SYNOPSIS
Human T-lymphotropic virus type 1 (HTLV-1) is associated with a number of human diseases. Its protease is essential for the virus replication, and similar to the human immunodeficiency virus type 1 (HIV-1) protease, it is a potential target for chemotherapy. The primary sequence of HTLV-1 protease is substantially longer compared to that of HIV-1 protease, and the role of the ten C-terminal residues is controversial. We have expressed C-terminally truncated forms of the HTLV-1 protease with and without N-terminal histidine tags. Removal of five C-terminal residues caused four- to fourtyfold decrease of the specificity constants, while the removal of an additional five C-terminal residues rendered the protease completely inactive. The addition of the N-terminal histidine tag dramatically decreased the activity of the HTLV-1 protease forms. Pull-down experiments carried out with histidine-tagged forms, gelfiltration experiments and dimerization assays provided the first unequivocal experimental data for the role of the C-terminal residues in dimerization of the enzyme. There is a hydrophobic tunnel on the surface of HTLV-1 protease close to the terminal ends that is absent in HIV-1 protease. This hydrophobic tunnel can accommodate the C-terminal extra residues of HTLV-1 protease, which was predicted to stabilize the dimer of the full-length enzyme and provides an alternative target site for protease inhibition.
Keywords: retroviral protease, HTLV-1, dimerization, C-terminal residues, gel filtration, pull down assay, molecular modeling
INTRODUCTION
Human T-lymphotropic virus type 1 (HTLV-1), a retrovirus similar to human immunodeficiency virus type 1 (HIV-1), has been etiologically associated with a number of diseases including adult T-cell leukemia/lymphoma (ATL) and tropical spastic paraparesis/HTLV-1 associated myelopathy (TSP/HAM) (reviewed in Ref. [1]). Approximately 10–20 million people are infected worldwide, and about 5 % of them develop one form of the diseases [2]. Currently there is no vaccine and no satisfactory therapy available to prevent infection or treat the patients [2]. Although active virus replication may not be required for the progression of all HTLV-1 related diseases, blocking viral replication with AZT or lamivudine can have a therapeutic effect [3]. Based on the successful clinical inhibitors of the protease (PR) of HIV-1, the PR of HTLV-1 is also a potential target for chemotherapy [4,5]. HTLV-1 PR, like HIV-1 PR, is a homodimeric aspartic protease, but each monomer comprises 125 residues, rather than 99 residues for HIV-1 PR (Fig. 1). Studies on the HTLV-1 PR have been recently reviewed [6–7]. HTLV-1 PR shares 28 % sequence identity with HIV-1 PR [8] and both the substrate specificity and inhibition profile of the two enzymes are substantially different [6,8,9–11].
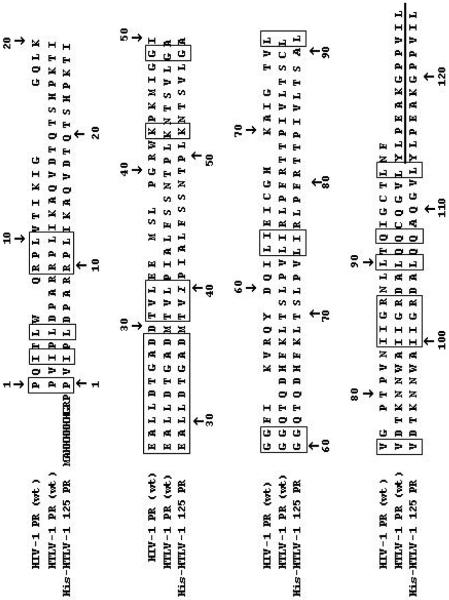
Figure 1. Sequence alignment of the HIV-1 (HXB2) and HTLV-1 (CS) proteases.
The identical residues are indicated by boxes. The sequence of the peptide reported to be inhibitory on the enzyme activity is underlined. The sequence of the longest histidine-tagged recombinant HTLV-1 protease form is also provided, where the stabilizing mutations (L40I, C90A, C109A) are in italics.
While the structures of several retroviral proteases were determined previously [12,13], the structure of a truncated HTLV-1 PR lacking 9 C-terminal residues was determined only recently [14]. Molecular models built for the protease suggested that the C-terminal residues 116–125 of the HTLV-1 PR would assume a flexible flanking region [6,8,15]. Nevertheless, the importance of the C-terminal residues for the activity and function of the HTLV-1 PR is controversial. The first study to address the potential role of the extended C-terminal region was performed by Hayakawa and coworkers [15]. They found that while the extreme five C-terminal residues were not required for autoprocessing from a larger precursor, the residues 115–120 were required for such activity [15]. Daenke and coworkers tested the inhibition effect of a decapeptide representing the last 12 residues of the HTLV-1 protease (shown underlined in Figure 1) and found that it inhibited the activity of the enzyme implying the involvement of the C-terminal extra residues in the enzyme dimerization [16]. However, since the nature of inhibition was not determined, the authors did not exclude the possibility that the peptide inhibits HTLV-1 PR by binding to the active site of the protease [16]. In contrast with these findings and suggestions, another study found the HTLV-1 enzyme form truncated C-terminally by 10 residues to have the same activity as the full length form in an in vitro assay system, although the observed catalytic constants were extremely low for both enzyme forms [17]. In another experiment, the 116-residue-long HTLV PR appeared to have 60 % activity compared to the full-length PR using a fluorometric substrate, although no kinetic parameters were published [14].
