Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression at the post-transcriptional level. They are involved in diverse biological processes, such as development, differentiation, cell proliferation and apoptosis. To study the role of miRNAs during pronephric kidney development of Xenopus, global miRNA biogenesis was eliminated by knockdown of two key components: Dicer and Dgcr8. These embryos developed a range of kidney defects, including edema formation, delayed renal epithelial differentiation and abnormal patterning. To identify a causative miRNA, mouse and frog kidneys were screened for putative candidates. Among these, the miR-30 family showed the most prominent kidney-restricted expression. Moreover, knockdown of miR-30a-5p phenocopied most of the pronephric defects observed upon global inhibition of miRNA biogenesis. Molecular analyses revealed that miR-30 regulates the LIM-class homeobox factor Xlim1/Lhx1, a major transcriptional regulator of kidney development. miR-30 targeted Xlim1/Lhx1 via two previously unrecognized binding sites in its 3′UTR and thereby restricted its activity. During kidney development, Xlim1/Lhx1 is required in the early stages, but is downregulated subsequently. However, in the absence of miR-30 activity, Xlim1/Lhx1 is maintained at high levels and, therefore, may contribute to the delayed terminal differentiation of the amphibian pronephros.
Keywords: Dgcr8, Dicer, Lhx1 (Lim1; Xlim1), Kidney, microRNA, Pronephros, Xenopus, Mouse
INTRODUCTION
The kidney is essential to clear waste products, to balance the concentration of body fluids and electrolytes and to reabsorb small molecules, such as amino acids, ions and water, to maintain blood homeostasis. It develops from the intermediate mesoderm via inductive processes that form three different and increasingly more complex kidney types: the pro-, meso- and metanephros (Saxén, 1987; Vize et al., 2003). The metanephros is the adult kidney of higher vertebrates, whereas the mesonephros is the adult kidney of amphibians and fish. The pronephros is of particular importance in aquatic animals during larval stages in order to maintain water homeostasis. The development of the three kidney forms is interconnected and both the meso- and metanephric kidney rely on the formation of the pronephros (Bouchard et al., 2002; Carroll and McMahon, 2003; Jones 2003; Dressler, 2006). Moreover, transcription factors, as well as markers of terminal differentiation, have a similar expression pattern and function during kidney development in human, mouse, zebrafish and Xenopus (Zhou and Vize, 2004; Drummond, 2005; Raciti et al., 2008).
MicroRNAs (miRNAs) are a class of ~22 nt, non-coding molecules expressed in multicellular organisms (Kloosterman and Plasterk, 2006; Bushati and Cohen, 2007; Stefani and Slack, 2008). The principal function of miRNAs is to regulate protein expression and mRNA stability by binding to complementary nucleotide sequences in the 3′UTRs. miRNAs are initially transcribed by RNA polymerase II as part of a much longer primary transcript (pri-miRNA) with a CAP structure and poly(A) tail. In the nucleus, the pri-miRNA is processed to a ~60 nt hairpin precursor miRNA (pre-miRNA) by the microprocessor complex, consisting of Drosha, an RNase III type endonuclease, and Dgcr8 (DiGeorge critical region 8)/Pasha, a double-stranded-RNA-binding protein. Once cleaved, pre-miRNAs are transported from the nucleus to the cytoplasm, where another RNase III enzyme, Dicer, cleaves pre-miRNA into the mature ~22 nt duplex miRNA. The mature miRNAs are then loaded into the RNA-induced silencing complex (RISC), which regulates binding of miRNAs to the 3′UTR of target mRNAs and induces translational inhibition or degradation. Target specificity is determined by the seed sequence (nucleotides 2 to 8 from the 5′ end of an miRNA) and is further strengthened by base pairing of flanking nucleotides (Brennecke et al., 2005; Bartel, 2009). The expression of miRNA and of its targets often show an inverse relationship (Farh et al., 2005; Stark et al., 2005). For example, expression levels of predicted targets for miR-1 are high in myoblasts, but are strongly reduced upon differentiation into myotubes, when expression of miR-1 is initiated (Chen et al., 2006).
The functions of several individual miRNAs have been characterized in detail, such as how lin-4 and let-7 control developmental timing in C. elegans and Drosophila, respectively (Lee et al., 1993; Wightman et al., 1993; Reinhart et al., 2000). However, many miRNAs belong to families with nearly identical seed sequences that can compensate for each other and make loss-of-function analyses difficult (Abbott et al., 2005; Stefani and Slack, 2008). For example, the zebrafish miR-430 family consists of five members that together control the transition from maternal to zygotic gene transcription (Chen, P. Y. et al., 2005; Giraldez et al., 2005; Giraldez et al., 2006). One approach to address the general role of miRNAs in development is to interfere with miRNA biogenesis. Mice lacking either Dicer (Dicer1) or Dgcr8 protein are embryonically lethal owing to defects in germ layer patterning (Bernstein et al., 2003; Wang et al., 2007). But tissue-specific conditional mutants have identified multiple roles for miRNAs during organogenesis (Stefani and Slack, 2008). In zebrafish, embryos lacking Dicer protein exhibit defects in the degradation of maternal transcripts as well as abnormal brain, somite and heart morphogenesis (Wienholds et al., 2003; Giraldez et al., 2005; Giraldez et al., 2006). However, little information is available regarding the role of miRNAs in kidney development. Expression analyses have identified several miRNAs that are expressed in the kidney (Sun et al., 2004; Naraba and Iwai, 2005; Kato et al., 2007) and conditional alleles eliminating Dicer from mouse podocytes have demonstrated that miRNAs are required for the maintenance of functional glomeruli (Harvey et al., 2008; Ho et al., 2008; Shi et al., 2008).
Here we extend the study of miRNAs to the pronephric kidney of Xenopus laevis. Using antisense morpholino oligomers against Dicer or Dgcr8 to inhibit miRNA biogenesis, we show that miRNAs are required for multiple facets of pronephros development, including patterning and terminal differentiation. We identify the miR-30 family as an essential player in this process. Knockdown of one family member, miR-30a-5p, phenocopies nearly all of the defects caused by the loss of Dicer or Dgcr8. Finally, we identified the transcription factor Xlim1/Lhx1 as an important target of miR-30 activity. This transcription factor is essential for kidney development in mouse and frog (Shawlot and Behringer, 1995; Carroll and Vize, 1999; Kobayashi et al., 2005) and we now demonstrate that miR-30 is required for its correct spatiotemporal expression during pronephros development.
MATERIALS AND METHODS
Embryo manipulations
Xenopus embryos obtained by in vitro fertilization were maintained in 0.1× modified Barth medium (Sive et al., 2000) and staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1994). Antisense morpholino oligomers (MOs) were obtained from GeneTools. The sequences of the antisense MOs used in this study were: Dicer-MO, 5′-TGCAGGGCTTTCATAAATCCAGTGA-3′; Dgcr8-MO, 5′-GGGCTACTTCCTCACACTCCTCCAT-3′; miR-30a5p-MO, 5′-AGCTTCCAGTCGAGGATGTTTACAG-3′; miR-34b-MO, 5′-ACAATCAGCTAACTACACTGCCTGA-3′; and Std-MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′. Antisense MOs were diluted to 1 mM. The Dgcr8-MO rescue construct pCS2-Dgcr8* was generated by PCR mutagenesis, introducing six nucleotide changes so that the construct was no longer targeted by Dgcr8-MO, but was translated into a protein of identical amino acid sequence. pCS2-Lhx1* was sub-cloned from pXH32 (a kind gift from Dr I. Dawid, NIH, USA), changing six nucleotides in the recognition site of Lhx1-MO and removing the entire 3′UTR. The pCS2-Dicer-GFP and pCS2-Dgcr8-GFP constructs were generated by PCR from pCS2-xDicer and pCS2-xDgcr8 and subcloned into pCS2-GFP. For synthetic mRNA, all plasmids were linearized with NotI and transcribed with SP6 RNA polymerase using the mMessage mMachine (Ambion).
