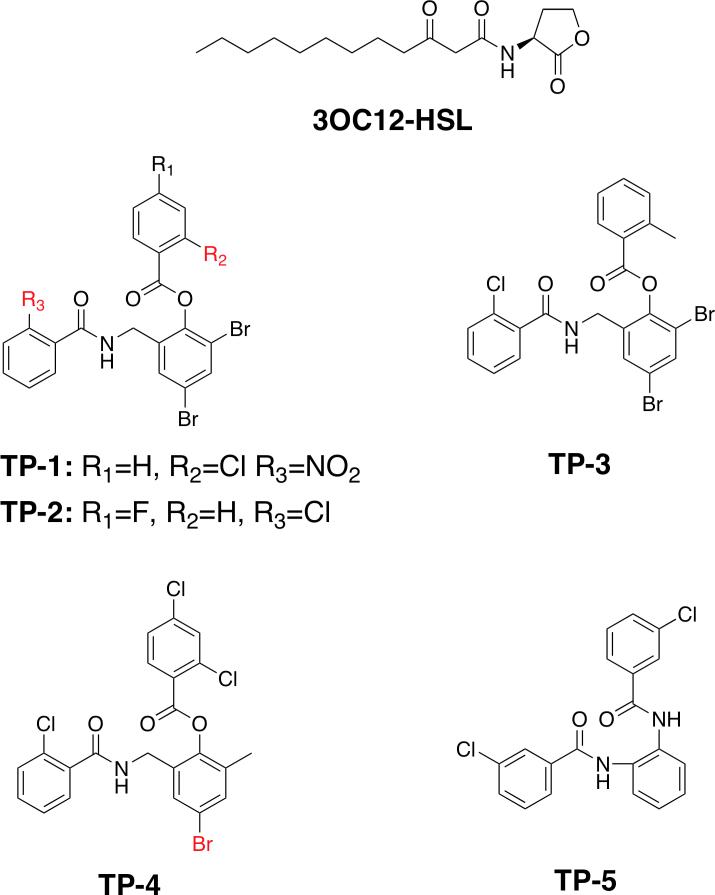
Abstract
The human pathogen Pseudomonas aeruginosa coordinates the expression of virulence factors using quorum sensing, a signaling cascade triggered by the activation of signal receptors by small molecule autoinducers. These homoserine lactone autoinducers stabilize their cognate receptors and activate their functions as transcription factors. As quorum sensing regulates the progression of infection and host immune resistance, significant efforts have been devoted towards the identification of small molecules that disrupt this process. Screening efforts have identified a class of triphenyl compounds that are structurally distinct from the homoserine lactone autoinducer, yet interact specifically and potently with LasR receptor to modulate quorum sensing (Muh et al., 2006a). Here we present the high-resolution crystal structures of the ligand-binding domain of LasR in complex with the autoinducer N-3-oxo-dodecanoyl homoserine lactone (1.4 Å resolution), and with the triphenyl mimics TP-1, TP-3, and TP-4 (to between 1.8-2.3 Å resolution). These crystal structures provide a molecular rationale for understanding how chemically distinct compounds can be accommodated by a highly selective receptor and provides the framework for the development of novel quorum sensing regulators, utilizing the triphenyl scaffold.
INTRODUCTION
The Gram-negative bacterium Pseudomonas aeruginosa is an opportunistic pathogen that targets immuno-compromised patients such as burn victims, neutropenic cancer, cystic fibrosis, and AIDS patients (Driscoll et al., 2007). P. aeruginosa infections kill several thousand of individuals each year and accounts for nearly 10% of all hospital-acquired nosocomial infections. The pathogen is notoriously resistant to most common antibiotics due to (a) the presence of a lipopolysaccharide-derived outer membrane permeability barrier (b) the actions of multidrug efflux pumps along with several chromosomally encoded antibiotic resistance genes, and (c) its tendency to manufacture and colonize surface-associated biofilms that further confer resistance (Hancock, 1998; Lambert, 2002). In addition to such intrinsic resistance, P. aeruginosa can also develop acquired resistance through horizontal gene transfer of antibiotic resistance determinants. The difficulty in treating Pseudomonas infections with antibiotics is illustrated by the fact that virtually all cystic fibrosis patients eventually become infected with a resistant strain that is untreatable (Gomez and Prince, 2007). The development of novel antimicrobials for the treatment of P. aeruginosa remains a global health concern.
Extensive research efforts over the past decade have established the quorum sensing signaling pathway as a viable target for the development of antimicrobials against P. aeruginosa (Bjarnsholt and Givskov, 2007; Clatworthy et al., 2007; Hentzer et al., 2003; Khmel and Metlitskaia, 2006). Quorum sensing is a strategy for intercellular communication that allows bacteria to coordinate their activity in response to population density (Waters and Bassler, 2005). The sensing mechanism is predicated upon the production of a small signal molecule called the autoinducer, which can diffuse through bacterial cell walls to allow for intercellular crosstalk (Fuqua et al., 1994). When the concentration exceeds a threshold value, the autoinducer binds its cognate receptor and activates the transcription of various genes. Pathogenic bacteria, including P. aeruginosa, use quorum sensing to evade the host immune response until they reach a threshold density, after which they activate the expression of virulence factors and participate in the maturation of biofilms (Clatworthy et al., 2007; Smith and Iglewski, 2003a).