To clarify the role of the C-terminal region in the activity and function of HTLV-1 PR, we have constructed C-terminally truncated forms of the wild type stabilized enzyme (HTLV-1 125 PR), shortened by 5- (HTLV-1 120 PR), 9- (HTLV-1 116 PR) and 10-residues (HTLV-1 115 PR). Furthermore, to test whether an N-terminal sequence extension with a His-tag could complement for the role of the C-terminal residues, as well as to facilitate pull-down dimerization experiments, N-terminally His-tagged proteases (His-HTLV-1 125 PR, His-HTLV-1 116 PR, His-HTLV-1 115 PR) were also constructed, expressed, purified to homogeneity, and studied for activity and dimerization. Using the pull-down, gelfiltration and kinetic studies, our work is the first to experimentally prove the importance of the C-terminal residues of HTLV-1 protease in the dimer stability of the enzyme.
MATERIALS AND METHODS
Construction of the shortened HTLV-1 protease forms
The clone coding for the stabilized HTLV-1 PR [11] was used as template for the mutagenesis. This enzyme contained Ala residues in place of cysteines, and a mutation (Leu40Ile) protecting the enzyme from autodegradation. Previous studies demonstrated that the activity of the stabilized enzyme is indistinguishable from that of the wild-type enzyme [11]. HTLV-1 120, 116 and 115 PR forms were generated by stop codon insertions using the QuikChange mutagenesis protocol (Stratagene) with the appropriate oligonucleotide pairs obtained from Genosys Sigma. Mutations were verified by DNA sequencing performed with ABI-Prism dye terminator cycle sequencing kit (Perkin-Elmer) and an Applied Biosystems Model 373A sequencer.
Construction of the His-tagged 10-residue shortened HTLV-1 protease
The DNA sequence of the wild-type, and 10-residue shortened HTLV-1 protease was amplified from the clone coding for HTLV-1 125 PR and HTLV-1 115 PR, respectively, by PCR using the following primer set: forward, 5'-CCT CCA GTT ATA CCG TTA GAT CCC GCC-3'; reverse, 5'-CGC GGA TCC TCA CAA GAT TAC AGG -3'. The PCR fragments were inserted into linearized pCR-Blunt (Invitrogen) vector. The plasmids were digested with StuI and BamHI restriction enzymes (New England Biolabs) to get the final protease fragments. The N-terminally His-tagged constructs were created by replacing the capsid coding sequence (originally cloned between StuI and BamHI restriction sites) of a recombinant HIV-1 capsid protein clone which had been created using pB6, a derivative of pET 11a that expresses N-His-tagged proteins (a gift from Dr. Carol Carter, Department of Molecular Genetics and Microbiology, S.U.N.Y. Stony Brook, USA). The pB6-derived plasmids were grown in DH5α E. coli cells and purified with QIAprep spin plasmid kit (Qiagen). For the construction of 9-residue shortened His-tagged HTLV-1 PR a stop codon was inserted into the His-HTLV-1 125 PR sequence using the QuikChange mutagenesis protocol (Stratagene) with oligonucleotide pairs used previously for generating HTLV-116 PR. Constructions were verified by DNA sequencing performed with ABI-Prism dye terminator cycle sequencing kit (Perkin-Elmer) and an Applied Biosystems Model 373A sequencer.
Enzyme purification and assays with oligopeptide substrates
Stabilized wild-type HTLV-1 PR was purified from inclusion bodies as described previously [11,18]. The C-terminally truncated forms were purified using the same procedure. The His-tagged HTLV-1 PR forms were purified by using two chromatography steps. The inclusion bodies were dissolved in 50 mM Tris-HCl, pH 8.0, 7.5 M guanidine-HCl, 5 mM EDTA, then loaded onto HisTrap column (Amersham-Pharmacia), which was equilibrated with 20 mM Na-phosphate, pH 7.4, containing 500 mM NaCl, 3M guanidine-HCl, 20 mM imidazole, 2% glycerol, 0.2% Triton X-100, and the PRs were eluted by using an increasing imidazole gradient (20–500 mM) in denaturing conditions. The eluted enzymes were purified further by reversed-phase HPLC as described previously [11]. The correct molecular weight of the purified HTLV-1 PRs was verified by using a 4000 Q TRAP mass spectrometer (Applied Biosystems). Folding of the enzymes was performed by dialysis of the HPLC fractions in large excess of 25 mM formic acid, pH 2.8, and subsequently in 50 mM sodium acetate buffer, pH 5.0, containing 100 mM NaCl. For the activity measurements and for purity check, folded proteases were concentrated using Amicon ultrafiltration devices (Amicon Ultra-15, 10 kDa cut-off). Protein concentration was determined by the Bradford spectrophotometric method (Bio-Rad). Purity of the samples was determined by SDS-polyacrylamide gel electrophoresis on 16% gels. 10–15 μg proteins were loaded per lane. Proteins were visualized by using Coomassie Brillant Blue staining after the electrophoretic separation. The protease assays were initiated by mixing of 5 μl (8 – 8500 nM) purified enzyme with 10 μl 2x incubation buffer A (0.5 M potassium phosphate buffer, pH 5.6, containing 10% glycerol, 2 mM EDTA, 10 mM dithiothreitol, 4 M NaCl) or buffer B (100 mM sodium acetate, pH 5.0, 0.2 M NaCl) and 5 μl 0.01 – 3.0 mM substrate. The synthesis and characterization of the oligopeptide substrates were described earlier [8,11,18]. The substrate concentration range for kinetic measurements was selected depending on the approximate Km values. The reaction mixture was incubated at 37 °C for one hour and the reaction was stopped by the addition of 180 μl 1 % trifluoroacetic acid (TFA), and an aliquot was injected onto a Nova-Pak C18 reversed-phase chromatography column (3.9 × 150 mm, Waters Associates, Inc.) using an automatic injector. Substrates and the cleavage products were separated using an increasing water-acetonitrile gradient (0–100%) in the presence of 0.05% TFA using a LaChrom HPLC system (Merck Hitachi). Cleavage products of PR-catalyzed hydrolysis for the peptides were identified by amino acid analysis. Kinetic parameters were determined by fitting the data obtained at less than 20% substrate hydrolysis to the Michaelis-Menten equation by using the SigmaPlot 8.02 program (Systat Software Inc). When Km was sufficiently high compared to the substrate concentrations used (e.g. in measurements performed at low ionic strength), the specificity constant (kcat/Km) was obtained by dividing the first-order rate constant (determined by linear regression) by the active enzyme concentration present in the reaction mixture. The standard deviation for the kcat/Km values was calculated according to Fenner [19]. The standard errors of the kinetic parameters were below 20 %. Active site titration of the wild-type and the 5-residue shortened PRs was performed using peptide KTKVL-r-VVQPK (IB268) where r represents a reduced peptide bond, as described for the wild-type enzyme [18].
