Abstract
arl13b was initially cloned as the novel cystic kidney gene scorpion (sco) in zebrafish and was shown to be required for cilia formation in the kidney duct. In mouse, a null mutant of Arl13b shows abnormal ultrastructure of the cilium and defective sonic hedgehog (Shh) signaling. Importantly, a recent study linked mutations in ARL13B to a classical form of Joubert syndrome (JS), an autosomal recessive disorder characterized by a distinctive cerebellar malformation. In this study, we analyzed the zebrafish arl13b (sco) mutant and gene products in detail. We first demonstrate that Arl13b is a protein that is highly enriched in the cilium and is required for cilia formation in multiple organs in zebrafish, and that knockdown of arl13b leads to multiple cilia-associated phenotypes. We additionally show that multiple regions of Arl13b are required for its localization to the cilium. By means of rescuing experiments with a series of deletion and point mutants, we further demonstrate that the ciliary localization is crucial for the in vivo function of Arl13b. Together, these results strongly support the hypothesis that JS-related disease (JSRD) is a ciliopathy, or a disease caused by ciliary defects, and that Arl13b functions mainly through the cilium.
Keywords: Arl13b, Scorpion, Cilium, Joubert syndrome, Kidney cyst, Laterality
INTRODUCTION
Joubert syndrome (JS) is an autosomal recessive disorder characterized by a malformation of the cerebellum, hypotonia, oculomotor apraxia, ataxia, abnormal breathing and mental retardation (for reviews, see Harris, 2007; Sharma et al., 2008). In addition to these classic manifestations, some individuals with JS have also presented abnormalities closely associated with ciliary defects, such as retinal dystrophy, polydactyly and familial juvenile nephronophthisis. Interestingly, a recent study found an association between mutations in the ARL13B gene and a classical form of JS (Cantagrel et al., 2008). ARL13B is a member of the ADP-ribosylation factor-like (ARL) family of small GTPases of the RAS superfamily. Consistent with a ciliary involvement in JS, mutations in Arl13b have been linked to cilia assembly and kidney cyst formation in model organisms. arl13b was cloned as the novel cystic kidney gene scorpion (sco) in zebrafish and was shown to be required for cilia formation in the kidney duct (Sun et al., 2004). In mouse, hennin (hnn) was identified as a null allele of Arl13b (Caspary et al., 2007). It was reported that hnn mutants have shorter cilia with a disrupted axonemal structure, and display phenotypes associated with defective sonic hedgehog (Shh) signaling.
On the surface, the association between ARL13B and JS is consistent with the hypothesis that Joubert syndrome and related disorders (JSRD) are ciliopathy disorders, or diseases caused by ciliary defects (Bielas et al., 2009; Cantagrel et al., 2008; Castori et al., 2005; Dixon-Salazar et al., 2004; Gleeson et al., 2004; Gorden et al., 2008; Harris, 2007; Helou et al., 2007; Jacoby et al., 2009; Sharma et al., 2008; Utsch et al., 2006; Wolf et al., 2007). However, phenotypical analysis of patients with ARL13B mutations and sequence analysis of the ARL13B gene in patients with non-classical forms of JS seem to suggest that mutations in ARL13B are restricted to the classical form of JS, with symptoms predominantly limited to brain structure and function (Cantagrel et al., 2008). In contrast to some well-established cilia-associated symptoms seen in other forms of JSRD, the symptoms of the classic form of JS have less clear functional connections to the cilium. At least two alternative models can be used to explain this observed specificity of involvement of ARL13B in JS. In the first model, the function of ARL13B is restricted to specific organs, such as the cerebellum, and hence mutations in ARL13B lead to phenotypes restricted to the brain. Alternatively, the identified alleles are partial loss-of-function alleles. Differential sensitivity of different organs to the reduction of ARL13B activity thus leads to restricted phenotypes. Distinguishing between these two possibilities will be informative for mutation screenings in JSRD patients.
To lay a foundation for a comprehensive understanding of Arl13b function, we analyzed the zebrafish arl13b (sco) mutant and gene products in detail in this study. We first show that Arl13b is a ciliary protein and is required for cilia formation in multiple organs, and that arl13b mutants display multiple cilia-associated phenotypes, including cystic kidney and body curvature. We also provide evidence that the knockdown of arl13b by a morpholino oligonucleotide additionally leads to left-right asymmetry defects and to phenotypes consistent with defective convergence extension (CE) movement during gastrulation.
Since Arl13b is highly enriched in the cilium, we searched for domains that are involved in trafficking it to the cilium. Our results suggest that both the small GTPase domain and the coiled-coil domain following the small GTPase domain are required for targeting Arl13b to the cilium.
If the cilium is the main site of function for Arl13b, the disruption of cilia targeting of Arl13b should disrupt most of its function in vivo. Through rescuing experiments with a series of deletion mutants, we show that disruption of the ciliary localization of Arl13b correlates with impairment of its rescuing capability for arl13b/scohi459 mutants. Together, these results strongly support the model that Arl13b mainly functions through the cilium and that JSRD is a ciliopathy. Our results also suggest that the involvement of Arl13b in cilia formation is systemic and that mutations in ARL13B might lead to typical ciliopathy with symptoms that are broader than the classical manifestations of JS. Our study also provides further support for the idea that rescuing experiments in zebrafish can be used both qualitatively and quantitatively in evaluating the functions of different alleles of a tested gene.
MATERIALS AND METHODS
Fish husbandry
Standard protocols were used for maintaining zebrafish colonies (Westerfield, 2000). Embryos were obtained through natural spawning. All lines were in a TAB background.
Histological analysis
Embryos were fixed in Bouin's fixative overnight at room temperature, washed three times with PBT (phosphate-buffered saline, pH 7.4, and 0.1% Tween-20), embedded in JB-4 resin from Polysciences and cut at 4 μm with a microtome. Slides were then stained with Hematoxylin and Eosin.
Cryosectioning of zebrafish embryos
Embryos were fixed in diluted formalin solution (1:4 in PBT) overnight at 4°C, washed in PBT, and then infiltrated with 10% and, subsequently, 25% sucrose solutions. Embryos were placed in Tissue-Tek OCT compound and solidified in a mold in isopentane placed in a liquid nitrogen bath. Sections (12 μm thick) were obtained using a cryostat before proceeding with immunohistochemistry.
Generation of arl13b constructs
The arl13b (sco) coding sequence was amplified via PCR from a zebrafish cDNA pool and was cloned into pCS2+. pCS2 scorpion tagged with egfp was generated via PCR cloning. gtpase::gfp, G28V::gfp, T35N::gfp and Δ(308-407)::gfp were generated using PCR-mediated cloning. sco::gfp, ΔCC::gfp, Δ(21-127)::gfp, Δ(E-rich)::gfp and Δ(394-407)::gfp were generated using the Gateway cloning vector kit (a kind gift from N. Lawson, University of Massachusetts Medical School, and C.-B. Chien, University of Utah) following a previously described strategy (Kwan et al., 2007).
In situ hybridization
Whole mount in situ hybridization was performed as described previously (Hauptmann and Gerster, 2000). Briefly, embryos were fixed in diluted formalin (1:2.7 in PBT) at room temperature for an hour or at 4°C overnight. Digoxigenin-UTP-labeled RNAs synthesized in vitro were used as probes. Alkaline phosphatase-coupled anti-digoxigenin (Roche) was used to localize hybridized probes. NBT/BCIP (Roche) was used as the chromogenic substrate to produce blue precipitates.
