Summary
Mitochondria are complex organelles whose dysfunction underlies a broad spectrum of human diseases. Identifying all the proteins resident in this organelle and understanding how they integrate into pathways represent major challenges in cell biology. Toward this goal, we performed mass spectrometry, GFP tagging, and machine learning to create a mitochondrial compendium of 1098 genes and their protein expression across 14 mouse tissues. We link poorly characterized proteins in this inventory to known mitochondrial pathways by virtue of shared evolutionary history. Using this approach we predict 19 proteins to be important for the function of complex I (CI) of the electron transport chain. We validate a subset of these predictions using RNAi, including C8orf38, which we further show harbors an inherited mutation in a lethal, infantile CI deficiency. Our results have important implications for understanding CI function and pathogenesis, and more generally, illustrate how our compendium can serve as a foundation for systematic investigations of mitochondria.
Introduction
Mitochondria are dynamic organelles essential for cellular life, death, and differentiation. Although they are best known for ATP production via oxidative phosphorylation (OXPHOS), they house myriad other biochemical pathways and are centers for apoptosis and ion homeostasis. Mitochondrial dysfunction causes over 50 diseases ranging from neonatal fatalities to adult onset neurodegeneration, and is a likely contributor to cancer and type II diabetes (DiMauro and Schon, 2003; Lowell and Shulman, 2005; Wallace, 2005). The 13 proteins encoded by the mitochondrial genome have been known since its sequencing (Anderson et al., 1981) and have been linked to a variety of maternally inherited disorders. However, there may be as many as 1500 nuclear-encoded mitochondrial proteins (Lopez et al., 2000), though less than half have been identified with experimental support. A complete protein inventory for this organelle across tissues would provide a molecular framework for investigating mitochondrial biology and pathogenesis.
Recent progress in defining the mitochondrial proteome has been driven by large-scale approaches, including mass spectrometry (MS) based proteomics in mammals (Forner et al., 2006; Foster et al., 2006; Johnson et al., 2007; Kislinger et al., 2006; Mootha et al., 2003a; Taylor et al., 2003) and yeast (Reinders et al., 2006; Sickmann et al., 2003), epitope tagging combined with microscopy in yeast (Huh et al., 2003; Kumar et al., 2002), and computation (Calvo et al., 2006; Emanuelsson et al., 2000; Guda et al., 2004). However each of these methods suffers from intrinsic technical limitations. MS-based approaches struggle with distinguishing genuine mitochondrial proteins from co-purifying contaminants, and published reports exhibit up to 41% false positive rates (Table S1). Additionally, these approaches tend to miss low abundance proteins or those expressed only in specific tissues or developmental states, and thus capture only 23-40% of known mitochondrial components (Table S1). Other experimental approaches such as epitope tagging are limited by the availability of cDNA clones, tag interference, and over-expression artifacts. While integrative machine-learning methods can be more comprehensive (Calvo et al., 2006; Jansen et al., 2003), they require subsequent experimental validation.
Here, we perform in-depth protein mass spectrometry, microscopy, and machine learning to construct a protein compendium of the mitochondrion. We perform MS-based proteomics on both highly purified and crude mitochondrial preparations to discover genuine mitochondrial proteins and distinguish them from contaminants based on enrichment. We integrate these MS data with six other genome-scale datasets of mitochondrial localization using a Bayesian framework and additionally perform the most extensive GFP tagging study focused on mammalian mitochondria. The resulting compendium consists of 1098 genes (Figure 1) and their protein expression across 14 mouse tissues. Although not complete, this represents the most comprehensive and accurate molecular characterization of the organelle to date.
Figure 1. Building a Compendium of Mitochondrial Proteins.
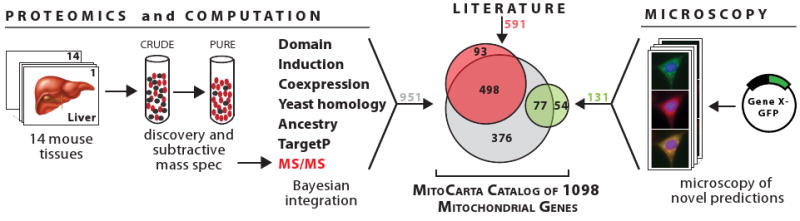
MitoCarta is a compendium of 1098 genes encoding proteins with strong support of mitochondrial localization. Each protein was determined to be mitochondrial by one or more of the following approaches: 1) an integrated analysis of seven genome-scale data sets, including in-depth proteomics of isolated mitochondria from 14 mouse tissues (gray circle), 2) large-scale GFP-tagging/microscopy (green circle), and 3) prior experimental support from focused studies (red circle). The union of genes from each approach comprises the MitoCarta compendium.
Our compendium provides a framework for identifying novel proteins within pathways resident in the mitochondrion. Here, we focus on complex I (CI) of the electron transport chain, a macromolecular structure composed of ∼45 subunits in mammals (Carroll et al., 2006). CI deficiency is the most common cause of rare, respiratory chain diseases (DiMauro and Schon, 2003) and has been implicated in Parkinson's disease (Schapira, 2008). Half of the patients with CI deficiency lack mutations in any known CI subunit, suggesting that yet unidentified genes crucial for maturation, assembly, or stability of CI are mutated in the remaining cases (Janssen et al., 2006). Multiple assembly factors for much smaller complexes IV and V have been identified in S. cerevisiae, and it is estimated that complex IV alone requires over 20 factors (Devenish et al., 2000; Fontanesi et al., 2006). However, the absence of CI in S. cerevisiae has impeded similar studies and, to date, only three CI assembly and maturation factors have been identified (Ogilvie et al., 2005; Saada et al., 2008; Vogel et al., 2005).
To systematically discover proteins essential for CI function, we apply the technique of phylogenetic profiling which uses shared evolutionary history to highlight functionally related proteins (Pellegrini et al., 1999). This approach was recently used to identify the CI assembly factor NDUFA12L using five yeast species (Ogilvie et al., 2005). We apply this approach more broadly to our mitochondrial protein inventory and report that 19 of these proteins share ancestry with a large subset of CI proteins. We validate several of these predictions in cellular models and additionally report that one of these genes, C8orf38, harbors a causative mutation in an inherited CI deficiency.
Together, these studies illustrate the utility of an expanded mitochondrial inventory in advancing basic and disease biology of the organelle. Our compendium, called MitoCarta, is freely available at www.broad.mit.edu/publications/MitoCarta.
