Abstract
A dominant allele at the D locus (also known as I in diploid potato) is required for the synthesis of red and purple anthocyanin pigments in tuber skin. It has previously been reported that D maps to a region of chromosome 10 that harbors one or more homologs of Petunia an2, an R2R3 MYB transcription factor that coordinately regulates the expression of multiple anthocyanin biosynthetic genes in the floral limb. To test whether D acts similarly in tuber skin, RT-PCR was used to evaluate the expression of flavanone 3-hydroxylase (f3h), dihydroflavonol 4-reductase (dfr) and flavonoid 3′,5′-hydroxylase (f3′5′h). All three genes were expressed in the periderm of red- and purple-skinned clones, while dfr and f3′5′h were not expressed, and f3h was only weakly expressed, in white-skinned clones. A potato cDNA clone with similarity to an2 was isolated from an expression library prepared from red tuber skin, and an assay developed to distinguish the two alleles of this gene in a diploid potato clone known to be heterozygous Dd. One allele was observed to cosegregate with pigmented skin in an F1 population of 136 individuals. This allele was expressed in tuber skin of red- and purple-colored progeny, but not in white tubers, while other parental alleles were not expressed in white or colored tubers. The allele was placed under the control of a doubled 35S promoter and transformed into the light red-colored cultivar Désirée, the white-skinned cultivar Bintje, and two white diploid clones known to lack the functional allele of D. Transformants accumulated pigment in tuber skin, as well as in other tissues, including young foliage, flower petals, and tuber flesh.
Introduction
Most natural variation in potato tuber color results from tissue-specific differences in the accumulation of anthocyanin pigments. An early study by Salaman (1910) in tetraploid potato (Solanum tuberosum L.) identified three loci—D (developer), R (red), and P (purple) that condition variation for color of potato tuber skin. R is required for the production of red anthocyanins, P is required for the production of purple anthocyanins, while D is required for the tissue-specific accumulation of anthocyanin pigments in tuber skin. Red and purple tuber anthocyanins have been shown to be derivatives of pelargonidin and delphinidin, respectively (Lewis et al. 1998; Naito et al. 1998; Rodriguez-Saona et al. 1998).
A parallel three-gene system, but using the gene symbols I (inhibitor), R, and P, was subsequently described in diploid potato by Dodds and Long (1955, 1956). With diploids P was shown to be epistatic to R. Thus, tubers with genotype I− R− P− or I− rr P− have purple tuber skin, those with genotype I− R− pp have red colored skin, and those with genotype ii − − are white. The similar phenotypes mediated by tetraploid D, R, and P and diploid I, R, and P imply that D is equivalent to I, tetraploid R is equivalent to diploid R, and tetraploid P is equivalent to diploid P (reviewed in De Jong 1991). To provide consistency, the original gene nomenclature of Salaman (1910) will be used throughout this paper.
D, R, and P (sensu Salaman) have previously been localized to chromosomes 10, 2, and 11, respectively (Gebhardt et al. 1989; van Eck et al. 1993, 1994). Subsequent mapping of many anthocyanin structural and regulatory genes in tomato gave rise to the suggestion that R corresponds to dihydroflavonol 4-reductase (dfr), that P codes for flavonoid 3′,5′-hydroxylase (f3′5′h), and that D encodes an R2R3 MYB transcription factor similar to Petunia an2 (De Jong et al. 2004). A combination of genetic cosegregation and transgenic evidence has confirmed that R encodes dfr (De Jong et al. 2003a; Zhang et al. 2009b) and that P encodes f3′5′h (Jung et al. 2005). R2R3 MYB genes have been shown to coordinately regulate the expression of multiple anthocyanin structural genes in several species, including maize, apple, and grape (Cone et al. 1986; Espley et al. 2007; Cutanda-Perez et al. 2009).
With this paper, we report that D is a transcriptional regulator of the anthocyanin biosynthetic pathway in tuber periderm and identify an R2R3 MYB domain gene that cosegregates with D and is expressed in red and purple, but not white, tuber skin.
Materials and methods
Plant material
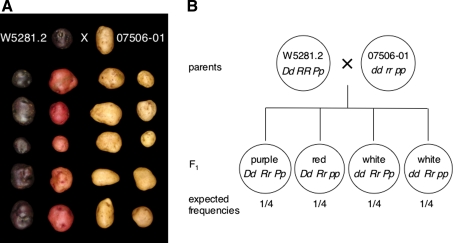
Diploid potato clones W5281.2 (female parent/purple tuber skin/genotype Dd RR Pp) (De Jong and Burns 1993) and 07506-01 (male parent/white tuber skin/genotype dd rr pp) (De Jong 1987; De Jong and Burns 1993) were crossed to generate an F1 population that segregates for tuber skin color (2 white:1 red:1 purple) (Fig. 1). Breeding clones with an NY designation were developed by the Cornell University potato breeding program. The tetraploid Cornell breeding clone Y83-5 (red tuber skin/white tuber flesh) was used to construct a cDNA library. In vitro plantlets of the cultivars Désirée and Bintje were obtained from the New York State Seed Farm in Lake Placid, NY, USA and the Potato Introduction Station in Sturgeon Bay, WI, USA, respectively.
Fig. 1.
Diploid population segregating for tuber skin color and schematic diagram illustrating allele segregation. a A cross between W5281.2 (female/purple tuber skin/genotype Dd RR Pp) and 07506-01 (male/white tuber skin/genotype dd rr pp) generated an F1 population that segregated for tuber skin color (76 purple:72 red:132 white). Parents are shown at the top, 20 representative progeny are shown below. b Schematic diagram of the F1 population showing parental and progeny genotypes and their predicted frequencies. D is required for color expression in tuber skin (red or purple), while R and P are required for red and purple color expression, respectively. Purple color is epistatic to red
cDNA library construction and screening
Total RNA was isolated from tuber skin of Y83-5 with a LiCl-based procedure (De Vries et al. 1988). Messenger RNA was then purified from total RNA with a PolyTract mRNA isolation system kit (Promega). Double stranded cDNA was synthesized with a ZAP cDNA synthesis kit (Stratagene) and cloned into a lambda DASH II vector (Stratagene). A fragment of the Petunia an2 gene was amplified with primers an2–5′ and an2–3′ (Quattrocchio et al. 1998), radioactively labeled, and then used as a probe to screen the Y83-5 cDNA library, as well as a cDNA library previously constructed from diploid W5281.2 (De Jong et al. 2003a). Hybridizing clones were plaque-purified and the inserts then excised to permit propagation in a pBlueScript SK(−) phagemid vector, following the manufacturer’s instructions. Inserts were sequenced using universal M13 vector primers.