Assay for rescue activity
mRNA was synthesized in vitro using the mMESSAGE mMACHINE kit (Ambion) following the manufacturer's instructions. If not stated otherwise, 265.5 pg mRNA for each construct was injected into zebrafish embryos. Embryos were scored for body curvature at 2 dpf (days post-fertilization), and for pronephric cysts at 4-5 dpf. For cilia rescue measurements, cilia were detected by immunostaining with anti-acetylated tubulin (clone 6-11B-1, Sigma-Aldrich), followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Pierce). Signal was detected via a color reaction using DAB (3,3′-diaminobenzidine, Sigma-Aldrich) as the chromogenic substrate.
Antibodies
A carboxy-terminal region of Sco (from amino acid 238) was produced as GST fusion protein in E. coli and purified following the manufacturer's protocol with the pGEX system (GE Healthcare LifeSciences). Purified antigen was injected into NZW rabbits by Covance. IgG was purified from serum with CM Affi-Gel blue gel following the instruction manual (Bio-Rad Laboratories). A region corresponding to amino acids 639-819 of Cdh17 was used to generate a polyclonal antibody following the same strategy as described above.
Morphometric measurements
A previously published protocol for measuring body gap angle was followed with minor modifications (Gerdes et al., 2007). Briefly, embryos at the 8-14 somite stage were selected randomly and side images were taken with a stereoscope. The centroid of the yolk was labeled using ImageJ. The angle of the lines connecting the centroid with the tip of the head and tip of the tail was measured using ImageJ.
Extension movement was followed using a previously published protocol with minor modifications (Sepich et al., 2000). Specifically, 0.5 nl of the lipophilic dye FM1-43 at 500 μg/ml was injected into the shield of embryos at the shield stage. Embryos with correctly labeled regions were selected, raised to the tailbud stage and imaged in side views. The anterior and the posterior borders of fluorescently labeled cells were then inspected and compared with the positions of the head and the tail.
Immunohistochemistry
A previously published protocol was followed (DiBella et al., 2009). Anti-acetylated tubulin (clone 6-11B-1, Sigma-Aldrich) was used at a dilution of 1:1000 to 1:10,000. Anti-γ-tubulin (T5326, Sigma-Aldrich) was used at 1:200. Anti-atypical PKC (Santa Cruz Biotechnology) was used at 1:50 to 1:200. Anti-Na+/K+ ATPase (α6F, DSHB) was used at 1:5. DAB reaction was used for detection of the HRP signal. For fluorescent signal, embryos were flat-mounted and visualized using a Zeiss Axioplan 2 microscope, a Zeiss Axiovert 200 equipped with a CARV II confocal imaging system, or a Zeiss LSM 510 Meta inverted laser scanning microscope. Images were cropped in Photoshop to create figures.
Protein extraction, immunoprecipitation and western analysis
Embryos were de-yolked and homogenized following a previously described protocol (Link et al., 2006) in TG buffer (PBS with 0.1% Triton X-100, 1% glycerol) with protease inhibitors (Roche). Lysates were cleared by microcentrifugation at top speed for 10 minutes and subsequently incubated with antibodies followed by protein A beads. Samples were eluted by boiling in sample buffer (2% SDS, 100 mM DTT and 2.5% β-mercaptoethanol), electrophoresed on a SDS-PAGE gel and transferred to nitrocellulose or PVDF membrane. Blots were incubated with primary antibody and HRP-conjugated secondary antibody, and signal was developed using the Western Lightning Detection Kit (PerkinElmer Life Sciences).
Electron microscopy
Zebrafish embryos were fixed in Karnovsky's Fixative at room temperature for one hour, washed in cacodylate buffer, post-fixed with Palade's osmium at room temperature for one hour, washed with cacodylate buffer, stained with Kellenberger, and taken through an ethanol dehydration series. Embryos were then infiltrated through a propylene oxide/Epon series, embedded in Epon and sectioned. Sections were post-stained with uranyl acetate and lead citrate, and viewed in a Tecnai 12 BioTwin electron microscope.
Morpholino injection
Morpholino oligonucleotides were purchased from Gene Tools and injected into zebrafish embryos at the one- to four-cell stage. The morpholino 5′-TTTCCCCCCTAAATGCTTTCACTGG-3′ was used to block the translation of arl13b.
High-speed videomicroscopy
A previously published protocol was followed (Kramer-Zucker et al., 2005). Embryos were incubated in 75 μM phenylthiourea (PTU) in embryo medium. On day 5, embryos were mounted in 6% methylcellulose containing 50 mM 2,3-butanedione monoxime (BDM) and 0.02% Tricaine. Images were taken using a 20×/0.4 DIC lens on a Nikon Eclipse TE2000-U microscope equipped with a Nikon Photometrics Coolsnap HQ camera under stream acquisition in Metamorph at 200 frames per second and analyzed in ImageJ.
Statistical analysis
Data from replicate experiments of body curvature and cyst rescue were examined for statistical heterogeneity using the replicated goodness-of-fit test (G-statistic). Data with heterogeneity G-values corresponding to P-values above 0.05 were considered to be homogeneous and were pooled, and a chi-squared test was performed on the pooled value with P-value shown in the figure; otherwise, the data replicate with the least significant P-value from a chi-squared test is shown. For cilia rescue, the data replicate with the least significant P-value from a chi-squared test is shown. For the rest of the experiments, Microsoft Excel was used to derive standard deviation and to perform Student's t-tests.
RESULTS
Arl13b (sco)hi459 mutants show similar phenotypes to ift mutants
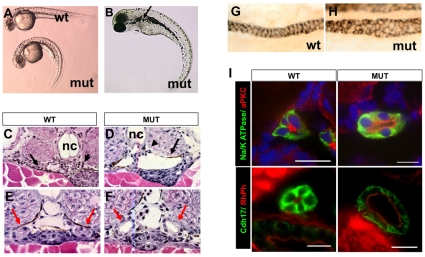
hi459 was originally isolated as a cystic kidney mutant in a large-scale insertional mutagenesis screen in zebrafish (Golling et al., 2002; Sun et al., 2004). Similar to multiple ift (intraflagellar transport) mutants isolated in the same screen, hi459 mutants exhibit body axis curvature and pronephric cysts (Fig. 1A-D). Cross sections reveal that the cysts are localized to the glomerular-tubular region of the nephron (Fig. 1C,D) as in the original definition of segments of the zebrafish pronephros, or the glomeruli-neck region in a more recent definition (Drummond et al., 1998; Wingert et al., 2007). For consistency and simplicity, we will use the original definition in the rest of the text. The pronephric duct also appears dilated in scohi459 mutants, as shown by histological sections (Fig. 1E,F) and by antibody staining with α6F, an antibody that recognizes a subunit of Na+/K+-ATPase enriched in the basolateral surface of the pronephric duct (Fig. 1G,H).
Fig. 1.
Phenotypes of scohi459 mutant zebrafish. (A) Side view of embryos at 36 hpf. (B) A mutant at 3 dpf. The arrow points to a cyst. (C,D) Cysts at the glomerular (arrowhead) and tubular (arrow) regions in cross-sections of mutant embryos at 50 hpf. (E-I) Increased size of the pronephric duct in mutant embryos as shown in cross-sections of the duct region (E,F red arrows) at 50 hpf. (G,H) The pronephric duct in side view in whole-mount embryos stained with α6F. (I) Cross sections of pronephric duct at 5 dpf. Anti-Na+/K+-ATPase and anti-Cdh17 are used as basolateral markers. Atypical PKC (aPKC) and rhodamine phalloidin (RhPh) are used as apical markers. DAPI is used as nuclei dye. mut, mutant; nc, notochord; wt, wild type. Scale bars: 10 μm.