For all injections, a total of 8 nl of MO solution was injected radially at the 2- to 4-cell stage into Xenopus embryos. For the rescue experiments, these injections were followed by a single injection of synthetic mRNA into the marginal zone. In the miR-30 reporter assays, a subset of embryos was injected with 160 fmol miRNA duplex (miR-30a-5p or miR-17) into the animal region at the 2-cell stage in the presence or absence of 3.2 pmol miR-30a5p-MO or miR-34b-MO. These embryos and uninjected embryos were then injected at the 4-cell stage with a total of 8 ng synthetic mRNA (pXEXβGal-Lhx1-3′UTR or pXEXβGal-miR-30a-5p) animally into two blastomeres. Embryos were processed at gastrula stage for lacZ staining using standard procedures.
In situ hybridization
In situ hybridizations were carried out as described previously (Belo et al., 1997). To generate antisense probes, plasmids were linearized and transcribed as follows: pGEM-T-Cadherin16, ApaI/SP6 (GenBank accession number GQ499200); pBC-ClC-K, EcoRI/T7 (Vize, 2003); pGEM-T-HNF1β, Asp718/T7 (Vignali et al., 2000); pSP64TS-Lim1, XhoI/T7 (Carroll et al., 1999); pSK-β1-Na/K-ATPase, EcoRI/T7 (Tran et al., 2007); pCMV-SPORT6-xNbc-1, SalI/T7 (Zhou and Vize, 2004); pSK-NCC, EcoRI/T7 (Tran et al., 2007); pSK-NKCC2, SmaI/T7 (Tran et al., 2007); pSK-Pax-2, XbaI/T7 (Carroll et al., 1999); pSK-Pax-8, BamHI/T7 (Carroll et al., 1999); pCMV-SPORT6-ROMK, EcoRI/T7 (Tran et al., 2007); pCMV-SPORT6-xSGLT1-K, SalI/T7 (Zhou and Vize, 2004); pGEM-T-Slc7a13, SalI/T7 (nt 862 to 1480 of GenBank accession number BC060020). The following LNA-modified degenerate and specific oligomers were synthesized by Integrated DNA Technologies and labeled with digoxigenin as previously described (Wienholds et al., 2005): miR-138, 5′-CGLNA-GCCLNA TGALNATTCLNAACALNAACALNACCALNAGCT-3′; miR-200 family, 5′-CRKCLNADTTLNAACCLNAMGRCLNAAGTLNARTTLNAA-3′; miR-30 family, 5′-SALNAGTLNASDRGLNAGATLNAGTTLNATACLNAA-3′; miR-30a-5p, 5′-CTTLNACCALNAGTCLNAGAGLNAGATLNAGTTLNA-TACLNAA-3′; miR-30b, 5′-AGCLNATGALNAGTGLNATAGLNAGATLNA-GTTLNA TACLNA A-3′; miR-30c, 5′-GCTGLNAAGALNAGTGLNATAGLNA-GATLNAGTTLNA TACLNAA-3′; miR-489, 5′-GCLNATGCLNACATLNA-ATALNATGTLNA GGTLNAGTCLNAATT-3′.
Histology, immunohistochemistry and TUNEL staining
For histological staining, Xenopus embryos were fixed in Bouin's Fixative, dehydrated, embedded in paraplast, sectioned at 7 μm, dewaxed, and stained with Hematoxylin and Eosin. For immunohistochemistry, embryos were fixed in Dent's fixative (4:1 methanol:DMSO). For whole-mount immunostaining, embryos were incubated overnight with 3G8 and 4A6 monoclonal antibodies (Vize et al., 1995) followed by incubation with a horseradish peroxidase-coupled anti-mouse IgG and developed using the ImmPACT DAB Kit (Vector Laboratories). For immunohistochemistry on sections (i.e. the proliferation analysis), the embryos were embedded in paraplast, sectioned at 25 μm and probed with anti-phospho-Histone H3 (Upstate Biotechnology). For TUNEL staining, Xenopus embryos were fixed in MEMFA, dehydrated, embedded in paraplast, sectioned at 10 μm and analyzed using the DeadEnd Colorimetric TUNEL System (Promega).
microRNA profiling
miRNA profiling was performed using the miRCURY LNA miRNA Arrays (Exiqon) consisting of all miRNA in all organisms as annotated in miRBase release 8.1. The results have been deposited to Gene Expression Omnibus (accession number GSE18609). For the candidate miRNA approach, total RNA was isolated using RNA STAT-60 (Tel-Test). RT-PCR was performed using a protocol described previously for miRNAs (Chen, C. et al., 2005) with minor modifications. The looped RT primer was modified to contain a random hexamer at the 3′ end (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACNNNNNN-3′). A common reverse primer (5′-GTGCAGGGTCCGAGGT-3′) and miRNA-specific forward primers (see Table S1 in the supplementary material) were used for PCR amplification and the products were analyzed on agarose gels.
miRNA reporter constructs and miRNA duplexes
The pXEXβGal-miR-30a-5p construct, containing a triplet of miR-30a-5p recognition sites (underlined in the oligomers), was designed as described (Giraldez et al., 2006) using the following four oligomers: miR-30a-5p-X, 5′-TCGAGAATCTAGATTCCAGTCGAGGATGTTTACATAGTA-3′; miR-30a-5p-Y, 5′-TTCCAGTCGAGGATGTTTACATAGTATTCCAGTCGAGGATGTTTACACTGCA-3′; miR-30a-5p-W, 5′-TCCTCGACTGGAATACTATGTAAACATCCTCGACTGGAATCTAGATTC-3′; and miR-30a-5p-Z, 5′-GTGTAAACATCCTCGACTGGAATACTATGTAAACA-3′. Oligomers were annealed and cloned first into pAcGFP (Clontech Laboratories) and then subcloned into pXEXβGal (a kind gift from R. Harland, University of California, Berkeley, CA, USA) using XbaI and Asp718. The pXEXβGal-Lhx1-3′UTR and pmir-GLO-Lhx1-3′UTR constructs were prepared by amplifying a part of the 3′UTR of Xlim1 (5′-GGGCTAGCTGTTAGGTGGTGCACAGGAC-3′, 5′-GAGGTACCCCTGTTTGGGTCTATGTAAATC-3′) and subcloning it into pXEXβGal using XbaI and Asp718 or into pmir-GLO (Promega) using NheI and SalI. pmir-GLO-Lhx1-3′UTR-mut was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene), changing the core of the two miR-30 binding sites from 5′-TGTTTAC-3′ to 5′-TCTATTC-3′.
For the synthetic miRNA duplexes, two RNA/DNA hybrid oligomers with the following sequences were synthesized (Integrated DNA Technologies) and annealed: miR-30a-5p duplex, 5′-rUrCrCrArGrUrCrGrArGrGrArUrGrUrUrUrArGrAAG-3′ and 5′-rUrGrUrArArArCrArUrCrCrUrCrGrArCrUrGrGrAAG-3′; miR-17 duplex, 5′-rArCrCrUrGrCrArCrUrGrUrArArGrCrArCrUrUrUrGTT-3′ and 5′-rCrArArArGrUrGrCrUrUrArCrArGrUrGrCrArGrGrUAG-3′.
