, i.e. resistant isolates/ 1,000 pd as described by Schwaber et al. [19, 20] and as RP. We used RD only for all the remaining analyses. The pitfall of RP, which is widely used and is easy to interpret, is that the number of resistant bacterial isolates depends on the number of susceptible isolates. If the number of susceptible isolates varies or changes over time, e.g. if some laboratories report contaminants, while others do not, or if screening procedures are introduced, these measures will all affect the RP. However, the resistance density only depends on the number of resistant isolates.
MRSA is by far the most resistant isolate with the highest burden of resistance (4.4 MRSA/1,000 pd), implying higher costs for isolation, decolonisation and the treatment of infections. Looking at changes over time in individual ICUs we saw no significant change in the RD of MRSA. The fact that the RD of MRSA increased in 18 and decreased in 12 ICUs might reflect the impact of different efforts undertaken by individual ICUs both with regard to infection control and antibiotic management. Especially the consumption of quinolones can have a significant impact on the MRSA resistance situation [21, 22]. A stable MRSA situation and a significant increase in third generation cephalosporin and ciprofloxacin resistant E. coli (2001–2004) confirm findings and trends seen earlier over a period of 3 years in SARI ICUs and described for MRSA in detail elsewhere [23, 24]. Comparison of the mean RD in 2004 with the mean RD in the years 2001 through 2003 showed a dramatic increase in the percentage of vancomycin resistant E. faecium. However, the increase in vancomycin resistant E. faecium was due to an outbreak in hospitals in South-West Germany (only two of the SARI ICUs were affected) and is a good example of the need to exercise care in the interpretation of pooled data [25].
Comparison of US-AUR data (2003) with German SARI data (2004) and Swedish STRAMA data (1999–2000) showed that resistance proportions for selected pathogens were highest in the US ICUs and lowest in Swedish ICUs [11]. However, interestingly, at 30%, resistance in P. aeruginosa to imipenem was highest in Swedish ICUs compared with 23.8% in German ICUs and 21.1% in US-American ICUs [7]. This might be due to the fact that while carbapenem consumption was 37.8 DDD/1,000 pd in interdisciplinary US AUR-ICUs and 84.1 in German SARI ICUs, it was 113 in Swedish STRAMA ICUs. The striking differences in prescribing preferences between one country and another should be taken into consideration and might be crucial for the success of possible interventions.
Thus, a Hawthorne effect, as described for nosocomial infection rates, i.e. reduction by observation alone, was not observed in either AUR or SARI. The 354% increase in vancomycin resistant E. faecium observed by SARI was caused by the regional outbreak, and ciprofloxacin resistant E. coli increased without an increase in quinolone ICUconsumption. Especially in the case of quinolones, hospital-wide and ambulatory care consumption influences the development of resistance. MacDougall et al. found that for quinolone resistant E. coli, total community quinolone use within a 16 km radius correlated significantly with resistance in the hospitals [26]. Therefore, campaigns for the appropriate use of antibiotics should ideally focus not only on ICUs, but on all those physicians prescribing them.
This study has some limitations. Selection bias cannot be excluded because participation in SARI was voluntary and ICUs encountering higher resistance rates would obviously be aware of the problem and wish to take part. Bias due to inclusion of all isolates must be taken into account, especially with respect to screening policies and outbreaks. In Germany, ICUs perform susceptibility testing in accordance either with NCCLS or DIN. Like other surveillance systems we did not differentiate between the methods used, which should be considered in the data analysis [27]. As data are laboratory and unit-based, they are prone to ecologic fallacy, i.e. improper inference about individual-level associations based on associations measured only at the aggregate level.
In conclusion, antibiotic consumption remained stable over a period of 4 years with a mean value of 1,321 DDD/1,000 pd. The burden of resistance in S. aureus and P. aeruginosa also remained stable, whereas resistance in third generation cephalosporin and ciprofloxacin resistant E. coli increased. The striking differences in the resistance situation and in antibiotic prescribing practices in different countries underline the need for national or even locally adapted guidelines on empiric antibiotic therapy. Further analysis of these differences will help improve the way in which antimicrobials are used in countries whose resistance situation is better than in others.