We followed the progression of ductal dilation by measuring the outer and inner perimeters of the duct on cross-sections. On 2 dpf, scohi459 mutants already showed increased inner perimeter along the entire length of the duct (see Table S1 in the supplementary material). On 4 dpf, both the inner and outer perimeters are significantly increased (see Table S1 in the supplementary material).
To evaluate the role of cell number in the dilation of the duct, we counted nuclei number in six consecutive cross-sections (4 μm sections) and used the average as an index of cell number in the circumference of the duct. On 2 dpf, there was a statistically significant increase in nuclei number in the mid-regions of the duct in mutant embryos (see Table S2 in the supplementary material). By 4 dpf, all regions in mutant embryos displayed a statistically significant increase in nuclei number (see Table S2 in the supplementary material), suggesting that an increase of cell number might contribute to the enlargement of the kidney duct in scohi459 mutants.
It was previously reported that Na+/K+-ATPase is mis-targeted to the apical side in some cyst-lining epithelial cells (Drummond et al., 1998; Wilson et al., 1991). We therefore performed immunostaining with anti-Na+/K+-ATPase in combination with anti-atypical PKC, which labels the apical side of the duct. Our results showed that in hi459 mutants, Na+/K+-ATPase is still absent from the apical side and atypical PKC remains exclusive to the apical side (Fig. 1I). We further labeled the duct with phalloidin, which stains filamentous actin enriched in the apical cortex, and anti-cadherin-17 (Cdh17) that labels the basolateral membrane of ductal cells. Consistently, no significant difference in the localization pattern of the two markers was detected in hi459 mutants (Fig. 1I). Thus, ductal cells in scohi459 mutants appear to maintain normal apicobasolateral polarity, at least at a gross level.
hi459 is a zygotic null allele of arl13b (sco)
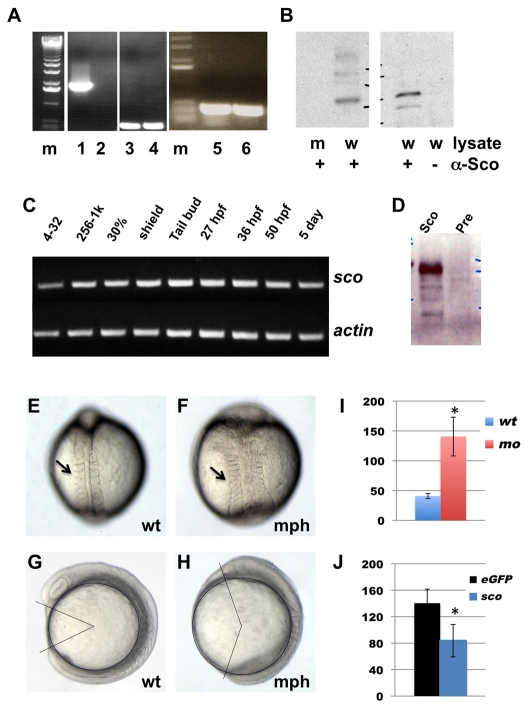
The proviral insertion in hi459 is located within the 5′-UTR region of the arl13b gene. RT-PCR with a pair of primers spanning the insertion site revealed a single band at the predicted size (Fig. 2A). This signal is completely abolished in samples from homozygous scohi459 mutants (Fig. 2A). However, transcript was detected in mutants using a pair of primers located 3′ to the insertion site (Fig. 2A). To verify that hi459 is a loss-of-function allele of arl13b, we injected arl13b mRNA into embryos from hi459 heterozygous carriers. Although six out of 24 uninjected embryos showed kidney cyst and body curvature, none of the 47 injected embryos were phenotypic. Genotyping experiments on 44 injected embryos verified the presence of seven homozygous mutant embryos in the injected sample, suggesting that the injected mRNA can rescue scohi459 mutants. This result is consistent with a previous report (Cantagrel et al., 2008). To analyze Arl13b protein products in scohi459 mutants, we generated a polyclonal antibody (anti-Sco) against the carboxy-terminal half of Arl13b (from amino acid 238). Immunoprecipitation (IP) and immunoblotting with anti-Sco revealed a band of the predicted size in wild-type samples, whereas this signal was completely absent both in scohi459 mutant samples and in wild-type lysates incubated with protein A beads alone (Fig. 2B), suggesting that the anti-Sco antibody is specific and that the production of Arl13b is completely disrupted in scohi459 mutants.
Fig. 2.
hi459 is a zygotic null allele of arl13b (sco). (A) arl13b transcripts in embryos at 5 dpf shown by RT-PCR of cDNA from hi459 mutant embryos (lanes 2, 4, 6) and wild-type siblings (lanes 1, 3, 5). m, size marker; lanes 1-2, RT-PCR with a pair of primers spanning the proviral insertion site in arl13b; lanes 3-4, RT-PCR with a pair of primers for beta-actin as loading controls; lanes 5-6, RT-PCR with a pair of primers 3′ to the proviral insertion site in arl13b. (B) Absence of Arl13b in hi459 mutant embryos at 5 dpf. A band of the predicted size can be seen in samples immunoprecipitated with anti-Sco from lysate of wild-type siblings (w), but not in the sample from hi459 mutant embryos (m), nor in the lysate precipitated with protein A beads alone. (C) RT-PCR time-course for arl13b expression. 4-32, four- to 32-cell stage; 256-1k, 256- to 1000-cell stage; 30%, 30% epiboly. (D) Presence of Arl13b in embryos at the 64-cell stage. Lysates from embryos at the 64-cell stage were subjected to immunoprecipitation using anti-Sco (Sco) or pre-immune IgG (Pre) and precipitated proteins detected by western analysis using anti-Sco. (E-H) Wild type (wt) and arl13b morphant (mph) embryos in dorsal view (E,F) and side view (G,H) at around the 8-somite stage. Arrows in E and F point to somites. In G and H, body gap angle was labeled using ImageJ. (I) Gap angle in degrees in wild type (wt, n=13) embryos and arl13b morphants (mo, n=14; *P<0.001). (J) Gap angle in degrees in arl13b morphants co-injected with eGFP (eGFP, n=13) or sco mRNA (sco, n=15; *P<0.001).
In zebrafish, the onset of zygotic transcription occurs at around the 512-cell stage (Westerfield, 2000). However, arl13b transcripts can be readily detected at the four- to 32-cell stage (Fig. 2C), which suggests that the arl13b transcript is supplied maternally. In addition, Arl13b protein can be readily detected in lysates of embryos at the 64-cell stage (Fig. 2D). Combined, these results suggest that hi459 represents a zygotic null allele of arl13b.
The maternal contribution of gene products of arl13b may mask an early function of Arl13b in hi459 mutants. However, translation-blocking morpholino oligonucleotides can block the translation of maternally deposited mRNA. We therefore designed a morpholino antisense oligonucleotide against a region within the 5′ UTR region of arl13b. Injection of 4 ng of this oligonucleotide into wild-type embryos caused phenotypes seen in scohi459 mutants, namely body curvature and kidney cyst formation. Importantly, phenotypes of morphants (knockdown animals generated by morpholino injection) could be rescued by the injection of arl13b mRNA, suggesting that the effect of this oligonucleotide is specific (see Fig. S1A in the supplementary material).
