Abstract
The recent emergence of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) marked a quantum step change in the biology and epidemiology of a major human pathogen. Various virulence determinants unique to CA-MRSA have been recently uncovered, shedding light on how these strains spread easily and sustainably among humans and frequently cause severe disease. The role of the Panton Valentine leukocidin (PVL) in CA-MRSA pathogenesis is a matter of much debate. While epidemiological data have suggested a role of PVL in the CA-MRSA disease process, recent data from relevant animal models suggest that PVL does not impact virulence of prevalent CA-MRSA strains. Identifying specialized pathogenic traits of CA-MRSA remains a challenge that will yield new diagnostic tools and therapeutic targets for drug and vaccine development.
Emergence of Community-associated Clones
Staphylococcus aureus is a major human pathogen and the leading cause of hospital-associated infections. S. aureus strains have developed resistance to almost all antibiotics, thereby complicating disease management. After first being reported in 1961, methicillin resistant Staphylococcus aureus (MRSA) has become endemic in hospitals worldwide, rendering the entire β-lactam class of antibiotics ineffective. Until recently, feral descendants of hospital-associated MRSA were the most common cause of sporadic disease in the community, frequently infecting persons with a history of recent hospitalization, or close contact with a person who had been hospitalized [3]. In contrast, community-associated MRSA (CA-MRSA) was first described in the late 1990s to cause severe and fatal infections in children without antecedent health care exposure [1, 2]. Since then, epidemiologically unassociated outbreaks of CA-MRSA have been reported throughout the world [6-24] (Box 1). The vast majority of CA-MRSA infections worldwide are caused by strains belonging to only 5 clonal lineages (Figure 1). CA-MRSA lineages differ markedly in genotypic and phenotypic characters from traditional hospital-associated MRSA lineages. Evidently, these five CA-MRSA strains are fit and well-adapted for dissemination in the community (Box 1).
Box 1. Community-associated Clones of MRSA.
Five CA-MRSA strains account for the vast majority of CA-MRSA disease worldwide.
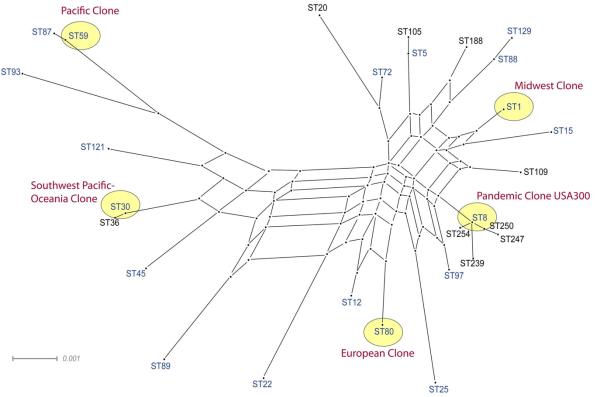
The first widely recognized CA-MRSA strain is commonly known as the Midwest Clone, coined after the region in the United States where it emerged [1, 2]. This clonal lineage, however, was prevalent in communities in Western Australia earlier [4]. According to multi-locus sequence typing (MLST) analysis, the Midwest Clone belongs to the sequence type ST1 clonal lineage (Figure 1) [5].
The second CA-MRSA strain—ST30 clonal lineage—is known as the Southwest Pacific/Oceania Clone, as it was implicated in localized community outbreaks in Australia, Greece, Mexico and United States [7-11].
The third CA-MRSA strain—ST80 clonal lineage—is known as the European Clone for causing endemic disease in many European communities [8, 14].
The fourth CA-MRSA strain—ST59 clonal lineage—is known as the Pacific Clone as it is endemic in the United States, Taiwan, and Vietnam [11-13].
The fifth CA-MRSA strain—ST8 clonal lineage—is better known as USA300, after the name of its unique pulsed-field gel electrophoresis profile [28]. Not seen before the year 2000, USA300 is now pandemic in communities across 38 U.S. states, Canada, and 9 European countries [11]. The pandemic clone USA300 has been implicated in unusually severe human diseases, including endocarditis, pneumonia, sepsis, and necrotizing fasciitis [16, 17, 29]. Remarkably, the introduction of USA300 into a new geographic area has often been associated with the displacement of locally endemic CA-MRSA strains belonging to ST1, ST30, ST59 and ST80 clonal lineages [22, 23, 32, 33]. In many locales, USA300 alone accounts for more than 50% of all disease caused by the entire S. aureus species [18], which points to the unique capacity of this clone to spread easily and sustainably among humans.
Figure 1.
Genetic diversity among S. aureus strains that have independently acquired type IV SCCmec (blue). The five predominant CA-MRSA clones (red) all have the lukPV gene operon encoding Panton Valentine leukocidin. The graph was constructed with Splitstree (version 4.8) using concatenated sequences of seven housekeeping gene fragments used in MLST for S aureus. The scale bar represents the evolutionary distance (number of substitutions per nucleotide position).
Here, we discuss recent advances in our understanding of CA-MRSA pathogenesis, focusing on the specialized pathogenic traits of CA-MRSA that are not typically found in traditional hospital-associated MRSA strains and methicillin susceptible S. aureus. Although numerous adhesins, toxins, and other important components of the staphylococcal background genome contribute to the disease process, a broad discussion of staphylococcal pathogenesis is beyond the scope of this review.