Dissection of the signal sensing pathway identifies three receptors in P. aeruginosa that respond to homoserine lactone autoinducers: LasR, QscR, and RhlR (Chugani et al., 2001; Ochsner and Reiser, 1995; Pearson et al., 1994). Among these, the LasR signal receptor is activated the earliest and regulates the expression of most virulence or biofilm production associated genes (including other quorum sensing receptors) and, thus, has been the focus of intense research efforts (Pearson et al., 1995; Smith and Iglewski, 2003a; Smith and Iglewski, 2003b). The LasR receptor consists of two independently folded protein domains: an amino terminal ligand binding domain and a carboxy-terminal DNA binding domain (Bottomley et al., 2007). As with other quorum-sensing signaling receptors, binding of the homoserine lactone autoinducer stabilizes LasR and promotes dimerization (Kiratisin et al., 2002) and the resultant LasR homodimer-N-3-oxo-dodecanoyl homoserine lactone autoinducer complex binds target DNA to activate gene transcription (Schuster et al., 2004).
Recent efforts provide compelling evidence that inhibition of the quorum sensing signaling pathway can attenuate the virulence of P. aeruginosa. For example, mice infected with P. aeruginosa strains with mutations in the quorum-sensing pathway show reduced mortality and a decrease in the spread of the pathogen throughout the body (Smith et al., 2002). The quorum sensing receptors have garnered significant research interest as a target for therapeutic intervention due to the ability to easily screen for small molecule inhibitors, and much of the interest is focused on the LasR receptor, as it is required for the function of all other acyl homoserine receptors. However, efforts to design mimics based on the scaffold of the N-3-oxo-dodecanoyl homoserine lactone are limited by the fact that LasR shows high specificity for its cognate autoinducer. For example, the autoinducer for the related signaling receptor TraR (N-3-oxo-octanoyl homoserine lactone) contains an acyl chain that is shorter by four carbon atoms but is otherwise identical in structure to the autoinducer for LasR (Piper et al., 1993; Zhang et al., 1993). However, despite their similar chemical structures, the two receptors are highly specific for their cognate autoinducers and these small molecules show no cross-functionality (Pearson et al., 1994). In addition, the homoserine lactone is unstable at pH levels above 7 and is targeted for hydrolysis by mammalian lactonases, limiting the efficacy of synthetic derivatives as suitable pharmaceuticals (Ozer et al., 2005; Yang et al., 2005).
Additional approaches towards the development of quorum sensing inhibitors have included characterization of natural product and high-throughput screening of small molecules. Recently, an in vitro screen utilizing a P. aeruginosa strain lacking the ability to produce acyl-homoserine lactones and carrying a yellow fluorescent protein marker under control of a LasR regulated promoter was subject to a library of 200,000 compounds, resulted in the identification of both inhibitors and activators of LasR-dependent signalling (Muh et al., 2006b). An intriguing finding from this screen was the identification of a novel triphenyl (TP) scaffold based compound that is shown to be a potent activator of LasR despite the lack of any appreciable chemical similarities to the acyl homoserine lactone autoinducer (Muh et al., 2006a) (Figure 1). This compound (TP-1) and its derivatives TP-2, TP-3, and TP-4 are shown to function directly through LasR, show binding affinities that are several orders of magnitude lower than for any other class of inhibitors, and shows no cross-reactivity with related signaling receptors including QscR, LuxR, or RhlR (Muh et al., 2006a).
Figure 1. Revised structures of the cognate and structurally distinct LasR ligands.
Chemical structures of the N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) and each of the five triphenyl mimics previously identified as LasR ligands (Muh et al., 2006a). The structures of TP-1 and TP-4 differ slightly from those reported previously, as determined by the presence of electron-rich halogen atoms in their respective co-crystal structures, and the revisions are noted in red.
The identification of these specific, potent small molecule activators of LasR, structurally distinct from the homoserine lactone autoinducer, serves as a linchpin towards the development of synthetic modulators of quorum sensing. In order to delineate the dictates of molecular recognition of these novel triphenyl compounds by LasR, we have determined the high resolution co-crystal structures of the ligand-binding domain of this signaling receptor bound to TP-1 and TP-3 (each to 1.8 Å resolution), and TP-4 (2.3 Å resolution). We also report the structure of the LasR ligand binding domain bound to the N-3-oxo-dodecanoyl homoserine lactone autoinducer to 1.4 Å resolution, which provides a highly accurate view of the native binary complex. This work presents the first structural characterization of any quorum sensing receptor bound to a structurally distinct ligand and provides insights into differential ligand recognition by a highly specific receptor. The molecular framework presented here may serve in the rational design of inhibitors to target the quorum sensing signal pathway in pathogenic P. aeruginosa.
RESULTS
Overall Fold of LasR and Interactions With Autoinducer
Initial attempts to produce co-crystals of full-length LasR bound to both small molecule ligands and to various synthetic oligonucleotides encompassing the target DNA binding sequence were unsuccessful. As prior analysis had established that the ligand binding domain, lacking the 60 carboxy-terminal residues, was stable in the presence of cognate acyl homoserine ligand and amenable to crystallization (Bottomley et al., 2007), we utilized a similar approach for our structural efforts. The validity of this approach is further confirmed by the fact that suitable quantities of soluble and stable protein could be heterologously produced in media supplemented with either the acyl homoserine lactone autoinducer or any of the triphenyl mimics. The 1.4 Å resolution resolution co-crystal structure of the LasR ligand binding domain (hereafter LasR)-N-3-oxo-dodecanoyl acyl homoserine lactone complex was determined by molecular replacement using the coordinates from the previously determined structure of the binary complex, determined to 1.8 Å resolution (Bottomley et al., 2007). All other structures were also determined by molecular replacement using coordinates from this higher resolution structure.