Results and Discussion
Discovery and Subtractive Proteomics of Mouse Mitochondria
As a first step toward establishing an experimentally supported inventory of mammalian mitochondrial proteins, we performed protein mass spectrometry on mitochondria from 14 diverse mouse organs (Figure 1). We designed our proteomic experiments in two phases in order to identify as many mitochondrial proteins as possible while systematically flagging co-purifying contaminants. In the discovery phase, we isolated highly purified mitochondria from cerebrum, cerebellum, brainstem, spinal cord, kidney, liver, heart, skeletal muscle, white adipose tissue, stomach, small intestine, large intestine, testis and placenta obtained from healthy C57BL/6 mice. Mitochondrial purity was assessed by western blots against selected mitochondrial and non-mitochondrial proteins, and intactness was verified by polarographic studies (data not shown) and electron microscopy (Figure 2A, S2). Each sample was separated by SDS-PAGE and then sectioned into 20 bands that were each analyzed by high performance, liquid chromatography tandem mass spectrometry (LC-MS/MS) using an LTQ Orbitrap Hybrid MS system. We captured 4.7 million tandem mass spectra and searched them against the mouse RefSeq protein database using stringent matching criteria, resulting in the confident identification of products from 3,881 genes (Table S3). The detected proteins are not biased by molecular weight, isoelectric point, or presence of transmembrane helices, but do show a slight bias against proteins whose transcripts exhibit low abundance (Figure S4). We estimate that we identify 85% of proteins within each sample (based on technical liver replicates), but we saturate detection of distinct proteins by sampling many tissues (Figure 2B). In total, we identify 88% of previously known mitochondrial proteins, including 93% of OXPHOS proteins.
Figure 2. Discovery Proteomics and Subtractive Proteomics of Isolated Mitochondria.
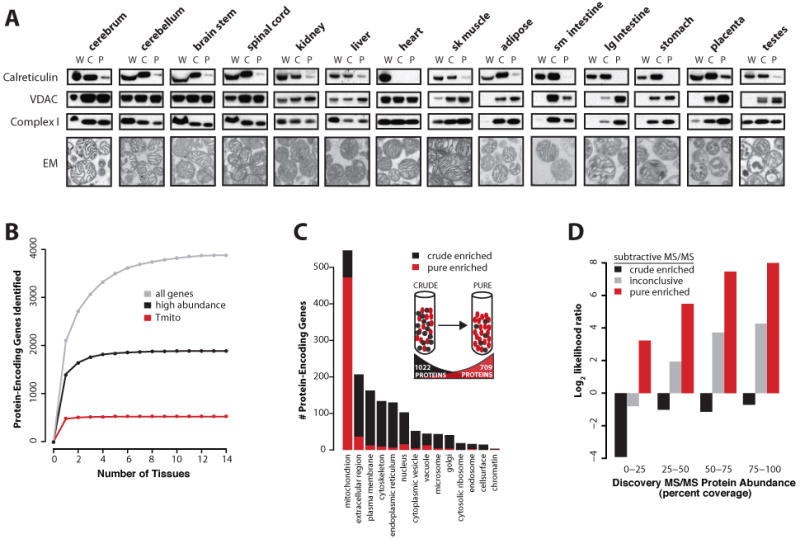
(A) Purification of mitochondria from 14 mouse tissues. Mitochondrial enrichment was tracked by the ratio of an ER protein (calreticulin) to mitochondrial proteins (VDAC and CI 8kDa subunit) at three stages of isolation (W, whole tissue lysate; C, crude mitochondrial extracts; P, purified mitochondrial extracts). Electron micrographs show intactness of the purified organelles.
(B) Saturation of protein identifications by discovery MS/MS is plotted for previously known mitochondrial proteins (Tmito), abundant proteins (>25% coverage), and all proteins.
(C) Gene Ontology annotations of proteins enriched in pure (red) or crude (black) mitochondrial samples based on subtractive MS/MS experiments. Inset: schematic overview of subtractive MS/MS method.
(D) Likelihood ratio of a protein being truly mitochondrial based on detection in discovery and subtractive MS/MS experiments.
In the subtractive proteomics phase, we applied in-solution LC-MS/MS on both crude and purified mitochondria from 10 of the above tissues. This approach is based on the observation that bona fide mitochondrial proteins should become enriched during the purification process, and likewise contaminants should become depleted (e.g., the loss of ER protein calreticulin in Figure 2A). This subtractive method is similar in concept to protein correlation profiling (Foster et al., 2006). Of the 2,565 gene products detected in either crude or pure samples, 1,022 were more abundant in crude samples (crude-enriched), 709 more abundant in purified samples (pure-enriched), and the remainder inconclusive (see Experimental Procedures). The crude-enriched set contained many plasma membrane and extracellular proteins (likely as precursors in the ER) whereas the pure-enriched set was almost exclusively mitochondrial, validating that the subtractive proteomics approach can aid in distinguishing genuine mitochondrial proteins from contaminants (Figure 2C).
We next combined the data from the discovery and subtractive phases in order to assign a probability that each protein detected by discovery MS/MS was truly mitochondrial. To do so, we compiled training sets comprised of 591 known mitochondria genes (Tmito) and 2519 non-mitochondrial genes (T∼mito), listed in Table S5. To avoid circularity, our curated Tmito list excludes mitochondrial proteins characterized solely by prior proteomic studies. Using our training data, we calculated the likelihood ratio that each protein is genuinely mitochondrial based on its discovery MS/MS protein abundance and its subtractive MS/MS enrichment (see Experimental Procedures and Figure S6). As shown in Figure 2D, the likelihood ratio quantifies the confidence that a protein detected by MS/MS is truly mitochondrial.
Integration of Mass Spectrometry Analysis with Genome-Scale Datasets
Our combination of discovery and subtractive proteomics is extremely powerful for discovering bona fide mitochondrial proteins, though this approach alone is not sufficiently sensitive or specific (Figure 3A). For example, these experiments miss proteins that are extremely low in abundance, lack tryptic peptides amenable to MS, or localize to mitochondria only under specific conditions. In order to approach a comprehensive mitochondrial inventory we need to integrate these data with other available information.
Figure 3. Data Integration and Validation by Microscopy.