Unique Genomic Contents
Based on clonal analysis (Figure 1), two prominent CA-MRSA strains from the United States were selected for further characterization to elucidate the molecular basis for inter-strain variations in epidemiologic characters, disease frequencies and disease severity. Genome sequencing of two CA-MRSA strains, the Midwest Clone MW2 (USA400) and the pandemic clone USA300 [7, 9-11, 25, 26], revealed that ~20% of the unique genomic contents of CA-MRSA strains are due to the horizontal acquisition of multiple mobile genetic elements, including prophages and pathogenicity islands, which are absent from the traditional hospital-associated MRSA strains COL, N315 and MRSA252 (Figure 2). The prophages and pathogenicity islands contained numerous specialized pathogenicity factors, such as enterotoxins and exoproteins that could allow CA-MRSA to evade or subvert host defenses [25, 26]. The MW2 strain, for example, contained 18 toxins not typically found in traditional hospital-associated MRSA strains [25]. Of these, superantigenic enterotoxin H, which is reported to have the highest binding affinity to major histocompatibility complex class II molecules ever measured for a staphylococcal enterotoxins [27], is uniquely found in MW2 and not other CA-MRSA strains or hospital-associated MRSA strains [6, 14].
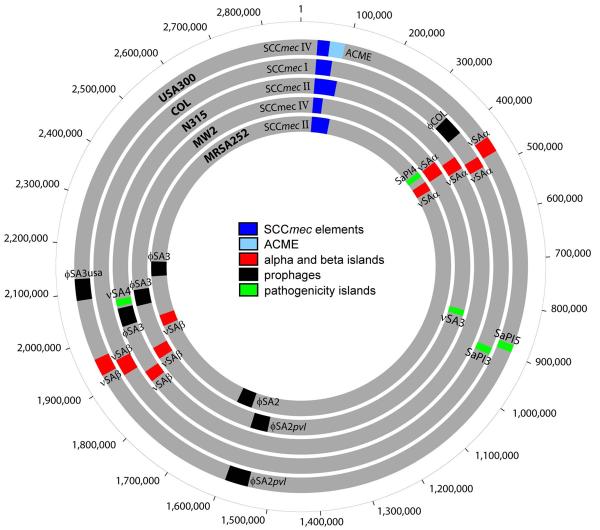
Figure 2.
Interstrain differences in gene content among methicillin resistant Staphylococcus aureus. Concentric circles represent the circular chromosomes of USA300, COL, N315, MW2, and MRSA252. The core chromosome, consisting of genes shared by all strains of S. aureus, is shown in gray. The accessory chromosome, consisting of genes acquired through horizontal acquisition of mobile genetic elements, includes the different allotypes of SCCmec (blue), ACME (light blue), pathogenicity islands (red or green), and prophages (black).
MW2 and USA300 have two mobile genetic elements in common: (1) the prophage ϕSA2pvl encoding the cytolytic toxin, Panton-Valentine leukocidin (PVL), and (2) the type IV staphylococcal chromosome cassette mec (SCCmec) element conferring resistance to the entire class of β-lactam antibiotics. Although PVL is found in sporadic CA-MRSA and methicillin susceptible S. aureus strains, it is epidemiologically linked only to the 5 predominant CA-MRSA clonal lineages widely associated with large disease outbreaks (see Figure 1). In comparison, the type IV SCCmec is present not only in the 5 predominant CA-MRSA clonal lineages but also almost all CA-MRSA strains causing sporadic disease in the community (Figure 1) [28, 30, 31]. The type IV SCCmec found in CA-MRSA strains is smaller in size than type I-III SCCmec elements found in traditional nosocomial strains (Figure 2), which could explain its extensive horizontal transfer among S. aureus lineages to generate the broad diversity of CA-MRSA strains (Figure 1). In addition, the genome sequence of the pandemic clone USA300 revealed the horizontal acquisition of the type I arginine catabolic mobile element (ACME) from the ubiquitous skin commensal S. epidermidis [26]. Type I ACME has been postulated to contribute to growth and survival of USA300 within the host, and promote colonization of the human skin [26]. ACME is integrated downstream of type IV SCCmec (Figure 2), utilizing the same cassette chromosome recombinases A and B (ccrAB) contained with SCCmec for mobilization and transfer [15-24, 34]. Type I ACME is a distinctive genetic feature of the USA300 pandemic clone [26, 34, 35], although ACME-like elements have been detected in sporadic MRSA [32-34].
Is PVL the major virulence determinant of CA-MRSA?