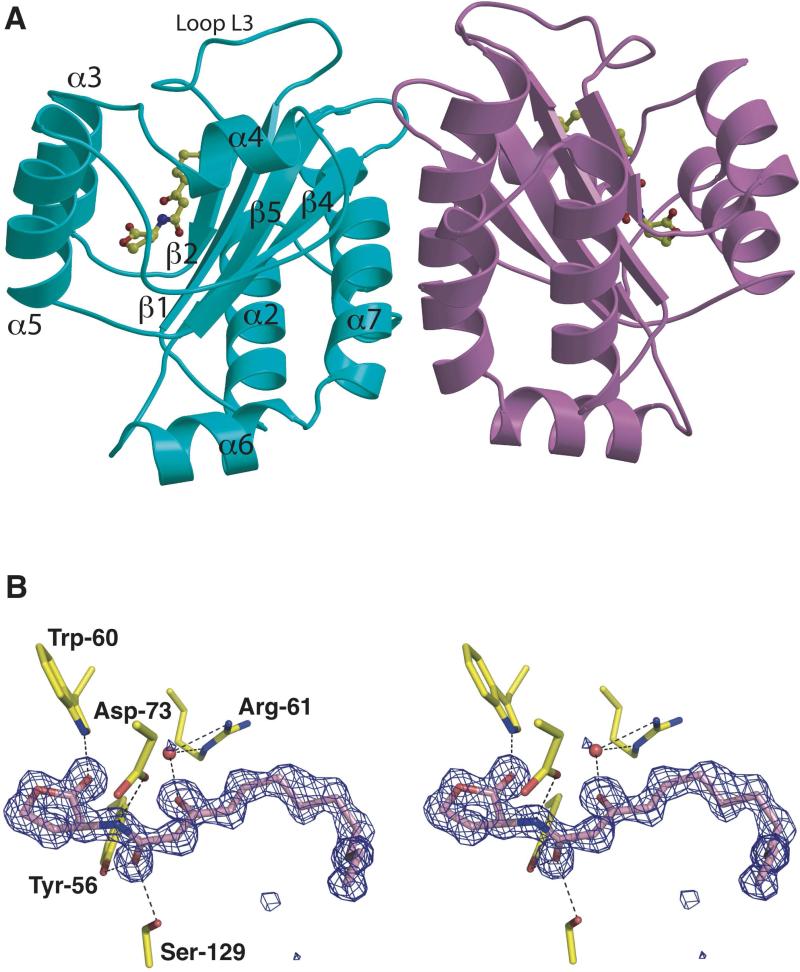
The overall fold of the LasR ligand binding domain presented here recapitulates the overall architecture of the same domain discerned from the prior structural analysis of its complex with acyl homoserine lactone autoinducer (Bottomley et al., 2007). Briefly, the ligand-binding domain consists of a five-stranded anti-parallel β-sheet flanked on both sides by two sets of three α-helices (Figure 2A). Consistent with previous biochemical and biophysical analysis, the LasR ligand binding domain exists as a homodimer in solution and in the 1.4 Å resolution structure of LasR-N-3-oxo-dodecanoyl homoserine lactone, there are two crystallographically independent copies of the complex present in the asymmetric unit. Dimerization is mediated by extensive polar and non-polar contacts with the central interface consisting of α-helix α6 and two loop regions.
Figure 2. Structure of LasR-homoserine lactone complex.
(A) Ribbon diagram showing the overall structure of the LasR-N-3-oxo-dodecanoyl homoserine lactone homodimer with one monomer colored in light blue and the second colored in purple. The homoserine lactone is shown in a ball-and-stick representation and secondary structure elements are noted on the light blue monomer. (B) Stereo-view of electron density maps calculated using Fourier coefficients Fobs – Fcalc with phases derived from the final refined model of the LasR-N-3-oxo-dodecanoyl homoserine lactone minus the coordinates of the ligand prior to one round of crystallographic refinement. The map is contoured at 4σ (blue mesh) and the final refined coordinates are superimposed with the protein residues colored in yellow and the ligand molecule colored in pink.
The ligand-binding pocket is formed between the β-sheet core and helices α3, α4, and α5. A large loop, L3 encompassing residues Leu-40 through Phe-51, covers the pocket and sequesters the ligand into the interior of the polypeptide and away from solvent. At the base of this pocket, several key residues engage in hydrogen bond interactions with the polar homoserine lactone head group of the autoinducer, including Tyr-56 (2.73 Å), Trp-60 (3.05 Å), Asp-73 (2.79 Å), and Ser-129 (2.69 Å) (Figure 2B). Each of these residues is strictly conserved between LasR, LuxR, and RhlR (with Thr-129 replacing the serine at this position in TraR), reflecting the fact that each of these receptor homologs can recognize cognate ligands with identical polar head groups.
At the other end of the ligand-binding pocket of LasR, a number of hydrophobic residues including Leu-36, Leu-40, Ile-52, Val-76 and Leu-125 engages the hydrophobic tail of the N-3-oxo-dodecanoyl homoserine lactone through van der Waals contacts (Figure S1). An important determinant of acyl chain interaction is mediated through interactions with Tyr-47 that is located in the L3 loop region that covers the ligand-binding pocket. The packing of Tyr-47 against the hydrocarbon chain of the autoinducer shields the ligand-binding pocket from bulk solvent, while reciprocally sequestering the nonpolar residues in the L3 loop into a favorable hydrophobic environment. The loss of this favorable interaction in the absence of the autoinducer would presumably result in loop flexibility, and subsequent exposure of the buried hydrophobic residues in this pocket to bulk solvent, resulting in protein aggregation. Similarly, decreasing the hydrocarbon chain length of the autoinducer would compromise the integrity of the favorable hydrophobic interactions resulting in an unstable complex. A comparison of the crystal structures of LasR and TraR (which utilizes a shorter N-3-oxo-hexanoyl homoserine lactone autoinducer) shows that the L3 loop is much shorter in TraR and does not cap the ligand-binding pocket (Figure S2) (Vannini et al., 2002; Zhang et al., 2002). Rather, a movement of sheets β1 and β2 towards helices α3 and α5 sequesters the hydrophobic pocket of TraR from bulk solvent. Such structural rearrangements at the acyl chain-binding region may account for the differential specificities of signaling receptors towards their cognate autoinducers.