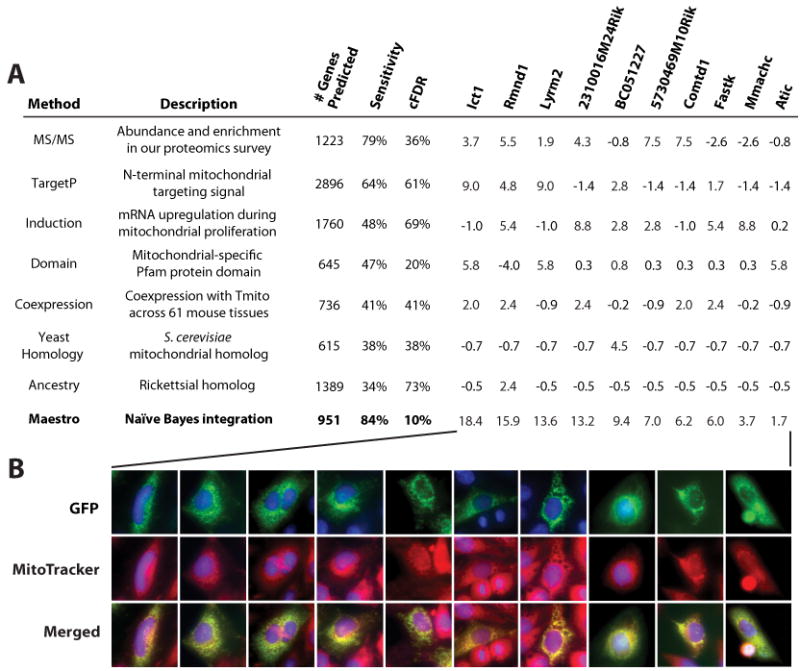
(A) Eight genome-wide methods for predicting mitochondrial localization, with sensitivity and corrected false discovery rates (cFDR) calculated from large training sets at predefined thresholds (Experimental Procedures). Rightmost columns show each method's log-likelihood score for a selection of mouse genes, which are summed to produce the Maestro log-likelihood of mitochondrial localization.
(B) Fluorescence microscopy images of 10 GFP-fusion constructs with clear mitochondrial localization, corresponding to examples in panel A. Images for all 131 constructs showing mitochondrial localization are available at www.broad.mit.edu/publications/MitoCarta.
We therefore combined our MS/MS results with six complementary computational, homology-based, and experimental techniques to determine likelihood of mitochondrial localization (Figure 3A and Experimental Procedures). Using the Maestro naïve Bayes framework we developed previously (Calvo et al., 2006), we used training data to convert each method's data values into log-likelihood scores of mitochondrial localization (Table S7). Since the seven methods are largely conditionally independent (Figure S8), we sum these individual log-likelihood scores into the combined Maestro score based on an independent probability model. Using Maestro, we systematically rank all mouse genes by their likelihood of mitochondrial localization (Table S5). We can assess accuracy at each score using a corrected false discovery rate statistic (cFDR), which accounts for the sizes of our training sets (see Experimental Procedures). At a Maestro score threshold of 4.56, corresponding to 10% cFDR, there are 951 mitochondrial gene predictions including 498/591 known mitochondrial genes (84% sensitivity). This Bayesian integration avoids overfitting the training data, as shown through 10-fold cross-validation (in rotation, training on 90% of the data and reserving 10% for testing) that achieves comparable 82% sensitivity at the same cFDR. As seen in Figure 3A, integration greatly increases prediction accuracy.
Large-Scale GFP-Microscopy of Mitochondrial Localization
We additionally undertook a large-scale microscopy study as a complementary experimental approach to confirm mitochondrial localization (Figure 1). We tested the human orthologs of our mouse predictions due to the availability of high quality clones from the human hORFeome v3.1 collection (Lamesch et al., 2007). We created C-terminus GFP-fusion constructs and visualized subcellular localization in HeLa cells by fluorescence microscopy. This method showed clear mitochondrial localization of 12/21 positive controls and none of 18 negative controls, indicating that this technique is specific but has limited sensitivity. We then tested 470 genes that lacked prior experimental support of mitochondrial localization. These candidates were selected from an interim Maestro analysis and have an estimated 59% cFDR based on our final Bayesian analysis. Of the 404 candidates successfully transfected, we identified 131 genes with clear mitochondrial localization (representatives shown in Figure 3B and the complete set available at www.broad.mit.edu/publications/MitoCarta). The success rate of this approach matches our estimated cFDR and sensitivity rates – thus validating our Bayesian integration. The 273 constructs without clear mitochondrial localization were less informative since it is possible that the GFP tag interfered with mitochondrial import, the wrong splice form was tested, or HeLa cells lacked necessary chaperones/modifiers.
MitoCarta: an Inventory of 1098 Genes Encoding the Mitochondrial Proteome and their Protein Expression across 14 Tissues
Combining our discovery and subtractive proteomics with computation, microscopy and previous literature, we defined a high-confidence mitochondrial compendium of 1098 genes, termed MitoCarta (Figure 1). This inventory is estimated to be over 85% complete and contain ∼10% false positives (see Supplemental Data). It contains 356 genes without previous mitochondrial annotation in Gene Ontology (GO) or MitoP2 (Prokisch et al., 2006) databases, and distinguishes itself from other catalogs by providing strong experimental support for 87% of genes based on: mass spectrometry (70%), GFP studies (12%), and/or literature curation (54%). We conservatively estimate that at least 85 of the MitoCarta proteins are also resident in other cellular locations, based on crossing MitoCarta with two organelle-based proteomic surveys shown in Table S9 (Foster et al., 2006; Kislinger et al., 2006).
The MitoCarta collection includes some notable components and highlights important regulatory features for the organelle. For example, the inventory includes several kinases, phosphatases, RNA-binding proteins and disease-related proteins (MMACHC, ATIC) not previously associated with the mitochondrion (Table S5B). Interestingly, as a collection the MitoCarta genes have significantly shorter UTRs and coding regions, and are more highly expressed, compared to all mouse genes (Figure S10). Their promoters tend to have CpG islands and lack TATA boxes, a feature shared with other “housekeeping” genes that may account for their higher expression (Carninci et al., 2006). Additionally MitoCarta promoters are enriched for the presence of eight conserved sequence motifs, including five known mitochondrial transcription factor binding sites and three novel elements (Figure S10).
In addition to expanding the number of known mitochondrial proteins, our inventory provides the opportunity to assess differences in mitochondrial protein expression across tissues (Figure 4A). We assessed the relative abundance of each MitoCarta protein across our 14 tissues using MS total peak intensity (see Experimental Procedures). This metric is highly reproducible across technical replicates (Figure S11) and correlates quite well with mRNA expression (see Supplemental Data). However, as our atlas contains only a single replicate per tissue, we note two caveats: first, it cannot be used to assess statistically significant differences in abundance across tissues; and second, due to stochastic sampling we estimate that we detect approximately 90% of proteins present in each tissue.
Figure 4. Mitochondrial Protein Expression Across 14 Mouse Tissues.