Gelfiltration of the purified proteases
Gelfiltration experiments were performed with HTLV-1 125 PR, 120 PR and 115 PR using a Superdex G-75 column (Amersham Pharmacia) equilibrated with 50 mM sodium-acetate, pH 5.0, containing 100 mM NaCl, with the flow rate of 0.5 ml/min. The experiments were performed using an AKTA Purifier instrument (Amersham-Pharmacia). Elution of the proteins was followed at 280 nm wavelength. For the HTLV-1 125 PR and 120 PR two consecutive runs were performed, in the first run 1.7–1.8 mg protein was loaded, and the monomer/dimer fractions were collected. Most of the injected proteins eluted at the exclusion volume, due to aggregation. The collected monomer/dimer fraction (10–15 μg) was reinjected and analyzed for the monomer/dimer ratio. For HTLV-1 115 PR purified refolded protease was directly injected to the column. The column was calibrated using a mixture of Bio-Rad gelfiltration standards (containing thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), vitamin B12 (1.35 kDa) and cytidine (243 Da) using the same buffer and flow rate.
Pull down assay of the HTLV-1 protease forms
Pull down assays were performed with a batch technique: 50 μl portions of nickel-charged ProBond resin (Invitrogen) were pipetted to Eppendorf tubes, then the resins were centrifuged down (3000 rpm) and the supernatant was removed. The gels were equilibrated for the assay with distilled water followed by 50 mM sodium-acetate, pH 5.0, containing 100 mM NaCl, 1% Triton-X100 and 5% glycerol. Excess of buffer was removed by centrifugation at 3000 rpm for 3 min. The HPLC purified HTLV-1 125, 120 and 115 proteases were refolded alone, or in the presence of equimolar amount of HPLC purified His-HTLV-1 125 PR. In another set, HPLC purified His-HTLV-1 116 PR was refolded alone or in the presence of equimolar amount of HTLV-1 115 protease. Based on Bradford assay and accurate volume measurement, same amounts of components in the pure or mixed samples were loaded onto the prepared nickel-chelate resins after the refolding process. Samples were let to stand on the resins for 5 min at room temperature then most of the unbound proteins were removed by centrifugation. The remaining unbound proteins were removed from the resins by repetitive washing with the equilibration buffer and centrifugation steps. The anchored proteins were eluted from the resins by adding 50 μl of 2x SDS loading buffer and heating for 5 min at 95 °C, then the tubes were cooled down and centrifuged. The supernatants were separated on 16% SDS-PAGE and stained with Comassie Brillant Blue. The intensity of the bands was determined using densitometric measurements performed with an AlphaImager 2200 gel documentation system (Alpha Innotech).
Determination of apparent Kd values of the HTLV proteases
Specific activity values were measured in duplicates as a function of dimeric enzyme concentration in 50 mM sodium-acetate buffer, pH 5.0, containing 100 mM NaCl using 1 mM KTKVF↓VVQPK oligopeptide substrate. Samples were incubated for 1 h at 37 °C, and processed for HPLC analysis as described for the protease above, as previously used to determine the Kd as described originally for HIV-1 protease [20]. Assuming that the active protease dimer is in rapid equilibrium with its inactive monomer, the following equation can be used to describe the activity of the enzyme [20]:
The apparent Kd values can be obtained by plotting the relative specific activities against the concentration of the HTLV-1 protease forms and fitting a curve using “Hyperbola single rectangular 2 parameters equation” setting of SigmaPlot 8.02 software.
Molecular Modeling
Crystal structures of HIV-1 PR (PDB code: 1K1T [21]) and HTLV-1 116 PR (PDB code: 2B7F [14]) complexed with inhibitors, and a molecular model of HTLV-1 125 PR were used for structural analysis. The nine residue long C-terminal tail of HTLV-1 125 PR was built manually into the crystal structure of the HTLV-1 116 PR using Sybyl software package (Tripos). Unfavorable interactions were removed by energy minimization (Kollmann all atom force field [22] 10 Simplex and 100 Powell iterations, 8 Angstrom cut-off). Lipophilic potentials on molecular surfaces were generated by MOLCAD modul of Sybyl. The model of HTLV-1 125 PR is available from the authors upon request.