Co-crystal Structure of the LasR-TP-1 Complex
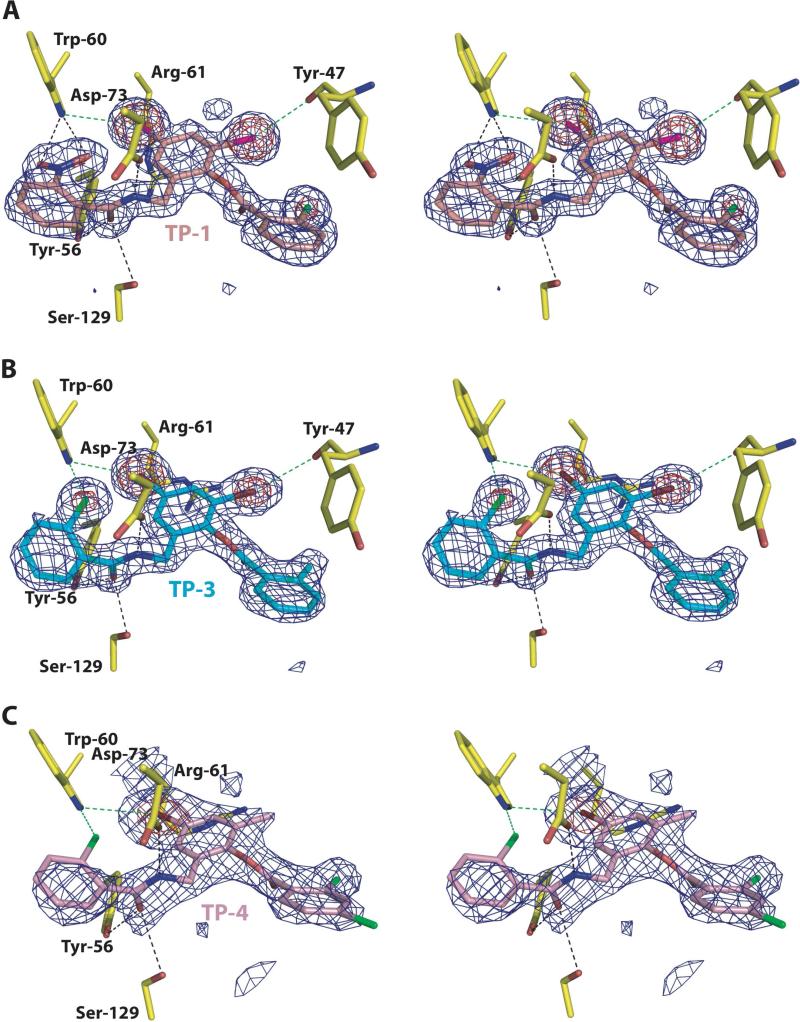
The structure of this binary complex was determined to 1.8 Å resolution by molecular replacement using the final refined coordinates of LasR from the acyl homoserine lactone complex structure. The structure of LasR is essentially identical to that of the acyl homoserine lactone complex with a root mean square deviation (RMSD) of 0.6 Å for main chain and 1.2Å Å for all atoms. Difference Fourier calculations show unambiguous electron density for the tri-phenyl mimic molecule at the ligand-binding site (Figure 3A).
Figure 3. Close-up view of the LasR-triphenyl mimic complexes.
Stereo-view of electron density maps calculated using Fourier coefficients Fobs – Fcalc with phases derived from the final refined model of a LasR-triphenyl mimic complex minus the coordinates of the ligand prior to one round of crystallographic refinement. (A) For the Lasr-TP-1 complex, the maps are contoured at 4σ (blue mesh) and 10σ (red mesh). The final refined coordinates are superimposed with the protein residues colored in yellow and the TP-1 ligand molecule colored in purple. (B) For the LasR-TP-3 complex, the maps are contoured at 4σ (blue mesh) and 10σ (red mesh). The final refined coordinates are superimposed with the protein residues colored in yellow and the TP-3 ligand molecule colored in light blue. (C) For the Lasr-TP-1 complex, the maps are contoured at 2.8σ (blue mesh) and 8σ (red mesh). The final refined coordinates are superimposed with the protein residues colored in yellow and the TP-4 ligand molecule colored in pink. Note the electron dense features that identify the location of the halogen substituents on each of the triphenyl scaffolds. Black dashed lines indicate hydrogen bond interactions and green dashed lines indicate halogen bond interactions.
The high quality of the electron density maps at this resolution, and the strong electron-rich feature of the chlorine atom suggest a slight inaccuracy in the previously reported chemical structure and that the chlorine atom on the first ring and the nitro functional group on the third ring should be transposed (Figure 1) (Muh et al., 2006a).