The epidemiological association between genetically diverse S. aureus strains carrying the pvl genes and fatal necrotizing pneumonia has renewed and intensified interest in understanding the biological role of PVL [36]. A PubMed search for articles on PVL published in 2002-2007 identified more than 250 articles noting an association between PVL and CA-MRSA disease outbreaks. Although compelling, epidemiological data alone is insufficient to establish whether PVL directly contributes to widespread dissemination of CA-MRSA clones [37, 38]. Of note, PVL-deficient variants of the predominant CA-MRSA strains also cause endemic community-associated disease [39]. For example, PVL-positive ST1 strains (MW Clone) are prevalent in the United States, but PVL-negative ST1 was earlier frequent in communities in Western Autralia [4]. Similarly, whereas PVL-positive ST59 are endemic in Taiwan and Vietnam, the vast majority of ST59 strains in the United States do not carry the PVL-harboring prophage [11-13, 29]. Despite the prevalence of PVL-deficient CA-MRSA strains, PVL is widely regarded as the primary virulence determinant driving the epidemic spread of the major CA-MRSA clones worldwide [14, 40, 41]. We summarize here our current understanding of the biochemistry, molecular biology, and general function of PVL in CA-MRSA pathogenesis.
PVL and its effects as purified toxin
PVL belongs to a family of similar bi-component leukocidal toxins (synergohymenotropic toxins) produced by staphylococci, with one component belonging to the F and one to the S class [42]. PVL is encoded by the lukPV operon encoding the LukF-PV and LukS-PV components on the ϕSLT, ϕPVL, ϕSA2MW, or ϕSA2usa phages [26, 42]. These β-barrel structured toxins form pores almost exclusively in leukocytes, via a mechanism similar to that used by α-hemolysin (α-toxin) [43]. Notably, α-toxin does not lyse neutrophils and the molecular underpinnings of the target cell specificity of these structurally very similar toxins are not completely understood [44]. Interestingly, early reports have concluded that PVL alone is not a very toxic substance [45], as intravenous injection of PVL in rabbits resulted in granulocytopenia followed by a marked granulocytosis, but was not lethal. Purified PVL also caused dermonecrotic lesions in rabbits [46], contributing to the hypothesis that this toxin plays a role in skin and soft tissue infections [42]. Recently, PVL has been shown to cause lethal necrotic lesions in the lungs of mice via nasal instillation of purified toxin [47].
Isogenic PVL+/PVL− CA-MRSA strains in abscess and sepsis models
The effects of deleting the lukPV operon in MW2 and USA300 genetic backgrounds were tested in a mouse abscess model, to reproduce the most common clinical disease associated with PVL-positive CA-MRSA disease [18]. No significant difference in weight loss, abscess size, or bacteria colony forming units per abscess were detected when using Crl:SKH1-hrBR hairless and BALB/c mice to compare the following isogenic strain pairs: 1) MW2Δpvl mutant, in which lukPV operon was deleted by allelic replacement with a spectinomycin resistance cassette, and the parental strain MW2; and 2) LACΔpvl mutant and the parental strain LAC (Los Angeles County), which is a prototypical USA300 strain [48, 49]. As bacteremia accounted for 65% of invasive CA-MRSA disease [24], the same isogenic strain pairs were compared in a CD1 Swiss mouse bacteremia model but no differences in lethality were observed, indicating that PVL is dispensable for CA-MRSA pathogenesis [48]. As rabbit and human PMNs are 10-100 times more sensitive than murine PMNs to the leukolytic effects of purified PVL [50, 51], a rabbit model of bacteremia was used to test the potential effects of PVL. Using both co-infection and single-strain infection designs in 117 outbred rabbits, no difference in lung, spleen or kidney infectivity were detected using the same MW2 and LAC isogenic strain pairs [52]. As the same mouse and rabbit models have been used successfully to demonstrate the role of other CA-MRSA virulence factors [53, 54], the null effect of PVL in these models strongly indicates that this toxin is not a major virulence factor of MW2 and USA300 strains.
PVL in experimental pneumonia: an exception?
Pneumonia is a rare disease caused by CA-MRSA [17, 24]. Using purified toxin or a laboratory strain of S. aureus that overproduced PVL via a multicopy plasmid, PVL was shown to impact mouse survival in a model of pneumonia using BALB/c mice [47]. It is of interest that when comparing isogenic S. aureus strains lysogenized with either wild-type ϕSLT or mutated ϕSLT in which the lukPV operon was deleted, no difference in mouse survival was found [47], indicating that PVL does not exhibit a lethal effect when expressed from a single transgene copy. In contrast, using isogenic Δpvl mutants in the MW2 and USA300 backgrounds, and when over-expressing PVL in S. aureus strain Newman, no significant contribution of PVL to lethal pneumonia was found using C57/black and BALB/c mice [49, 55]. Passive immunization with anti-PVL immune sera also failed to protect mice against challenge with USA300 in the murine pneumonia model [56], indicating that PVL is not necessary for the pathogenesis of pulmonary disease.
Isogenic PVL+/PVL− CA-MRSA strains in the interaction with human neutrophils
Neutrophils are the main human leukocyte type to eliminate invading bacteria and the main cellular target of PVL [42]. As animal models can only partially reproduce how pathogens cause disease in humans, the null effect of PVL in the mouse and rabbit models may be due to a specific interaction of PVL with human neutrophils. Surprisingly, culture supernatants prepared from MW2 and USA300 clinical strains and their Δpvl isogenic mutants did not exhibit differences in capacity for pore formation or lysis of human neutrophils, even though PVL was secreted in abundance into the supernatants of the parental strains [48]. Thus, despite a detailed understanding of the molecular mechanistic basis of the leukolytic activities of purified PVL [57], the relevance of this toxin in the context of host-pathogen interactions remains unclear, particularly because CA-MRSA strains also secrete other exotoxins with much more potent leukolytic activities than PVL (see the section on PSMs below).