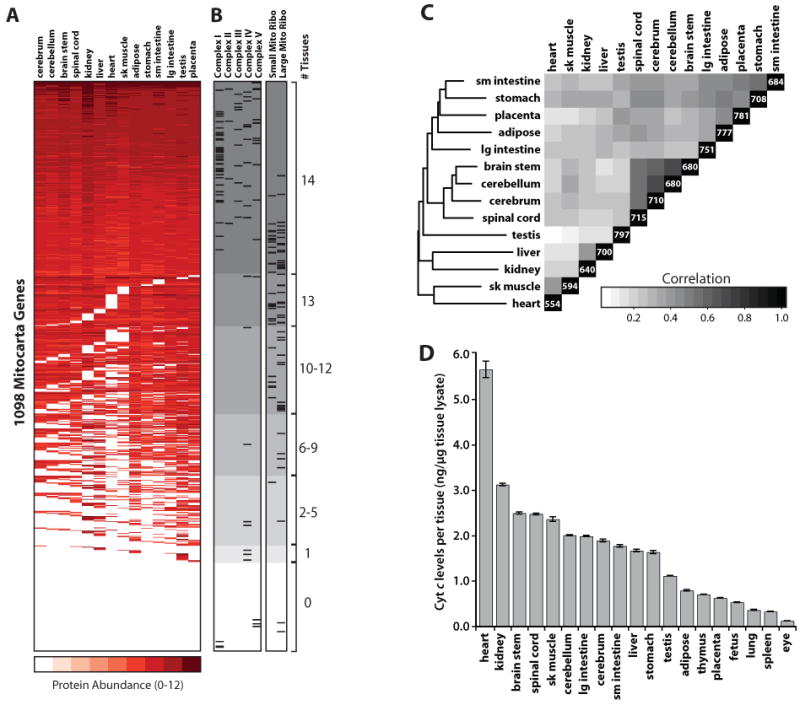
(A) Heatmap of protein abundance, measured by log10 (total MS peak intensity), for 1098 MitoCarta genes across 14 tissues. Genes are ordered by number of tissues and total intensity. White background indicates genes whose protein product was not detected by MS/MS, but are mitochondrial based on prior annotation, computation, or microscopy.
(B) Tissue-distribution of proteins within selected pathways. Tick marks indicate locations of corresponding proteins within (A), and gray shading indicates the total number of tissues in which the protein was detected (0-14).
(C) Correlation matrix of MitoCarta proteins detected by MS/MS in each tissue, clustered hierarchically. Counts on diagonal indicate number of MitoCarta proteins identified by MS/MS.
(D) Mitochondrial quantity per tissue, assessed by ELISA measurements of cytochrome c from whole tissue lysates.
We utilize this protein atlas to investigate the differences in mitochondrial pathways between tissues. We find that approximately 1/3 of MitoCarta genes are core mitochondrial components present across all sampled tissues, including most OXPHOS subunits and the TCA cycle (Figure 4B). However, most MitoCarta genes show some degree of tissue specificity (Figure 4A). Interestingly, these include much of the mitochondrial ribosome and half the subunits of complex IV, several of which have previous verification of tissue-specific expression (Huttemann et al., 2003). Additionally, the enzymes of the ketogenesis and urea cycle pathways are expressed in a broader set of tissues than expected, including brain and placenta (Figure S12). Typically, we find that mitochondria express an average of ∼760 unique gene products per tissue (range 554-797, Figure 4C), with pairs of tissues typically sharing ∼75% of proteins (range 63-88%). Moreover, using a cytochrome c ELISA, we estimate that mitochondrial quantity varies by a remarkable 30-fold amongst a panel of 19 tissues (Figure 4D). Together these analyses reveal the tissue diversity of mitochondrial quantity and composition, and demonstrate how our compendium can serve as a resource for future investigations into tissue-specific mitochondrial biology.
Identifying Complex I Associated Proteins Through Phylogenetic Profiling
The expanded mitochondrial compendium also provides an opportunity to discover novel components for pathways resident in the organelle. Nearly 300 genes—26% of our inventory—have no association with a GO biological process. To associate a subset of these with known pathways, we perform phylogenetic profiling, which uses shared evolutionary history to identify functionally related proteins (Pellegrini et al., 1999). This approach is likely to be particularly applicable to the mitochondrion, given its unique evolutionary history of descending from a Rickettsia-like endosymbiont early in eukaryotic evolution (Andersson et al., 1998).
To explore the utility of phylogenetic profiling for mitochondria, we first identified homologs of mouse MitoCarta proteins in 500 fully sequenced species (Figure 5A, Table S13). We find that 75% of present-day mitochondrial components have clear bacterial ancestry (BlastP expect < 1e-3) and that 57% have bacterial best-bidirectional orthologs, which is more than three-fold higher than that of all mouse proteins (Figure 5C). The phylogenetic profiles confirm that functionally-related mitochondrial proteins tend to have similar evolutionary histories. For example, most proteins involved in fatty acid metabolism, the citric acid cycle, and folate metabolism have ancient origins (Figure 5B). Conversely, the mitochondrial protein import machinery and mitochondrial carriers are more recent innovations (Figure 5B). Thus, it may be possible to use shared evolutionary history to associate unannotated MitoCarta proteins with known pathways.
Figure 5. Ancestry of Mitochondrial Proteins.
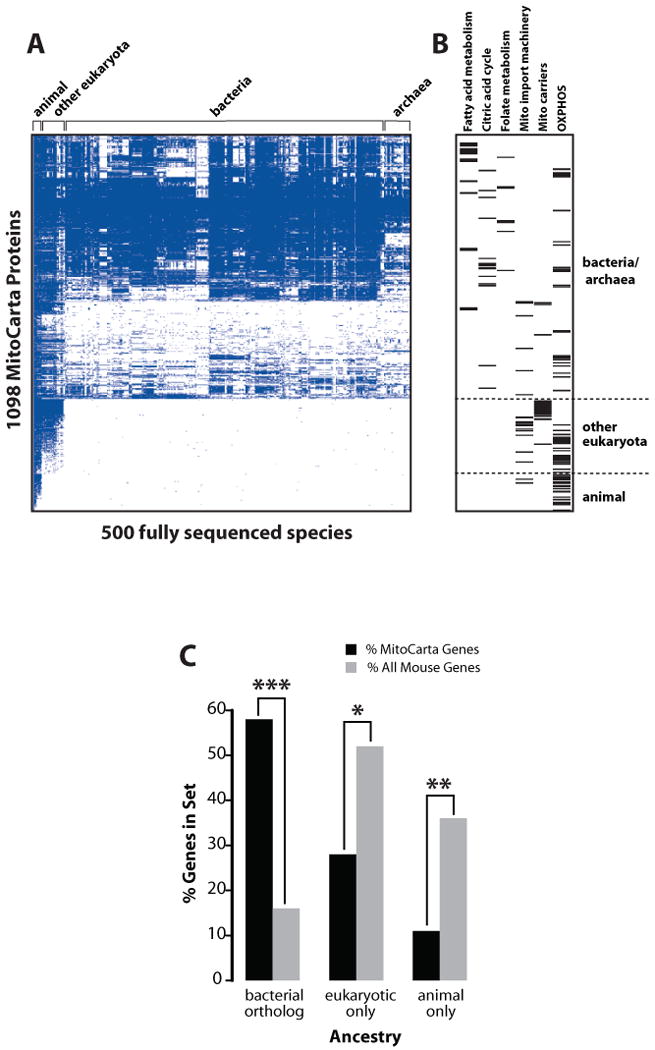
(A) Presence/absence matrix for the 1098 MitoCarta proteins across 500 fully sequenced organisms. Blue squares indicate homology of the mouse protein (row) to a protein within a target species (column).