Within the co-crystal structure, the TP-1 mimic participates in interactions with LasR similar to those engaged by the autoinducer. The four residues that participate in hydrogen bond interactions with the polar head group of the autoinducer similarly engage atoms near the first aromatic ring of the TP-1 mimic: Tyr-56-O17 (2.72 Å), Trp-60-O18 (3.07 Å), Trp-60-O17 (3.24 Å), Asp-73-N8 (2.70 Å), and Ser-129-O17 (3.06 Å) (shown as black dashed lines in Figure 3A). The geometry and stereochemistry of these hydrogen bond interactions are similar to those observed in the interactions with the autoinducer. The first aromatic ring participates in face-to-face π-stacking interaction with Trp-88 and an edge-to-face interaction with Phe-101 (Burley and Petsko, 1985).
The second and third aromatic rings of the TP-1 mimic occupy a location similar to that of the long chain hydrocarbon of the autoinducer. In order to accommodate the meta bromine substituents on the second ring, Arg-61, which faces towards the ligand-binding pocket in the autoinducer co-crystal structure, is displaced out towards bulk solvent. Each of the bromine atoms participate in halogen bonds that fix the orientation of the molecule and interact with the carbonyl oxygen of Tyr-47 (3.52 Å) and Nε1 of Trp-60 (3.69 Å) (shown as green dashes lines in Figure 3A). Despite the slightly larger than expected distances, these halogen bonds associated with an aromatic compound are expected to stronger than those with aliphatic molecules (Auffinger et al., 2004). In addition, the ortho location of an oxygen atom to one of the bromines is expected to have a further polarizing effect on the halogen. The second aromatic ring of the TP-1 mimic also participates in a face-to-face π-stacking interaction with the side chain of Tyr-64, which is further enhanced by the additive, dispersive effects of the multiple bromine substitutions on the π electrons of the ring (Ringer et al., 2006). A modest movement of the Tyr-64 in the TP-1 co-crystal structure results in a better alignment of the two conjugate aromatic systems to maximize the stacking interactions. Lastly, the third aromatic ring of the TP-1 mimic packs in the ligand-binding pocket in an orientation similar to that observed for the trajectory of the final four carbons of the hydrocarbon tail of the autoinducer. The planarity of this third ring results in a more compact packing arrangement within the pocket. Consequently, the side chain of Tyr-47 is shifted about 3 Å towards the pocket when TP-1 is bound, relative to its position with bound autoinducer.
Co-crystal Structure of the LasR-TP-3 Complex
The 1.8 Å resolution co-crystal structure of the LasR-triphenyl mimic 3 (TP-3) complex was similarly determined by molecular replacement. Difference Fourier calculations show unambiguous electron density for TP-3 and the orientation of the ligand was further confirmed by the electron rich features of the halogen atoms (Figure 3). The interactions between LasR and TP-3 are similar to those observed for TP-1, consistent with the similar properties of this molecule as an autoinducer mimic. The position of the second and third aromatic rings of the triphenyl mimics in the ligand-binding pocket aligns well, and TP-3 utilizes similar hydrophobic and π-stacking interactions in the interaction with LasR.
The interactions between the first aromatic of TP-3 and LasR are similar to those observed for TP-1, except that a chlorine atom replaces the nitro group substituent. This results in a loss of one of the two hydrogen bonds between the nitro oxygen and a torsional movement of this ring in order to maximize a halogen bond between the chlorine atom and Nε1 of Trp-60 (3.24 Å). This movement compromises the favorable π-stacking interactions observed between the first ring of TP-1 and Phe-101 and Trp-88. This loss of a favorable hydrogen bond and π-stacking interactions in the LasR-TP-3 structure is consistent with the higher IC50 value and lower activation effect of this triphenyl mimic relative to that for TP-1 (Muh et al., 2006a).
Co-crystal Structure of the LasR-TP-4 Complex
Unambiguous density corresponding to triphenyl mimic 4 (TP-4) could be observed in electron density maps calculated using difference Fourier coefficients on the 2.3 Å resolution structure of the LasR-TP-4 complex. Despite the lower quality of the electron density map at this resolution, strong electron density on one of the substituents of the second aromatic ring (at least 10 σ above background) is consistent with a bromine atom and suggest another minor inaccuracy in the previously reported chemical structure (Figure 1) (Muh et al., 2006a). The orientation of the three aromatic rings of TP-4 is similar to those observed for each of the other triphenyl mimics in the co-crystal structures reported here. As with the TP-3 ligand, the chlorine substituent on the first aromatic ring results in a loss of a hydrogen bond and compromises the π-stacking interactions with Phe-101 and Trp-88. Additionally, the loss of the two halogens on the second aromatic ring weakens the dispersive effects on the π electrons, resulting in weaker face-to-face π-stacking interaction with the side chain of Tyr-64 (Burley and Petsko, 1985). The cumulative effects of each of these alterations results in a weaker interaction of LasR with TP-4, consistent with the highest IC50 value and lowest activation effect for this mimic relative to the other two under investigation here (Muh et al., 2006a).
DISCUSSION
Despite the limited sequence identity between these molecules, each of the four quorum sensing signaling receptors (LasR, TraR, LuxR and RhlR) recognize similar cognate ligands, each of which have identical head groups but differing chain lengths (Fuqua and Greenberg, 2002; Fuqua et al., 2001). Residues involved in intermolecular hydrogen bond interactions with the head group of the autoinducer are conserved, providing a molecular rationale for the conservation of this interaction in spite of the low sequence similarity between these receptors (Bottomley et al., 2007; Vannini et al., 2002; Zhang et al., 2002). For each of these receptors, the length of the acyl chain dictates the specificity for interaction with the autoinducer. For each ligand/receptor pair, specificity for the cognate ligand is regulated by the size and hydrophobicity of the acyl chain-binding pocket, rather than by detailed side chain interactions. In the case of LasR, the long hydrocarbon acyl chain is accommodated into a correspondingly larger pocket that is then capped off by the L3 loop to shield this pocket from bulk solvent. Small molecule compounds mimics of the N-3-oxo-dodecanoyl homoserine lactone autoinducer must be able to both meet the hydrogen bonding requirements with Tyr-56. Trp-60, Asp-73 and Ser-129 and satisfy the constraints of the acyl chain-binding pocket to induce the proper configuration of the L3 loop. Each of the tri-phenyl compounds under study here can meet both of these requirements, making them suitable mimics of the autoinducer, in spite of the lack of overall chemical similarities.