Cloning and analysis of genomic DNA
Primers Stan2-F4 (5′GTAATAAGGAAAGGTTCATGGACT) and Stan2-R (5′AAAATTCATCCCAACCACCATCAC) were designed against the predominant sequence class (Stan2) isolated from the Y83-5 cDNA library and used to amplify the majority of the corresponding gene from W5281.2. The resulting PCR products were cloned into the vector pGEM-T (Promega), sequenced, and examined for polymorphism. Two haplotypes were observed. To determine whether either haplotype cosegregated with tuber skin color, primers Stan2-ClaI-F2 (5′GTGATTATGTCATCCAAAAGTTTATAG) and Stan2-ClaI-R1 (5′GAATTTCTGAGGTTGAGGTCTTA) were designed to flank a polymorphic Cla I restriction site within the second intron. Products were digested with Cla I after amplification, separated on a 1% agarose gel, and visualized by staining with ethidium bromide. The same primers were used to evaluate the allelic composition of tetraploid cultivars and breeding clones. Here, primer Stan2-ClaI-F2 was radioactively labeled with 33P prior to PCR, and undigested products were separated on a 5% denaturing acrylamide sequencing gel.
Primers Stan3-F1 (5′GGTTCATGGACTGAACAAGAAGATTT) and Stan3-R1 (5′ATTCAGTCAGATTCTAATATAAGTACC) were designed against the less abundant cDNA sequence class (Stan3) and used to amplify the majority of one allele from W5281.2. Comparing the sequence of this allele to the cDNA previously isolated from Y83-5 revealed a polymorphic (CTC)n simple sequence repeat in the third exon. Primers Stan3-F4 (5′CAGCAAACGATGTGAAGAAC) and Stan3-R4 (5′TGTAGATGAGGATTAGGACGAG) were designed to flank this repeat and then used to test whether either Stan3 alleles of W5281.2 cosegregated with tuber skin color.
RT-PCR analysis of gene expression
A schematic diagram of part of the anthocyanin biosynthetic pathway, illustrating metabolic intermediates and enzymes, is presented in Fig. 2. Primers F3H-F1 (5′AACAAGTTTTATTAGGGATGAA) and F3H-R1 (5′CACGCCAGTCTTGAACCA) were designed against conserved regions of two published potato flavanone 3-hydroxylase (f3h) cDNA sequences (GenBank accessions AY102035 and BE339992). Primers potDFR1 and potDFR2 amplify an internal fragment of dihydroflavonol 4-reductase (dfr) and have been described previously (De Jong et al. 2003a). Primers F35H-F2 (5′GTTGTTGCCTCTACCCCTAAT) and F35H-R2 (5′AACTTTTCATCCCTTTTTCAAT) were designed against the sequence of a potato cDNA clone isolated from a W5281.2 cDNA library using Petunia flavonoid 3′,5′-hydroxylase (f3′5′h) as a probe (Jung et al. 2005). Primers Stan2-F1 (5′GAGAGGTGACTTTGATTG) and Stan2-R1 (5′CAGTATATTTGCCCACCATTG) were used to study expression of Stan2. Primers Stan3-F2 (5′GAGACTTCATAAACTCTTAGGCAATAG) and Stan3-R2 (5′TATGTAGATGAGGATTAGGACGAG) were used to study expression of Stan3. All five primer pairs flank an intron so that amplification products generated from genomic and cDNA templates can readily be distinguished. Primers 18S-F1 (5′TGCCAGTAGTCATATGCTTGTCTC) and 18S-R1 (5′AGCCCGGTATTGTTATTTATTGTC) were designed against potato 18S ribosomal RNA. This primer pair was used to control for the success of RT-PCR when characterizing W5281.2 and 07506-01 and their progeny. When evaluating expression of Stan2 in tuber skin of selected cultivars and breeding clones, a QuantumRNA™ 18S internal standard (Ambion) was used to control for the success of RT-PCR, following the manufacturer’s instructions.
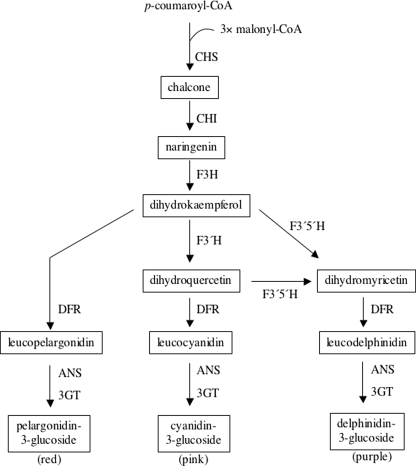
Fig. 2.
Schematic diagram of the anthocyanin biosynthetic pathway, adapted from Holton and Cornish (1995). Condensation of one molecule of p-coumaroyl CoA with three molecules of malonyl CoA is catalyzed by chalcone synthase. The chalcone is then processed along the pathway in a series of enzymatic reactions to form three basic types of anthocyanins—pelargonidin-3-glucoside, cyanidin-3-glucoside, and delphinidin-3-glucoside—after which varying additional modifications ultimately give rise to red, pink, and purple anthocyanin pigments, respectively. Some of the genes encoding pathway enzymes are: chs (chalcone synthase), chi (chalcone isomerase), f3h (flavanone 3-hydroxylase), f3′h (flavonoid 3′-hydroxylase), f3′5′h (flavonoid 3′,5′-hydroxylase), dfr (dihydroflavonol 4-reductase), ans (anthocyanidin synthase), and 3gt (flavonoid 3-glucosyltransferase). Metabolic intermediates are shown in boxes
For each independent experiment evaluating anthocyanin structural gene expression, total RNA was isolated with an RNeasy Plant Mini Kit (Qiagen) from the tuber skin (the surface tissue that could be peeled off with forceps) of parents W5281.2 and 07506-01, two progeny with red skin, two progeny with white skin, and two progeny with purple skin. The identity of the red-, white- and purple-skinned progeny varied between experiments. The experiment was performed four times, so that a total of eight red, eight white, and eight purple progenies were examined. RNA was isolated from young tubers, one to three cm in diameter, from plants that had not yet senesced. Total RNA from each clone was treated with RQ1 RNase-free DNase (Promega) to eliminate genomic DNA prior to reverse transcription. RNA was then converted into cDNA using random primers and a reverse transcription system kit (Promega). Approximately 70 ng of cDNA was then used as a template for PCR, employing the following thermal profile: 1 min at 94°C, followed by 35 cycles of [15 s at 94°C, 15 s at 53°C, and 1 min at 72°C], and a final 10 min incubation at 72°C.
Testing which alleles of Stan2 are expressed
Primers Stan2-F4 and Stan2-ClaI-R1 were used to amplify a fragment containing a polymorphic Alu I restriction site to determine which Stan2 alleles of W5281.2 and 07506-01 are expressed in the skin of colored progeny. Complementary DNA, prepared in the same way as for the RT-PCR analyses described above, was amplified from a total of eight red-skinned and seven purple-skinned progenies. PCR products were then digested with Alu I.