RESULTS
The sequence alignment of HTLV-1 and HIV-1 PRs, based on crystal structures, is shown in Fig. 1 for the wild-type proteases and for the His-HTLV 125 PR. All the other constructs used in this paper contained shortened versions of this sequence or no His-tag (starting with the first Pro residue). The 125 residue-length protease (HTLV-1 125 PR) was previously characterized in detail [8,11,18]. Based on the active site titrations, this form folded completely, and efficiently processed the oligopeptide substrates at high ionic strength (Table 1). At lower ionic strength the enzyme exhibited an approximately thirty to eightyfold reduction in the specificity constants (Table 1). The beneficial effect of ionic strength, similar to that for HIV-1 PR [23], on the catalytic activity of HTLV-1 PR has been reported earlier [24]. To determine the kinetic capability of the C-terminally shortened enzyme forms without or with an N-terminal His-tag, these proteins were expressed and purified to homogeneity, as demonstrated by SDS-polyacrylamide gel electrophoresis (Fig. 2). These purified concentrated enzymes (at least 1.7 mg/ml final concentration) were used for further assays. The 5-residue truncated form (HTLV-1 120 PR) also folded very efficiently (98 % based on active site titration performed on the freshly prepared and refolded enzyme), nevertheless, it showed substantially lower Km and kcat values, resulting in a thirty to fourtyfold decrease in the specificity constants at high ionic strength but only four- to sevenfold decrease at low ionic strength (Table 1). Small but detectable activity was observed for the HTLV-1 116 PR form at high ionic strength. While the wild-type and the HTLV-1 120 PR were able to efficiently cleave various substrates under various conditions at 25–2500-fold dilutions, no activity was observed for the HTLV-1 115 PR, even when it was used in the concentrated form (data not shown). The N-terminal histidine tag dramatically affected the kinetic parameters of the HTLV-1 PR forms. Only the His-HTLV-1 125 PR showed activity on various substrates at high ionic strength, while it was inactive at low salt concentration. Its Km values were higher and kcat values were much lower as compared to those obtained with HTLV-1 125 PR, resulting in extremely low specificity constants (Table 1). These results suggest that the C-terminal residues are important for efficient substrate hydrolysis, and the N-terminal extra sequence with the His tag is not favorable for the enzyme activity. Interestingly, N-terminal extension of HIV-1 PR also had an unfavorable effect on the activity of the enzyme [25].
Table 1.
Kinetic parameters determined for various HTLV-1 protease formsa
| Buffer A_____________ | Buffer B_ | ||||
|---|---|---|---|---|---|
| Enzyme form | Substrate | Km(mM) | kcat (s-1) | kcat/Km(mM-1s-1) | kcat/Km(mM-1s-1) |
| HTLV-1 125 PR | KTKVL↓VVQPK | 0.063 ± 0.004b | 10.0 ± 0.2b | 158.7 ± 10.6b | 5.41 ± 0.17c |
| KTKVF↓VVQPK | 0.049 ± 0.006b | 16.4 ± 0.9b | 334.7 ± 44.9b | 4.16 ± 0.06c | |
| His-HTLV-1 125 PR | KTKVL↓VVQPK | 0.088 ± 0.013 | 0.0020 ± 0.0001 | 0.022 ± 0.003 | No cleavaged |
| KTKVF↓VVQPK | 0.116 ± 0.014 | 0.021 ± 0.001 | 0.180 ± 0.023 | No cleavaged | |
| HTLV-1 120 PR | KTKVL↓VVQPK | 0.034 ± 0.003 | 0.150 ± 0.004 | 4.4 ± 0.4 | 0.81 ± 0.03c |
| KTKVF↓VVQPK | 0.014 ± 0.001 | 0.160 ± 0.001 | 11.7 ± 0.4 | 1.03 ± 0.08c | |
| HTLV-1 116 PR | KTKVL↓VVQPK | No cleavaged | No cleavaged | ||
| KTKVF↓VVQPK | Residual activitye | No cleavaged | |||
Measurements were performed at least in triplicate sets providing values within the experimental errors. A representative set is given the table. No activity was observed for His-HTLV-1 116 PR, HTLV-1 115 PR and His-HTLV-1 115 PR forms in buffer A or in buffer B.
These data are taken from Ref. (8).
These values were determined using first-order kinetic conditions ([S] << Km)
No hydrolysis was observed neither at low nor at high protease concentration.
Residual activity was observed when the substrate was incubated with 3.1 μsM protease in assay conditions.
Figure 2. SDS-polyacrylamide gel electrophoresis of the expressed HTLV-1 protease forms.
Samples are indicated below the gel using the following abbreviations: H125: His-HTLV-1 125 PR; H116: His-HTLV-1 116 PR; H115: HTLV-1 115 PR; 125: HTLV-1 125 PR; 120: HTLV-1 120 PR; 116: HTLV-1 116 PR; 115: HTLV-1 115 PR; M, unstained molecular mass marker from Fermentas. The gel was stained with Coomassie Brillant Blue to visualize the proteins.