For the miRNA expression constructs, pri-miRNA precursors including ~200 bp flanking sequences (to facilitate proper folding and cleavage to the mature miRNAs) were amplified by PCR and subcloned into pCS2 to generate pCS2-miR-30a-5p (5′-GCTTTGCAGTTTACAGAATGT-3′ and 5′-CCTCGAGTCTGTGGTTTTGAAGTTGTTT-3′) and pCS2-miR-17 (5′-GAACTTCTGGCTATTGGCTCCTC-3′ and 5′-CCTCGAGCACGCAGCACCAGCAG-3′). For the luciferase assays, HEK 293T cells were transfected using Lipofectamine 2000 (Invitrogen) with 50 ng of the pmir-GLO constructs and 1 μg of the miRNA vectors and processed using the Dual-Luciferase Reporter Assay (Promega) after 48 hours.
RESULTS
Global inhibition of miRNA biogenesis
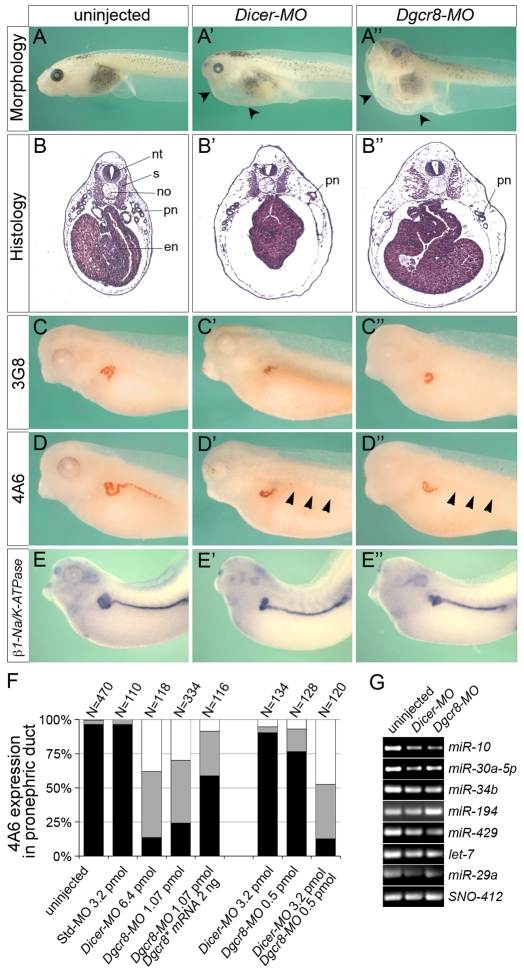
miRNAs undergo multiple steps of post-transcriptional processing and are non-functional until the mature miRNA is loaded into the RISC complex (Kloosterman and Plasterk, 2006; Bushati and Cohen, 2007; Stefani and Slack, 2008). As a first step to explore the overall role of miRNAs during pronephric kidney development, we depleted two key proteins in this process, Dicer and Dgcr8, using antisense morpholino oligomers (MOs). Xenopus embryos were injected at the 2- to 4-cell stage in all four blastomeres with different amounts of Dicer-MO or Dgcr8-MO to determine the sub-optimal doses that allowed survival until tailbud stage. Interestingly, the two antisense MOs did not behave identically. Injection of a total of 6.4 pmol Dicer-MO permitted reasonable survival of the embryos, whereas only a 6-fold lower amount of Dgcr8-MO (1.07 pmol) was tolerated. As shown in Fig. 1A-A″, the Dicer-MO- and Dgcr8-MO-injected embryos at stage 43 were generally malformed, displayed eye defects and developed edema, a condition often caused by an impaired osmoregulatory function of the pronephros (Howland, 1916). Histological sections at stage 42 showed an enlarged body cavity, malformations of the endoderm, a reduced neural tube, disintegrating somites and a reduction of the pronephric tubules (Fig. 1B-B″ and data not shown). Although these morphants had a wide range of defects, we focused in this study on the development of the pronephric kidney.
Fig. 1.
Inhibition of miRNA biogenesis results in pronephric abnormalities. (A-E″) Xenopus embryos were injected with 6.4 pmol Dicer-MO or 1.07 pmol Dgcr8-MO and compared with uninjected sibling embryos by morphology at stage 43 (A-A″), histology with Hematoxylin and Eosin at stage 42 (B-B″), immunostaining with 3G8 and 4A6 at stage 40 (C-D″), and by whole-mount in situ hybridization for β1-Na/K-ATPase at stage 39 (E-E″). Arrowheads indicate edema formation (A′,A″) and the loss of 4A6 staining in duct (D′,D″). en, endoderm; no, notochord; nt, neural tube; pn, pronephros; s, somites. (F) Quantification of 4A6 staining in the pronephric duct, comparing uninjected control embryos with embryos injected with a standard control MO (Std-MO), Dicer-MO, Dgcr8-MO, a combination of Dicer-MO and Dgcr8-MO, or Dgcr8-MO together with Dgcr8* mRNA at stage 40. The graph represents the summary of at least three independent experiments. The number (N) of embryos analyzed is indicated above the bars. Black, normal expression; gray, partial expression; white, no expression. (G) RT-PCR analysis of multiple miRNAs after Dicer and Dgcr8 knockdown at stage 35. SNO-412 served as loading control.
To address whether specific pronephric malformations could be detected, embryos were examined at stage 40 before the onset of gross morphological abnormalities and the formation of edema. The pronephros was visualized by immunostaining with two antibodies, 3G8 and 4A6, that specifically stain the proximal tubules or the distal tubules and the pronephric duct, respectively (Vize et al., 1995). In agreement with the histology, both Dicer-MO- and Dgcr8-MO-injected embryos displayed a reduced area of 3G8-positive proximal tubules (Fig. 1C-C″). Interestingly, 4A6 staining was detected in the distal tubules, but was absent in the pronephric duct (Fig. 1D-D″). This was not due to a general disappearance of the pronephric duct, as whole-mount in situ hybridization showed expression of the pan-pronephros marker β1-Na/K-ATPase throughout the tubules and duct (Fig. 1E-E″). The absence of 4A6 staining was probably a result of delayed terminal differentiation of the renal epithelial cells in the pronephric duct, as Dgcr8-MO-injected embryos gradually recovered 4A6 staining at later stages (see Fig. S2 in the supplementary material).
The highly similar pronephric defect caused by targeting of two independent proteins involved in miRNA biogenesis (Dicer and Dgcr8) suggested that this phenotype was due to the lack of miRNAs. It was not observed in embryos injected with a standard control morpholino (Std-MO, Fig. 1F and data not shown). Moreover, the efficacy of the Dicer-MO and the Dgcr8-MO was demonstrated using GFP-fusion protein constructs that contained the MO binding sites (see Fig. S1 in the supplementary material). Whereas Xenopus embryos injected animally with 8 ng mRNA encoding Dicer-GFP or Dgcr8-GFP displayed GFP expression at late gastrula stage, this fluorescence was lost upon co-injection with Dicer-MO or Dgcr8-MO, respectively.
To further underscore the specificity of the knockdowns, a rescue experiment was performed using a Dgcr8* mRNA that has six nucleotide mismatches in the recognition site of the Dgcr8-MO and is resistant to its activity. Injection of synthetic Dgcr8* mRNA into all four blastomeres of a 4-cell stage embryo resulted in severe developmental defects and did not allow us to perform rescue experiments (data not shown). Thus, we decided to use a strategy similar to the one previously described by Tran et al. (Tran et al., 2007). Xenopus embryos were injected at the 2-cell stage radially with Dgcr8-MO. Subsequently, a subset of these embryos was injected with Dgcr8* mRNA into a single blastomere at the 4-cell stage. Embryos were grown until stage 40 and processed for whole-mount immunostaining with 4A6. As described above, 4A6 staining was lost in the pronephric duct upon injection of Dgcr8-MO, but was now recovered upon co-injection of Dgcr8* mRNA (Fig. 1F). Unfortunately, we could not perform a similar rescue experiment for the Dicer-MO as we were not successful in isolating a functional allele of Xenopus Dicer.
