Abstract
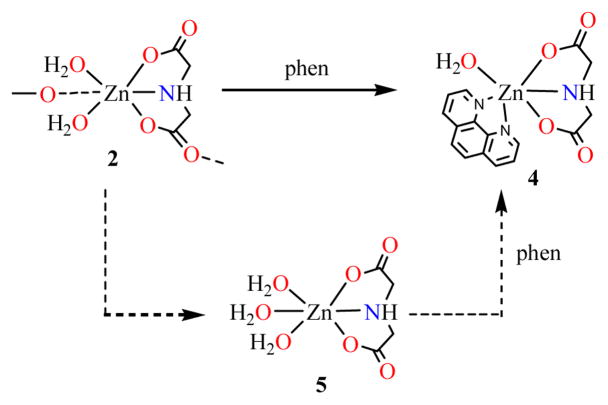
Three novel zinc coordination polymers (NH4)n[Zn(Hida)Cl2]n (1), [Zn(ida)(H2O)2]n (2), [Zn(Hida)2]n·4nH2O (3) (H2ida = iminodiacetic acid) and a monomeric complex [Zn(ida)(phen)(H2O)]·2H2O (4) (phen=1,10-phenanthroline) have been synthesized and characterized by X-ray diffraction methods. 1 and 2 form one-dimensional (1-D) chain structures, whereas 3 exhibits a three-dimensional (3-D) diamondoid framework with an open channel. The mononuclear complex 4 is extended into a 3-D supramolecular architecture through hydrogen bonds and π-π stacking. Interestingly, cyclic nonplanar tetrameric water clusters are observed that encapsulated in the 3-D lattice of 4. Based on 1H and 13C NMR observations, there is obvious coordination of complex 2 in solution, while 1 and 3 decompose into free iminodiacetate ligand. Monomer [Zn(ida)(H2O)3] (5) is considered as a possible discrete species from 2. These coordination polymers can serve as good molecular precursors for zinc oxide.
Keywords: Zinc, Iminodiacetate, Coordination polymer, NMR, ZnO nanoparticles, Photoluminescence
1. Introduction
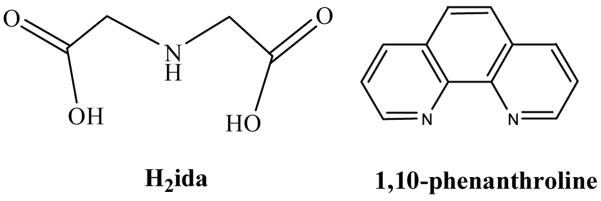
Iminodiacetic acid (H2ida = iminodiacetic acid) (Chart 1) is employed as in immobilized metal affinity chromatography (IMAC) purification of proteins and nucleic acids as a typical metal chelating agent. Transition metal ions like zinc, nickel and copper are immobilized on Sephadex with chelating groups of iminodiacetic acid in polymeric form, which are allowed to interact with proteins and nucleic acids in the mobile phase [1–7]. More attention has been paid on the interactions of the zinc ion with aminocarboxylic acids, due to the role in maintaining the structural integrity of the metalloenzyme and determining the binding ability of substrate and proteins [8–12]. Moreover, as the solid precursors, zinc carboxylate species are technologically useful in the preparation of Zn-containing semiconductor materials [13–18]. In the other aspect, dicarboxylate ligands have been used for the construction of coordination polymers like thiodiacetate [H2tda= S(CH2COOH)2], oxydiacetate [H2oda= O(CH2COOH)2], and iminodiacetate[H2ida= HN(CH2COOH)2] [19], which contain different coordination of monodentate, bridging bidentate and bridging tridentate modes [20]. At present, complex formation between zinc and iminodiacetate has been investigated by spectroscopic and structural methods [21–28]. In a neutral solution, monomeric diiminodiacetato zinc K2[Zn(ida)2]·3H2O is isolated as a dominant species However, little concern is focused on the formation of coordination polymer between zinc and iminodiacetate. Herein, we report the interactions of zinc and iminodiacetic acid in acidic solution, which results in the isolations and characterizations of three novel coordination polymers (NH4)n[Zn(Hida)Cl2]n (1), [Zn(ida)(H2O)2]n (2), [Zn(Hida)2]n·4nH2O (3) and a monomeric mixed ligand complex [Zn(ida)(phen)(H2O)]·2H2O (4). The structure of monomer complex 4 is isomorphous with a mixed ligand copper(II) complex, [Cu(ida)(phen)(H2O)]·1.5H2O [29]. However, unprecedented cyclic nonplanar tetrameric water clusters are observed that encapsulated in the 3-D lattice of 4. The intermediate in the formation of coordination polymers 1 – 3 and the solution NMR spectra were investigated. Moreover, the complexes were used as molecular precursors for the thermal decomposition of ZnO. The resulted nanoparticles were studies by photoluminescence spectra, XRD patterns and SEM images.