To determine the apparent dimer stability of the HTLV-1 125 and HTLV-1 120 PR forms, an activity-based assay originally used for HIV-1 PR [20]. The assay was performed in low ionic strength condition (Buffer B). As fully formed dimer should have a constant specific activity, the decrease of specific activity by decreasing enzyme concentration is considered to be due to the less efficient dimerization, hence, the enzyme concentration corresponding to the 50% loss in enzyme activity represents the apparent dissociation constant Kd [20]. Substantial differences were observed in the apparent dimer stability of the HTLV-1 125 PR (Kdapp. = 491 nM) and HTLV-1 120 PR (Kdapp. > 3000nM), as shown in Fig. 3. These results indicated that the removal of the C-terminal 5 residues substantially decreased the apparent dimer stability of the enzyme. A substantial decrease of the specific activity was observed for the HTLV-1 125 PR above 2500 nM concentration (data not shown), and a similar effect was found for the HTLV-1 120 PR above 7000 nM (data not shown). This effect was predicted to be due to the high aggregation tendency of HTLV-1 protease, as observed in the gelfiltration experiments (see below) and is in a good agreement with our observations of aggregation tendencies during crystallization of HTLV-1 PR forms.
Figure 3. Determination of the apparent dissociation constant for HTLV-1 protease forms.
Relative specific activities were measured for the HTLV-1 125 PR (upper panel) and for the HTLV-1 120 PR (lower panel) as a function of dimeric enzyme concentration at a final substrate (KTKVFVVQPK) concentration of 1 mM, in 50 mM sodium acetate buffer (pH 5.0), 100 mM NaCl using an HPLC assay. The obtained apparent Kd values were 491 ± 119 nM for HTLV-1 125 PR and > 3000 nM for HTLV-1 120 PR.
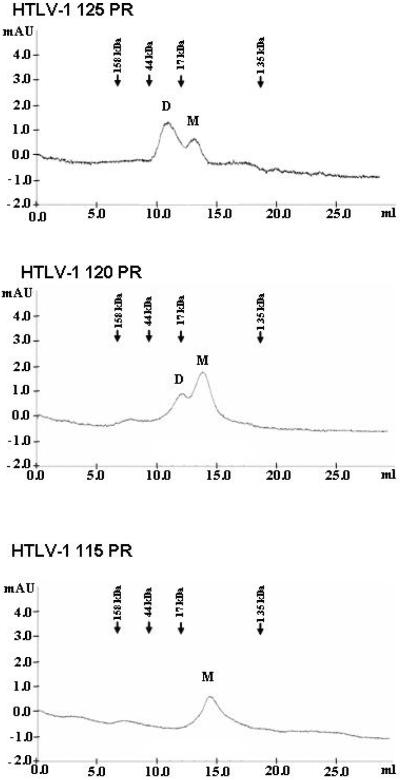
To further characterize the effect of truncation on the dimer stability as well as dimerization capability of the enzymes, gelfiltration experiments were performed using the same buffer conditions as utilized for the dimerization assay. As shown in Fig. 4, while the wild type HTLV-1 125 PR appeared to be mostly in the dimeric form under the assay conditions of gelfiltration, the equilibrium was shifted towards the monomeric form for the HTLV-1 120 PR, while the HTLV-1 115 PR was completely in the monomeric form, suggesting that the elimination of the 10 residue-long C-terminal flanking sequence detrimental for the dimerization capability of the enzyme in low ionic strength conditions.
Figure 4. Gelfiltration of the HTLV-1 protease forms.
Gelfiltration of the indicated HTLV-1 protease forms was performed on a Superdex G75 column in 50 mM sodium-acetate buffer, pH 5.0, containing 100 mM NaCl (Buffer B in Table 1), at 0.5 ml/min flow rate. The position of the eluted monomeric (M) and dimeric (D) forms is indicated. Elution volume for the standard proteins used for the calibration of the column under identical conditions is indicated above the chromatograms. Based on the calibration, the apparent molecular weights of the eluted proteins are: HTLV-1 125 PR dimer: 28 kDa, monomer: 12 kDa; HTLV-1 120 PR dimer: 24 kDa, monomer: 11 kDa; HTLV-1 115 PR monomer: 10 kDa.
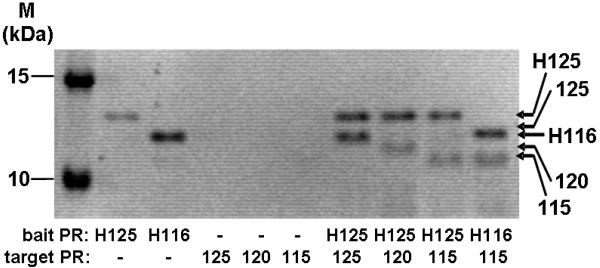
As the HTLV-1 115 PR and the His-tagged HTLV-1 PRs were not active at low ionic strength (Table 1), a pull-down assay was performed to obtain more information about the effect of the C-terminal residues of the HTLV-1 PR on dimerization. While the HTLV-1 125 PR formed heterodimer with the His tagged HTLV-1 125 PR with close to 1:1 molar ratio, the HTLV-1 120 and 115 PRs associated significantly weaker with the His-HTLV-1 125 PR (Figure 5). Interestingly, the dimerization capability of the HTLV-1 115 PR was similar with the His-HTLV-1 116 and His-HTLV-1 25 PRs.
Figure 5. Pull-down assay of the HTLV-1 protease forms.
SDS-PAGE of the pull-down assay samples is shown. M, unstained molecular mass marker from Fermentas; Protease samples are indicated below the gel using the same abbreviations as in Figure 2. Densitometric evaluation of gels from four independent experiments provided the following ratios: 125/H125: 84.9 ± 10; 120/H125: 37.2 ± 13.4; 115/H125: 37.3 ± 12; 115/H116: 43.1 ± 18.7.