Transformation
A Stan2 777 cDNA isolated from W5281.2 (GenBank accession AY841129) appeared to contain all but the first two codons of the open reading frame when compared to Stan2 816. The Stan2 777 cDNA was amplified with primers 777 Sac I adaptor F (5′ GAGCTCatgagtACTCCTATGATGTGTA) and 777 Sac I adaptor R (5′ GAGCTCCTAATTAAGTAGATTCCATATATC) using a low error-rate polymerase (Platinum Pfx DNA polymerase; GIBCO-BRL). The 5′ portions of the primers were extended to include Sac I restriction sites (underlined). The forward primer also added the missing two codons (lowercase) to the open reading frame. Amplification products were digested with Sac I and ligated into the Sac I site of transformation vector pPS1 (Huang and Mason 2004). This vector contains a doubled CaMV promoter and tobacco etch virus translational enhancer sequence to enhance transgene expression. The construct was partially sequenced to ensure that no undesirable mutations had been introduced during PCR and confirm that the insert was in the desired sense orientation. The transformation construct was then introduced into Agrobacterium tumefaciens strain LBA4404. Transformation of the cultivar Désirée, the cultivar Bintje, and two diploid progeny (dd Rr pp and dd Rr Pp) from a cross between W5281.2 and 07506-01 was performed as previously described (Jung et al. 2005).
Thin layer chromatography
Approximately 0.1 g of tuber skin was macerated in a 1.5 mL tube containing 200 μL of 99% ethanol/1% HCl (v:v) and then centrifuged to pellet insoluble material. Repeated applications of 1 μL aliquots were spotted onto cellulose thin layer chromatography plates (Eastman Kodak 6064), allowed to dry, and then developed with butanol/2 N HCl (1:1 v:v) (Dodds and Long 1955).
Sequence alignment
Multiple sequence alignment was performed with MegAlign software (Lasergene, DNAStar, Inc.).
Results
Coordinate regulation of three anthocyanin structural genes in tuber skin
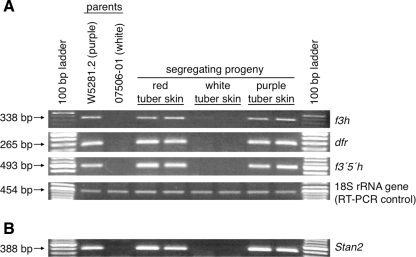
The potato D locus is required for the tissue-specific accumulation of red or purple anthocyanins in tuber skin. Several genes that control similar tissue-specific accumulation of anthocyanins in model plant systems (Petunia, maize, Arabidopsis) are known to be transcription factors, which act by coordinately regulating the expression of multiple structural genes (Holton and Cornish 1995; Mol et al. 1998). Diploid W5281.2 has purple tuber skin and is heterozygous Dd, while diploid 07506-01 has white skin and is homozygous recessive dd (Fig. 1). Progeny of a cross between W5281.2 and 07506-01 segregate 1:1 for the presence or absence of anthocyanins in tuber skin (Fig. 1). To test the hypothesis that D encodes a transcription factor, we examined the expression of three anthocyanin structural genes—f3h, dfr, and f3′5′h—in the tuber skin of W5281.2 and 07506-01, as well as in selected progeny, by RT-PCR (Fig. 3a). All three structural genes were found to be expressed in W5281.2 and in all colored progeny, while no expression of dfr and f3′5′h, and only weak expression of f3h, were detected in 07506-01 or in any white progeny.
Fig. 3.
Expression patterns of three anthocyanin structural genes (f3h, dfr, and f3′5′h) as well as Stan2, a candidate gene for D. a Expression of three anthocyanin structural genes in tuber periderm was examined by RT-PCR in parents W5281.2 and 07506-01 as well as in two progeny with red skin, two progeny with white skin, and two progeny with purple skin. Expression of 18S rRNA was evaluated as a control for RT-PCR. PCR product size and the corresponding gene name are denoted on the left and right of each gel, respectively. b Expression of Stan2, a candidate gene for the D locus, was examined by RT-PCR in the same parents and progeny as in a
Isolation of candidate genes for D and cosegregation with tuber skin color
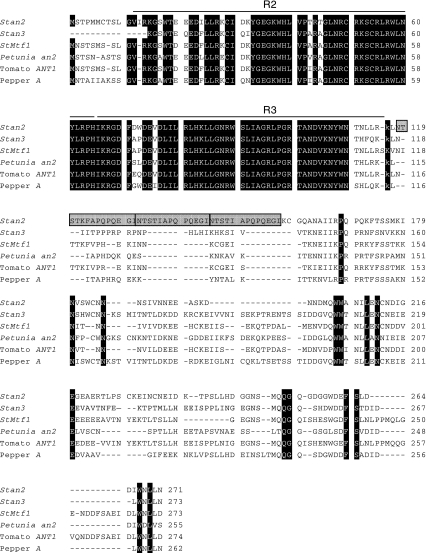
The Petunia R2R3 MYB domain gene an2 coordinately regulates the expression of multiple anthocyanin genes in the floral limb (Quattrocchio et al. 1993). Tomato harbors at least two tightly linked homologs of an2 that map to the same region of chromosome 10 where D is located in potato (De Jong et al. 2004). Reasoning that potato homolog(s) of an2 expressed in tuber skin might correspond to D, a cDNA library constructed from tuber skin of Y83-5, a red-colored tetraploid breeding clone, was screened with an an2 probe. Thirteen positively hybridizing clones were isolated and sequenced. Eleven cDNA clones were derived from one gene, which we designated Stan2 for S olanum t uberosum anthocyanin 2, while the other two cDNA clones were derived from a second gene, which we designated Stan3. One of the Stan2 cDNA clones (sequence deposited as GenBank accession AY841127) appeared, based on comparison with an2, to code for the entire open reading frame and was predicted to encode a protein of 271 amino acids. An interesting feature of the Y83-5 Stan2 allele (hereafter designated Stan2 816) was a perfect duplication of 39 nucleotides (coding for 13 amino acids) in the third exon (Fig. 4). Neither of the Stan3 cDNA clones were full-length (partial cDNA sequence has been deposited as accession AY841131). The predicted gene products of both Stan2 and Stan3 share extensive sequence similarity in their amino-terminal regions with several other known regulators of the anthocyanin pathway, including potato StMtf1 (Rommens et al. 2008), Petunia an2 (Quattrocchio et al. 1999, GenBank accession AF146702), tomato ANT1 (Mathews et al. 2003), and pepper A (Borovsky et al. 2004) (Fig. 4). Sequence conservation is most evident in the amino-terminal R2 and R3 MYB domains. Relatively few amino acid residues are absolutely conserved in the second half of these proteins (Fig. 4).
Fig. 4.