When injected with 6 ng of this morpholino, morphants showed a severely shortened body axis at 1 dpf (see Fig. S1B in the supplementary material). Further analysis revealed wider somites and a shorter body axis at around the 8-somite stage (Fig. 2E,F), phenotypes that are suggestive of defective CE movement during gastrulation. To quantify this phenotype, we measured body gap angle (Gerdes et al., 2007), which is defined as the angle between lines connecting the center of the yolk with the tips of the head and the tail. In wild-type embryos, the gap angle averaged 41±4°, whereas in arl13b morphants this angle was increased to 141±32° (Fig. 2G-I, P<0.001). This effect of the arl13b morpholino was specific to the depletion of arl13b, because co-injection of arl13b mRNA could rescue the gap angle from 140±21° to 84±26° (Fig. 2J, P<0.001). To measure cell movement during gastrulation more directly, we labeled the shield with the lipophilic dye FM1-43 at the shield stage (see Fig. S1C in the supplementary material) and examined the position of labeled cells at the tailbud stage. Results suggest that the anterior and posterior end of labeled cells closely followed the positions of the head and the tail (Fig. S1D,E in the supplementary material), consistent with the notion that CE movement during gastrulation is a driving force to generate distance between the head and the tail. Together, these data suggest that the depletion of arl13b at early embryonic stage leads to phenotypes that are consistent with defects in CE movement during gastrulation.
Arl13b is localized to the cilium and is required for cilium assembly in multiple organs
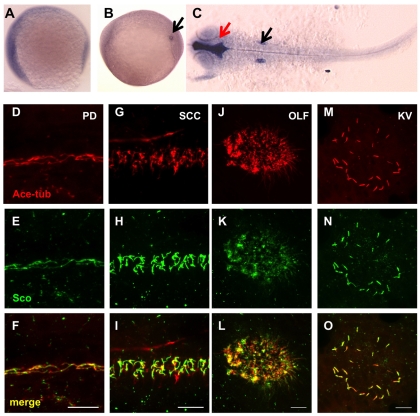
We analyzed the expression pattern of arl13b during early zebrafish development. Whole-mount in situ hybridization revealed that the arl13b message was widely distributed (Fig. 3A-C). At the 8-somite stage, arl13b transcript showed an occasional enrichment in the Kupffer's vesicle. At 25 hpf, arl13b expression was ubiquitous with enrichment in brain ventricles and otic vesicles, two regions with a high density of cilia.
Fig. 3.
Gene products of arl13b are widely distributed. (A-C) In situ hybridization for arl13b on embryos at the 1-somite stage (A), the 8-somite stage (B) and 25 hpf (C). A and B are side views and C is a dorsal view, all with anterior to the left. In B, the transcript is enriched in the Kupffer's vesicle (KV, arrow). In C, enrichment in the brain ventricle (red arrow) and the otic vesicle (black arrow) can be seen. (D-O) Arl13b is highly enriched in cilia in multiple organs as shown by whole-mount immunostaining of embryos at 2 dpf (D-L) and the 10-somite stage (M-O). The cilia marker anti-acetylated tubulin is shown in red, while anti-Sco is shown in green. (D-F) The pronephric duct (PD). (G-I) The spinal cord canal (SCC). (J-L) The olfactory placode (OLF). (M-O) The KV. Scale bars: 10 μm.
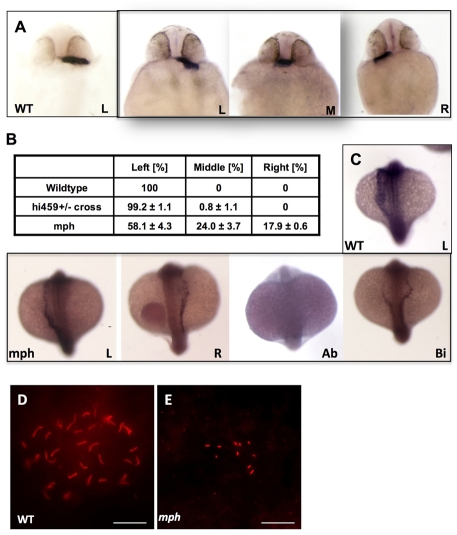
To investigate how Arl13b might be involved in preventing cyst formation, we examined the subcellular localization of this protein in zebrafish via immunostaining with the anti-Sco antibody. Results show that Arl13b is localized to cilia in the pronephric duct (Fig. 3D-F), consistent with previous reports on Arl13b in mouse and in cultured cells (Cantagrel et al., 2008; Caspary et al., 2007; Hori et al., 2008). To verify the specificity of anti-Sco, we tested whether the ciliary localization detected by anti-Sco was reduced in embryos with less Arl13b. Because we had previously shown that cilia formation itself is affected in scohi459 mutants (Sun et al., 2004), we injected the above mentioned arl13b morpholino oligonucleotide at a low dose to allow cilia formation, and stained the embryos with anti-Sco and anti-acetylated tubulin. Results showed that the signal of Arl13b in cilia is greatly reduced in morphants, while the acetylated tubulin signal remains (Fig. 4A,B). Thus anti-Sco is highly specific for immunostaining. Using this antibody, we detected the presence of Arl13b on cilia in all of the organs we examined, including the neural tube, the olfactory placode and Kupffer's vesicle (KV; Fig. 3G-O).
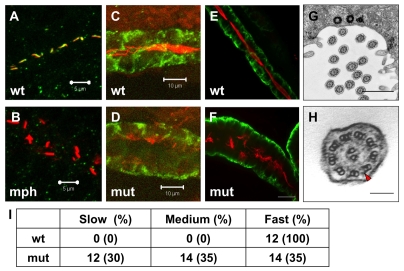
Fig. 4.
Cilia in scohi459 mutant embryos. (A,B) Confocal projections showing the pronephric duct in a wild-type embryo (A) and an arl13b morphant (B) at the 20-somite stage, stained with anti-acetylated tubulin (red) and anti-Sco (green). Signal in B is overexposed. Scale bars: 5 μm. (C,D) Confocal projections showing cilia defect (stained with anti-acetylated tubulin, red) in the pronephric duct (stained with anti-Cdh17, green) in a scohi459 mutant embryo (D), compared with a wild-type sibling (C), at 33 hpf. Scale bars: 10 μm. (E,F) Confocal projection showing cilia (stained with anti-acetylated tubulin, red) in the pronephric duct (stained with anti-Cdh17, green) in embryos at 5 dpf. Scale bar: 20 μm. (G,H) Electron micrographs showing the ultrastructure of cilia in the scohi459 mutant at 5 dpf. H shows an enlarged cross section; red arrowhead indicates a dynein arm. Scale bars: 1 μm in G; 100 nm in H. (I) Cilia motility in scohi459 mutant and wild-type siblings at 5 dpf. Cilia with a beating frequency of between 36 and 55 Hz are categorized as fast, whereas cilia with frequency of between 21 and 35, and between 5 and 20 are categorized as medium and slow, respectively. mph, morphant; mut, mutant; wt, wild type.
Using immunostaining and chromogenic reaction, we previously showed that cilia formation in the pronephric duct is affected in scohi459 mutants (Sun et al., 2004). In this study, we analyzed cilia formation in scohi459 mutants in more detail using fluorescence microscopy. At 33 hpf, we detected significant reduction of cilia in the pronephric duct of scohi459 mutant embryos (Fig. 4C,D). Interestingly, we did detect a significant number of cilia later at 5 dpf (Fig. 4E,F). Instead of being tightly bundled together as tufts, as in wild-type siblings, cilia appeared disorganized in mutant embryos. By contrast, in ift mutants, cilia are able to form during early embryonic stages but degenerate with time (Tsujikawa and Malicki, 2004) (Y. Cao and Z.S., unpublished), suggesting that the role of Arl13b in cilia formation may not be identical to that of IFT proteins.