(B) Ancestry of MitoCarta proteins from selected groups. Tick marks indicate location of proteins within (A).
(C) Comparison of MitoCarta protein ancestry to all mouse proteins, considering only best-bidirectional hits. P values based on hypergeometric distribution with Bonferroni multiple hypothesis correction: *p = 6e-64, **p = 4e-78, ***p = 2e-232.
We focused this strategy on identifying factors essential to respiratory chain complex I (CI) because of its prominent role in energy metabolism and disease. Currently, there are only three known assembly factors for this large, macromolecular complex, though clinical data suggest that there are many unidentified factors needed for its assembly and activity (Janssen et al., 2006). These factors likely reside in the mitochondrion, and thus our MitoCarta compendium aids in prioritizing candidates. Additionally, the evolutionary history of CI across 5 yeast species has recently been proven useful in identifying the assembly factor NDUFA12L, supporting this phylogenetic approach (Gabaldon et al., 2005; Ogilvie et al., 2005).
In order to establish a broader phylogenetic profile for CI, we first built a rooted phylogenetic tree of 42 eukaryotes (Figure 6C, Experimental Procedures). This tree is robust to different phylogenetic reconstruction methods, except for some positioning uncertainty of three deep branching protist species (see Supplemental Data). We observed that a set of 15 CI proteins are not only absent from several yeast species, but are ancestral bacterial subunits that have been independently lost at least four times in eukaryotic evolution (Figure 6A, Table S14). It is probable that the species that lost CI also lost the proteins required for its assembly and function. Only 19 other MitoCarta proteins share this profile and now represent strong candidates for functional association with CI (Figure 6B). These 19 MitoCarta proteins, termed COPP (Complex One Phylogenetic Profile), as well as an expanded set with weaker phylogenetic signatures, are listed in Table S14. The COPP set includes two well-studied proteins involved in branched chain amino acid degradation (Ivd, Mccc2), and four proteins involved in lipid breakdown (Dci, Phyh, Amacr, AF397014), which raises the intriguing hypothesis of an association between these pathways and complex I activity.
Figure 6. Identification of Complex I Associated Proteins through Phylogenetic Profiling.
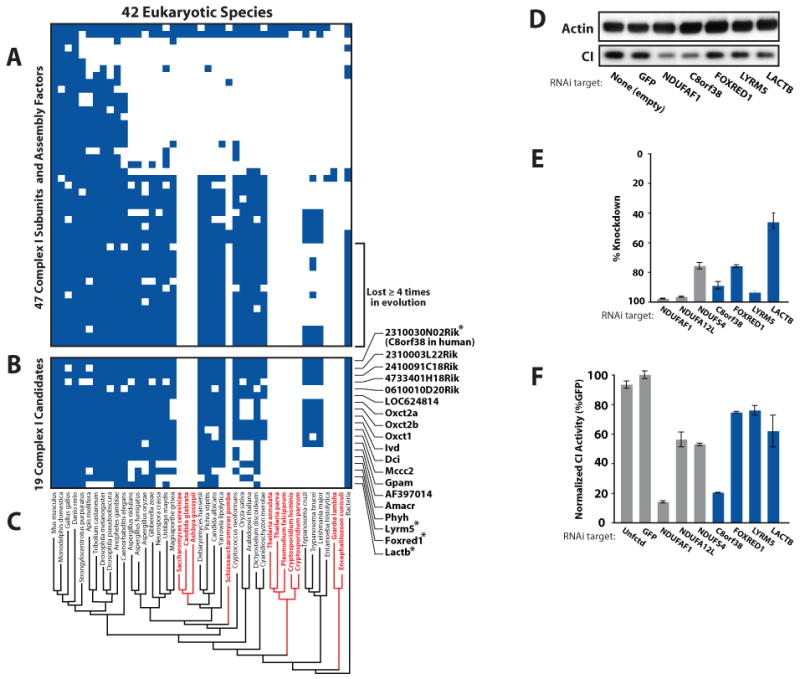
(A) Presence/absence matrix for 44 respiratory chain CI subunits and 3 assembly factors across 42 eukaryotic species. Blue squares indicate homology of the mouse protein (row) to a protein in a target species (column).
(B) MitoCarta proteins matching the phylogenetic profile of the subset of CI subunits lost independently at least four times in evolution. Asterisks indicate candidates tested by RNAi in (D-F).
(C) Reconstructed phylogenetic eukaryotic tree, with red text indicating species that have lost CI.
(D) Effect of candidate knockdown on CI levels in human fibroblasts. Immunoblots of actin and a CI subunit from whole cell lysates were performed following lentiviral-mediated delivery of an empty vector or hairpins targeted against GFP (negative control), NDUFAF1 (known CI assembly factor) and four CI candidates.
(E) Percent knockdown of mRNA expression achieved for controls (gray bars) or CI candidates (blue bars) as measured by real-time qPCR.
(F) CI activity assays from fibroblast lysates (as in D) for controls (gray bars) and four candidates (blue bars). Error bars represent the range of duplicate assays.
We tested four of our COPP genes for an involvement in CI activity by creating stable knockdowns in human fibroblasts using lentiviral-mediated RNAi (Root et al., 2006). Given that we are interested in the clinical relevance of these predictions, we chose to test the human orthologs of our mouse candidates. We achieved ≥ 80% knockdown of 3 COPP genes and 50% knockdown of the fourth, as measured by quantitative real-time PCR (Figure 6E). We next assessed both CI abundance, using immunoblots against a CI subunit, and CI activity, using immunocapture-based activity assays (see Experimental Procedures and Figure S15). Knockdown of C8orf38 showed the strongest reduction of both CI abundance and activity, comparable to the known CI assembly factor NDUFAF1 (Figure 6D-F). These data strongly suggest that C8orf38, which previously had no prior association to any biological process or subcellular location, is crucial for activity and/or assembly of endogenous CI. The other three candidate knockdown lines showed 20-40% reduction of CI activity (Figure 6F) with variable effects on CI abundance (Figure 6D). The moderate reduction of CI activity does not offer definitive evidence of association with CI, however we note that the CI activity assay measures only the NADH dehydrogenase activity, which may still be largely intact even if other modules of CI are improperly assembled. Thus we experimentally validate the importance of a one COPP gene, show suggestive evidence for three other COPP genes, and prioritize more than one dozen additional proteins for future studies of complex I.