Alignment of the predicted amino acid sequences of Stan2 and Stan3 with those of known regulators of the anthocyanin biosynthetic pathway in the Solanaceae. Amino acids conserved across the six peptides—Stan2, Stan3, potato StMtf1, Petunia an2, tomato ANT1, and pepper A—are highlighted in black. The Stan2 816 allele shown contains an almost perfect tandem repeat (highlighted in gray) immediately after the R3 MYB domain: 12 of 14 residues at positions 118–131 are identical to 12 of 13 amino acids at positions 132–144. In addition, Stan2 816 residues 145–157 are a perfect duplication of residues 132–144
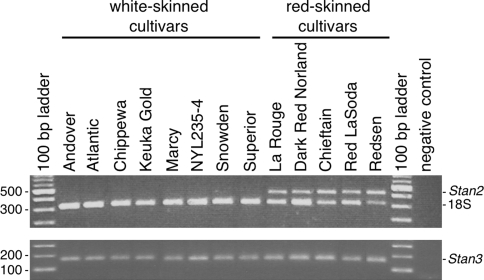
Subsequent RT-PCR analyses with RNA template isolated from 13 cultivars revealed that Stan2 was expressed in the skin of red-colored, but not white-colored, cultivars (Fig. 5). In addition, Stan2 was not expressed in the tuber flesh of any of the same 13 clones (data not shown). Stan3, on the other hand, was expressed in the skin of both red- and white-skinned clones (Fig. 5), as well as in the tuber flesh of all these cultivars (data not shown). Thus, of the two candidate genes, Stan2 exhibited an expression pattern that was more consistent with the possibility that it corresponds to D.
Fig. 5.
Expression of Stan2 and Stan3 in the periderm of red- and white-skinned potato cultivars. Expression was evaluated by RT-PCR, using mRNA template prepared from the periderm of eight white-skinned and five red-skinned cultivars. For Stan2 a QuantumRNA™ 18S internal standard was used to control for the success of RT-PCR. The gene names are indicated on the right side of each gel. The sizes of selected molecular weight markers, in base pairs, are indicated at left. The negative control was an amplification performed without cDNA template
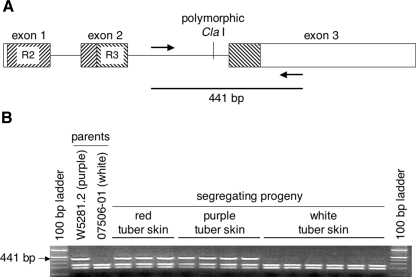
Genomic DNA of both alleles of Stan2 and Stan3 from diploid W5281.2 was amplified, cloned, and sequenced. Neither Stan2 allele from W5281.2 (GenBank accessions AY841130 and AY842506) exhibited the perfect 39 bp/13 amino acid duplication observed in Stan2 816. One of the W5281.2 alleles, designated Stan2 777, lacked a Cla I restriction site within the second intron (Fig. 6a). Evaluation of 73 colored and 63 white-skinned progeny from a cross between W5281.2 and 07506-01 revealed that Stan2 777 cosegregated perfectly with colored skin. Results from a subset of progeny are shown in Fig. 6b. One of the two alleles of Stan3 from W5281.2 (Genbank accession AY842507) also cosegregated perfectly with colored skin in the same 136 progeny (data not shown), demonstrating that Stan2, Stan3, and D are all tightly linked.
Fig. 6.
Structure of Stan2 and cosegregation of this gene with tuber skin color. a The Stan2 gene contains three exons, indicated with boxes, and two introns, indicated with horizontal lines. R2 and R3 MYB DNA-binding domains are indicated with diagonal hatch lines. The position of a polymorphic Cla I restriction site, which differentiates the two alleles of this gene in W5281.2, is also shown. Arrows indicate binding sites of oligonucleotide primers Stan2-ClaI-F2 and Stan2-ClaI-R1 (see “Materials and Methods”). b Primers diagrammed in a were used to amplify genomic DNA from parents W5281.2 and 07506-01 as well as from segregating progeny. Products were digested with Cla I and then electrophoresed on a 2% agarose gel. The Stan2 777 allele yields a 441 bp fragment with this assay and was observed to cosegregate with colored tuber skin
Allele-specific expression
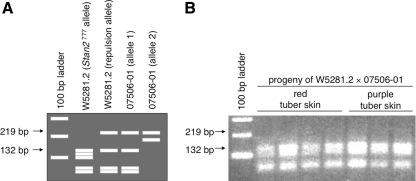
Like the structural genes dfr and f3′5′h, RT-PCR analyses revealed that Stan2 was expressed in the tuber skin of colored parent W5281.2 and in colored progeny, but not in the tuber skin of white-skinned parent 07506-01 or in white progeny (Fig. 3b). To determine which allele(s) of Stan2 are expressed in colored progeny, portions of the genomic DNA of both alleles of Stan2 were also sequenced from 07506-01. Sequence comparison indicated that Stan2 777 could be differentiated from all other parental alleles by virtue of a polymorphic Alu I restriction site in the first exon (Fig. 7a). RT-PCR followed by Alu I digestion demonstrated that the only allele expressed in tuber periderm of W5281.2 and colored progeny was Stan2 777 (Fig. 7b).
Fig. 7.