Does PVL have a gene regulatory effect?
Recently, a pronounced global gene regulatory effect has been ascribed to PVL [47], with the regulatory changes reminiscent of disrupting the accessory gene regulator agr [58]. For example, expression of PVL in a laboratory S. aureus strain background lysogenized with ϕSLT appeared to result in a very strong up-regulation of protein A compared to the isogenic strain lysogenized with a mutated ϕSLT in which the lukPV operon was deleted, suggesting that PVL regulates the expression of protein A via an unknown mechanism [47]. Protein A not only plays a classical role in immune evasion by binding to the Fc part of human IgG but also causes inflammatory effects in mouse lungs by interacting with TNFR1 on airway epithelial cells [59]. The interrelated effects of PVL and protein A were thought to be central in causing the overwhelming inflammation and necrosis of the mouse lungs [47]. However, misinterpretation of the microarray data due to the apparent lack of confirmatory experiments might have led to the model in which PVL plays a role in global gene regulation [47]. The possibility of spontaneous mutations or polar effects could have been ruled out by genetic complementation analysis, which however was not done for these microarray experiments [47]. Further transcriptional analysis using the SH1000 strain (a laboratory strain deficient in the rsbU component of the alternative sigma factor sigB, complemented with a functional rsbU copy) did not provide appropriate external validation, as it was compared to a strain that was not isogenic, containing both the lukPV operon and the rest of the phage ϕSLT [47]. In contrast, no effects of PVL on global gene regulation and protein expression were detected in three independent CA-MRSA strains belonging to MW2 and USA300 backgrounds and their Δpvl isogenic mutants (Diep et al., unpublished). While the reasons underlying these extreme differences will need to be investigated further, the failure to replicate the regulatory effects of PVL in clinically relevant CA-MRSA strains suggests that unintended genetic perturbations—not ruled out by appropriate controls—may have caused the dramatic gene regulatory differences reported in that study [47].
Taken together, it is clear that PVL has no impact on CA-MRSA pathogenesis when the lukPV operon is expressed via its own promoter at single gene copy. Only purified PVL at high concentrations or over-expression of the lukPV operon via a multicopy plasmid may cause lung inflammation and necrosis. Notwithstanding the compelling epidemiological linkage between PVL and CA-MRSA strains, is there anything overlooked when analyzing the contribution of PVL to the pathogenesis of CA-MRSA? For example, PVL may be produced in abundance in the human host, perhaps in the context of antibiotic-induced bacterial SOS response that could result in prophage induction and enhanced expression of phage-encoded PVL [60, 61]. Also, PVL may harm human cells other than leukocytes in a yet undiscovered interaction, which in this case would have to be human-specific, as mouse and rabbit infection models clearly do not show a PVL effect. Future research on the biological relevance of PVL is cautiously warranted. Nonetheless, attributing enhanced CA-MRSA pathogenicity to PVL alone ignores the possible contributions of numerous other genetic determinants unique to these epidemic strains (Figure 2) [25, 26, 53, 62].
The linkage of SCCmec and ACME
MRSA strains contain distinct allotypes of the SCCmec element. In the absence of antibiotic selection pressure, SCCmec is thought to reduce the biological fitness of MRSA. The type IV SCCmec element found in CA-MRSA strains is smaller in size than the type I-III SCCmec elements found in hospital-associated MRSA strains. Thus, it may impose only a slight cost to fitness, as it does not encode resistance determinants other than mecA. When comparing isogenic strains with different allotypes of the SCCmec element, type I SCCmec but not type IV SCCmec was found to impose a fitness cost to the bacterial host in terms of decreased in vitro growth rate and cell yield per mole of ATP consumed [63]. A highly discriminating rabbit co-infection model was used to further demonstrate that a USA300 clinical strain and its isogenic mutant with precise deletion of the type IV SCCmec exhibited no difference in infectivity of vital rabbit organs [34]. Taken together, these data strongly indicate that type IV SCCmec does not engender a biological fitness cost. Accordingly, stable populations of CA-MRSA strains have emerged worldwide by transfer of type IV SCCmec into genetically diverse S. aureus strains (Figure 1) [5, 64]. In contrast, the burden that other SCCmec allotypes (type I-III) impose on bacterial fitness (i.e. decreased in vitro growth rate [63]) has likely restricted traditional hospital-associated strains bearing these elements from spreading into the community. Hospital-associated MRSA seems to depend critically on high rates of antibiotic use in hospitals to overcome the fitness burden associated with the presence of antibiotic resistance elements.