Acknowledgments
SARI is part of the SIR (Spread of Nosocomial Infections and Resistant Pathogens) research network supported by the Federal Ministry of Education and Research. This study was supported by a grant from the Federal Ministry of Education and Research (01Kl 9907).
References
- 1.Fridkin SK: Increasing prevalence of antimicrobial resistance in intensive care units. Crit Care Med 2001; 29: N64–N68. [DOI] [PubMed]
- 2.Kollef MH, Fraser VJ: Antibiotic resistance in the intensive care unit. Ann Intern Med 2001; 134: 298–314. [DOI] [PubMed]
- 3.Harbarth S, Samore MH: Antimicrobial resistance determinants and future control. Emerg Infect Dis 2005; 11: 794–811. [DOI] [PMC free article] [PubMed]
- 4.Council recommendation on the prudent use of antimicrobial agents in human medicine. Official Journal of the European Communities (2002/77/EC), L 34-13-L 34/16, 2002.
- 5.World Health Organisation: WHO global strategy for containment of antimicrobial resistance. 2001 WHO/CDS/CSR/ DRS/2001.2.
- 6.Fridkin SK, Lawton R, Edwards JR, Tenover FC, McGowan JE Jr, Gaynes RP: Monitoring antimicrobial use and resistance: comparison with a national benchmark on reducing vancomycin use and vancomycin-resistant enterococci. Emerg Infect Dis 2002; 8: 702–707. [DOI] [PMC free article] [PubMed]
- 7.Hanberger H, Erlandsson M, Burman LG, Cars O, Gill H, Lindgren S, Lennart N, Olsson-Liljequist B, Walther SM: High antibiotic susceptibility among bacterial pathogens in Swedish ICUs. Report from a nation-wide surveillance program using TA90 as a novel index of susceptibility. Scan J Infect Dis 2004; 36: 24–30. [DOI] [PubMed]
- 8.Meyer E, Jonas D, Schwab F, Rueden H, Gastmeier P, Daschner FD: Design of a surveillance system of antibiotic use and bacterial resistance in German intensive Care units (SARI). Infection 2003; 31: 208–215. [DOI] [PubMed]
- 9.Meyer E, Schwab F, Jonas D, Rueden H, Gastmeier P, Daschner FD: Surveillance of antimicrobial use and antimicrobial resistance in intensive care units (SARI): 1. Antimicrobial use in German intensive care units. Intensive Care Med 2004; 30: 1089–1096. [DOI] [PubMed]
- 10.Walther SM, Erlandsson M, Burman LG, Cars O, Gill H, Hoffman M, Isaksson B, Kahlmeter G, Lindgren S, Nilsson L, Olsson-Liljequist B, Hanberger H: Antibiotic prescription practices, consumption and bacterial resistance in a cross section of Swedish intensive care units. Acta Anaesthesiol Scand 2002; 46: 1075–1081. [DOI] [PubMed]
- 11.National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control 2004; 32: 470–485. [DOI] [PubMed]
- 12.Fridkin SK, Steward CD, Edwards JR, Pryor ER, McGowan JE Jr, Archibald LK, Gaynes RP, Tenover FC: Surveillance of antimicrobial use and antimicrobial resistance in United States hospitals: project ICARE phase 2. Project Intensive Care Antimicrobial Resistance Epidemiology (ICARE) hospitals. Clin Infect Dis 1999; 29: 245–252. [DOI] [PubMed]
- 13.Gastmeier P, Geffers C, Sohr D, Dettenkofer M, Daschner F, Ruden H: Five years working with the German nosocomial infection surveillance system (Krankenhaus Infektions Surveillance System). Am J Infect Control 2003; 31: 316–321. [DOI] [PubMed]
- 14.WHO Collaborating centre for Drug Statistics Methodology: Guidelines For ATC Classification and DDDs Assignment. WHO Collaborting Centre, Oslo, Norway, 2004.