At the center of the cilium is a microtubule axoneme, which comprises nine doublet (A and B) microtubules at the periphery and, mostly in motile cilia, a pair of microtubules in the center. The mouse hnn mutant, which is a null allele of Arl13b, shows incomplete B microtubules in the axoneme (Caspary et al., 2007). To test whether similar defects can be detected in scohi459 mutants, we performed transmission electron microscopy on embryos fixed at 5 dpf. Results indicate that the ultrastructure of cilia appears intact (Fig. 4G,H).
We further examined cilia motility using high-speed videomicroscopy. Cilia in scohi459 mutants are still able to beat. However, compared with wild-type siblings, the beating pattern in mutants is less coordinated: whereas all analyzed cilia in wild-type embryos beat with a frequency of between 36 and 55 Hz, cilia in mutant embryos beat with a frequency ranging from 5-55 Hz (Fig. 4I, see Movies 1 and 2 in the supplementary material).
Functional cilia in the KV are essential for establishing the left-right (LR) asymmetry of the body plan, as are cilia in the mouse node (Essner et al., 2005; Essner et al., 2002). However, we failed to detect any LR defects in scohi459 mutants. One possibility is that the maternal contribution of arl13b transcript is sufficient to support the role of this gene in the KV. To test this hypothesis, we used the above mentioned morpholino oligonucleotide to block the translation of arl13b mRNA. Indeed, injection of 4 ng of this oligonucleotide leads to randomization of left-right asymmetry, as shown by the position of the heart tube at 1 dpf (Fig. 5A). In control embryos, 99.2% displayed a left-sided heart tube. By contrast, in morphants 17.9% had a right-sided heart tube, and in 24% the heart tube remained in the middle (Fig. 5B). To analyze the mechanism of LR asymmetry disruption by arl13b knockdown, we examined the expression pattern of southpaw, which encodes a nodal related protein that is expressed in the left lateral plate mesoderm in wild-type embryos (Long et al., 2003). southpaw is the earliest laterality marker and is essential for the establishment of left-right asymmetry in zebrafish. Our results indicate that the southpaw expression pattern is abnormal in arl13b morphants (Fig. 5C). In a representative experiment, whereas 90% of the control embryos show left-sided expression of southpaw, only 65% of the morphants show such a pattern, the rest show right-sided (6.5%), bilateral (11.1%) or absent (17.5%) expression, suggesting that Arl13b functions early in the establishment of laterality, not far from the KV. We further examined organogenesis of the KV, and cilia formation in the KV in morphants at the 8-10 somite stage. Results indicated that although formation of the KV was not affected (data not shown), cilia in the KV were either very sparse or completely absent in morphants, compared with those in wild-type embryos and in embryos injected with a control morpholino (Fig. 5D,E). Furthermore, cilia length in morphants was significantly shorter (1.37±0.27 μm, P<0.001) than in age-matched wild-type embryos (3.01±0.33 μm) or control morphants (3.07±0.32 μm).
Fig. 5.
arl13b is required for proper left-right asymmetry. (A) In situ hybridization for cardiac myosin light chain (cmlc2) showing the heart tubes of wild-type and morphant embryos at 26 hpf. (B) Tabulation of heart position from three independent experiments. (C) In situ hybridization for southpaw on wild-type and morphant embryos at the 18- to 20-somite stage, in dorsal view with head to the top. (D,E) KV in a wild-type embryo (D) and a morphant (E). Scale bars: 20 μm. Ab, absent; Bi, bilateral; L, left-sided; M, middle; mph, morphant; R, right-sided; wt, wild type.
Together, these results suggest that the ciliary localization of Arl13b and its function in cilia formation are systemic, rather than kidney or brain specific.
Multiple regions of Arl13b are required for its ciliary localization
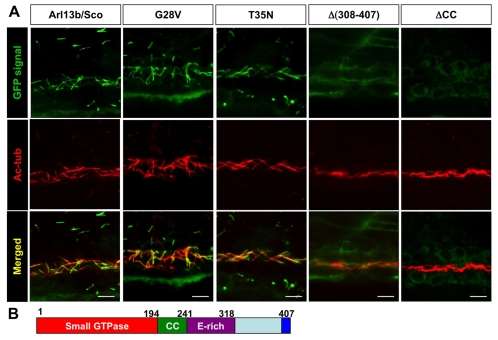
One striking feature of Arl13b localization is that it is almost exclusively localized to the cilium, whereas many other ciliary proteins including IFT proteins can be found in other compartments of the cell in significant quantities (Follit et al., 2006; Qin et al., 2004). We therefore sought to isolate cilia-targeting sequences in Arl13b. We first overexpressed a full-length Arl13b with an eGFP tag at the carboxy-terminus by injecting mRNA transcribed in vitro into wild-type embryos at the one- to four-cell stage. We collected embryos at around 24 hpf and performed immunostaining with anti-acetylated tubulin as the cilium marker. Results showed that the GFP signal was enriched in the cilium of the pronephric duct (Fig. 6A), recapitulating the subcellular localization pattern of endogenous Arl13b. Importantly, injected embryos appeared morphologically normal. To test whether the tagged protein was functional, we injected mRNA into embryos from heterozygous carriers of hi459. In uninjected embryos, 25% (n=24) were phenotypic with body curvature and kidney cysts, whereas no (n=23) injected embryos showed such phenotypes, verifying that the tagged protein is functional in vivo.
Fig. 6.
Multiple regions, but not the small GTPase activity, of Arl13b are required for its targeting to the cilium. (A) Images show side views of the pronephric duct in embryos injected with mRNA encoding eGFP-tagged versions of Arl13b indicated above. eGFP signal is shown in green, while anti-acetylated tubulin (Ac-tub) is labeled in red. CC, coiled-coil domain. Scale bars: 10 μm. (B) Schematic of the structure of Arl13b. Numbers are amino acid coordinates.
Arl13b is unique among the Arl family of small GTPases in that it contains a relatively extensive carboxy-terminal sequence that makes up more than half of the protein. Immediately after the small GTPase domain is a highly conserved coiled-coil domain (residues 194-241, simplified as CC), followed by a glutamate-rich region (residues 241-318, simplified as E-rich), and a region at the very C terminus that is also highly conserved in Arl13b among multiple species (Fig. 6B). To determine the role of these regions in the ciliary localization of Arl13b, we made a series of deletion constructs of Sco-eGFP. We injected mRNA transcribed in vitro into embryos and inspected the subcellular localization pattern of eGFP at 1 dpf. Contrary to our expectation of a single cilia-targeting sequence, multiple regions were required for the ciliary localization of Sco (Fig. 6A, Table 1). Deletion of either the small GTPase domain, or the coiled-coil domain, abolishes the ciliary localization of Arl13b. By contrast, deletion of either the glutamate-rich region or the C-terminal 14 residues [Δ(394-407)] failed to disrupt the localization of Arl13b to the cilium. Together, these results suggest that more than one region is essential for targeting Arl13b to cilia.
Table 1.
Summary of localization and rescue result
| Cilia localization | BC/cyst rescue | Cilia rescue | |
| Full-length Sco | + | + | + |
| GTPase [Δ(196-407)] | − | − | NA |
| Δ(21-127) | − | − | NA |
| G28V | + | − | − |
| T35N | + | − | − |
| ΔCC [Δ(195-243)] | − | − | − |
| Δ(E-rich) [Δ(244-318)] | + | + | NA |
| Δ(308-407) | − | + | + |
| Δ(394-407) | + | + | NA |
Detailed information on cilia rescue results are shown in Fig. S1 in thesupplementary material.
NA, not analyzed; CC, coiled-coil domain.