Only one allele of Stan2 is expressed in progeny with colored skin. Primers Stan2-F4 and Stan2-ClaI-R1 were used to amplify cDNA prepared from red- and purple-skinned progeny of the cross between W5281.2 and 07506-01. Amplification products were then digested with Alu I and separated on a 2% agarose gel to determine which W5281.2 and 07506-01 alleles are transcribed. a Predicted fragment sizes. Complementary DNA of Stan2 777 was predicted to give restriction fragments of 132, 119, 100, 52, and 44 bp. Based on genomic DNA sequence, the other allele of W5281.2 and one allele of 07506-01 were predicted to give fragments of 219, 132, 52, and 44 bp, while the remaining allele of 07506-01 was predicted to give fragments of 219 and 186 bp. b Actual fragment sizes observed. Panel shows an ethidium-bromide-stained gel and demonstrates that only Stan2 777 was expressed
Transgenic expression
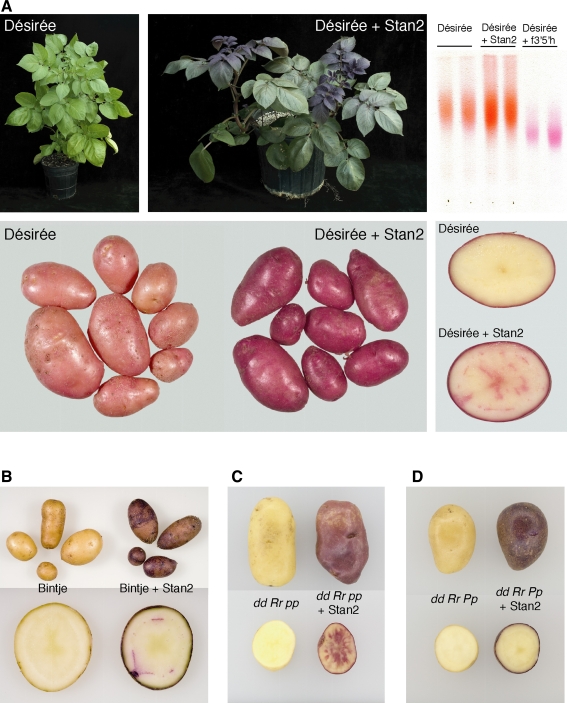
To test directly whether Stan2 777 can regulate anthocyanin biosynthesis, the open reading frame of this allele (see “Materials and Methods”) was placed into binary vector pPS1, behind a doubled CaMV 35S promoter and tobacco etch translational enhancer, and transformed into the light red-skinned cultivar Désirée. Désirée was chosen for transformation because it is relatively easy to transform and its tuber color demonstrates that it possesses a functional allele at the R locus. As the CaMV 35S promoter is known to direct expression in many plant tissues (Sunilkumar et al. 2002), we examined transformed plants to see if anthocyanins accumulated in any tissues that are not normally pink or red in color. Callus of Stan2 777-transformed Désirée was discernibly more pigmented than that of empty vector-transformed or untransformed Désirée (data not shown). Four independent transformants were selected for further analysis. The youngest foliage of each transformant was reddish-purple in color (Fig. 8a). Tuber skin of the same transgenic lines was deep red in color, while tuber flesh exhibited weak pigmentation in the vascular ring and central pith (Fig. 8a). The foliage of untransformed Désirée, as well as that of empty vector-transformed Désirée, was green (Fig. 8a and data not shown). Control tubers displayed light red skin and white flesh (Fig. 8a and data not shown). The flowers of the four transgenic lines were also dark red in color, in contrast to the faint pink color of untransformed Désirée (data not shown).
Fig. 8.
Transformation of the cultivars Désirée and Bintje, and two progeny from a cross between W5281.2 and 07506-01, with Stan2. The open reading frame of the R2R3 MYB domain allele Stan2 777 was placed into the vector pPS1 under the control of a doubled 35S promoter and introduced via Agrobacterium-mediated transformation. Tubers were harvested from mature plants and photographed several days later. a Foliage, tubers, and tuber flesh of cultivar Désirée, as well as corresponding tissues from a Désirée plant transformed with Stan2 777. Top right thin layer chromatography of anthocyanin pigments isolated from tuber skin of untransformed Désirée, Désirée transformed with Stan2 777, and Désirée transformed with a functional allele of f3′5′h (Jung et al. 2005). Pigment samples were applied near the bottom of the TLC plate shown and developed with a solvent comprised butanol/2 N HCl. The orange color and faster migration of the sample prepared from Désirée and Désirée transformed with Stan2 777 is characteristic of derivatives of pelargonidin, while the purple color and slower migration in the remaining two lanes is characteristic of derivatives of petunidin (Dodds and Long 1955). b Tuber and tuber flesh of Bintje, as well as corresponding tissues from Bintje transformed with Stan2 777. c Tuber and tuber flesh of a diploid clone with genotype dd Rr pp, as well as corresponding tissues from the same clone transformed with Stan2 777. d Tuber and tuber flesh of a diploid clone with genotype dd Rr Pp, as well as corresponding tissues from the same clone transformed with Stan2 777
Thin layer chromatography showed that although Désirée tubers transformed with Stan2 777 contained more anthocyanin, the anthocyanin profile was qualitatively indistinguishable from that of untransformed Désirée (Fig. 8a). These results demonstrate that Stan2 777 can regulate anthocyanin synthesis and that tissue non-specific overexpression leads to increased anthocyanin accumulation in multiple plant tissues.
The same construct used to transform Désirée was also introduced into the white-skinned cultivar Bintje. Periderm of seven (out of seven) transgenic Bintje lines was purple in color, while tuber flesh exhibited weak pigmentation in the vascular ring (Fig. 8b). Tuber periderm and flesh of Bintje transformed with an empty vector remained white (data not shown). The purple coloration observed in transgenic Bintje suggests that Bintje carries a functional allele at P.
For a more direct complementation test, two white-skinned progeny from the cross between W5281.2 and 07506-01 were selected and transformed with the same construct used for Bintje and Désirée. Based on marker and phenotypic analyses (this paper; De Jong et al. 2003a; Jung et al. 2005), one daughter clone was known to have genotype dd Rr pp (white skin, white tuber sprouts), while the other daughter clone had genotype dd Rr Pp (white skin, purple tuber sprouts). Seven (out of seven) transgenic lines derived from the dd Rr pp clone exhibited red coloration in tuber skin and tuber flesh (Fig. 8c), while one (out of three) transgenic line derived from the dd Rr Pp clone produced tubers with purple skin and no apparent pigmentation in tuber flesh (Fig. 8d).
Allelic composition in tetraploid cultivars and breeding clones
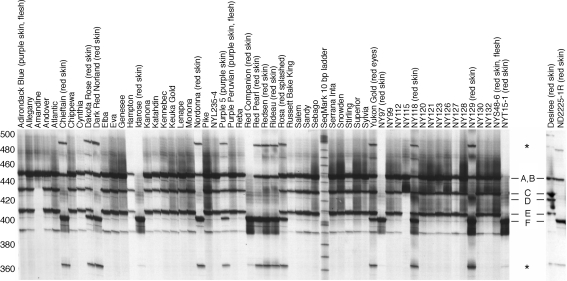
To assess the allelic diversity of Stan2 in tetraploid germplasm, a portion of the second intron and third exon was amplified from 60 cultivars and breeding clones. The products were then separated at high resolution on a 5% denaturing acrylamide gel. Products of six different sizes were observed (Fig. 9). The fastest migrating band, approximately 400 bp in size and labeled ‘F’ in Fig. 9, was found in 14 of 15 red-skinned/white-fleshed cultivars and breeding clones as well as in ‘Purple 5’, a purple-skinned/white-fleshed clone. The same band was absent in all 39 white-skinned clones tested. Cloning and sequencing band F from Idarose and NY97 revealed that it corresponded to Stan2 816. Amplification of a plasmid containing genomic DNA of Stan2 777 yielded a product that comigrated with band B (data not shown). Although the third exon of Stan2 816 is 39 bp longer than the third exon of Stan2777, it nevertheless gives rise to a smaller PCR product with this assay because the second intron of Stan2 816 (genomic DNA sequence deposited as accession AY841128) is 82 nucleotides shorter than that of Stan2 777. Band F was also observed in one cultivar with red tuber eyes (Yukon Gold) and one cultivar (Rosa) with a red ‘splashed’ phenotype (red eyes and areas of red skin around the eyes). The only red-skinned/white-fleshed cultivar tested that did not contain Stan2 816 was Désirée. Désirée was also unique in displaying a fragment (labeled ‘D’ in Fig. 9) that was absent in all 59 other clones.