Another argument for the specificity of the phenotype was the cooperativity of Dicer and Dgcr8. Whereas sub-optimal doses of the two antisense MOs (3.2 pmol Dicer-MO or 0.5 pmol Dgcr8-MO) did not affect 4A6 staining, the combination of both resulted in a more-than-additive reduction in 4A6 staining in the pronephric duct (Fig. 1F). Finally, semi-quantitative RT-PCR at stage 35 showed that the levels of multiple mature miRNAs were reduced, but - as expected from the suboptimal doses of the MOs - not completely eliminated (Fig. 1G). Based on the identical results of the two MOs, we decided to primarily use the Dgcr8-MO for the remainder of the study.
The pronephros is organized along its proximal-distal axis in a manner that is highly similar to the metanephric nephron (Zhou and Vize, 2004; Tran et al., 2007; Raciti et al., 2008). To address whether inhibition of miRNA biogenesis would affect this organization, Dgcr8-MO-injected and uninjected control embryos were cultured until stage 39 and processed for whole-mount in situ hybridization. A panel of segment-specific terminal differentiation genes (Fig. 2H) showed that in the Dgcr8 knockdown, all pronephric segments were present, but the expression domains of individual markers were affected (Fig. 2I). The domains of the proximal tubule markers SGLT1-K and Slc7a13 were reduced (Fig. 2A,A′ and data not shown). NKCC2, a marker for the intermediate tubules and part of the distal tubule, was less convoluted and was extended posteriorly (Fig. 2B,B′). The expression of NBC1 in the distal tubule was shorter and more abutted to the proximal tubule (Fig. 2C,C′). ClC-K, Cadherin-16 and ROMK, three genes expressed in the intermediate and distal tubule as well as in the pronephric duct, showed a dramatic reduction in the anterior part of their expression domains (Fig. 2D-F′). Only NCC, a marker for the distal tubule and the pronephric duct, appeared relatively unchanged (Fig. 2G,G′). Similar results were observed in embryos injected with the Dicer-MO (see Fig. S3 in the supplementary material).
Fig. 2.
Inhibition of miRNA biogenesis affects patterning of the pronephros. (A-G′) Whole-mount in situ hybridization for markers of terminal pronephros differentiation on uninjected and Dgcr8-MO-injected Xenopus embryos at stage 39. (A,A′) SGLT1-K; (B,B′) NKCC2; (C,C′) NBC1; (D,D′) ClC-K; (E,E′) Cadherin-16; (F,F′) ROMK; (G,G′) NCC. Arrowheads indicate a reduction in the intermediate and distal tubular domain. (H,I) Schematics illustrating the different pronephric regions and their corresponding marker gene expression (H) and the phenotype of Dicer-MO- or Dgcr8-MO-injected embryos showing a reduction in the proximal and intermediate tubules as well as in distal tubule DT1 (I).
Together, these data suggested that miRNAs play an important role during pronephros development. Interestingly, they do not seem to regulate the early inductive events, but rather later phases, such as the outgrowth and morphogenesis of the individual tubule segments and the timing of pronephric duct differentiation.
Screening for miRNAs expressed in the kidney
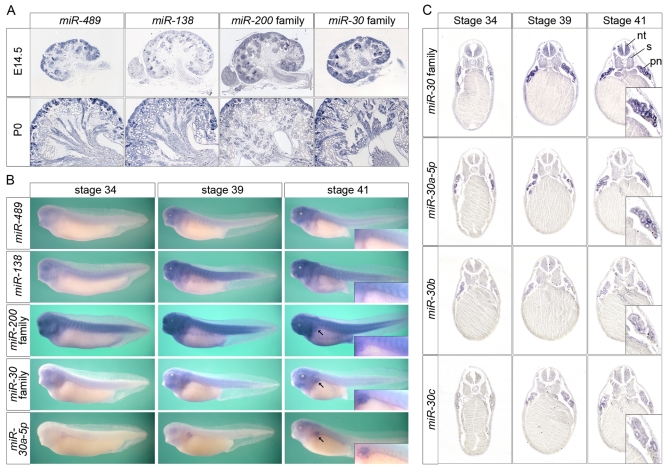
Next, we set out to identify those miRNAs that could exert this regulatory function in pronephros development, pursuing two parallel approaches. First, mouse metanephric kidneys from embryonic day (E) 14.5 and postnatal day (P) 1 were screened using the miRCURY LNA Array miRNA profiling service from Exiqon to identify not only miRNAs present in the kidney, but also those that undergo differential regulation during development (see Fig. S4A in the supplementary material). Second, candidates for kidney-enriched miRNAs were selected from previous publications on human, mouse, rat and zebrafish (Sun et al., 2004; Naraba and Iwai, 2005; Wienholds et al., 2005) and re-analyzed by RT-PCR, comparing four organs of adult mice and different stages of Xenopus development (see Fig. S4B in the supplementary material). From these two data sets, several miRNAs/miRNA families were selected for subsequent expression profiling. In situ hybridization on mouse kidney sections at E14.5 and P0 and Xenopus embryos at stages 34, 39 and 41 were performed using locked nucleic acid (LNA)-modified probes (Fig. 3 and data not shown). To accelerate the screen, miRNA families were initially studied using degenerate LNA oligomers. Individual family members were only analyzed once expression of the family was confirmed in the kidney.
Fig. 3.
Kidney expression of miRNAs. (A) In situ hybridizations of mouse kidney sections at E14.5 and P0 using LNA-modified oligonucleotide probes. miRNA families were studied using degenerate probes that recognize all family members. (B) Whole-mount in situ hybridization of Xenopus embryos at stages 34, 39 and 41. Arrows indicate pronephros. Insets show magnified view of pronephros. (C) Transverse section of Xenopus embryos after whole-mount in situ hybridization with probes recognizing the entire miR-30 family or individual members. Insets show enlargements of the pronephros.
Although many of the candidates were expressed in the kidney, the miR-30 family (miR-30a-5p, miR-30b, miR-30c-1, miR-30c-2, miR-30d and miR-30e) was the most striking. In particular, its expression in Xenopus was rather specific for the pronephros in comparison with other miRNAs, such as the miR-200 family, that were strongly expressed in various tissues besides the pronephros (Fig. 3B). Therefore, individual members of the miR-30 family were analyzed using three LNA-modified probes: miR-30a-5p (which cross-reacts with miR-30d and miR-30e), miR-30b and miR-30c (which recognizes miR-30c-1 and miR-30c-2). All family members showed a very similar expression pattern, with miR-30a-5p appearing slightly earlier than the other two (Fig. 3C and data not shown). They were strongly expressed in the pronephros and weakly in neural tube, somites and head structures (Fig. 3B,C and data not shown). No staining was detected before stage 30 (data not shown) and pronephric expression was first detected around stage 34.