We then tested whether more subtle manipulations of the small GTPase domain would disrupt the ciliary localization of Arl13b. Members of the Ras superfamily of small GTPases contain a P loop (GXXXXGKS/T), which is required for guanine nucleotide binding (Lu et al., 2001). Substitution of the first G with V frequently leads to reduced GTP hydrolysis and thus generates a dominant-active protein (Lu et al., 2001). By contrast, mutants with a substitution of N for the T/S site have a higher affinity for GDP and function as a dominant-negative protein (Fan et al., 2004; Kobayashi et al., 2009; Lu et al., 2001). We introduced these two mutations into the corresponding sites of Arl13b and tested their in vivo effect on trafficking to cilia by expressing eGFP fusion constructs as described above. Results showed that both mutated forms of Arl13b still localized to the cilium (Fig. 6A). However, neither mutant showed detectible function when overexpressed in vivo (see below). Whether these point mutations affected the activity of the small GTPase domain as predicted is currently unclear.
Ciliary localization is required for in vivo function of Arl13b
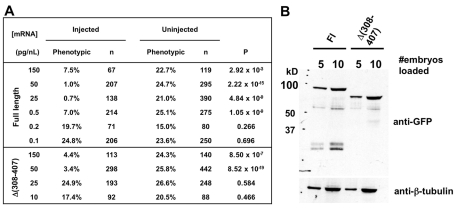
To investigate whether ciliary localization is essential for the in vivo function of Arl13b, we tested deletion constructs for their ability to rescue the scohi459 mutant. We injected mRNA into embryos from heterozygous carriers of hi459 and observed for the development of body axis curvature and pronephric cyst phenotypes. Surprisingly, although failing to traffic to the cilium in a significant quantity, Δ(308-407) was able to rescue the scohi459 mutant. When injected with 265.5 pg of Δ(308-407) mRNA, only 4.4% of embryos showed body curvature and kidney cysts (Tables 1, 2). It is possible that a small percentage of the overexpressed Δ(308-407) was able to enter cilium and carry out its essential function. To test this hypothesis, we injected progressively smaller amounts of mRNA encoding either full-length Arl13b or Δ(308-407). Results showed that whereas 0.885 pg full-length arl13b mRNA was able to rescue scohi459 mutants, 88.5 pg of Δ(308-407) mRNA was required to rescue scohi459 mutants to a significant extent (Fig. 7A). To rule out the possible involvement of protein stability or expression level in this assay, we performed western blot analysis of full-length Arl13b and Δ(308-407) in lysates obtained at 1 dpf from embryos injected with same quantities of mRNAs encoding either protein. Results indicated that both proteins were expressed at similar levels (Fig. 7B). Together, these results suggest that the in vivo activity of Δ(308-407) is greatly reduced, correlating with its drastically reduced ciliary localization. The rest of the deletion proteins showed complete correlation between ciliary localization and rescuing capability. Specifically, Δ(E-rich) and Δ(394-407) were able to rescue scohi459 mutants, whereas GTPase, ΔCC, and Δ(21-127) failed to do so (Tables 1, 2).
Table 2.
Body curvature and cyst rescue activity of different alleles
| Injected | Uninjected | ||||
| Allele | % Ph | n | % Ph | n | P |
| Fl | 7.5 | 67 | 22.7 | 119 | 2.9×10−3 |
| GTPase | 22.2 | 221 | 22.3 | 386 | 1.0 |
| Δ(21-127) | 23.6 | 161 | 26.3 | 194 | 0.4 |
| G28V | 22.1 | 289 | 21.2 | 364 | 0.7 |
| T35N | 22.2 | 221 | 24.2 | 260 | 0.5 |
| ΔCC | 23.8 | 223 | 21.9 | 269 | 0.5 |
| Δ(E-rich) | 2.3 | 261 | 24.1 | 286 | 1.70×10−16 |
| Δ(308-407) | 4.4 | 113 | 24.3 | 140 | 8.5×10−7 |
| Δ(394-407) | 0.0 | 76 | 23.2 | 112 | 1.6×10−6 |
Ph, phenotypic; n, number of embryos analyzed; Fl, full length.
Fig. 7.
Localization and functions of Δ(308-407) Arl13b in vivo. (A) arl13b::gfp mRNA retains detectable rescue activity at a quantity 100-fold less than that of Δ(308-407)::gfp mRNA. (B) Arl13b::GFP and Δ(308-407)::GFP are expressed at similar levels when the same amount of mRNA is injected.
We also tested whether the putative dominant mutants described above were functional in vivo. Despite their normal localization pattern shown above, both G28V and T35N failed to rescue either the body curvature or the cystic kidney phenotype of scohi459 mutants (Tables 1, 2). In addition to morphological phenotypes, we further tested G28V and T35N for their ability to rescue cilia formation in scohi459 mutants. We immunostained embryos with anti-acetylated tubulin, performed a chromogenic reaction and observed signal at a low magnification. This assay reveals a global picture of cilia formation in the kidney duct. In wild-type embryos, bundles of cilia in the duct appear as a tight line running between the yolk and the somites, with occasional gaps representing areas of cells with a single cilium. In scohi459 mutant embryos, only spotted signals could be detected (see Fig. S2A,B in the supplementary material). Injection of either G28V or T35N mRNA failed to rescue this phenotype (Fig. S2C in the supplementary material). It is perplexing that the putative dominant-active Arl13b failed to rescue either morphological phenotypes or cilia formation. Moreover, overexpression of either of the two putative dominant constructs did not lead to any obvious morphological phenotypes (data not shown). The mechanism for the lack of function of these two mutants awaits further investigation.
DISCUSSION
The role of Arl13b in cilia assembly and implications for human diseases
The cell surface organelle the cilium is almost ubiquitously present on vertebrate cells. Not surprisingly, structural or functional defects in this organelle have been linked to an expanding list of human diseases, ranging from polycystic kidney disease to polydactyly (Badano and Katsanis, 2006; Sharma et al., 2008). The ‘molar tooth sign’ (MTS) in brain MRI scans resulting from structural abnormalities in the cerebellar vermis is a hallmark of classical JS (Gleeson et al., 2004; Harris, 2007; Sharma et al., 2008). As MTS can be found in patients with additional symptoms associated with ciliary defects, it is thought that ciliary defects might be the underlying mechanism of JSRD defects (Cantagrel et al., 2008; Castori et al., 2005; Dixon-Salazar et al., 2004; Gleeson et al., 2004; Gorden et al., 2008; Harris, 2007; Helou et al., 2007; Sharma et al., 2008; Utsch et al., 2006; Wolf et al., 2007). The findings that mutations in ARL13B are associated with JS and that Arl13b is required for cilia formation support this model. A lingering question is what causes different combinations of the spectrum of symptoms in different JS patients. Because, so far, mutations in human ARL13B are limited to the classical form of JS, we sought to determine whether the function of the zebrafish homolog is also restricted. Our results show that Arl13b is localized to the cilium and is required for cilia formation in multiple regions outside of the brain. Additionally, a mutated form of Arl13b retains residual activity. We further showed that defective Arl13b leads to phenotypes in multiple organs. Together, these data support a hypothesis that the involvement of ARL13B may not be limited to classical JS and that ARL13B could be a candidate gene for other cilia-related diseases.
Morpholino knockdown of arl13b leads to additional phenotypes that are consistent with defective CE movement during gastrulation. The specificity of the observed phenotype is verified by its rescue by the injection of arl13b mRNA. This result is consistent with the involvement of multiple cilia-related genes in the planar cell polarity (PCP) pathway that regulates CE movement (Aanstad et al., 2009; Kishimoto et al., 2008; Ross et al., 2005). However, a recent report suggests that a maternal-zygotic mutant of ift88oval does not display any obvious CE phenotype (Huang and Schier, 2009). It is possible that cilia themselves may not be directly involved in PCP and that genes involved in cilia formation might play additional, separate roles in PCP. Alternatively, a less likely scenario is that the maternal zygotic ift88oval mutant is not a complete null mutant and that residual undetected cilia are present in this mutant. Additional maternal-zygotic mutants of ift genes will further clarify this issue.