Chart 1.
The structures of H2ida and 1,10-phen.
2. Experimental
2.1. Materials and physical measurements
The pH value was measured by potentiometric method with a digital PHB-8 pH meter. Infrared spectra were recorded as Nujol mulls between KBr plates using a Nicolet 360 FT-IR or a Bruker Vertex 70v spectrometer. Elemental analyses were performed using EA 1110 elemental analyzer. 1H NMR and 13C NMR spectra were measured on a Bruker Avance AV-400 MHz resonance spectrometer with D2O solvent using DSS (sodium 2,2-dimethyl-2-silapentane-5-sulfonate) as an internal reference at room temperature. The thermogravimetric analyses (TG) were collected with a Netzsch TG 209 F1 instrument with a 10 °C/min from 30 to 900 °C in flowing nitrogen atmosphere. Scanning electron micrograph (SEM) images were obtained with a LEO-1530 electron microscope equipped with an energy dispersive X-ray spectrometer (EDS). The X-ray powder diffraction patterns (XRD) were recorded on a Philips X’Pert PRO diffractometer, operated at 40 kV and 30 mA using a Cu target tube.
2.2 Synthesis of complexes 1–4
2.2.1. (NH4)n[Zn(Hida)Cl2]n (1) and [Zn(ida)(H2O)2]n (2)
Zinc chloride (0.68 g, 5.0 mmol) was reacted with iminodiacetic acid (0.67 g, 5.0 mmol) in 20.0 ml water with continuously stirring. The pH of the solution was adjusted to 2.0 by the addition of ammonium hydroxide (2.0 M). The reaction mixture was filtered and kept at room temperature for one week. Colorless block crystals of 1 were obtained, washed with water and ethanol, and dried in air. Yield: 0.64 g (45%). Anal. Found: C, 17.18; H, 3.45; N, 9.56. Calc. for C4H10Cl2N2O4Zn: C, 16.77; H, 3.52; N, 9.78. IR (KBr, cm−1): νas(C=O) 1638vs, 1622vs, 1547s; νs(C=O) 1418vs, 1368s, 1315vs. 1H NMR δH (400 MHz, D2O, ppm, decomposed): 3.71 (s, CH2); 13C NMR δC(D2O, ppm, decomposed): 173.45 (CO2), 51.16 (CH2). In a similar condition, the pH value of the reaction mixture was adjusted to 5.0 with the addition of ammonium hydroxide (2.0 M). The mixture was left standing for several days to give a colorless crystalline product of 2 in a yield of 0.39 g (34%). Anal. Found: C, 20.91; H, 3.79; N, 6.00 Calc. for C4H9NO6Zn: C, 20.66; H, 3.90; N, 6.02. IR (KBr, cm−1):νas(C=O) 1583vs; νs(C=O) 1473m, 1438m, 1403s. 1H NMRδH (400 MHz, D2O, ppm): 3.65 (s, CH2); 13C NMR δC(D2O, ppm): 181.48 (CO2), 55.79 (CH2).
2.2.2. [Zn(Hida)2]n·4nH2O (3)
Zinc nitrate hexahydrate (1.49 g, 5.0 mmol) was reacted with iminodiacetic acid (1.34 g, 10.0 mmol) in 20.0 ml water with continuously stirring. The pH of the solution was adjusted to 2.0 by the addition of ammonium hydroxide. The reaction mixture was filtered and kept at room temperature for one week. Colorless block crystals of 3 were obtained, washed with water and ethanol, and dried in air. Yield: 1.27 g (31.6%). Anal. Found: C, 24.14; H, 4.81; N, 6.96. Calc. for C8H20N2O12Zn: C, 23.92; H, 5.01, N, 6.97. IR (KBr, cm−1):νas(C=O) 1664vs;νs(C=O) 1422vs, 1378vs, 1312m. 1H NMR δH (400 MHz, D2O, ppm, decomposed): 3.68 (s, CH2); 13C NMR δC (D2O, ppm, decomposed): 173.84 (CO2), 51.48(CH2).