A Mutation in C8orf38 Causes an Inherited Complex I Deficiency in Humans
The 19 MitoCarta COPP genes identified above represent strong candidates for genes underlying clinical CI deficiency. We used these candidates in combination with homozygosity mapping to search for a causative gene mutation in two siblings (female and male) with severe isolated CI deficiency, born to first cousin Lebanese parents (Figure 7A). The siblings presented at 10 and 7 months, respectively, with focal right hand seizures, decreased movement and strength, ataxia and evolving rigidity. Both had persistent lactic acidosis and neuroimaging was consistent with Leigh syndrome. The affected girl had isolated CI deficiency in muscle, liver and fibroblasts with normal or elevated activities of other complexes and citrate synthase (Figure 7B). She died at 34 months of age from a cardiorespiratory arrest following admission to hospital with pneumonia. The affected boy had an isolated CI defect confirmed in fibroblasts and is currently 22 months of age.
Figure 7. Discovery of a C8orf38 Mutation in an Inherited Complex I Deficiency.
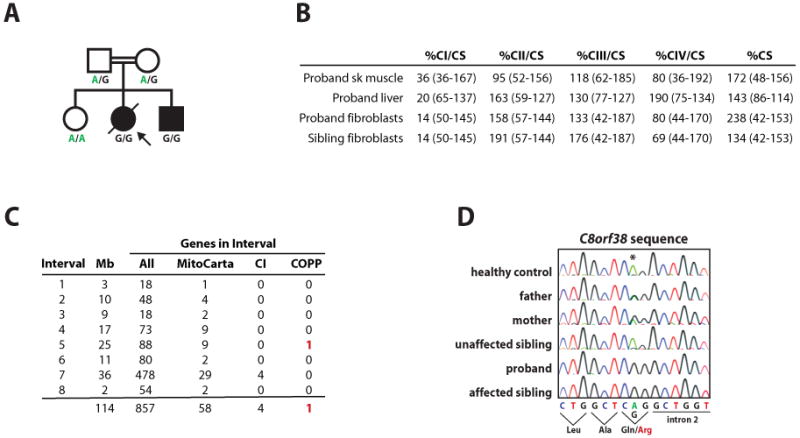
(A) Pedigree from a consanguineous Lebanese family with two children affected by Leigh syndrome and complex I deficiency. Letters beneath each family member represent the genotype for a c.296A>G mutation in C8orf38. Proband indicated by arrow.
(B) Respiratory chain enzyme activities, standardized against the mitochondrial matrix marker enzyme citrate synthase, expressed as percentages of the mean value (normal ranges in parentheses). The final column lists citrate synthase activities (relative to total protein) as % of normal control mean (see Experimental Procedures).
(C) Results of homozygosity mapping using DNA from family members in (A). Eight intervals of homozygosity shared by the affected siblings but not the parents or unaffected sibling are listed along with the number of genes in various categories for each interval (CI, known complex I genes; COPP, Complex One Phylogenetic Profiling candidates).
(D) Sequence traces of C8orf38 from each family member in (A) and one healthy control demonstrating homozygosity for a c.296A>G mutation in both affected siblings.
Since the underlying molecular defect is likely a recessive mutation, we performed homozygosity mapping on DNA isolated from the five family members and identified eight chromosomal regions of homozygosity shared only by the affected siblings (Figure 7C and Experimental Procedures). Collectively, these regions contain 857 genes, including 4 CI structural subunits and one COPP gene: C8orf38 (Figure 7C). Sequencing of two CI structural subunit genes showed no mutations, however sequencing of C8orf38 (NM_152416) revealed a c.296A>G mutation in exon 2 that segregated with the disease in the family (Figure 7D). This mutation causes a predicted Gln99Arg substitution in a residue fully conserved across vertebrates, and may also cause a splicing defect due to its position at the 3′ end of exon 2 (Figure 7D). This mutation was not present in EST databases, SNP databases, or in 100 Lebanese chromosomes tested. The localization of C8orf38 to the mitochondrion, its RNAi phenotype of CI deficiency (Figure 6F), and the segregating C8orf38 mutation at a highly conserved residue together strongly establish that C8orf38 is a human CI disease gene.
Conclusion
We have constructed a high quality compendium of mitochondrial proteins, used comparative genomics to predict roles for unannotated proteins in CI biology, and validated these predictions using cellular models and human genetics. Our inventory of 1098 mitochondrial genes and their protein expression across 14 tissues represents the most comprehensive characterization of the organelle to date and provides a framework for addressing major questions in mitochondrial biology.
We leveraged our compendium to discover proteins essential for proper complex I activity. Despite CI's critical importance in energy production and broad role in rare and common human disease, many aspects of its structure, assembly and activity are poorly understood. Through phylogenetic profiling, we identified 19 additional genes likely to be associated with CI, most notably C8orf38, which we further show is mutated in an inherited CI deficiency. C8orf38 was first shown to be mitochondrial in this study and was not previously associated with any biological function. The domain structure of C8orf38 suggests involvement in phytoene metabolism, potentially implicating it in branched chain lipid metabolism along with other COPP proteins Phyh, Amacr, and AF397014. The remaining COPP genes are now prime candidates for other CI deficiencies, and may help unravel the assembly and maturation program for CI.
In addition to fueling the discoveries we present here, the MitoCarta inventory can be used immediately in other disease related projects. As we have demonstrated in the current report, the mitochondrial compendium can help highlight specific candidates within linkage regions of any Mendelian mitochondrial disease. MitoCarta can also help elucidate the pathogenesis of common degenerative diseases, which have recently been associated with declining mitochondrial gene expression and rising reactive oxygen species production (Houstis et al., 2006; Mootha et al., 2003b; Schon and Manfredi, 2003). Importantly, MitoCarta can also serve as a foundation for basic mitochondrial biology. The orchestrated transcription, translation, and assembly of the mitochondrial components, encoded by two genomes, into functioning, tissue-specific organelles is a remarkable feat about which much remains unknown. Our protein compendium provides a framework with which these tissue-specific programs can be deciphered.
Experimental Procedures
Protein mass spectrometry
Discovery phase
Mitochondria were isolated from C57BL/6 mouse tissues by Percoll density gradient purification (see Supplemental Data for complete details), and assessed for purity with antibodies against Calreticulin (Calbiochem), VDAC1 (Abcam), and an 8 KDa CI subunit (Mitosciences). To further demonstrate purity, a more extensive set of organelle marker antibodies were used for a subset of the mitochondrial preparations (Figure S2). Each sample was size separated by 4-12% bis-Tris gradient SDS-PAGE, separated into 20 gel slices and then reduced, alkylated, and subjected to in-gel tryptic digestion. Extracted peptides from each slice were analyzed by reversed-phase LC-MS/MS using an LTQ-Orbitrap (Thermo Scientific). Data dependent MS/MS were collected in the LTQ for the top ten most intense ions observed in the Orbitrap survey scan, using dynamic exclusion to exclude re-sampling peaks recently selected for tandem MS/MS (within 60s intervals). MS/MS spectra were filtered for spectral quality, pooled from all 14 tissues, and searched against the RefSeq mouse protein database using the Spectrum Mill MS Proteomics Workbench. We required proteins to have ≥2 unique peptides detected, with at least one peptide that distinguished the matching gene from all other mouse Entrez genes. Data were aggregated at the gene level, using the highest MS values for any splice form. Abundance was measured by coverage (percent of amino acids with MS evidence) for cross-protein comparisons, and by total peak intensity (the sum of MS peak areas for all sequence identified peptides matching a protein) for cross-tissue comparisons.