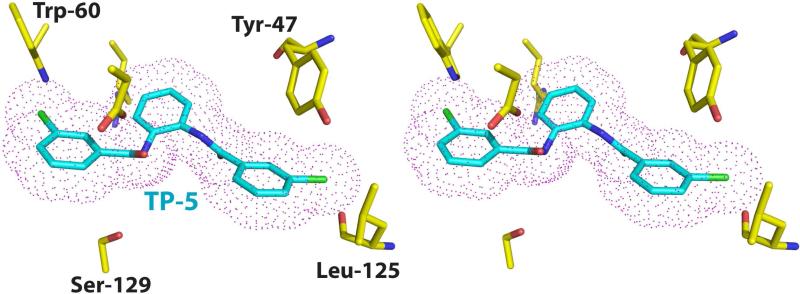
Among the several tri-phenyl mimics identified and characterized, TP-5 is of particular interest as, despite the similarities to the other tri-phenyl compounds, this molecule functions as an antagonist rather than an agonist of LasR function (Muh et al., 2006a). Elucidation of the mechanism for this agonist to antagonist switch should provide a valuable framework for the development of effective inhibitors of LasR. Unfortunately, we have not been able to determine the structure of the LasR-TP-5 due to instability of this complex, resulting in LasR aggregation. The TP-5 ligand is easily dissociated from the receptor, despite persistent efforts to ensure a stoichiometric excess of the ligand during cell growth and in all solution used in protein purification. In spite of a lack of structural data for the LasR-TP-5 complex, the similarity in chemical structures between TP-5 and TP-3 allows modeling of the LasR-TP-5 complex based on our experimentally determined structure of the LasR-TP-3 complex. Manual docking of the TP-5 ligand based on the position of TP-3 reveals that the chlorine atom on the third aromatic ring of TP-5 would result in a steric clash with the main chain carbonyl of Leu-125, as the chlorine-oxygen distance of 2.2 Å is shorter than the sum of the van der Waal's radii for the two atoms. Consequently, the β sheet that harbors Leu-125 would be pushed away from the ligand-binding pocket, resulting in an aberrant conformation of LasR. Disruption of the native conformation of the LasR receptor may account for the inhibitory activity of TP-5, and may explain the instability of this complex in our experiments. However, in the absence of structural data, we can not rule out the possibility that TP-5 may bind to LasR in an entirely different manner from that observed for the other triphenyl mimics.
The triphenyl mimics have been demonstrated to be specific for LasR and show no cross-reactivity to other other structural similar quorum sensing receptors, such as QscR or TraR (Muh et al., 2006a). A superposition of the crystal structure of TraR with the LasR-TP-1 structure provides a molecular rationale for this specificity. While the triphenyl mimics can be suitably accommodated into the ligand binding site of LasR, steric clashes between TraR residues Leu-40, Ile-42, and Ile-47, and the third ring of the mimic would preclude binding of any triphenyl compound. Notably, these residues are all located in the L3 loop. The uncharacteristically longer L3 loop in LasR allows for the accommodation of the third ring of the triphenyl compounds, whereas the shorter loops of TraR (and QscR) would clash with this group and preclude effective binding.
The efficacy of the different triphenyl mimics to activate LasR quorum-sensing controlled reporter genes correlates directly with the ability of these compounds to maintain hydrogen bonding interactions observed in the LasR-N-3-oxo-dodecanoyl homoserine lactone complex. Notably, comparison of the co-crystal structures of LasR bound to its cognate ligand against those bound to triphenyl mimics suggests a critical role for hydrogen bonding interactions with Trp-60. For example, the triphenyl mimics TP-1 and TP-3 make near identical interactions with the receptor but the two hydrogen bonds between Trp-60 and the nitro group on TP-1 are replaced with s single halogen bond with a chlorine atom located at the equivalent position on TP-3. The fact that quorum activation with the TP-3 ligand is lower than that with TP-1 may be a consequence of the weaker donor-acceptor interactions between the triphenyl mimic and Trp-60 in the receptor. In our modeled structure of the LasR-TP-5 complex, the halogen bond between the chlorine atom on TP-5 and Trp-60 has poor stereochemistry, and would be expected to be even weaker than for TP-3. This may explain, in part, why TP-5 is an inhibitor, rather than an activator, of LasR signaling.