Type IV SCCmec is physically linked to type I ACME in the pandemic clone USA300 (Figure 2), suggesting that selection for antibiotic resistance and pathogenicity may be interconnected [34]. Precise deletion of ACME in a USA300 clinical isolate resulted in attenuated pathogenicity or fitness when compared to the parental strain in the rabbit co-infection model, thereby providing evidence that ACME contributes to pathogenesis [34]. Two gene clusters identified in ACME, arc and opp-3, could confer the phenotypes of interest. As L-arginine is a substrate for nitric oxide production, depletion of L-arginine by arginine deiminase system (arc) could inhibit nitric oxide production, a molecule used in both the innate and adaptive immune responses against bacterial infections [65]. L-arginine catabolism could also be important for ATP production and pH homeostasis on the acidic human skin (pH 4·2–5·9) [66]. opp-3 belongs to the ABC transporter family, members of which have a wide variety of functions, including peptide nutrient uptake, quorum sensing, pheromone transport, chemotaxis, eukaryotic cell adhesion, binding of serum components, and resistance to antimicrobial peptides. Thus, in community settings largely devoid of antibiotic selection pressure, ACME may enhance the growth, survival and dissemination of USA300, thus allowing for the genetic “hitchhiking” of SCCmec. In turn, SCCmec protects against exposure to β-lactam antibiotics, further enhancing rapid dissemination of USA300 into health care settings with high rates of antibiotic use.
The SCCmec-ACME linkage alone, however, is likely not sufficient to bestow the epidemic properties of USA300. USA300 variants lacking the SCCmec-ACME composite island are also prevalent among methicillin susceptible S. aureus causing sporadic disease in the United States [18, 67], suggesting a substantial contribution of the genomic background to the epidemic properties of USA300 [26, 29]. It is also of interest that ACME-like elements have been found in MRSA isolates in association with other SCCmec elements, including type IV SCCmec in ST1, type II SCCmec in ST5 and ST59, and type V SCCmec in ST97 strains [32-34]. The associated SCCmec allotypes II, IV, and V, contained genes encoding CcrAB recombinases essential for mobilization and integration of ACME into a chromosomal attachment site found at the terminus of SCCmec [34, 68]. ACME-like elements found in ST5 and ST59 strains differed significantly in gene content from the type I ACME found in USA300, including the lack of the opp-3 operon that could contribute to virulence [34]. Although it appears that these ACME-like elements are found in MRSA strains causing sporadic disease, it remains to be determined whether they contribute to bacterial pathogenesis or fitness.
Phenol-soluble modulins and other toxins
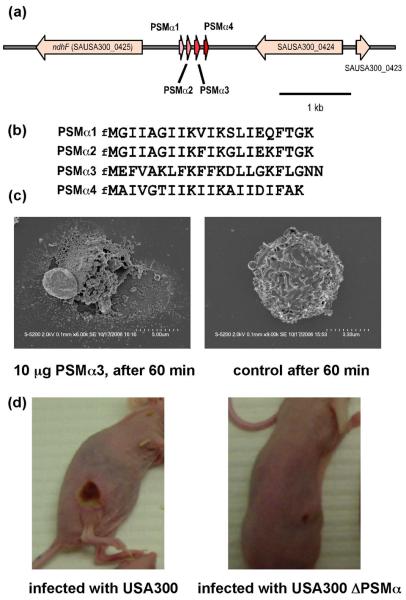
While S. aureus produces many molecules with the capacity to lyse human leukocytes, it has remained obscure which molecules are mainly responsible for neutrophil lysis in vivo and the pronounced lytic activity seen in CA-MRSA. Recently, novel cytolytic peptides have been found in S. aureus, the α-type phenol-soluble modulins (PSMs), which are encoded in an operon on the genomes of all sequenced S. aureus strains (Figure 3a). The α-helical and amphipathic α-type PSMs are secreted in a yet undiscovered fashion as the primary translation products carrying an N-terminal N-formyl methionine (Figure 3b). They have pronounced in vitro and in vivo leukocidal (Figure 3c), in addition to pro-inflammatory and chemotactic activities [53]. However, whereas hospital-associated MRSA often lack PSM production or produce PSMs only at reduced levels, PSMs are expressed at considerable levels in CA-MRSA. Notably, over-expression of α-type PSMs in a hospital-associated MRSA strain caused leukocidal activity equal to that observed in CA-MRSA strains, indicating that expression of these peptides is the main cause for the extreme difference in lytic activity between CA-MRSA and hospital-associated MRSA and a possible major contributor to the pronounced pathogenic potential of CA-MRSA strains. In fact, a dramatic influence of the α-type PSMs on the infectivity of CA-MRSA was demonstrated using the bacteremia and abscess mouse infection models (Figure 3d) [53].
Figure 3.
The α-type phenol-soluble modulins (PSMs). (a), arrangement of the α-type PSM genes in the S. aureus genome (shown for strain USA300). (b), amino acid sequences of α-type PSMs. (c), α-type PSMs cause neutrophil lysis. Representative images of neutrophils incubated with PSMα3 (10 μg) or buffer after 60 min of incubation. (d), α-type PSMs are crucial for CA-MRSA skin and soft tissue infection. Representative images of dermonecrosis formed by USA300 wild-type and isogenic PSMα operon deletion strains.