Subtractive phase
Matched crude and highly purified mitochondria were collected from 10 tissues. Sample proteins were reduced, alkylated and then digested with trypsin in-solution. MS/MS spectra were obtained and searched as above, but proteins required only ≥1 peptide spectra, since these results affected only proteins detected via discovery MS/MS. Proteins found only in crude extracts, or found at ≥ twofold higher peak intensity in crude extracts compared to pure were considered crude-enriched (and similarly for pure-enriched).
Data combination
Proteins were assigned integrated MS/MS scores using the likelihood ratio L(d,s)=P(d,s|Tmito)/P(d,s|T∼mito) where d is the discovery MS/MS abundance level (coverage), s is the subtractive MS/MS enrichment category, and Tmito and T∼mito are training sets. See Supplemental Data for complete details.
Mouse and human datasets
Mouse RefSeq Release 20 proteins were mapped to 23,640 NCBI Entrez gene identifiers (ftp.ncbi.nih.gov/gene/DATA/, 12/12/2006), excluding proteins mapped to non-reference assemblies or to pseudogenes (Entrez annotation, 6/21/07). Human-mouse orthologs were obtained from Homologene (ftp.ncbi.nih.gov/pub/HomoloGene, 1/26/2007). Training sets (Table S5) included Tmito: 591 genes with mitochondrial annotations from MitoP2 or Gene Ontology (GO) databases, that were manually curated for experimental evidence of mitochondrial localization in mammals, excluding genes with support solely from large scale proteomics surveys; T∼mito: all 2519 genes with GO subcellular localization annotations (type “inferred by direct assay”), excluding mitochondrial and uninformative categories (Calvo et al., 2006). Protein domains from Pfam (ftp.sanger.ac.uk/pub/databases/Pfam, 11/22/2006) were identified using HMMER (expect parameter=0.1, trusted threshold cutoffs).
Integration of genome-scale data sets
Seven methods for determining mitochondrial localization were integrated using the Maestro naïve Bayes classifier (Calvo et al., 2006). Training sets (Tmito and T∼mito) were used to convert each of the individual feature scores (s1..s7) into a log-likelihood ratio, defined as log2[P(s1..s7| Tmito) / P(s1..s7| T∼mito)]. For transcript or protein level scores, the gene inherited the highest score of any splice form. The scores for the seven genomic features were calculated at predefined ranges (see Table S7) as follows (see Supplemental Data for details):
Proteomics: one of 12 categories shown in Figure 2D, or NA if not detected
Targeting sequence: TargetP v1.1 confidence score (Emanuelsson et al., 2000)
Protein domain: categorical score (M+, M-, M±, NA) representing presence of a protein domain that is exclusively mitochondrial, exclusively non-mitochondrial, ambiguous, or not present in any annotated SwissProt eukaryotic protein.
Yeast homology: 1 if the best S. cerevisiae homolog (BlastP expect < 1e-3, coverage >50% of longer gene) is mitochondrial (Saccharomyces Genome Database, 12/27/06), 0 otherwise
Ancestry: BlastP expect value from R. prowazekii homolog, or NA if expect > 1e-3
Coexpression: N50 score (number of Tmito genes found within the gene's 50 nearest transcriptional co-expression neighbors) within the GNF1M atlas of 61 mouse tissues (Su et al., 2004)
Induction: fold-change of mRNA expression in cellular models of mitochondrial proliferation (overexpression of PGC-1α in mouse myotubes) compared to controls (Calvo et al., 2006; Mootha et al., 2004)
The corrected false discovery rate was used to assess accuracy of predictions since the sizes of the training sets Tmito and T∼mito do not match our prior expectation of the proportion of mitochondrial to non-mitochondrial cellular proteins (Calvo et al., 2006). We define cFDR = (1 − SP) / (1 − SP + SN × Oprior), where TP, TN, FP, FN represent true/false positives and negatives, specificity SP = TN / (TN + FP), sensitivity SN = TP / (TP + FN), and Oprior = 1500/21000.
To compare performance of each method (Figure 3A), we chose the following thresholds: MS/MS pure-enriched, or inconclusive with coverage > 25%; TargetP ≥ 1; Induction ≥ 1.5; Domain M+; Coexpression ≥ 5; Yeast Homology 1; Ancestry ≤ 1e-3; Maestro ≥ 4.56.
Epitope tagging with GFP and microscopy
cDNAs from the Human Orfeome collection (Lamesch et al., 2007) were cloned into the C-terminal GFP vector pcDNA6.2/C-EmGFP-DEST (Invitrogen). Approximately 4 ×103 HeLa cells were seeded in 100 μL of medium (DMEM with 10% FBS, 1× GPS) in 96-well imaging plates (Falcon) 24 h before transfection using Lipofectamine LTX (Invitrogen). 48 h post transfection, cells were stained with medium containing 50 nM MitoTracker Red CMXRos and 1:1000 diluted Hoechst 33258 (Molecular Probes), washed, fixed, and imaged (see Supplemental Data). Mitochondrial localization was determined by overlap of GFP and MitoTracker signals.
Cytochrome c ELISA assays
Fresh mouse tissues were prepared in ice-cold PBS (see Supplemental Data). Following homogenization, tissue lysates were resuspended in PBS containing 0.5% Triton X-100 detergent and protease inhibitors (Roche) and spun at maximum speed in a table top centrifuge set to 4°C for 30 minutes. Supernatant was drawn off, flash frozen in liquid nitrogen and stored at -80°C until use. Cytochrome c levels were measured in duplicate using an ELISA kit (Quantikine) following the manufacturer's protocol.