Fig. 9.
An allele of Stan2 common to most red-skinned cultivars but absent in white-skinned clones. A portion of the Stan2 gene was amplified from 60 tetraploid cultivars and breeding clones with primers Stan2-ClaI-F2 and Stan2-ClaI-R1 and then separated on a 5% denaturing acrylamide gel. Six different fragments, labeled A through F, were observed. Relative to the SeqMark 10 base pair ladder, band A migrated at 447, band B at 445, band C at 430, band D at 423, band E at 408, and band F at 402 bp. The bands marked by asterisks appear to result from recombination during PCR. Amplification of individual plasmids containing genomic DNA corresponding to Stan2 777 and Stan2 816 gave rise to bands B and F, respectively, while amplification of a mixture of these two plasmids gave the two expected bands as well as the bands marked with an asterisk (data not shown). Recombination during PCR has been reported by others (Meyerhans et al. 1990; Judo et al. 1998)
Three potato clones with pigmented skin and pigmented flesh (Adirondack Blue, Purple Peruvian, NYS48-6) also did not contain Stan2 816 (Fig. 9).
Bands C and E were either both present or both absent in all clones tested, implying that the underlying alleles are tightly linked, possibly as (part of) a tandem array. Band E was 39 nucleotides shorter than band A, suggesting that the alleles underlying bands A and E differ in copy number of the same 39 bp region that is duplicated in Stan2 816.
Discussion
D is required for the accumulation of anthocyanin pigments in tuber skin. Several genes known to control tissue-specific accumulation of anthocyanins in model systems are transcription factors that regulate the expression of multiple anthocyanin biosynthetic genes, e.g., the MYB domain gene an2 of Petunia (Quattrocchio et al. 1999) and the basic helix-loop-helix gene R of maize (Consonni et al. 1997). To test the hypothesis that D might also act to regulate the activity of multiple genes, the expression of three unlinked structural genes (f3h, dfr, and f3′5′h) were evaluated in a potato population segregating for pigmented skin. All three were expressed in progeny with colored tuber skin, while dfr and f3′5′h were not expressed, and f3h was only weakly expressed, in progeny with white tuber skin. Two candidates for D (Stan2 and Stan3) were isolated from a potato cDNA library prepared from pigmented tuber skin, using Petunia an2 as a hybridization probe. Both candidates cosegregated with D, but only Stan2 was expressed specifically in potatoes with colored skin. When placed under the control of a doubled 35S promoter and transformed into the light red-skinned variety Désirée, the white-skinned variety Bintje, and two white-skinned diploids, Stan2 directed increased accumulation of anthocyanins in tuber skin, as well as in foliage. These results collectively support the hypothesis that Stan2 corresponds to D. The difference between dominant and recessive alleles of D appears to be at the level of transcription, as functional alleles were transcribed while non-functional alleles were not. We did not observe any premature stop codons or splice-site alterations in the non-expressed alleles sequenced.
In petunia flowers the structural gene f3h is expressed independently from anthocyanin regulatory genes (Quattrocchio et al. 1993, 1998), while in potato tubers f3h appears to be regulated, at least in part, by the D locus (Fig. 4). The differential regulation of f3h between these two closely related species, albeit in different tissues, is interesting. Lewis et al. (1998) observed a small but significant difference (p < 0.05) in flavonoid levels (other than anthocyanins) between purple/red-skinned and white-skinned tuber classes, with colored skin containing higher concentrations of flavonoids than white skin. Increased expression of f3h in colored potato skin may be responsible for this difference, although further study is needed to test this relationship more rigorously.
The function of Stan3 remains unknown at present. It may be a pseudogene, as we were unable to obtain a full-length cDNA, or it may regulate a function unrelated to anthocyanin production. Testing the function of individual members of closely related gene families has always been challenging, although the artificial miRNA approach outlined by Schwab et al. (2006) and Warthmann et al. (2008) may provide a way to do so.
In tetraploid potato, additional gene symbols have been assigned to loci that direct anthocyanin expression in tuber periderm (E) (Lunden 1937, 1960) or tuber phelloderm (the R locus of Lunden 1960, 1974). We consider it most likely that Lunden’s E and R are simply alleles of D, although further experimentation will nevertheless be required to confirm/disprove this. Another locus that has been described at the tetraploid level is M. When present together with E, M has been reported to restrict tuber pigment to the areas around the tuber eyes (Howard 1970), which could explain why the cultivars Yukon Gold and Rosa both appear to possess a functional allele of D (Fig. 9), yet have pigment restricted to areas around the tuber eye.
It has recently been reported that application of the rare-earth metal Cerium (Ce [IV]) to potato cell tissue cultures induces many anthocyanin structural genes (chs, f3h, f3′5′h, dfr and 3gt) and increases anthocyanin production (Qineng et al. 2006). Since these parallel the phenotypes mediated by D, Cerium may exert its effects by activating expression of D or a closely related gene, or by substituting for R2R3 MYB gene function.
The completely sequenced genome of Arabidopsis thaliana codes for approximately 126 R2R3 MYB domain genes (Stracke et al. 2001; Yanhui et al. 2006). Two of these, AtMYB75 (also known as Pap1) (Kranz et al. 1998; Romero et al. 1998; Borevitz et al. 2000) and AtMYB90 (Pap2) (Kranz et al. 1998; Borevitz et al. 2000), are known to regulate anthocyanin synthesis. These two genes, as well as two other R2R3 MYB genes, AtMYB113 and AtMYB114, are closely related in sequence (Stracke et al. 2001). Three of these genes—AtMYB113, AtMYB114, and AtMYB90—are tightly linked in a tandem array on chromosome 1, while AtMYB75 is located approximately 3.5 Mbp away on the same chromosome. The cosegregation of bands C and E in Fig. 9 suggests that at least one haplotype of the D locus contains a tandem array of two or more R2R3 MYB domain genes. Previous studies have shown that potato loci conditioning tissue-specific accumulation of anthocyanins are tightly linked to each other and to D. These include B (where a hierarchy of alleles is required for pigmentation of seed spot, nodal band, floral abscission layer, and tuber eyebrow), F (required for anthocyanin expression in flowers), Ow (pigmentation of ovary wall), Pf (pigmentation of tuber flesh), Pw (pigmentation of leaf whorl), Ul (pigmentation of the underside of leaves), Pd (pigmentation of the dorsal side of leaves), and Pv (pigmentation of the ventral side of leaves) (Kessel and Rowe 1974; De Jong 1987, 1991; Dodds and Long 1956; De Jong and Rowe 1972; Garg et al. 1981). One or more of these classical loci may correspond to bands C and/or E, or other bands, of Fig. 9.