- 15.DIN 58940-4 Medical microbiology — Methods for the determination of susceptibility of pathogens (except mycobacteria) to antimicrobial agents — part 4: evaluation classes of the minimum inhibitory concentration. Beuth Verlag, Berlin, 1998, pp 1–11.
- 16.National Committee for Clinical Laboratory Standards: Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th edn. Approved Standard M7-A4. NCCLS, Wayne, PA, 1997.
- 17.Fridkin SK, Hill HA, Volkova NV, Edwards JR, Lawton RM, Gaynes RP, McGowan JE Jr: Temporal changes in prevalence of antimicrobial resistance in 23 US Hospitals. Emerg Infect Dis 2002; 8: 697–701. [DOI] [PMC free article] [PubMed]
- 18.DANMAP 2004: Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark, 2005, pp 26–31. ISSN 1600-2032.
- 19.Schwaber MJ, De Medina T, Carmeli Y: Epidemiological interpretation of antibiotic resistance studies — what are we missing? Nat Rev Microbiol 2004; 2: 979–983. [DOI] [PubMed]
- 20.Cornaglia G, Hryniewicz W, Jarlier V, Kahlmeter G, Mittermayer H, Stratchounski L, Baquero F: European recommendations for antimicrobial resistance surveillance. Clin Microbiol Infect 2004; 10: 349–383. [DOI] [PubMed]
- 21.Charbonneau P, Parienti JJ, Thibon P, Ramakers M, Daubin C, du Cheyron D, Lebouvier G, Le Coutour X, Leclercq R: Fluoroquinolone use and methicillin-resistant Staphylococcus aureus isolation rates in hospitalized patients: a quasi experimental study. Clin Infect Dis 2006; 42: 778–784. [DOI] [PubMed]
- 22.Weber SG, Gold HS, Hooper DC, Karchmer AW, Carmeli Y: Fluoroquinolones and the risk for methicillin-resistant Staphylococcus aureus in hospitalized patients. Emerg Infect Dis 2003; 9: 1415–1422. [DOI] [PMC free article] [PubMed]
- 23.Meyer E, Schwab F, Gastmeier P, Jonas D, Rueden H, Daschner FD: Methicillin-resistant Staphylococcus aureus in German Intensive Care Units during 2000–2003: data from Project SARI (Surveillance of Antimicrobial Use and Antimicrobial Resistance in Intensive Care Units). Infect Control Hosp Epidemiol 2006; 27: 146–154. [DOI] [PubMed]
- 24.Meyer E, Schwab F, Jonas D, Ruden H, Gastmeier P, Daschner FD: Temporal changes in bacterial resistance in German intensive care units, 2001–2003: data from the SARI (surveillance of antimicrobial use and antimicrobial resistance in intensive care units) project. J Hosp Infect 2005; 60: 348–352. [DOI] [PubMed]
- 25.Robert-Koch Institute: On the emergence and spread of glycopeptide resistant enterococci. Epidemiol Bull 2005; 17/2005: 149–155.
- 26.MacDougall C, Powell JP, Johnson CK, Edmond MB, Polk RE: Hospital and community fluoroquinolone use and resistance in Staphylococcus aureus and Escherichia coli in 17 US hospitals. Clin Infect Dis 2005; 41: 435–440. [DOI] [PubMed]
- 27.Jones ME, Draghi DC, Thornsberry C, Karlowsky JA, Sahm DF, Wenzel RP: Emerging resistance among bacterial pathogens in the intensive care unit — a European and North American Surveillance study (2000–2002). Ann Clin Microbiol Antimicrob 2004; 3: 1–11. [DOI] [PMC free article] [PubMed]