Knockdown of miR-30a-5p activity
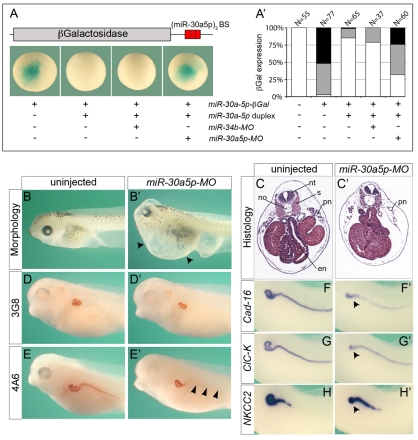
Next, we sought to determine whether the miR-30 family contributed to the kidney phenotype observed in embryos injected with Dicer-MO or Dgcr8-MO. We targeted the miR-30 family by antisense MOs similar to those used in recent reports in zebrafish (Flynt et al., 2007; Eberhart et al., 2008). Since miR-30a-5p was the earliest expressed family member (Fig. 3B,C), we designed a miR-30a-5p-specific antisense MO (miR-30a5p-MO). This MO most likely also targeted miR-30d and miR-30e as they differ by only one nucleotide from miR-30a-5p. A second antisense MO was designed in an identical fashion, but directed against an unrelated miRNA, miR-34b (miR-34b-MO), to serve as an internal control. To test the efficacy of miR-30a5p-MO, the following strategy was used. Xenopus embryos were injected in the animal pole at the 2-cell stage with 160 fmol miR-30a-5p duplex in the presence or absence of either 3.2 pmol miR-30a5p-MO or miR-34b-MO. At the 4-cell stage, these embryos, as well as uninjected control embryos, were injected with 4 ng of a β-galactosidase reporter containing three copies of an optimal miR-30a-5p binding site in its 3′UTR. At gastrula stage, embryos were processed for lacZ staining. As shown in Fig. 4A,A′, the miR-30a-5p duplex downregulated the activity of the β-galactosidase reporter. Co-injection of miR-30a5p-MO reversed this effect and lacZ expression was regained. This was specific, as it was not observed when miR-34b-MO was injected.
Fig. 4.
miR-30a-5p knockdown and global inhibition of miRNA biogenesis have very similar kidney phenotypes. (A) To determine the specificity of miR-30a5p-MO, three consecutive miR-30a-5p binding sites (BS) were introduced in the 3′UTR of a lacZ reporter. Xenopus embryos were injected with the reporter in the presence or absence of miR-30a-5p duplex and miR-30a5p-MO and processed for lacZ staining at gastrula stage. Injections with miR-34b-MO served as a specificity control. (A′) Quantification of the lacZ staining in embryos: white, no staining; gray, partial staining; black, strong staining. The number of embryos analyzed in three different experiments is indicated above the bars. (B-H′) Xenopus embryos injected with miR-30a5p-MO and uninjected controls were analyzed by morphology (B,B′), histology (C,C′), immunohistochemistry with 3G8 and 4A6 (D-E′), and whole-mount in situ hybridization for Cadherin-16 (Cad-16; F,F′), ClC-K (G,G′) and NKCC2 (H,H′). Arrowheads indicate the presence of edema (B′), loss of 4A6 staining in the pronephric duct (E′), and the shortening of the tubule segments IT1, IT2 and DT1 (F′,G′,H′). en, endoderm; no, notochord; nt, neural tube; pn, pronephros; s, somites.
Next, to analyze the role of miR-30a-5p during pronephros development, Xenopus embryos were injected with 3.2 pmol miR-30a5p-MO at the 2- to 4-cell stage and cultured until late tailbud stage. Interestingly, these embryos exhibited pronephric defects very similar to those observed in Dicer-MO- or Dgcr8-MO- injected embryos. miR-30a5p morphants developed severe edema after stage 43 and exhibited reduced pronephric tubules when examined by histology (Fig. 4B-C′). Moreover, immunohistochemistry with 4A6 and 3G8, as well as whole-mount in situ hybridization with the same panel of marker genes as in Fig. 2, revealed reduced proximal tubules, delayed differentiation of the pronephric duct and defects in the organization of the individual tubular segments (Fig. 4D-H′ and data not shown). The similarity of the two phenotypes (Dicer-MO or Dgcr8-MO and miR-30a5p-MO) was striking, but clearly specific. Microinjections targeting another kidney miRNA (miR-34-MO) did not result in any of these pronephric defects (data not shown). Unfortunately, a rescue experiment turned out to be unfeasible because the synthetic miR-30a-5p duplex was insufficiently stable to persist until late stages of development (data not shown).
These data demonstrated that the miR-30 family is important for pronephros development and that its inhibition phenocopies most of the pronephric defects observed upon global inhibition of miRNA biogenesis.
miR-30 regulates proliferation
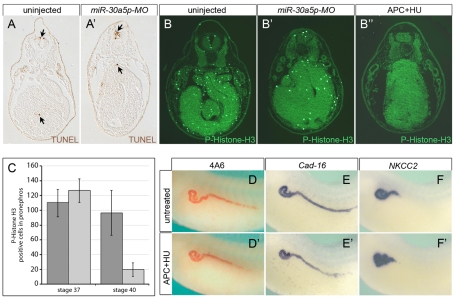
One important function of miRNAs is to regulate the balance between proliferation and apoptosis (Chivukula and Mendell, 2008). Such an imbalance could therefore be responsible for certain aspects of the miR-30a5p-MO phenotype (e.g. the reduced pronephric tubules). However, TUNEL staining did not detect an increase in apoptosis in the pronephros at stage 40 (Fig. 5A,A′). By contrast, mitotic cells positive for phospho-Histone H3 were dramatically decreased (Fig. 5B,B′,C). Inhibition of proliferation was observed only at stage 40, and not at stage 37 (Fig. 5C) when changes in gene expression were already detectable (data not shown). To address the contribution of decreased proliferation to the phenotype, uninjected Xenopus embryos were treated with aphidicolin and hydroxyurea from stage 33/34 until stage 40 to abolish all cell divisions (Harris and Hartenstein, 1991). Even though these embryos were devoid of mitotic cells (Fig. 5B″), they did not exhibit any differences in the pattern of 4A6 staining or in Cadherin-16 and NKCC2 expression (Fig. 5D-F′). This suggested that although proliferation is clearly affected by knockdown of miR-30 activity, it was not responsible for the phenotypic changes seen with miR-30a5p-MO.
Fig. 5.
Apoptosis and proliferation do not contribute to the kidney phenotype. (A,A′) TUNEL staining of transverse sections from uninjected and miR-30a5p-MO-injected Xenopus embryos at stage 39. TUNEL-positive cells appear brown and are indicated by arrows. Note that no significant differences were observed by analyzing many serial sections of the pronephros. Several embryos and multiple independent experiments were analyzed. (B-B″) Immunofluorescence analysis of proliferation with an anti-phospho-Histone H3 antibody comparing transverse sections of uninjected control embryos, embryos injected with miR-30a5p-MO and embryos treated with aphidicolin and hydroxyurea (APC+HU) at stage 40. (C) Quantification of phospho-Histone H3-positive cells in the pronephros of uninjected (dark gray) and miR-30a5p-MO-injected (light gray) embryos at stages 37 and 40. Consecutive sections covering the entire pronephric tubular area were counted. Numbers are the average of at least four different embryos from two independent experiments. Error bars represent s.d. (D-F′) Immunostaining and in situ hybridization of untreated and APC+HU-treated embryos with 4A6 (D,D′), Cadherin-16 (Cad-16; E,E′) and NKCC2 (F,F′). Note the absence of any patterning differences, even though proliferation was completely inhibited.