Several genes or quantitative trait loci that influence tissue-specific anthocyanin expression in other Solanceous species—such as pepper A, tomato ag, and eggplant fap 10.1—map to regions comparable to D in their respective genomes (reviewed in De Jong et al. 2004), suggesting that all code for similar MYB domain genes. Indeed evidence was recently presented that pepper A, which conditions anthocyanin expression in the foliage, flower, and immature fruit, encodes a gene related to an2 (Borovsky et al. 2004). The precedents of an2 and A, and now D, strongly suggest that evolution and subsequent selection for novel tissue-specific expression patterns of R2R3 MYB domain genes in the Solanaceae is responsible for much of the anthocyanin-mediated, visually conspicuous phenotypic variation that is so prevalent in this economically important and diverse plant family.
Evaluation of tetraploid germplasm with a simple PCR assay revealed that the Stan2 816 allele was common to 14/15 red-skinned cultivars and absent in all white-skinned cultivars tested. The unique size of this allele should make it simple to track in potato breeding programs, e.g., for the purpose of developing homozygous (quadruplex) parental lines. In maize, the expression level of transcriptional regulators of anthocyanin biosynthesis is directly correlated with the amount of anthocyanin produced (Patterson et al. 1993; Hollick et al. 2000). Consistent with this, overexpression of Stan2 in transgenic Désirée increased the amount of anthocyanin in tuber skin. De Jong et al. (2003b) and Salaman (1910) have previously postulated that dosage of functional alleles at D correlates with the intensity of red tuber color. Figure 9 suggests, but does not prove, that four clones—IdaRose, Red Companion, NY97 and NYT15-1—are quadruplex for the 816 allele, as the PCR assay only amplified band F from each of them. It is thus of interest to note that NY97 and NYT15-1 are two of the deepest red-skinned clones ever developed by the Cornell University breeding program. With the availability of Stan2 816 sequence it should now be possible to test whether dosage of this allele, or other functional alleles, influence intensity of tuber color.
We have previously presented evidence that a functional allele at R, also required for red tuber color, is present in many white-skinned potatoes (De Jong et al. 2003a). In the current study, we found no white potatoes that possess Stan2 816. Taken together these observations suggest that white-skinned cultivars are most frequently white because they lack a functional allele of D, not because they lack a functional allele of R (or P).
One curious result in the present study was that the flesh of transgenic tubers of Désirée, Bintje, and two progeny from the cross between W5281.2 and 07506-01 was not completely red or purple. In diploids, pigmented flesh is mediated by Pf, a locus tightly linked to D (De Jong 1987). The pigmented flesh trait exhibits variable expressivity, i.e., it is not unusual in a single segregating cross to observe some progeny with completely colored flesh, some progeny with only traces of pigmentation, and other progeny with a wide range of intermediate phenotypes. Désirée, Bintje and the two transformed diploid clones may have a genetic background that limits the extent to which flesh can be pigmented. Alternatively the 35S promoter used to drive Stan2 expression may not function well in all tuber tissues. Sung et al. (1994) reported that GUS marker gene expression in mature tubers from a plant transformed with a 35S:GUS construct was higher in tuber vasculature than in other tuber tissues, although GUS was expressed in most tissues from young tubers. Rommens et al. (2008) recently transformed Bintje with another Stan2-like MYB gene, StMtf1, and observed similar, variable tuber flesh coloration, when the gene was expressed under the control of two alternative constitutive promoters (ubiquitin-7 and GBSS). The combined results from the current and Rommens et al. (2008) studies suggest that, at least in Bintje, MYB gene expression is not sufficient for complete coloration of tuber flesh. Quantitative trait loci that influence the degree of tuber flesh pigmentation have recently been mapped on chromosomes 5, 8, and 9 (Zhang et al. 2009a); allelic configurations at these, and/or other unknown loci, may influence the tuber flesh phenotype when Stan2 is constitutively expressed.
Acknowledgments
We thank H. De Jong for providing diploid clones and reviewing the manuscript, K. Paddock and M. Compton for plant maintenance, and K. Loeffler for photography. This work was supported in part by federal Hatch funds provided to W.D.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- dfr
Dihydroflavonol 4-reductase
- f3′5′h
Flavonoid 3′,5′-hydroxylase
- f3h
Flavanone 3-hydroxylase
- Stan2
Solanum tuberosum anthocyanin 2
- Stan3
Solanum tuberosum anthocyanin 3
References
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovsky Y, Oren-Shamir M, Ovadia R, De Jong W, Paran I. The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor Appl Genet. 2004;109:23–29. doi: 10.1007/s00122-004-1625-9. [DOI] [PubMed] [Google Scholar]
- Cone KC, Burr FA, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni G, Ronchi A, Pilu R, Gavazzi G, Dellaporta SL, Tonelli C. Ectopic anthocyanin pigmentation in maize as a tool for defining interactions between homologous regulatory factors. Mol Gen Genet. 1997;256:265–276. doi: 10.1007/s004380050569. [DOI] [PubMed] [Google Scholar]
- Cutanda-Perez M-C, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L. Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plaant Mol Biol. 2009;69:633–648. doi: 10.1007/s11103-008-9446-x. [DOI] [PubMed] [Google Scholar]
- De Jong H. Inheritance of pigmented tuber flesh in cultivated diploid potatoes. Am Pot J. 1987;64:337–343. doi: 10.1007/BF02853595. [DOI] [Google Scholar]
- De Jong H. Inheritance of anthocyanin pigmentation in the cultivated potato: a critical review. Am Pot J. 1991;68:585–593. doi: 10.1007/BF02853712. [DOI] [Google Scholar]
- De Jong H, Burns VJ. Inheritance of tuber shape in cultivated diploid potatoes. Am Pot J. 1993;70:267–283. doi: 10.1007/BF02849314. [DOI] [Google Scholar]
- De Jong H, Rowe PR. Genetic markers in inbred clones of cultivated diploid potatoes. Potato Res. 1972;15:200–208. doi: 10.1007/BF02361807. [DOI] [Google Scholar]
- De Jong WS, De Jong DM, De Jong H, Kalazich J, Bodis M. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.) Theor Appl Genet. 2003;107:1375–1383. doi: 10.1007/s00122-003-1395-9. [DOI] [PubMed] [Google Scholar]
- De Jong WS, De Jong DM, Bodis M. A fluorogenic 5′ nuclease (TaqMan) assay to assess dosage of a marker tightly linked to red skin color in autotetraploid potato. Theor Appl Genet. 2003;107:1384–1390. doi: 10.1007/s00122-003-1420-z. [DOI] [PubMed] [Google Scholar]