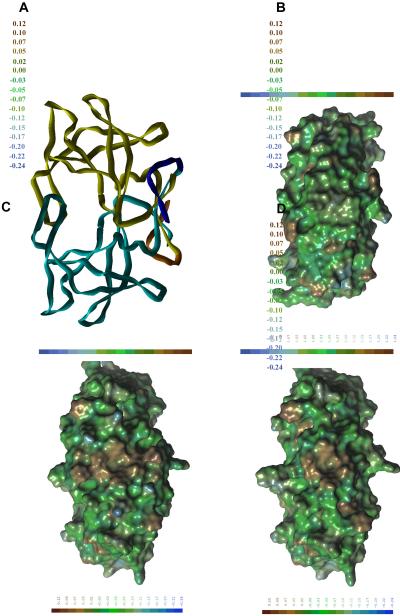
To obtain a feasible explanation for our results, we analyzed the nature of the C-terminal extension in HTLV-1 PR (Fig. 1) as well as created a molecular model for HTLV-1 125 PR. Various surface representations of HIV-1 and HTLV-1 116 PRs showed substantial differences between hydrophobicities of the C-terminal regions of the two enzymes (Fig. 6 A and B). The more hydrophilic HIV-1 PR also showed a more compact structure in this region (Fig. 6A), while residues of the terminal beta-sheet and the Arg77-Pro84 loop of the HTLV-1 116 PR formed a groove (Fig 6B). Based on the experimental results, we predicted that this hydrophobic groove[a tunnel is an enclosed hole – in the figure it looks like a surface groove] may be filled by the extra C-terminal residues which may preferentially interact with the hydrophobic “outer” surface of the terminal beta-sheet. To obtain a molecular model for the full-length protease, we have incorporated the 9 missing residues into the crystal structure of the HTLV-1 116 PR in such a way that this interaction may occur. The structural requirements of this step and the characteristics of PEAKGPPVIL fragment (residues 116–125) were in good agreement: the PEAKGPP part was dominated by Pro, Gly and charged residues with strong turn-forming propensity, while residues of the terminal Val-Ile-Leu part had hydrophobic and beta-sheet-forming characters. The C-terminal residues were predicted to form two new “strands” for the terminal beta-sheet (Fig 6C), however they were stabilized not by H-bonds of the peptide backbone, rather by hydrophobic interactions of the side-chains. Interestingly, cellular aspartic proteases also have six strands in their corresponding beta-sheet region, but the topologies are completely different (data not shown). The presence of the additional C-terminal residues in the full-length protease model resulted in a substantial decrease in size of the hydrophobic patch, although it did not disappear completely (Fig. 6D).
Figure 6. Structural representations of the HIV-1 and HTLV-1 proteases.
(A) Lipophilic potential of HIV-1 PR X-ray structure on Connolly-type molecular surface. (B) Lipophilic potential of HTLV-1 116 PR X-ray structure on Connolly-type molecular surface. (C) Ribbon representation of HTLV-1 125 PR. Residues 1–116 (X-ray structure) are shown in cyan (monomer A) and yellow (monomer B), while residues 117–125 (model) are shown in blue (monomer A) and orange (monomer B). The structure is shown in a view perpendicular to that of Figures A, B and D. (D) Lipophilic potential of HTLV-1 125 PR model on Connolly-type molecular surface.
DISCUSSION
Based on structure-based sequence comparison, HTLV-1 PR appears to contain a 10 residue-long C-terminal flanking sequence, as shown in Fig. 1. The C-terminal extension is a characteristic feature of the proteases belonging to the deltaretrovirus family. While some other proteases also appear to contain C-terminally extended sequences, none of the proteases for which the 3-dimensional structure is available contain such C-terminal extension [12] including the newly determined structure of HTLV-1 116 PR [14]. The importance of this C-terminal flanking sequence is controversial. In case of bovine leukemia virus (BLV) PR, another deltaretrovirus, the C-terminal flanking region appeared to be dispensable for the activity of purified recombinant protease [25], while at lease five out of the ten C-terminal flanking amino acid residues were required for self-processing activity of HTLV-1 PR [15]. Inhibition by a peptide representing the C-terminal 12 amino acid residue-long region of the protease also suggested the importance of this region in the enzyme dimerization [16]. However, recently a purified, N-terminally His-tagged but C-terminally 10 residues shortened HTLV-1 PR was described, which showed an activity equivalent to that of the full-length His-tagged protease [17]. Surprisingly, both the wild-type and the shortened enzyme showed very low catalytic efficiency (0.004–0.006 s−1 kcat values) compared to the catalytically active HTLV-1 125 PR and HTLV-1 120 PR forms, described in this paper. The HTLV-1 116 PR used for crystallization appeared to have 60 % activity of that of the full length protease, but no kinetic data were provided [14], therefore the folding/activity level of this form is uncertain. In our hands the HTLV-1 116 PR appeared to have residual catalytic activity (Table 1). We have speculated that the activity of the His-HTLV-1 115 PR described by Herger et al. [17] might be due to the presence of N-terminal His-tag, therefore, we have also expressed the HTLV-1 115 PR with an N-terminal histidine tag, but this form appeared to be completely inactive in our assay systems. Furthermore, the N-terminally His-tagged HTLV-1 116 PR did not show any activity (data not shown) while the activity of the His-HTLV-1 125 PR was substantially lower than that of the HTLV-1 125 PR (Table 1).