Multiple regions of Arl13b are required for its ciliary localization
The cilium is a narrow cell surface organelle protruding from the cell surface into its milieu. Mounting evidence suggests that the cilium is a specific compartment of the cell with distinct protein compositions. The mechanism by which ciliary proteins are targeted to the cilium remains enigmatic. Nonetheless, ciliary targeting motifs have been shown to be sufficient to target proteins to the cilium (Geng et al., 2006; Nasser and Landfear, 2004; Snapp and Landfear, 1999; Tam et al., 2000). For example, an N-terminal RVxP motif in Pkd2 has been shown to be both necessary and sufficient for targeting membrane proteins to the cilium (Geng et al., 2006). However, the conservation of these motifs seems to be restricted to proteins of similar structures. Because Arl13b is highly enriched in the cilium and because we are not aware of any reported cilia-targeting motif in proteins with a small GTPase domain, we sought to identify a cilia-targeting motif in Arl13b. Instead, our result indicates that multiple regions in Arl13b are essential for its ciliary localization. Conceivably, different regions of Arl13b could be involved in different steps of the trafficking process, and therefore are required for the ciliary localization of this protein. Alternatively, multiprotein interaction might be essential for the targeting of Arl13b to the cilium and different regions of Arl13b might be involved in protein-protein interactions.
Ciliary localization of Arl13b is essential for its function
There is much evidence to suggest that Arl13b/Sco functions through the cilium. First, Arl13b is highly enriched in the cilium and is required for cilium formation. Second, scohi459 mutants show almost identical phenotypes to those of known ciliary mutants. Our result that there is a strong correlation between the ciliary localization of different deletion constructs and their rescuing capability provides further support for this model.
Although our results suggest that the physical presence of the small GTPase domain is essential for the ciliary localization of Arl13b, the role of its GTPase activity is currently unclear. Both G28V, which is predicted to be trapped in a GTP-bound form, and T35N, which is predicted to bind to GDP with higher affinity, are still able to traffic to the cilium. However, both G28V and T35N mutant constructs failed to rescue the scohi459 mutants. Moreover, overexpression of neither of the two point mutants led to obvious phenotypes, suggesting that the functional regulation of the small GTPase domain might be different from that of classic small GTPases and more complex than what we had imagined. Indeed, a prominent difference of Arl13b is the conserved glycine at amino acid position 71, whereas almost all known Arfs and Arls contain a glutamine at the corresponding site. How the GTPase activity of Arl13b is regulated remains enigmatic.
Zebrafish can be used quantitatively to evaluate the activity of different alleles of tested genes
With the advance of sequencing and genotyping techniques, human genetic variations are being detected at an increasingly rapid rate. In the meantime, the ability to ascertain a causal relationship between a genotype and a phenotype remains limited. Furthermore, many genetic variations may not totally disrupt the function of affected genes. There is consequently an urgent need to develop new methods for rapid analysis of gene function both qualitatively and quantitatively. Many zebrafish homologs of human disease genes, when mutated, lead to phenotypes that can be directly compared to human symptoms. For example, defects in the zebrafish homolog of the human autosomal dominant polycystic kidney disease (ADPKD) gene PKD2 lead to kidney cyst formation in fish (Sun et al., 2004). Conversely, arl13b was initially identified as a cystic kidney gene in zebrafish and later associated with JS in humans (Cantagrel et al., 2008; Sun et al., 2004). In addition, morpholino oligonucleotides can be used very effectively and on a relatively high-throughput scale in zebrafish to disrupt gene function (Nasevicius and Ekker, 2000), and recently a zinc-finger technique has been used successfully for targeted mutagenesis (Foley et al., 2009; Meng et al., 2008; Yan et al., 2009). Our result that Δ(308-407) reduced rescuing capability, together with previous publications using zebrafish to evaluate the functions of variants of human genes (Leitch et al., 2008; Weber et al., 2008), support the idea that similar experiments can be used to evaluate gene function quantitatively. Together, these features make zebrafish an excellent model system in which to analyze the in vivo functions of human disease genes.
Supplementary Material
Acknowledgements
We thank members of the Sun lab and members of the Yale Center for PKD Research for helpful discussions, the Reinke lab and the Cooley lab for use of their microscopes, Nicole Semanchik for superb technical assistance, SueAnn Mentone for assistance on histology and Christoph Rahner for assistance on EM. N.A.D. was partly supported by a Genetics training grant from the NIH (GENTG#5, T32 GM007499). RO1 DK069528 and P50 DK057328 (project #3) from the NIDDK, and a research grant from the PKD Foundation to Z.S. provided the rest of the support. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/23/4033/DC1
References
- Aanstad P., Santos N., Corbit K. C., Scherz P. J., Trinh, le A., Salvenmoser W., Huisken J., Reiter J. F., Stainier D. Y. (2009). The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol. 19, 1034-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J. L., Katsanis N. (2006). Life without centrioles: cilia in the spotlight. Cell 125, 1228-1230 [DOI] [PubMed] [Google Scholar]
- Bielas S. L., Silhavy J. L., Brancati F., Kisseleva M. V., Al-Gazali L., Sztriha L., Bayoumi R. A., Zaki M. S., Abdel-Aleem A., Rosti R. O., et al. (2009). Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 9, 1032-1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V., Silhavy J. L., Bielas S. L., Swistun D., Marsh S. E., Bertrand J. Y., Audollent S., Attie-Bitach T., Holden K. R., Dobyns W. B., et al. (2008). Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 83, 170-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C. E., Anderson K. V. (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell 12, 767-778 [DOI] [PubMed] [Google Scholar]
- Castori M., Valente E. M., Donati M. A., Salvi S., Fazzi E., Procopio E., Galluccio T., Emma F., Dallapiccola B., Bertini E. (2005). NPHP1 gene deletion is a rare cause of Joubert syndrome related disorders. J. Med. Genet. 42, e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella L. M., Park A., Sun Z. (2009). Zebrafish Tsc1 reveals functional interactions between the cilium and the TOR pathway. Hum. Mol. Genet. 18, 595-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar T., Silhavy J. L., Marsh S. E., Louie C. M., Scott L. C., Gururaj A., Al-Gazali L., Al-Tawari A. A., Kayserili H., Sztriha L., et al. (2004). Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 75, 979-987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond I. A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., Schier A. F., Neuhauss S. C., Stemple D. L., Zwartkruis F., Rangini Z., et al. (1998). Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125, 4655-4667 [DOI] [PubMed] [Google Scholar]
- Essner J. J., Vogan K. J., Wagner M. K., Tabin C. J., Yost H. J., Brueckner M. (2002). Conserved function for embryonic nodal cilia. Nature 418, 37-38 [DOI] [PubMed] [Google Scholar]
- Essner J. J., Amack J. D., Nyholm M. K., Harris E. B., Yost H. J. (2005). Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247-1260 [DOI] [PubMed] [Google Scholar]