- De Jong WS, Eannetta NY, De Jong DM, Bodis M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet. 2004;108:423–432. doi: 10.1007/s00122-003-1455-1. [DOI] [PubMed] [Google Scholar]
- De Vries S, Hoge H, Blisseling T (1988) Isolation of total and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoort RA, Verma DPS (eds) Plant molecular biology manual, vol B6, pp 1–13
- Dodds KS, Long DH. The inheritance of colour in diploid potatoes. I. Types of anthocyanidins and their genetic loci. J Genet. 1955;53:136–149. doi: 10.1007/BF02981517. [DOI] [Google Scholar]
- Dodds KS, Long DH. The inheritance of colour in diploid potatoes. II. Three-factor linkage group. J Genet. 1956;54:27–41. doi: 10.1007/BF02981699. [DOI] [Google Scholar]
- Espley RV, Hellen RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg KC, Tiwari SP, Sharma KP. Inheritance of leaf pigmentation in dihaploid-Phureja hybrids of potato. J Ind Pot Assn. 1981;8:31–34. [Google Scholar]
- Gebhardt C, Ritter E, Schachtschabel U, Walkemeier B, Uhrig H, Salamini F. RFLP analysis and linkage mapping in Solanum tuberosum. Theor Appl Genet. 1989;78:65–75. doi: 10.1007/BF00299755. [DOI] [PubMed] [Google Scholar]
- Hollick JB, Patterson GI, Asmundsson IM, Chandler VL. Paramutation alters regulatory control of the maize pl1 locus. Genet. 2000;154:1827–1838. doi: 10.1093/genetics/154.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard HW. Genetics of the potato. Great Britain: Logos Press Limited; 1970. [Google Scholar]
- Huang Z, Mason HS. Conformational analysis of hepatitis B surface antigen fusions in an Agrobacterium-mediated transient expression system. Plant Biotechnol J. 2004;2:241–249. doi: 10.1111/j.1467-7652.2004.00068.x. [DOI] [PubMed] [Google Scholar]
- Judo MSB, Wedel AB, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CS, Griffiths HM, De Jong DM, Cheng S, Bodis M, De Jong WS. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theor Appl Genet. 2005;110:269–275. doi: 10.1007/s00122-004-1829-z. [DOI] [PubMed] [Google Scholar]
- Kessel R, Rowe PR. Inheritance of two qualitative traits and a proposed genetic map for their linkage group in diploid potatoes. Potato Res. 1974;17:283–295. doi: 10.1007/BF02360322. [DOI] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Paz-Ares J, Weisshaar B. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Lewis CE, Walker JRL, Lancaster JE, Sutton KH. Determination of anthocyanins, flavonoids and phenolic acids in potatoes. I: Coloured cultivars of Solanum tuberosum L. J Sci Food Agric. 1998;77:45–57. doi: 10.1002/(SICI)1097-0010(199805)77:1<45::AID-JSFA1>3.0.CO;2-S. [DOI] [Google Scholar]
- Lunden AP. Arvelighetsunderskelser i potet. (Inheritance studies in the potato) Meld Norg Landbr. 1937;17:1–156. [Google Scholar]
- Lunden AP. Some more evidence of autotetraploid inheritance in the potato (Solanum tuberosum) Euphytica. 1960;9:225–234. doi: 10.1007/BF00022227. [DOI] [Google Scholar]
- Lunden A. Inheritance of tuber and flower colour in the potato (Solanum tubersosum, L.) Meld Norg Landbr (Scientific Reports of the Agricultural University of Norway) 1974;53:1–8. [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, Wagner DR. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell. 2003;15:1689–1703. doi: 10.1105/tpc.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A, Vartanian J, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Plant Sci. 1998;3:212–217. doi: 10.1016/S1360-1385(98)01242-4. [DOI] [Google Scholar]
- Naito K, Umemura Y, Mori M, Sumida T, Okada T, Takamatsu N, Okawa Y, Hayashi K, Saito N, Honda T. Acylated pelargonidin glycosides from a red potato. Phytochem. 1998;47:109–112. doi: 10.1016/S0031-9422(97)00520-7. [DOI] [Google Scholar]
- Patterson GI, Thorpe CJ, Chandler VL. Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genet. 1993;135:881–894. doi: 10.1093/genetics/135.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qineng L, Qing Y, Huawen Z. Effects of Cerium on accumulation of anthocyanins and expression of anthocyanin biosynthetic genes in potato cell tissue cultures. J Rare Earth. 2006;24:479–484. doi: 10.1016/S1002-0721(06)60147-6. [DOI] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes R. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JNM, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998;13:475–488. doi: 10.1046/j.1365-313X.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Saona LE, Giusti MM, Wrolstad RE. Anthocyanin pigment composition of red-fleshed potatoes. J Food Sci. 1998;63:458–465. doi: 10.1111/j.1365-2621.1998.tb15764.x. [DOI] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313X.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Rommens CM, Richael CM, Yan H, Navarre DA, Ye J, Krucker M, Swords K. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotech J. 2008;6:870–886. doi: 10.1111/j.1467-7652.2008.00362.x. [DOI] [PubMed] [Google Scholar]
- Salaman RN. The inheritance of colour and other characters in the potato. J Genet. 1910;1:7–46. doi: 10.1007/BF02981567. [DOI] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant J. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Sung S-K, Choi S-B, Jeon J-S, Park MC, Lee K-W. Expression patterns of CaMV 35S promoter-GUS in transgenic potatoes and their clonal progenies. J Plant Biol. 1994;37:17–25. [Google Scholar]
- Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS. Developmental and tissue-specific expression of CaMV promoter in cotton as revealed by GFP. Plant Mol Biol. 2002;50:463–474. doi: 10.1023/A:1019832123444. [DOI] [PubMed] [Google Scholar]
- Van Eck HJ, Jacobs JME, van Dijk J, Stiekema WJ, Jacobsen E. Identification and mapping of three flower colour loci of potato (S. tuberosum) by RFLP analysis. Theor Appl Genet. 1993;86:295–300. doi: 10.1007/BF00222091. [DOI] [PubMed] [Google Scholar]
- Van Eck HJ, Jacobs JME, Van Den Berg PMMM, Stiekema WJ, Jacobsen E. The inheritance of anthocyanin pigmentation in potato (Solanum tuberosum L.) and mapping of tuber skin colour loci using RFLPs. Hered. 1994;73:410–421. doi: 10.1038/hdy.1994.189. [DOI] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Herve P. Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE. 2008;3:e1829. doi: 10.1371/journal.pone.0001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Yunping S, Li Z, Xiaohui D, Jingchu L, Xing-Wang D, Zhangliang C, Hongya G, Li-Jia Q. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jung CS, De Jong W. Genetic analysis of pigmented tuber flesh in potato. Theor Appl Genet. 2009;119:143–150. doi: 10.1007/s00122-009-1024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng S, De Jong D, Griffiths H, Halitschke R, De Jong W. The potato R locus codes for dihydroflavonol 4-reductase. Theor Appl Genet. 2009;119:931–937. doi: 10.1007/s00122-009-1100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]