miR-30 regulates Xlim1/Lhx1 activity
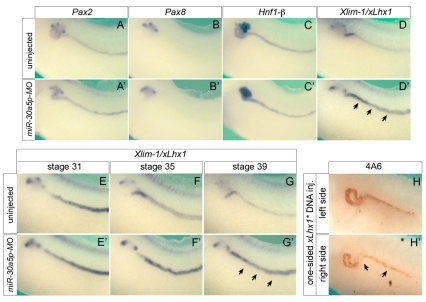
All the genes analyzed so far are markers for terminal differentiation in the pronephros. Next, we tested whether miR-30a5p-MO affected upstream transcription factors. Xenopus embryos injected with miR-30a5p-MO and uninjected controls were processed for in situ hybridization with Pax2, Pax8, Hnf1-β and Xlim1 at stage 39. As shown in Fig. 6A-C′, the expression levels of Pax2, Pax8 and Hnf1-β mRNA were unchanged, even though their domains reflected the altered pronephros architecture, as seen with terminal differentiation markers such as β1-Na/K-ATPase (Fig. 1E-E″). By contrast, Xlim1 mRNA was elevated in the pronephric duct (Fig. 6D,D′). Loss of miRNA activity not only results in increased translation, but can also increase mRNA stability (Giraldez et al., 2006), suggesting that Xlim1 could be a direct target of miR-30 activity. In Xenopus, Xlim1 is expressed in the pronephric anlage as early as stage 13 (Taira et al., 1994; Carroll and Vize, 1999), whereas the miR-30 family is first detected at stage 34 (Fig. 3B,C). If miR-30 directly downregulates Xlim1, the loss of miR-30 should stabilize Xlim1 mRNA levels as soon as miR-30 normally becomes expressed in the pronephros. Indeed, comparison of miR-30a5p-MO-injected embryos with uninjected sibling controls at stages 31, 35 and 39 showed that the expression of Xlim1 was nearly indistinguishable at stage 31, but was maintained at higher levels in embryos with reduced miR-30 activity as early as stage 35 (Fig. 6E-G′).
Fig. 6.
Xlim1 is regulated by miR-30a-5p. (A-D′) Whole-mount in situ hybridization for Pax2 (A,A′), Pax8 (B,B′), Hnf1-β (C,C′) and Xlim1 (D,D′), comparing uninjected and miR-30a5p-MO-injected Xenopus embryos at stage 39. (E-G′) Whole-mount in situ hybridization for Xlim1 mRNA of uninjected and miR-30a5p-MO-injected Xenopus embryos at stages 31, 35 and 39. Note that Xlim1 expression is maintained at a higher level in miR-30a5p-MO-injected embryos after stage 35, when endogenous expression of miR-30a-5p can first be detected (arrows in D′,G′). (H,H′) Immunohistochemistry with 4A6 of stage 40 embryos injected with 4 pg pCS2-Lhx1* DNA into one blastomere at the 4-cell stage, comparing the left with the right side. Arrows indicate changes in 4A6 staining. Ectopic expression of Lhx1 mRNA transcribed from the pCS2-Lhx1* was confirmed by in situ hybridization (data not shown).
Increased Xlim1 expression could be part of the miR-30a5p-MO phenotype. To test this, we elevated Xlim1 levels without changing miR-30 activity by a one-sided injection with 4 pg pCS2-Lhx1* DNA. This construct contains an optimized Kozak sequence and lacks the entire 3′UTR. Xenopus embryos were injected at the 2-cell stage, cultured until stage 40 and processed for 4A6 immunostaining. Fifty-five percent of the embryos (n=98) displayed various degrees of change in 4A6 staining upon comparison of the uninjected and DNA-injected sides (Fig. 6H,H′ and see Fig. S5 in the supplementary material). This result agreed with our hypothesis that increased levels of Xlim1 interfere with terminal differentiation of the pronephros.
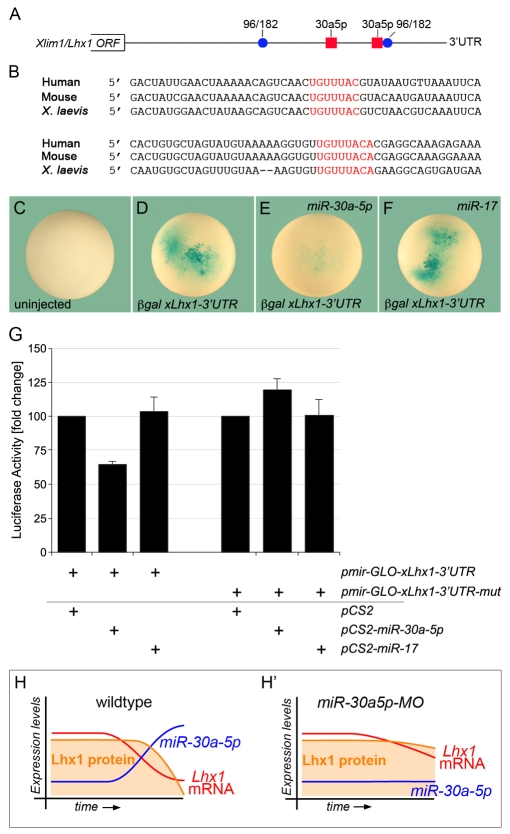
To test whether Lhx1 mRNA is a direct target of the miR-30 family, we first performed an in silico analysis to detect putative miRNA binding sites. Unfortunately, the 3′UTR of Lhx1 had not been described. So we initially used a prediction algorithm based on genomic sequences (DIANA microT version 3.0). This approach identified potential miRNA binding sites in mouse Lhx1 including two strong miR-30 binding sites (Fig. 7A). Moreover, these sites were present in authentic Lhx1 transcripts, as they were found in ESTs of human, mouse and Xenopus, as well as in the previously uncharacterized 3′UTR of a full-length Xenopus Xlim1 cDNA clone (Taira et al., 1992) (GenBank accession number GQ485551). As shown in Fig. 7B, the two miR-30 binding sites were completely conserved.
Fig. 7.
Xlim1/Lhx1 is a direct target of miR-30a-5p-mediated repression. (A) In silico analysis of putative miRNA binding sites in the 3′UTR of mouse Lhx1 using DIANA microT (version 3.0 using strict threshold conditions). Blue circles represent predicted miRNA binding sites and the red boxes indicate two strong miR-30 binding sites in the Lhx1 3′UTR. (B) Sequence alignment showing the conservation of the miR-30 binding sites in human, mouse and Xenopus Xlim1/Lhx1. The miR-30 seed sequences are indicated in red. (C-F) β-galactosidase reporter assay of Xenopus embryos injected with pXEXβGal-Lhx1-3′UTR mRNA in the presence or absence of a miR-30a-5p or a miR-17 duplex at gastrula stage. (G) Luciferase reporter assay of HEK 293T cells transfected with pmir-GLO-xLhx1-3′UTR and pmir-GLO-xLhx1-3′UTR-mut in the presence of pCS2, pCS2-miR-30a-5p or pCS2-miR-17. Values were corrected for the expression of Renilla luciferase and calculated as fold change compared with the pCS2 control. Multiple independent experiments were averaged and s.d. is indicated. (H,H′) Model for the regulation by miR-30 of Lhx1 expression. During kidney development, Lhx1 mRNA and protein levels gradually decline, whereas miR-30a-5p levels increase (H). Upon injection of the miR-30a5p-MO, Lhx1 levels persist, impairing terminal differentiation (H′).
To address the functionality of these two sites, the 3′UTR of Xenopus Xlim1 was subcloned downstream of the lacZ reporter. Synthetic mRNA was injected into the animal region of Xenopus embryos in the absence or presence of a synthetic miR-30a-5p or miR-17 duplex and processed for lacZ staining at gastrula stage. Injection of the reporter construct alone showed strong lacZ staining, but was significantly reduced upon co-injection of the miR-30a-5p duplex (Fig. 7C-F). This effect was specific, as injection of a miR-17 duplex that does not have a predicted binding site within the 3′UTR did not alter the lacZ staining.
Next, in order to quantify the miR-30a-5p effects, HEK 293T cells were transfected with a dual luciferase reporter construct harboring the 3′UTR of Xlim1 (pmir-GLO-xLhx1-3′UTR) in the presence or absence of expression constructs expressing miR-30a-5p or miR-17 under the control of the CMV promoter (pCS2-miR-30a-5p, pCS2-miR-17). As shown in Fig. 7G, miR-30a-5p reduced luciferase expression by 35%. miR-17 did not have any effect. Moreover, when both miR-30 binding sites in the 3′UTR of Xlim1 were mutated (pmir-GLO-xLhx1-3′UTR-mut), the repression by miR-30a-5p was lost. Thus, repression of Xlim1 by miR-30 was mediated by the two miR-30 binding sites identified in its 3′UTR.