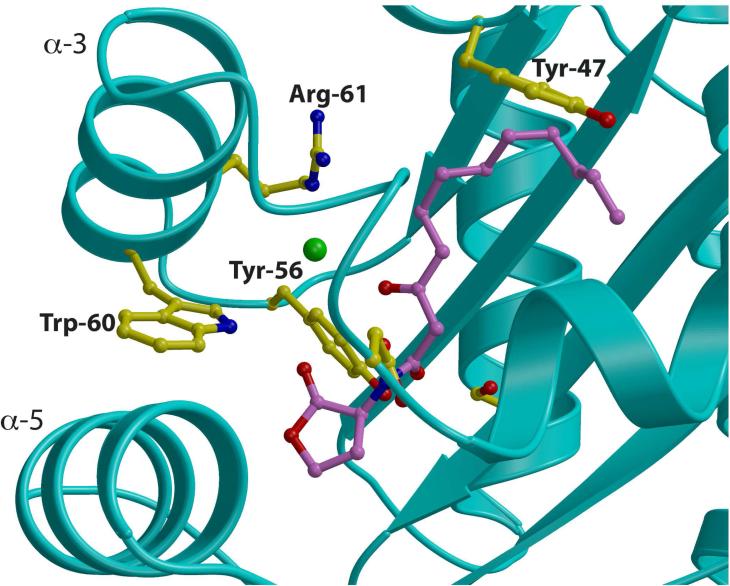
Within the LasR receptor ligand binding domain, Trp-60 is situated on helix α3 and serves to buttress helix α5 through van der Waals contact with Phe-101, Ala-108 and Leu-110 on α5 (Figure 5). While there are no significant conformational differences near Trp-60 between the various triphenyl mimic, the correlation between hydrogen bonding through Trp60 and quorum activation/inhibition suggests that the local structure around Trp-60 may be involved in the process of transcription activation, perhaps through contacts with RNA polymerase or other transcriptional factor. Additional efforts are needed to elucidate the mechanism of differential activation, with the goal of developing efficient inhibitors of LasR that show low toxicity.
Figure 5. Role of Trp-60 in Receptor Stability.
Close-up view of the ligand-binding site in the LasR-N-3-oxo-dodecanoyl homoserine lactone structure showing the orientation of Trp-60 relative to helices α3 and α5. Residues that participate in ligand binding or direct the trajectory of the ligand are shown as yellow ball-and-stick and the homoserine lactone is shown as purple ball-and-stick.
SIGNIFICANCE
Pseudomonas aeruginosa is an opportunistic pathogen that kill several thousand of individuals each year and accounts for nearly 10% of all hospital-acquired infections. Treatment of P. aeruginosa infections with traditional antibiotics is limited by an intrinsic resistance to a large range of antibiotics and by its facile acquisition of additional resistance determinants. As disruption of the quorum sensing signaling pathway is shown to attenuate the virulence of P. aeruginosa, significant research efforts have focused on the identification of compounds that target quorum sensing. However, synthetic chemical efforts to design mimics based on this structural scaffold have been limited by the high specificity of the LasR signaling receptor for its cognate autoinducer. High-throughput screening efforts have identified a novel class of structurally distinct compounds that can interact specifically and potently with LasR receptor to modulate quorum sensing (Muh et al., 2006a). The crystal structures of the LasR ligand-binding domain bound to its cognate autoinducer, and to each of the triphenyl mimics TP-1, TP-3, and TP-4 provide the molecular bases for understanding how the chemically distinct triphenyl compounds can be accommodated by a highly selective receptor. These studies establish a framework for further development of quorum sensing modulators based on the triphenyl scaffold.
EXPERIMENTAL PROCEDURES
Materials
Genomic DNA for Pseudomonas aeruginosa was purchased from the American Tissue Culture Collection. The N-3-oxo-dodecanoyl homoserine lactone was purchased from Cayman Chemicals (Ann Arbor, Michigan) and the triphenyl mimics were purchased from ChemBridge Corporation (San Diego, CA).
Cloning, protein expression and purification
Full details are presented in the Supplemental Methods section.
Crystallization and Data Collection
As attempts to produce co-crystals of full-length LasR complex bound to various synthetic oligonucleotides encompassing the target DNA binding sequence were unsuccessful, we focused on co-crystallization efforts on the ligand binding domain in the presence of cognate ligand or mimics. For crystallization of the LasR-N-3-oxo-dodecanoyl acyl homoserine lactone, LasR-TP-1 and LasR-TP-3 complexes, 1.5 μl of protein (8 mg/ml) was mixed with 1.5 μl precipitant solution containing 80 mM calcium acetate, 40 mM HEPES pH=7.3, 3mM dithiothreitol, and 16% polyethylene glycol 4,000. For crystallization of the LasR-TP-4 complex, 1.5 μl of protein (8 mg/ml) was mixed with 1.5 μl precipitant solution containing 100 mM ammonium sulfate, 50 mM MES pH=5.6, 3mM dithiothreitol, and 20% polyethylene glycol 8,000. The mixture drop was equilibrated over a well containing the same precipitant solution at either 8o or 20oC and crystals reached their maximum size after 2-4 days. Crystals of each complex were stepwise equilibrated with the crystallization media supplemented with incremental concentrations of ethylene glycol, up to a final concentration of 30%, prior to vitrification in liquid nitrogen.
Structure Determination and Refinement
Data from each crystal was collected to their respective resolution limit, utilizing a Mar 300 CCD detector (LS-CAT, Sector 21 ID-D, Advanced Photon Source, Argonne, IL). Data were indexed and scaled using the HKL2000 package (Otwinowski et al., 2003). The structures of each complex were determined by molecular replacement (McCoy, 2007), using the refined coordinates of the previously reported 1.8 Å resolution structure of the LasR ligand binding domain-N-3-oxo-dodecanoyl acyl homoserine lactone complex (Bottomley et al., 2007) without any water molecules or bound ligands. For each structure, iterative model building was carried out was using XtalView (McRee, 1999) and further improved by rounds of refinement with REFMAC5 (Murshudov et al., 1997; Murshudov et al., 1999). Cross-validation used 5% of the data in the calculation of the free R factor (Kleywegt and Brunger, 1996). The appropriate ligand was built into difference Fourier electron density maps after the free R factor for the protein model fell below 30%. Solvent molecules were added towards the latter stages of model building and refinement. For each of the structures, stereochemistry of the model was monitored throughout the course of refinement using PROCHECK (Laskowski et al., 1996). Data collection and refinement statistics are given in Table 1. The refined coordinates have been deposited in the PDB with identification numbers XX1 (LasR- N-3-oxo-dodecanoyl acyl homoserine lactone), XX2 (LasR-TP-1), XX3 (LasR-TP-3), XX4 (LasR-TP-4).