- Fan Y., Esmail M. A., Ansley S. J., Blacque O. E., Boroevich K., Ross A. J., Moore S. J., Badano J. L., May-Simera H., Compton D. S., et al. (2004). Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 36, 989-993 [DOI] [PubMed] [Google Scholar]
- Foley J. E., Yeh J. R., Maeder M. L., Reyon D., Sander J. D., Peterson R. T., Joung J. K. (2009). Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN). PLoS ONE 4, e4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J. A., Tuft R. A., Fogarty K. E., Pazour G. J. (2006). The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell 17, 3781-3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., Somlo S. (2006). Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 119, 1383-1395 [DOI] [PubMed] [Google Scholar]
- Gerdes J. M., Liu Y., Zaghloul N. A., Leitch C. C., Lawson S. S., Kato M., Beachy P. A., Beales P. L., DeMartino G. N., Fisher S., et al. (2007). Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39, 1350-1360 [DOI] [PubMed] [Google Scholar]
- Gleeson J. G., Keeler L. C., Parisi M. A., Marsh S. E., Chance P. F., Glass I. A., Graham J. M., Jr., Maria B. L., Barkovich A. J., Dobyns W. B. (2004). Molar tooth sign of the midbrain-hindbrain junction: occurrence in multiple distinct syndromes. Am. J. Med. Genet. A 125, 125-134; discussion 117 [DOI] [PubMed] [Google Scholar]
- Golling G., Amsterdam A., Sun Z., Antonelli M., Maldonado E., Chen W., Burgess S., Haldi M., Artzt K., Farrington S., et al. (2002). Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31, 135-140 [DOI] [PubMed] [Google Scholar]
- Gorden N. T., Arts H. H., Parisi M. A., Coene K. L., Letteboer S. J., van Beersum S. E., Mans D. A., Hikida A., Eckert M., Knutzen D., et al. (2008). CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet. 83, 559-571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. C. (2007). Genetic complexity in Joubert syndrome and related disorders. Kidney Int. 72, 1421-1423 [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Gerster T. (2000). Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 137, 139-148 [DOI] [PubMed] [Google Scholar]
- Helou J., Otto E. A., Attanasio M., Allen S. J., Parisi M. A., Glass I., Utsch B., Hashmi S., Fazzi E., Omran H., et al. (2007). Mutation analysis of NPHP6/CEP290 in patients with Joubert syndrome and Senior-Loken syndrome. J. Med. Genet. 44, 657-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y., Kobayashi T., Kikko Y., Kontani K., Katada T. (2008). Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem. Biophys. Res. Commun. 373, 119-124 [DOI] [PubMed] [Google Scholar]
- Huang P., Schier A. F. (2009). Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development 136, 3089-3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby M., Cox J. J., Gayral S., Hampshire D. J., Ayub M., Blockmans M., Pernot E., Kisseleva M. V., Compere P., Schiffmann S. N., et al. (2009). INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 9, 1027-1031 [DOI] [PubMed] [Google Scholar]
- Kishimoto N., Cao Y., Park A., Sun Z. (2008). Cystic kidney gene seahorse regulates cilia-mediated processes and Wnt pathways. Dev. Cell 14, 954-961 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Hori Y., Ueda N., Kajiho H., Muraoka S., Shima F., Kataoka T., Kontani K., Katada T. (2009). Biochemical characterization of missense mutations in the Arf/Arl-family small GTPase Arl6 causing Bardet-Biedl syndrome. Biochem. Biophys. Res. Commun. 381, 439-442 [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker A. G., Olale F., Haycraft C. J., Yoder B. K., Schier A. F., Drummond I. A. (2005). Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132, 1907-1921 [DOI] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099 [DOI] [PubMed] [Google Scholar]
- Leitch C. C., Zaghloul N. A., Davis E. E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R. A., Eyaid W., Banin E., et al. (2008). Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 40, 443-448 [DOI] [PubMed] [Google Scholar]
- Link V., Shevchenko A., Heisenberg C. P. (2006). Proteomics of early zebrafish embryos. BMC Dev. Biol. 6, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S., Ahmad N., Rebagliati M. (2003). The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130, 2303-2316 [DOI] [PubMed] [Google Scholar]
- Lu L., Horstmann H., Ng C., Hong W. (2001). Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 114, 4543-4555 [DOI] [PubMed] [Google Scholar]
- Meng X., Noyes M. B., Zhu L. J., Lawson N. D., Wolfe S. A. (2008). Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26, 695-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216-220 [DOI] [PubMed] [Google Scholar]
- Nasser M. I., Landfear S. M. (2004). Sequences required for the flagellar targeting of an integral membrane protein. Mol. Biochem. Parasitol. 135, 89-100 [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D. R., Geimer S., Cole D. G., Rosenbaum J. L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. J., May-Simera H., Eichers E. R., Kai M., Hill J., Jagger D. J., Leitch C. C., Chapple J. P., Munro P. M., Fisher S., et al. (2005). Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat. Genet. 37, 1135-1140 [DOI] [PubMed] [Google Scholar]
- Sepich D. S., Myers D. C., Short R., Topczewski J., Marlow F., Solnica-Krezel L. (2000). Role of the zebrafish trilobite locus in gastrulation movements of convergence and extension. Genesis 27, 159-173 [DOI] [PubMed] [Google Scholar]
- Sharma N., Berbari N. F., Yoder B. K. (2008). Chapter 13 ciliary dysfunction in developmental abnormalities and diseases. Curr. Top Dev. Biol. 85, 371-427 [DOI] [PubMed] [Google Scholar]
- Snapp E. L., Landfear S. M. (1999). Characterization of a targeting motif for a flagellar membrane protein in Leishmania enriettii. J. Biol. Chem. 274, 29543-29548 [DOI] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G. J., Cole D. G., Miller M. S., Hopkins N. (2004). A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085-4093 [DOI] [PubMed] [Google Scholar]
- Tam B. M., Moritz O. L., Hurd L. B., Papermaster D. S. (2000). Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. J. Cell Biol. 151, 1369-1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa M., Malicki J. (2004). Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 42, 703-716 [DOI] [PubMed] [Google Scholar]
- Utsch B., Sayer J. A., Attanasio M., Pereira R. R., Eccles M., Hennies H. C., Otto E. A., Hildebrandt F. (2006). Identification of the first AHI1 gene mutations in nephronophthisis-associated Joubert syndrome. Pediatr. Nephrol. 21, 32-35 [DOI] [PubMed] [Google Scholar]
- Weber S., Taylor J. C., Winyard P., Baker K. F., Sullivan-Brown J., Schild R., Knuppel T., Zurowska A. M., Caldas-Alfonso A., Litwin M., et al. (2008). SIX2 and BMP4 mutations associate with anomalous kidney development. J. Am. Soc. Nephrol. 19, 891-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book: a guide for the laboratory use of zebrafish (Danio rerio): University of Oregon Press; [Google Scholar]
- Wilson P. D., Sherwood A. C., Palla K., Du J., Watson R., Norman J. T. (1991). Reversed polarity of Na(+) -K(+) -ATPase: mislocation to apical plasma membranes in polycystic kidney disease epithelia. Am. J. Physiol. 260, F420-F430 [DOI] [PubMed] [Google Scholar]
- Wingert R. A., Selleck R., Yu J., Song H. D., Chen Z., Song A., Zhou Y., Thisse B., Thisse C., McMahon A. P., et al. (2007). The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 3, 1922-1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M. T., Saunier S., O'Toole J. F., Wanner N., Groshong T., Attanasio M., Salomon R., Stallmach T., Sayer J. A., Waldherr R., et al. (2007). Mutational analysis of the RPGRIP1L gene in patients with Joubert syndrome and nephronophthisis. Kidney Int. 72, 1520-1526 [DOI] [PubMed] [Google Scholar]
- Yan Z., Sun X., Engelhardt J. F. (2009). Progress and prospects: techniques for site-directed mutagenesis in animal models. Gene Ther. 5, 581-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.