To further characterize the role of the C-terminal extension in HTLV-1 PR, we have determined the apparent Kd values of the HTLV-1 125 and 120 PRs using a specific activity-based method. The shorter enzyme appeared to have dramatically lower dimer stability (Fig. 3). The importance of the C-terminal residues in dimerization also supported by the result of gelfiltration experiments, which showed that the HTLV-1 115 PR appeared only in monomeric form in low ionic strength conditions (Fig. 4). An aggregation effect was observed for the HTLV-1 125 PR when the protein concentration was increased (Fig. 3), an effect also seen for HTLV-1 120 PR at threefold higher protein concentration (data not shown). This phenomenon partly explains the sustained failure of the crystallization of the full length HTLV-1 PR and the fact that HTLV-1 116 PR was favorably crystallized, although only in the presence of a ligand that is expected to strongly facilitate dimerization [14]. These results suggest that the C-terminal 10 residues may also have a critical role mediating the in vitro aggregation of the protease.
The results of the pull-down assay further confirmed our findings that the C-terminal extension of the HTLV-1 PR has a critical role in dimerization in the absence of ligand that may also be a prerequisite for sufficient catalytic efficiency. There is no structural evidence [evidence is always singular] for how these extended residues can contribute to the dimerization. Retroviral PR dimers are held together by an intricate H-bond network at the catalytic aspartates called the “fireman's grip” as well as by the beta sheets formed by C- and N-terminal residues [12]. The two regions are not independent: in case of HIV protease, the residues above the beta sheet form a hydrophobic cluster which interacts with the side chain of the Thr residues after the catalytic aspartates. The side chains below the beta sheet are hydrophilic and they interact with the bulk solvent. Unlike in HIV-1 PR, the residues of the beta sheet exposed to the water are hydrophobic in HTLV-1 PR, which might explain the high aggregation tendency of the enzyme. Furthermore, it also provides a surface for interaction with the residues of the extreme C-terminus, which may contribute to the dimer stability of the full length HTLV-1 protease. However, the exact way in which these residues contribute to the dimer stability of HTLV-1 PR requires further structural studies. We have created a molecular model for the positioning of the C-terminal residues. This model explains the importance of these residues in the dimerization process and does not contradict the observed high aggregation and the low crystal-forming propensities of the full length HTLV-1 PR. The flexibility of the C-terminal tail may allow the dimer to switchbetween a more compact and a more open structure, in which the tail does not interact with the body of the PR, which may interfere in the crystallization processes. It is important to note that the C-terminal tail of the HTLV-1 protease was also found to be critical for the infectivity of recombinant HTLV-1 virions: while the 5-residue truncation already decreased infectivity, elimination of the 10 residues practically abolished infectivity in a wild-type PR sequence background (M. Mitchell and D. Derse, personal communication). Therefore the proposed extended dimerization interface of HTLV-1 PR may provide an additional target site for design of inhibitors against this enzyme.
ACKNOWLEDGMENTS
We thank Tamas Sperka for the mass spectrometry analyses and Szilvia Petö for technical assistance. The research was supported in part by the Hungarian OTKA T43482, F35191, GVOP 3.2.1. 0149/3.0 and by the National Institute of Health grants GM62920, AIDS-FIRCA TW01001.
Abbreviations used
- HTLV-1
Human T-lymphotropic virus type 1
- HIV-1
human immunodeficiency virus type 1
- ATL
adult T-cell leukemia/lymphoma
- TSP/HAM
tropical spastic paraparesis/HTLV-1 associated myelopathy
- PR
protease
- BLV
bovine leukemia virus
REFERENCES
- 1.Johnson JM, R. Harrod R, Franchini G. Molecular biology and pathogenesis of the human T-cell leukemia/lymphotropic virus type-1. Int. J. Exp. Path. 2001;82:135–147. doi: 10.1046/j.1365-2613.2001.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bangham CR. HTLV-1 infections. J. Clin. Pathol. 2000;53:581–586. doi: 10.1136/jcp.53.8.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazarbachi A, Hermine O. Treatment of adult T-cell leukaemia/lymphoma: current strategy and future perspectives. Virus Res. 2001;78:78–92. doi: 10.1016/s0168-1702(01)00286-6. [DOI] [PubMed] [Google Scholar]
- 4.Gustchina A, Jaskolski M, Wlodawer A. Lessons learned fighting HIV can be applied to anti-cancer drug design. Cell Cycle. 2006;5:463–464. doi: 10.4161/cc.5.5.2490. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen JT, Zhang M, Kumada HO, Itami A, Nishiyama K, Kimura T, Cheng M, Hayashi Y, Kiso Y. Truncation and non-natural amino acid substitution studies on HTLV-I protease hexapeptidic inhibitors. Bioorg. Med. Chem. Lett. 2008;18:366–370. doi: 10.1016/j.bmcl.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 6.Shuker SB, Mariani VL, Herger BE, Dennison KJ. Understanding HTLV- I protease. Chem. Biol. 2003;10:373–380. doi: 10.1016/s1074-5521(03)00104-2. [DOI] [PubMed] [Google Scholar]