Table 1.
Data collection, phasing and refinement statistics
| LasR-3OC12 | LasR-TP1 | LasR-TP3 | LasR-TP4 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | P21 | C2 |
| Cell dimensions | ||||
| a, b, c (Å) | 54.4, 67.6, 54.3 | 54.2, 84.5, 156.4 | 54.2, 85.2, 76.6 | 153.1, 82.4, 90.5 |
| α, β, γ (°) | 90.0, 114.9, 90.0 | 90.0, 95.9, 90.0 | 90.0, 96.4, 90.0 | 90.0, 126.0, 90.0 |
| Resolution (Å)* | 50-1.4 (1.45-1.4) | 50-1.8 (1.86-1.8) | 50-1.8 (1.86-1.8) | 50-2.3 (2.38-2.3) |
| Rsym (%) | 2.9 (8.5) | 5.4 (19.2) | 3.7 (18.2) | 4.6 (36.5) |
| I /σ (I) | 37.1 (17.7) | 19.6 (4.7) | 27.8 (5.3) | 23.1 (2.4) |
| Completeness (%) | 93.3 (87.1) | 97.9 (91.6) | 97.3 (88.2) | 96.3 (80.4) |
| Redundancy | 4.8 (4.7) | 2.8 (2.7) | 3.2 (2.9) | 3.7 (3.1) |
| Refinement | ||||
| Resolution (Å) | 20.0-1.4 | 25.0-1.8 | 25.0-1.8 | 25.0-2.3 |
| No. reflections | 61,343 | 120,893 | 59,151 | 37,071 |
| Rwork / Rfree | 16.8/20.7 | 19.9/24.7 | 20.3/24.9 | 22.8/28.6 |
| Number of atoms | ||||
| Protein | 2,586 | 10,500 | 5,196 | 6,358 |
| Ligand | 42 | 248 | 116 | 150 |
| Water | 457 | 1,497 | 542 | 83 |
| B-factors | ||||
| Protein | 17.32 | 21.31 | 25.91 | 34.49 |
| Ligand | 13.98 | 18.85 | 26.87 | 58.2 |
| Water | 30.93 | 34.21 | 36.81 | 32.29 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.010 | 0.007 | 0.009 | 0.019 |
| Bond angles (°) | 1.39 | 1.09 | 1.12 | 1.79 |
Highest resolution shell is shown in parenthesis.
Supplementary Material
Figure 4. Model of the LasR-TP-5 interaction.
Stereo-view of a model structure of the LasR-TP-5 complex derived from the experimentally determined structure of the LasR-TP-3 complex. The protein residues are shown in yellow and the TP-5 ligand is shown in cyan. A van der Waals surface for the ligand is superimposed and shown as pink dots. Note the steric clash between the backbone carbonyl of Leu-125 and a chlorine atom on the TP-5 ring.
ACKNOWLEDGEMENTS
We thank Brian Bae for assistance with data collection and Keith Brister and staff at LS-CAT (21-BM at Argonne National Labs, APS) for facilitating data collection. Supported in part by grants to S.K.N. from the NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no competing interests.
REFERENCES
- Auffinger P, Hays FA, Westhof E, Ho PS. Halogen bonds in biological molecules. Proc Natl Acad Sci U S A. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, Givskov M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos Trans R Soc Lond B Biol Sci. 2007;362:1213–1222. doi: 10.1098/rstb.2007.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley MJ, Muraglia E, Bazzo R, Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MI, Prince A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr Opin Pharmacol. 2007;7:244–251. doi: 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Hancock RE. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin Infect Dis. 1998;27(Suppl 1):S93–99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmel IA, Metlitskaia AZ. [Quorum sensing of genes expression--perspective drug target against bacterial pathogenicity]. Mol Biol (Mosk) 2006;40:195–210. [PubMed] [Google Scholar]
- Kiratisin P, Tucker KD, Passador L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J Bacteriol. 2002;184:4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Brunger AT. Checking your imagination: applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- Lambert PA. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95(Suppl 41):22–26. [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Muh U, Hare BJ, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, Greenberg EP. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc Natl Acad Sci U S A. 2006a;103:16948–16952. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006b;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- Ozer EA, Pezzulo A, Shih DM, Chun C, Furlong C, Lusis AJ, Greenberg EP, Zabner J. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol Lett. 2005;253:29–37. doi: 10.1016/j.femsle.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Ringer AL, Sinnokrot MO, Lively RP, Sherrill CD. The effect of multiple substituents on sandwich and T-shaped pi-pi interactions. Chemistry. 2006;12:3821–3828. doi: 10.1002/chem.200501316. [DOI] [PubMed] [Google Scholar]
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci U S A. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Harris SG, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002;184:1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003a;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J Clin Invest. 2003b;112:1460–1465. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. Embo J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Yang F, Wang LH, Wang J, Dong YH, Hu JY, Zhang LH. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 2005;579:3713–3717. doi: 10.1016/j.febslet.2005.05.060. [DOI] [PubMed] [Google Scholar]
- Zhang L, Murphy PJ, Kerr A, Tate ME. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.