Together, these data make a compelling argument that gene expression in the kidney must be tightly controlled to allow proper pronephric kidney development and that the miR-30 miRNA family plays an important role in this regulation.
DISCUSSION
miRNAs are involved in a variety of biological processes and the number of isolated or predicted miRNAs continues to increase. However, the in vivo significance of only a few miRNAs has been elucidated so far (Stefani and Slack, 2008). We now reveal the role of the miR-30 family during pronephric kidney development in Xenopus. Knockdown of miR-30a-5p by antisense MOs resulted in a complex phenotype characterized by a delay in differentiation, shortening of individual nephron segments and reduced proliferation. Intriguingly, these changes were very similar to the kidney phenotype observed when global miRNA maturation was inhibited using Dicer-MO or Dgcr8-MO. This argues that the miR-30 family has a central role in nephrogenesis. Such an interpretation finds support in three recent studies of miRNAs in podocytes that show that Dicer is required for miR-30 expression (Harvey et al., 2008; Ho et al., 2008; Shi et al., 2008). Nevertheless, the central role of miR-30 is also puzzling. Our expression profiling identified several other miRNAs present in the kidney of frog and mouse, including the miR-200 family, miR-489 and miR-138. It is likely that they too will have important functions in the kidney, but they might promote different aspects of kidney development that were not addressed with the panel of markers used in this study. Indeed, a parallel study in our laboratory has identified the miR-17 family as a crucial regulator of polycystic kidney disease genes in Xenopus and mouse (our unpublished results).
A second interesting aspect of this study was that global inhibition of miRNA biogenesis did not interfere with the induction of the pronephros. The initial expression of transcription factors patterning the intermediate mesoderm (such as Pax2, Pax8, Xlim1 and Hnf1- β) was unaffected (Fig. 6 and data not shown). Defects only became apparent when these transcription factors are downregulated to allow terminal differentiation of the pronephros (Carroll et al., 1999). At this junction, miRNAs might provide some plasticity. Indeed, several miRNAs, such as the miR-200 family, are crucial during the epithelial-mesenchymal transition, when a differentiated epithelial cell is converted into a mesenchymal cell type (Cano and Nieto, 2008). Interestingly, miR-200 was also identified during our expression profiling as a kidney-enriched miRNA and it will be interesting to address their function on E-Box genes, such as Zeb1 and Zeb2, during kidney development.
One of the most challenging aspects in studying miRNAs is the prediction and verification of genes regulated by a given miRNA (Bartel, 2009). In this study, we identified the LIM-class homeobox gene Xlim1/Lhx1 as a target of miR-30 activity. This argument is based on four observations. (1) The 3′UTR of human, mouse and Xenopus Lhx1 contains two highly conserved miR-30 binding sites. (2) miRNA reporter assays showed that this 3′UTR was repressed by miR-30a-5p and this depended on the presence of these two binding sites. (3) Downregulation of Xlim1 mRNA in the Xenopus pronephros coincided with the appearance of miR-30 expression. (4) Overexpression of Xlim1 mimicked the loss of 4A6 staining observed in the miR-30 knockdown. Based on these data, we hypothesize that Xlim1 mRNA is an important target of miR-30 activity and sharpens the time window of Xlim1 expression in the kidney (Fig. 7H,H′). In the presence of miR-30a-5p, the expression of Xlim1 starts to decline at about stage 35 (Fig. 7H). However, in the absence of miR-30a-5p activity, this window of Xlim1 expression is extended, contributing to the delayed terminal differentiation of the renal epithelial cells (Fig. 7H′).
The observation that Xlim1/Lhx1 is under miRNA control is even more important because it is a central player in kidney development and exhibits a dynamic expression pattern that needs to be tightly controlled (Dressler, 2006). In Xenopus, Xlim1 mRNA is initially found throughout the pronephric tubules and duct. As tubules begin to lumenize, expression becomes restricted to the nephrostomes and the late distal tubule (Carroll et al., 1999; Tran et al., 2007). Similarly, in mouse, Lhx1 is expressed in the nephric duct, the tips of the ureteric buds, the pretubular aggregates, the comma- and S-shaped bodies and even the podocytes of the immature glomerulus (Barnes et al., 1994; Fujii et al., 1994; Kobayashi et al., 2005). Moreover, all these individual domains are important for proper kidney development and tubular morphogenesis (Kobayashi et al., 2005; Pedersen et al., 2005). An miRNA-based regulation provides the possibility to fine-tune Xlim1/Lhx1 expression independently of transcriptional control. Based on the data presented here, the miR-30 family constitutes such a regulator. It controls terminal differentiation of the Xenopus pronephros by modulating Xlim1. Because miR-30 expression appears at around stage 34, it cannot control all phases of pronephric expression of Xlim1. The early regulation might be accomplished by other miRNAs. Our analysis of the Lhx1 3′UTR identified two additional miRNA binding sites for miR-96/182 (Fig. 7A), an miRNA family that, based on microarray data (data not shown), is expressed during early mouse kidney development.
Obviously, Xlim1/Lhx1 will not be the only target of miR-30 activity. In silico analyses suggest many other potential targets. The osmoregulatory transcription factor NFAT5 (TonEBP) (Ho, 2006) has three conserved miR-30 binding sites. Similarly, the mRNAs of a large number of solute carrier (SLC) proteins (He et al., 2009) have miR-30 binding sites in their 3′UTR. These proteins are involved in renal homeostasis. In the future, it will be interesting to see whether loss of miR-30 activity in the adult has consequences for kidney function. This might bring forth examples of dual-function miRNAs: although they direct patterning during early development, they are later required to maintain organ function.