- 7.Daenke S. In: Handbook of Proteolytic Enzymes. 2nd ed. Barrett AJ, Rowlings ND, Woessner JF, editors. Elsevier Science Ltd.; London: pp. 166–169. [Google Scholar]
- 8.Tözsér J, Zahuczky G, Bagossi P, Louis JM, Copeland TD, Oroszlan S, Harrison RW, Weber IT. Comparison of the substrate specificity of the human T-cell leukemia virus and human immunodeficiency virus proteinases. Eur. J. Biochem. 2000;267:6287–6295. doi: 10.1046/j.1432-1327.2000.01714.x. [DOI] [PubMed] [Google Scholar]
- 9.Hruskova-Heidingsfeldova O, Blaha I, Urban J, Strop P, Pichova I. Substrates and inhibitors of human T-cell leukemia virus type 1 (HTLV-1) proteinase. Leukemia. 1997;11(Suppl 3):45–46. [PubMed] [Google Scholar]
- 10.Ding YS, Rich, Ikeda RA. Substrates and inhibitors of human T-cell leukemia virus type I protease. Biochemistry. 1998;37:17514–17518. doi: 10.1021/bi982004a. [DOI] [PubMed] [Google Scholar]
- 11.Louis JM, Oroszlan S, Tözsér J. Stabilization from autoproteolysis and kinetic characterization of the human T-cell leukemia virus type 1 proteinase. J. Biol. Chem. 1999;274:6660–6666. doi: 10.1074/jbc.274.10.6660. [DOI] [PubMed] [Google Scholar]
- 12.Wlodawer A, Gustchina A. Structural and biochemical studies of retroviral proteases. Biochim. Biophys. Acta. 2000;1477:16–34. doi: 10.1016/s0167-4838(99)00267-8. [DOI] [PubMed] [Google Scholar]
- 13.Dunn BM, Goodenow MM, Gustchina A, Wlodawer A. Retroviral proteases. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-reviews3006. REVIEWS3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Laco GS, Jaskolski M, Rozycki J, Alexandros J, Wlodawer A, Gustchina A. Crystal structure of human T-cell leukemia virus protease, a novel target for anti-cancer drug design. Proc. Natl. Acad. Sci. USA. 2005;102:18332–18337. doi: 10.1073/pnas.0509335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa T, Misumi Y, Kobayashi M, Yamamoto Y, Fujisawa Y. Requirement of N- and C-terminal regions for enzymatic activity of human T-cell leukemia virus type I protease. Eur. J. Biochem. 1992;206:919–925. doi: 10.1111/j.1432-1033.1992.tb17001.x. [DOI] [PubMed] [Google Scholar]
- 16.Daenke S, Schramm HJ, Bangham CRM. Analysis of substrate cleavage by recombinant protease of human T cell leukaemia virus type 1 reveals preferences and specificity of binding. J. Gen Virol. 1994;75:2233–2239. doi: 10.1099/0022-1317-75-9-2233. [DOI] [PubMed] [Google Scholar]
- 17.Herger BE, Mariani VL, Dennison K, Shuker SB. The 10 C-terminal residues of HTLV-I protease are not necessary for enzymatic activity, Biochem. Biophys. Res. Commun. 2004;320:1306–1308. doi: 10.1016/j.bbrc.2004.06.087. [DOI] [PubMed] [Google Scholar]
- 18.Kádas J, Weber IT, Bagossi P, Miklóssy G, Boross P, Tözsér J. Narrow substrate specificity and sensitivity towards ligand binding site mutations of human T-cell leukemia virus protease. J. Biol. Chem. 2004;279:27148–27157. doi: 10.1074/jbc.M401868200. [DOI] [PubMed] [Google Scholar]
- 19.Fenner G. Das Genauigkeitsmaß von Summen, Differenzen, Produkten und Quotienten der Beobachtungsreichen. Naturwissenschaften. 1931;19:310. [Google Scholar]
- 20.Jordan SP, Zugay J, Darke PL, Kuo LC. Activity and dimerization of human immunodeficiency virus protease as a function of solvent composition and enzyme concentration. J. Biol. Chem. 1992;267:20028–20032. [PubMed] [Google Scholar]
- 21.Mahalingam B, Boross P, Wang YF, Louis JM, Fischer CC, Tozser J, Harrison RW, Weber IT. Combining mutations in HIV-1 protease to understand mechanisms of resistance. Proteins. 2002;48:107–116. doi: 10.1002/prot.10140. [DOI] [PubMed] [Google Scholar]
- 22.Weiner SJ, Kollman PA, Nguyen DT, Case DA. An All Atom Force Field for Simulations of Proteins and Nucleic Acids. J. Comp. Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 23.Wondrak EM, Louis JM, Oroszlan S. Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. FEBS Lett. 1991;280:344–346. [Google Scholar]
- 24.Bagossi P, Kádas J, Miklóssy G, Boross P, Weber IT, Tözsér J. Development of a microtiter plate fluorescent assay for inhibition studies on the HTLV-1 and HIV-1 proteinases. J. Virol. Methods. 2004;119:87–93. doi: 10.1016/j.jviromet.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Louis JM, Wondrak EM, Kimmel AR, Wingfield PT, Nashed NT. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 1999;274:23437–23442. doi: 10.1074/jbc.274.33.23437. [DOI] [PubMed] [Google Scholar]
- 26.Precigoux G, Geoffre S, Leonard R, Llido S, Dautant A, d'Estaintot BL, Picard P, Menard A, Guillemain B, Hospital M. Modelling, synthesis and biological activity of a BLV proteinase, made of (only) 116 amino acids. FEBS Lett. 1993;326:237–240. doi: 10.1016/0014-5793(93)81798-5. [DOI] [PubMed] [Google Scholar]