2.2.3. [Zn(H2O)(ida)(phen)]·2H2O (4)
The procedure was similar to that for complex 2. Reaction of zinc chloride (0.68 g, 5.0 mmol), iminodiacetic acid (0.67 g, 5.0 mmol) and 1,10-phenanthroline (1.01 g, 5.0 mmol) at pH 7.0 results in the isolation of 4 in a yield of 0.99 g (46%). The complex was also prepared from the addition of 1,10-phenanthroline (1.01 g, 5.0 mmol) to the aqueous solution of [Zn(H2O)2(ida)]n (2) (1.16 g, 5.0 mmol) in 78% yield (1.67 g). Anal. Found: C, 44.28; H, 4.36; N, 9.74. Calc. for C16H19N3O7Zn: C, 44.62; H, 4.44; N, 9.76. IR (KBr, cm−1): νas(C=O) 1625vs, 1597vs, 1518m;νs(C=O) 1426m, 1387s, 1297m.
2.3. X-ray crystallography
Data collections of compounds 1–4 was performed on an Oxford Gemini S Ultra system with graphite monochromate Mo Kα radiation (λ = 0.7107 Å) at 173 K. An absorption correction was applied by using the program CrysAlis (multi-scan) [30]. The structures were solved using the WinGX package [31] and refined by full-matrix least-squares procedures with anisotropic thermal parameters for all the non-hydrogen atoms using SHELXL-97 [32]. Hydrogen atoms were included and located from difference Fourier maps but not refined anisotropically. Crystal data collections and refinement parameters are summarized in Table 1. Selected bond lengths and bond angles are listed in Supplementary material (see Table S1).
Table 1.
Crystal data summaries of intensity data collections and structure refinements for (NH4)n[Zn(Hida)Cl2]n (1), [Zn(H2O)2(ida)]n (2), [Zn(Hida)2]n·4n H2O (3), [Zn(H2O)(ida)(phen)]·2H2O(4).
| (1) | (2) | (3) | (4) | |
|---|---|---|---|---|
| Empirical formula | C4H10Cl2N2O4Zn | C4H9NO6Zn | C8H20N2O12Zn | C16H19N3O7Zn |
| Formula weight | 286.41 | 232.49 | 401.63 | 430.71 |
| Crystal color | colorless | colorless | colorless | colorless |
| Crystal size (mm) | 0.38 × 0.10 × 0.08 | 0.42 × 0.13 × 0.10 | 0.40 × 0.40 × 0.20 | 0.35 × 0.12 × 0.10 |
| Temperature (K) | 173(2) | 173(2) | 173(2) | 173(2) |
| λ (Mo Kα) (Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Crystal system | monoclinic | orthorhombic | orthorhombic | triclinic |
| Space group | P 21/n | P c 21/b | C 2 2 2 | P 1̄ |
| a (Å) | 7.3123(1) | 5.2512(1) | 12.951(2) | 6.6205(3) |
| b (Å) | 16.1727(4) | 9.7224(3) | 17.855(2) | 10.6527(5) |
| c (Å) | 8.4717(2) | 14.2052(4) | 6.6597(8) | 11.5500(6) |
| α (°) | 95.192(4) | |||
| β (°) | 107.561(2) | 91.936(4) | ||
| γ (°) | 92.193(4) | |||
| V (Å3) | 955.17(4) | 725.24(3) | 1540.0(3) | 810.07(7) |
| Z | 4 | 4 | 4 | 2 |
| Dcalc (g·cm−3) | 1.992 | 2.129 | 1.732 | 1.766 |
| Abs. coefficient (mm−1) | 3.115 | 3.379 | 1.661 | 1.566 |
| F000 | 576 | 472 | 832 | 444 |
| Reflections collected/unique/Rint | 7834/3147/0.0373 | 4066/2093/0.0024 | 3901/2290/0.0186 | 7716/3130/0.0222 |
| Data/restraints/parameters | 3147/3/130 | 2093/7/121 | 2290/7/125 | 3130/9/262 |
| θ range (°) | 2.82 – 32.51 | 3.55 – 32.50 | 4.39 – 32.59 | 2.73 – 26.00 |
| Goodness-of-fit on F2 | 0.968 | 0.935 | 1.020 | 1.141 |
| Final R indices[I > 2(I)] | R1a = 0.0302 | R1a =0.0237 | R1a =0.0314 | R1a = 0.0326 |
| wR2b = 0.0575 | wR2b =0.0504 | wR2b =0.0869 | wR2b =0.0891 | |
| R indices (all data) | R1a = 0.0468 | R1a = 0.0296 | R1a = 0.0344 | R1a = 0.0387 |
| wR2b = 0.0579 | wR2b = 0.0513 | wR2b =0.0877 | wR2b = 0.0908 | |
| Largest diff. peak/hole (e.Å−3) | 0.573/−0.582 | 0.392/−0.528 | 0.929/−0.888 | 0.876/−0.883 |
R1 = Σ||Fo| − |Fc||/Σ(|Fo|).