The results obtained with the PSMs indicate that we will have to consider differential gene expression between CA-MRSA and HA-MRSA strains to explain differences in virulence. For example, it is known that α-hemolysin, a strong pro-inflammatory and pore-forming toxin, may differ considerably in expression among different strains. Although α-hemolysin does not lyse neutrophils, it has recently been shown to have a key impact on the outcome of necrotizing pneumonia by CA-MRSA [56, 69]. Finally, as these toxins and the PSMs are under control of the global regulator agr and there is strong expression of the agr regulatory molecule RNAIII in CA-MRSA strains, whereas hospital-associated MRSA strains are often agr-negative, there may be a key role of this virulence regulator in CA-MRSA disease that remains to be investigated [53].
Concluding Remarks
In recent years perhaps no infectious disease problem has generated as much interest as the epidemic spread of CA-MRSA strains. In 2005, 18,650 deaths were estimated in patients with invasive MRSA disease in the United States, which was comparable to the number of deaths attributed to HIV/AIDS [24, 70]. The pandemic clone USA300 has contributed more than any other strain to the dramatic increase in MRSA disease because it is unusually virulent and especially well-adapted for transmission in the community. A frequently overlooked attribute of USA300 pathogenicity is its enhanced transmission between susceptible hosts. The precise mechanism involved in transmission is poorly understood, although specific bacterial factors, such as ACME, may facilitate colonization of new niches within the host [26]. Recent studies further indicate that the molecular basis of CA-MRSA pathogenicity resides not solely in the horizontal acquisition of novel pathogenicity determinants but also in the differential regulation of effector molecules encoded by all S. aureus genomes. Specifically, the α-type PSMs exemplify the importance of understanding the regulatory mechanisms underlying increased expression of virulence determinants in CA-MRSA strains [53]. The current state of knowledge regarding CA-MRSA pathogenesis is summarized in Figure 4. It should be noted that the prototypical strains MW2 and USA300 have been used exclusively for studies of CA-MRSA pathogenesis, although the proposed virulence mechanisms may not be present or active in other CA-MRSA strains. The specific pathogenic traits of other prevalent CA-MRSA strains, including ST30, ST59 and ST80, will need to be identified and their relative contributions to the disease process elucidated in order to prioritize targets for drug and vaccine development.
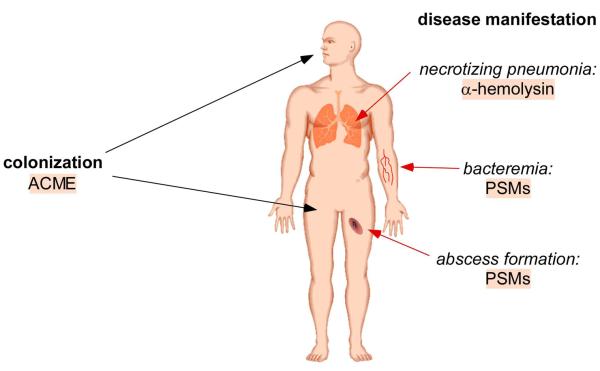
Figure 4.
Molecular basis of CA-MRSA infection. The exceptional success of CA-MRSA strains in the community is likely due to enhanced virulence and colonization. In addition to an enhanced capacity for nasal carriage [11], CA-MRSA strains are also commonly carried on the skin, thereby facilitating skin-to-skin spread and even sexual transmission [71-75]. Using isogenic mutant strains in CA-MRSA, 3 factors have a proven impact on CA-MRSA pathogenesis, as determined in animal models. PSMs (α-type) are crucial for abscess formation and bacteremia, the most widely found manifestations of CA-MRSA disease. α-toxin has a dramatic impact on the development of necrotizing pneumonia, one of the rarer and more dramatic diseases CA-MRSA may cause. Whether PSMs have a role in necrotizing pneumonia, or α-hemolysin in abscess formation and bacteremia of CA-MRSA, has not been tested yet. Both factors are widely found in S. aureus strains. However, CA-MRSA strains may differ from HA-MRSA strains in the expression of these factors, as shown for PSMs. The mobile genetic element ACME is thought to enhance colonization and survival on the skin and is only found in the USA300 background. USA300 CA-MRSA strains with ACME outcompete isogenic strains without ACME in a rabbit infection model.