Phylogenetic profiling
Homologs of mouse proteins within 500 fully sequenced species (Table S13) were defined by BlastP expect < 1e-3. Mouse genes with ≤1 bacterial homologs were called “eukaryotic innovations”. We built a rooted phylogenetic tree of 42 eukaryotic species and a bacterial outgroup (E. coli) using PhyML (Guindon and Gascuel, 2003) (JTT matrix, 4 substitution rate categories) based on ClustalW multiple alignments of 6 well-conserved mouse proteins (Rps16, Ak2, Drg1, Dpm1, Cct7, Psmc3) that were concatenated and manually edited to remove regions of poor alignment. COPP genes were identified using the following profile: absent in 11 species (S. pombe, A. gossypii, C. glabrata, S. cerevisiae, C. hominis, C. parvum, P. falciparum 3D7, T. annulata, T. parva, G. lamblia, E. cuniculi), present in a bacterial genome, present in ≥ 1 plant-like species (A. thaliana, O. sativa, D. discoideum, C. merolae) and present in ≥ 2 other yeasts (Y. lipolytica, C. albicans, P. stipitis, D. hansenii), where presence was defined by BlastP expect < 1e-3. See Supplemental Data and Table S14 for full details.
Complex I abundance and activity assays
Lentiviral vectors (pLKO.1) encoding short hairpin sequences were obtained from the Broad RNAi Consortium (TRC) (Root et al., 2006). These vectors were transfected with a packaging plasmid (pCMV-dR8.91) and VSV-G envelope plasmid (pMD2.G) into 293T cells using Fugene (Roche) following TRC protocols (www.broad.mit.edu/genome_bio/trc/publicProtocols.html). Virus-containing medium was harvested 24 and 48 hours post transfection. Approximately 30,000 MCH58 human fibroblasts were seeded onto 24-well plates the day prior to infection. To infect cells, 150 μl of virus-containing medium mixed with 350 μl of low antibiotic medium containing 8μg/ml polybrene was added to each well and the plate spun at 2250 rpm for 90 minutes at 37°C. Post spin, medium was replaced with DMEM (5% FBS, 1× GPS) for 12-24 hours and then switched to DMEM with 2 μg/ml puromycin for 1-2 weeks for selection of stably infected cells. RNA was extracted from each cell line (Qiagen RNAeasy) and used for 1st strand cDNA synthesis (Invitrogen). Knockdown efficiency was then assessed using real-time PCR (ABI Taqman Assays) using HPRT as an endogenous control. For immunoblot analysis of CI and actin, 10 μg of cleared whole cell lysate was separated on a 4-12% gel (Invitrogen) and transferred to pvdf membrane. Membranes were probed with antibodies against β-actin (Sigma) and an 8kDa CI subunit (Mitosciences). CI activity assays were performed on 15 μg of cell lysate using immunocapture-based assays following the manufacturer's protocol (Mitosciences). Results were scanned using a BioRad GS-800 scanner and analyzed with Quantity One software.
Mitochondrial enzyme assays
Respiratory chain complexes I, II, III and IV plus the mitochondrial marker enzyme citrate synthase were assayed in skeletal muscle and liver homogenates and in enriched fibroblast mitochondrial preparations by spectrophotometric methods as described previously (Kirby et al., 1999; Rahman et al., 1996). Respiratory chain enzyme assays measured NADH:coenzyme Q1 reductase (CI), succinate:coenzyme Q1 reductase (CII), decylbenzylquinol:cytochrome c reductase (CIII) and cytochrome c oxidase (CIV). Enzyme activities were expressed as a ratio relative to citrate synthase and then as a percentage of normal control mean value.
Homozygosity mapping
DNA from five family members was analyzed using Affymetrix GeneChip Mapping 50K XbaI SNP arrays. Loss of heterozygosity regions were detected using Affymetrix software (GDAS v.3.0.2.8, CNAT v.2.0.0.9 and IGB v.4.56).
Supplementary Material
Acknowledgments
We thank J. Jaffe, K. Clauser, P. Matsudaira, for advice; D. Arlow, S. Silver, O. Goldberger, T. Gilbert, and T. Hirozane-Kishikawa for technical assistance; M. McKee for performing electron microscopy; E. A. Shoubridge for providing MCH58 cell lines; and A. Ting, D. Altshuler, and J. Hirschhorn for comments on the manuscript. Electron microscopy was performed in the Microscopy Core of the Center for Systems Biology, which is supported by an Inflammatory Bowel Disease Grant DK43351 and a Boston Area Diabetes and Endocrinology Research Center Award DK57521.This work was supported by a Principal Research Fellowship from the Australian NHMRC awarded to D.R.T., and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences, an Early Career Award from the Howard Hughes Medical Institute, a Charles E. Culpeper Scholarship in Medical Science, and a grant from the National Institutes of Health (GM077465) awarded to V.K.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- Devenish RJ, Prescott M, Roucou X, Nagley P. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim Biophys Acta. 2000;1458:428–442. doi: 10.1016/s0005-2728(00)00092-x. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. The New England journal of medicine. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of molecular biology. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. American journal of physiology. 2006;291:C1129–1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) Journal of molecular biology. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- Guda C, Fahy E, Subramaniam S. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20:1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huttemann M, Jaradat S, Grossman LI. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb--the counterpart to testes-specific cytochrome c? Molecular reproduction and development. 2003;66:8–16. doi: 10.1002/mrd.10327. [DOI] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science (New York, NY. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA. Mitochondrial complex I: structure, function and pathology. Journal of inherited metabolic disease. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Johnson DT, Harris RA, French S, Blair PV, You J, Bemis KG, Wang M, Balaban RS. Tissue heterogeneity of the mammalian mitochondrial proteome. American journal of physiology. 2007;292:C689–697. doi: 10.1152/ajpcell.00108.2006. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology. 1999;52:1255–1264. doi: 10.1212/wnl.52.6.1255. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427–3440. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science (New York, NY. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003a;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003b;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. The Journal of clinical investigation. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokisch H, Andreoli C, Ahting U, Heiss K, Ruepp A, Scharfe C, Meitinger T. MitoP2: the mitochondrial proteome database--now including mouse data. Nucleic Acids Res. 2006;34:D705–711. doi: 10.1093/nar/gkj127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Blok RB, Dahl HH, Danks DM, Kirby DM, Chow CW, Christodoulou J, Thorburn DR. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Annals of neurology. 1996;39:343–351. doi: 10.1002/ana.410390311. [DOI] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. Journal of proteome research. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- Saada A, Edvardson S, Rapoport M, Shaag A, Amry K, Miller C, Lorberboum-Galski H, Elpeleg O. C6ORF66 is an assembly factor of mitochondrial complex I. American journal of human genetics. 2008;82:32–38. doi: 10.1016/j.ajhg.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet neurology. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- Schon EA, Manfredi G. Neuronal degeneration and mitochondrial dysfunction. The Journal of clinical investigation. 2003;111:303–312. doi: 10.1172/JCI17741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- Vogel RO, Janssen RJ, Ugalde C, Grovenstein M, Huijbens RJ, Visch HJ, van den Heuvel LP, Willems PH, Zeviani M, Smeitink JA, et al. Human mitochondrial complex I assembly is mediated by NDUFAF1. The FEBS journal. 2005;272:5317–5326. doi: 10.1111/j.1742-4658.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