Supplementary Material
Acknowledgements
We thank Drs S. El-Dahr, J. Larraín, T. Obara, M. Oelgeschläger and E. Pera, as well as all members of the laboratory, for critically reviewing the manuscript and for helpful discussions; Dr E. Jones for the 4A6 and 3G8 antibodies; and Drs I. Dawid, R. Harland, R. Vignali, P. Vize and the NIBB/NIG/NBRP Xenopus laevis EST project for plasmids. This work was supported by a grant from NIH/NIDDK (#5R21DK077763-03) to O.W. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/23/3927/DC1
References
- Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P., Horvitz H. R., Ambros V. (2005). The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, 403-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J. D., Crosby J. L., Jones C. M., Wright C. V., Hogan B. L. (1994). Embryonic expression of Lim-1, the mouse homolog of Xenopus Xlim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev. Biol. 161, 168-178 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belo J. A., Bouwmeester T., Leyns L., Kertesz N., Gallo M., Follettie M., De Robertis E. M. (1997). Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech. Dev. 68, 45-57 [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim S. Y., Carmell M. A., Murchison E. P., Alcorn H., Li M. Z., Mills A. A., Elledge S. J., Anderson K. V., Hannon G. J. (2003). Dicer is essential for mouse development. Nat. Genet. 35, 215-217 [DOI] [PubMed] [Google Scholar]
- Bouchard M., Souabni A., Mandler M., Neubuser A., Busslinger M. (2002). Nephric lineage specification by Pax2 and Pax8. Genes Dev. 16, 2958-2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B., Cohen S. M. (2005). Principles of microRNA-target recognition. PLoS Biol. 3, e85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S. M. (2007). microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175-205 [DOI] [PubMed] [Google Scholar]
- Cano A., Nieto M. A. (2008). Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition. Trends Cell Biol. 18, 357-359 [DOI] [PubMed] [Google Scholar]
- Carroll T. J., Vize P. D. (1999). Synergism between Pax-8 and lim-1 in embryonic kidney development. Dev. Biol. 214, 46-59 [DOI] [PubMed] [Google Scholar]
- Carroll T. J., McMahon A. P. (2003). Overview: the molecular basis of kidney development. In The Kidney: From Normal Development to Congenital Disease (ed. Vize P. D., Woolf A. S., Bard J. B. L.), pp. 343-376 Amsterdam: Academic Press; [Google Scholar]
- Carroll T. J., Wallingford J. B., Vize P. D. (1999). Dynamic patterns of gene expression in the developing pronephros of Xenopus laevis. Dev. Genet. 24, 199-207 [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula R. R., Mendell J. T. (2008). Circular reasoning: microRNAs and cell-cycle control. Trends Biochem. Sci. 33, 474-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G. R. (2006). The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22, 509-529 [DOI] [PubMed] [Google Scholar]
- Drummond I. A. (2005). Kidney development and disease in the zebrafish. J. Am. Soc. Nephrol. 16, 299-304 [DOI] [PubMed] [Google Scholar]
- Eberhart J. K., He X., Swartz M. E., Yan Y. L., Song H., Boling T. C., Kunerth A. K., Walker M. B., Kimmel C. B., Postlethwait J. H. (2008). MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat. Genet. 40, 290-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K. K., Grimson A., Jan C., Lewis B. P., Johnston W. K., Lim L. P., Burge C. B., Bartel D. P. (2005). The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310, 1817-1821 [DOI] [PubMed] [Google Scholar]
- Flynt A. S., Li N., Thatcher E. J., Solnica-Krezel L., Patton J. G. (2007). Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 39, 259-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Pichel J. G., Taira M., Toyama R., Dawid I. B., Westphal H. (1994). Expression patterns of the murine LIM class homeobox gene lim1 in the developing brain and excretory system. Dev. Dyn. 199, 73-83 [DOI] [PubMed] [Google Scholar]
- Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., Hammond S. M., Bartel D. P., Schier A. F. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833-838 [DOI] [PubMed] [Google Scholar]
- Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., Enright A. J., Schier A. F. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75-79 [DOI] [PubMed] [Google Scholar]
- Harris W. A., Hartenstein V. (1991). Neuronal determination without cell division in Xenopus embryos. Neuron 6, 499-515 [DOI] [PubMed] [Google Scholar]
- Harvey S. J., Jarad G., Cunningham J., Goldberg S., Schermer B., Harfe B. D., McManus M. T., Benzing T., Miner J. H. (2008). Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J. Am. Soc. Nephrol. 19, 2150-2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Vasiliou K., Nebert D. W. (2009). Analysis and update of the human solute carrier (SLC) gene superfamily. Hum. Genomics 3, 195-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J., Ng K. H., Rosen S., Dostal A., Gregory R. I., Kreidberg J. A. (2008). Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J. Am. Soc. Nephrol. 19, 2069-2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N. (2006). Intracellular water homeostasis and the mammalian cellular osmotic stress response. J. Cell. Physiol. 206, 9-15 [DOI] [PubMed] [Google Scholar]
- Howland R. B. (1916). On the effect of removal of the pronephros of the amphibian embryo. Proc. Natl. Acad. Sci. USA 2, 231-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. A. (2003). Molecular control of pronephric development: an overview. In The Kidney: From Normal Development to Congenital Disease (ed. Vize P. D., Woolf A. S., Bard J. B. L.), pp. 93-117 Amsterdam: Academic Press; [Google Scholar]
- Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J. J., Natarajan R. (2007). MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 104, 3432-3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W. P., Plasterk R. H. (2006). The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441-450 [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Kwan K. M., Carroll T. J., McMahon A. P., Mendelsohn C. L., Behringer R. R. (2005). Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development 132, 2809-2823 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854 [DOI] [PubMed] [Google Scholar]
- Naraba H., Iwai N. (2005). Assessment of the microRNA system in salt-sensitive hypertension. Hypertens. Res. 28, 819-826 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. (1994). Normal Table of Xenopus laevis New York: Garland Publishing; [Google Scholar]
- Pedersen A., Skjong C., Shawlot W. (2005). Lim 1 is required for nephric duct extension and ureteric bud morphogenesis. Dev. Biol. 288, 571-581 [DOI] [PubMed] [Google Scholar]
- Raciti D., Reggiani L., Geffers L., Jiang Q., Bacchion F., Subrizi A. E., Clements D., Tindal C., Davidson D. R., Kaissling B., et al. (2008). Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 9, R84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R., Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901-906 [DOI] [PubMed] [Google Scholar]
- Saxén L. (1987). Organogenesis of the Kidney Cambridge, UK: Cambridge University Press; [Google Scholar]
- Shawlot W., Behringer R. R. (1995). Requirement for Lim1 in head-organizer function. Nature 374, 425-430 [DOI] [PubMed] [Google Scholar]
- Shi S., Yu L., Chiu C., Sun Y., Chen J., Khitrov G., Merkenschlager M., Holzman L. B., Zhang W., Mundel P., et al. (2008). Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J. Am. Soc. Nephrol. 19, 2159-2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M., Harland R. M. (2000). Early Development of Xenopus laevis: A Laboratory Manual Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R. B., Cohen S. M. (2005). Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123, 1133-1146 [DOI] [PubMed] [Google Scholar]
- Stefani G., Slack F. J. (2008). Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219-230 [DOI] [PubMed] [Google Scholar]
- Sun Y., Koo S., White N., Peralta E., Esau C., Dean N. M., Perera R. J. (2004). Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 32, e188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira M., Jamrich M., Good P. J., Dawid I. B. (1992). The LIM domain-containing homeo box gene Xlim-1 is expressed specifically in the organizer region of Xenopus gastrula embryos. Genes Dev. 6, 356-366 [DOI] [PubMed] [Google Scholar]
- Taira M., Otani H., Jamrich M., Dawid I. B. (1994). Expression of the LIM class homeobox gene Xlim-1 in pronephros and CNS cell lineages of Xenopus embryos is affected by retinoic acid and exogastrulation. Development 120, 1525-1536 [DOI] [PubMed] [Google Scholar]
- Tran U., Pickney L. M., Ozpolat B. D., Wessely O. (2007). Xenopus Bicaudal-C is required for the differentiation of the amphibian pronephros. Dev. Biol. 307, 152-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali R., Poggi L., Madeddu F., Barsacchi G. (2000). HNF1(beta) is required for mesoderm induction in the Xenopus embryo. Development 127, 1455-1465 [DOI] [PubMed] [Google Scholar]
- Vize P. D. (2003). The chloride conductance channel ClC-K is a specific marker for the Xenopus pronephric distal tubule and duct. Gene Expr. Patterns 3, 347-350 [DOI] [PubMed] [Google Scholar]
- Vize P. D., Jones E. A., Pfister R. (1995). Development of the Xenopus pronephric system. Dev. Biol. 171, 531-540 [DOI] [PubMed] [Google Scholar]
- Vize P. D., Carroll T. J., Wallingford J. B. (2003). Induction, development and physiology of the pronephric tubules. In The Kidney: From Normal Development to Congenital Disease (ed. Vize P. D., Woolf A. S., Bard J. B. L.), pp. 19-50 Amsterdam: Academic Press; [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Koudijs M. J., van Eeden F. J., Cuppen E., Plasterk R. H. (2003). The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35, 217-218 [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H. R., Kauppinen S., Plasterk R. H. (2005). MicroRNA expression in zebrafish embryonic development. Science 309, 310-311 [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855-862 [DOI] [PubMed] [Google Scholar]
- Zhou X., Vize P. D. (2004). Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric kidney tubules. Dev. Biol. 271, 322-338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.