REFERENCES
- 1.Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. Jama. 1998;279:593–8. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 2.CDC Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus: Minnesota and North Dakota, 1997-1999. MMWR Morb Mortal Wkly rep. 1999;37:2658–2662. [PubMed] [Google Scholar]
- 3.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178–82. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 5.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. Jama. 2003;290:2976–84. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 7.Nimmo GR, Schooneveldt J, O'Kane G, McCall B, Vickery A. Community acquisition of gentamicin-sensitive methicillin-resistant Staphylococcus aureus in southeast Queensland, Australia. J Clin Microbiol. 2000;38:3926–31. doi: 10.1128/jcm.38.11.3926-3931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aires de Sousa M, Bartzavali C, Spiliopoulou I, Sanches IS, Crisostomo MI, de Lencastre H. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J Clin Microbiol. 2003;41:2027–32. doi: 10.1128/JCM.41.5.2027-2032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson DA, Kearns AM, Holmes A, et al. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet. 2005;365:1256–8. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- 10.Velazquez-Meza ME, Aires de Sousa M, Echaniz-Aviles G, et al. Surveillance of methicillin-resistant Staphylococcus aureus in a pediatric hospital in Mexico City during a 7-year period (1997 to 2003): clonal evolution and impact of infection control. J Clin Microbiol. 2004;42:3877–80. doi: 10.1128/JCM.42.8.3877-3880.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan ES, Diep BA, Charlebois ED, et al. Population Dynamics of Nasal Strains of Methicillin-Resistant Staphylococcus aureus—and Their Relation to Community-Associated Disease Activity. J Infect Dis. 2005;192:811–818. doi: 10.1086/432072. [DOI] [PubMed] [Google Scholar]
- 12.Huang YC, Su LH, Wu TL, Lin TY. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream isolates from a teaching hospital in Northern Taiwan. J Clin Microbiol. 2006;44:2268–70. doi: 10.1128/JCM.00776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang CT, Nguyen DT, Ngo TH, et al. An outbreak of severe infections with community-acquired MRSA carrying the Panton-Valentine leukocidin following vaccination. PLoS ONE. 2007;2:e822. doi: 10.1371/journal.pone.0000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandenesch F, Naimi T, Enright MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–84. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazakova SV, Hageman JC, Matava M, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med. 2005;352:468–75. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- 16.Miller LG, Perdreau-Remington F, Rieg G, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–53. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 17.Francis JS, Doherty MC, Lopatin U, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–7. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 18.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 19.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med. 2006;144:309–17. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert M, MacDonald J, Gregson D, et al. Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness or incarceration. Cmaj. 2006;175:149–54. doi: 10.1503/cmaj.051565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hota B, Ellenbogen C, Hayden MK, Aroutcheva A, Rice TW, Weinstein RA. Community-Associated Methicillin-Resistant Staphylococcus aureus Skin and Soft Tissue Infections at a Public Hospital: Do Public Housing and Incarceration Amplify Transmission? Arch Intern Med. 2007;167:1026–33. doi: 10.1001/archinte.167.10.1026. [DOI] [PubMed] [Google Scholar]
- 22.Pan ES, Diep BA, Charlebois ED, et al. Population Dynamics of Nasal Strains of Methicillin-Resistant Staphylococcus aureus--and Their Relation to Community-Associated Disease Activity. J Infect Dis. 2005;192:811–8. doi: 10.1086/432072. [DOI] [PubMed] [Google Scholar]
- 23.Larsen A, Stegger M, Goering R, Sorum M, Skov R. Emergence and dissemination of the methicillin resistant Staphylococcus aureus USA300 clone in Denmark (2000-2005) Euro Surveill. 2007;12 [Google Scholar]
- 24.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 25.Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 26.Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson H, Bjork P, Dohlsten M, Antonsson P. Staphylococcal enterotoxin H displays unique MHC class II-binding properties. J Immunol. 1999;163:6686–93. [PubMed] [Google Scholar]
- 28.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-Field Gel Electrophoresis Typing of Oxacillin-Resistant Staphylococcus aureus Isolates from the United States: Establishing a National Database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2006;193:1495–503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 30.Okuma K, Iwakawa K, Turnidge JD, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–94. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coombs GW, Pearson JC, O'Brien FG, Murray RJ, Grubb WB, Christiansen KJ. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg Infect Dis. 2006;12:241–7. doi: 10.3201/eid1202.050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC. Epidemiologic Distribution of the Arginine Catabolic Mobile Element (ACME) Among Selected Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Isolates. J Clin Microbiol. 2007 doi: 10.1128/JCM.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellington MJ, Yearwood L, Ganner M, East C, Kearns AM. Distribution of the ACME-arcA gene among methicillin-resistant Staphylococcus aureus from England and Wales. J Antimicrob Chemother. 2008;61:73–7. doi: 10.1093/jac/dkm422. [DOI] [PubMed] [Google Scholar]
- 34.Diep BA, Stone GG, Basuino L, et al. The ACME and SCCmec Linkage: Convergence of Virulence and Resistance in the USA300 Clone of Methicillin Resistant Staphylococcus aureus. J Infect Dis. 2007 doi: 10.1086/587907. accepted for publication. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy AD, Otto M, Braughton KR, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1327–32. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillet Y, Issartel B, Vanhems P, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–9. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 37.Chambers HF. Community-associated MRSA--resistance and virulence converge. N Engl J Med. 2005;352:1485–7. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- 38.Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O'Connell B, Coleman DC. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol. 2007;45:2554–63. doi: 10.1128/JCM.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, McClure JA, Elsayed S, Tan J, Conly JM. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J Infect Dis. 2008;197:195–204. doi: 10.1086/523763. [DOI] [PubMed] [Google Scholar]
- 40.Tristan A, Ferry T, Durand G, et al. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J Hosp Infect. 2007;65(Suppl 2):105–9. doi: 10.1016/S0195-6701(07)60025-5. [DOI] [PubMed] [Google Scholar]
- 41.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest. 2007;87:3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 42.Szmigielski S, Prevost G, Monteil H, Colin DA, Jeljaszewicz J. Leukocidal toxins of staphylococci. Zentralbl Bakteriol. 1999;289:185–201. doi: 10.1016/s0934-8840(99)80105-4. [DOI] [PubMed] [Google Scholar]
- 43.Joubert O, Voegelin J, Guillet V, et al. Distinction between Pore Assembly by Staphylococcal alpha-Toxin versus Leukotoxins. J Biomed Biotechnol. 2007;2007:25935. doi: 10.1155/2007/25935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valeva A, Walev I, Pinkernell M, et al. Transmembrane beta-barrel of staphylococcal alpha-toxin forms in sensitive but not in resistant cells. Proc Natl Acad Sci U S A. 1997;94:11607–11. doi: 10.1073/pnas.94.21.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodin AM. Staphylococcal leukocidin. In: Montje TC, Kadis S, Ajl SJ, editors. Microbial toxins. Vol. 3. Academic Press Inc.; New York: 1970. pp. 327–355. [Google Scholar]
- 46.Ward PD, Turner WH. Identification of staphylococcal Panton-Valentine leukocidin as a potent dermonecrotic toxin. Infect Immun. 1980;28:393–7. doi: 10.1128/iai.28.2.393-397.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labandeira-Rey M, Couzon F, Boisset S, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–3. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 48.Voyich JM, Otto M, Mathema B, et al. Is Panton-Valentine Leukocidin the Major Virulence Determinant in Community-Associated Methicillin-Resistant Staphylococcus aureus Disease? J Infect Dis. 2006;194:1761–70. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 49.Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. 2008 doi: 10.1086/592053. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szmigielski S, Jeljaszewicz J, Wilczynski J, Korbecki M. Reaction of rabbit leucocytes to staphylococcal (Panton-Valentine) leucocidin in vivo. J Pathol Bacteriol. 1966;91:599–604. doi: 10.1002/path.1700910237. [DOI] [PubMed] [Google Scholar]
- 51.Gladstone GP, van Heyningen WE. Staphylococcal leukocidins. Br J Exp Pathol. 1957;38:123–37. [PMC free article] [PubMed] [Google Scholar]
- 52.Diep BA, Palazzolo-Ballance AM, Basuino L, et al. Panton-Valentine leukocidin does not impact virulence of community-associated MRSA. 2008 submitted. [Google Scholar]
- 53.Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 54.Diep BA, Stone GG, Basuino L, et al. The ACME and SCCmec Linkage: Convergence of Virulence and Resistance in the USA300 Clone of Methicillin Resistant Staphylococcus aureus. J Infect Dis. 2008 doi: 10.1086/587907. in press. [DOI] [PubMed] [Google Scholar]
- 55.Wardenburg JB, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–6. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 56.Wardenburg JB, Schneewind O. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med. 2008;205:287–94. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Genestier AL, Michallet MC, Prevost G, et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest. 2005;115:3117–27. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–49. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 59.Gomez MI, Lee A, Reddy B, et al. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med. 2004;10:842–8. doi: 10.1038/nm1079. [DOI] [PubMed] [Google Scholar]
- 60.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195:202–11. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 61.Dumitrescu O, Boisset S, Badiou C, et al. Effect of antibiotics on Staphylococcus aureus producing Panton-Valentine leukocidin. Antimicrob Agents Chemother. 2007;51:1515–9. doi: 10.1128/AAC.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burlak C, Hammer CH, Robinson MA, et al. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9:1172–90. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bachi B, Cook GM. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother. 2007;51:1497–9. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–34. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moncada S, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med. 1993;329:2002–12. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- 66.Ehlers C, Ivens UI, Moller ML, Senderovitz T, Serup J. Females have lower skin surface pH than men. A study on the surface of gender, forearm site variation, right/left difference and time of the day on the skin surface pH. Skin Res Technol. 2001;7:90–4. doi: 10.1034/j.1600-0846.2001.70206.x. [DOI] [PubMed] [Google Scholar]
- 67.McCaskill ML, Mason EO, Jr., Kaplan SL, Hammerman W, Lamberth LB, Hulten KG. Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections. Pediatr Infect Dis J. 2007;26:1122–7. doi: 10.1097/INF.0b013e31814536e0. [DOI] [PubMed] [Google Scholar]
- 68.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–55. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bubeck Wardenburg J, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–4. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bancroft EA. Antimicrobial resistance: it's not just for hospitals. Jama. 2007;298:1803–4. doi: 10.1001/jama.298.15.1803. [DOI] [PubMed] [Google Scholar]
- 71.Miller LG, Diep BA. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:752–60. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- 72.Lee NE, Taylor MM, Bancroft E, et al. Risk factors for community-associated methicillin-resistant Staphylococcus aureus skin infections among HIV-positive men who have sex with men. Clin Infect Dis. 2005;40:1529–34. doi: 10.1086/429827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cook HA, Furuya EY, Larson E, Vasquez G, Lowy FD. Heterosexual transmission of community-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2007;44:410–3. doi: 10.1086/510681. [DOI] [PubMed] [Google Scholar]
- 74.Cohen AL, Shuler C, McAllister S, et al. Methamphetamine use and methicillin-resistant Staphylococcus aureus skin infections. Emerg Infect Dis. 2007;13:1707–13. doi: 10.3201/eid1311.070148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diep BA, Chambers HF, Graber CJ, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 2008;148:249–57. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]