wR2 =Σ[w(Fo2 −Fc2)2]/Σ[w(Fo2)2]1/2.
3. Results and discussion
3.1. Synthesis
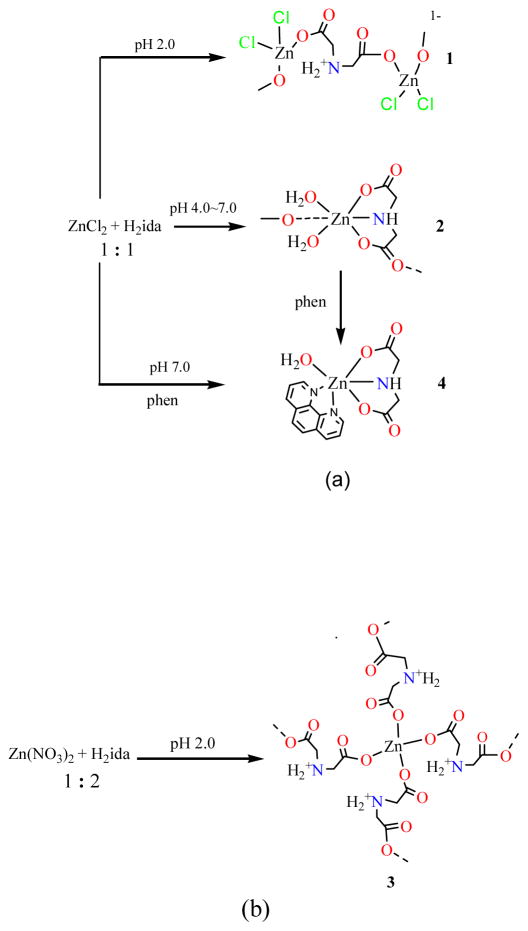
It has been showed that self-assembly processes of zinc coordination polymers are highly affected by several factors such as the ligand’s nature, medium, metal-ligand ratio, pH value, and counterion [33]. The syntheses and transformation of zinc iminodiacetato coordination polymers are outlined in Scheme 1, which illustrates the sensitivity of the reactions to pH and the metal-to-ligand molar ratio in aqueous solution. The formations of 1 and 2 show a pH dependent reaction pattern. The [ZnL2] core (L = Cl or H2O) is a dominant fragment for the construction of the complexes. Compound 1 (NH4)n[Zn(Hida)Cl2]n was prepared from the reaction of zinc chloride and iminodiacetic acid at pH 2.0, while the increase of pH value resulted in the formation of [Zn(ida)(H2O)2]n (2) in weak acidic solution between pH 4.0 ~ 7.0 (Scheme 1a).
Scheme 1.
Syntheses and transformation of zinc iminodiacetato complexes at various pH values and molar ratios.
From Scheme 1b, the reaction between zinc nitrate and iminodiacetic acid in ZnII/H2ida ratio of 1:2 at pH 2.0 gave a novel 3-D coordination polymer [Zn(Hida)2]n·4H2O (3). Therefore, at lower pH value of 2.0, tetracoordinated zinc species 1 and 3 are both obtained. Formation of hydrogen iminodiacetato zinc complex can be accomplished by the controlling low pH value and temperature, in which the bridging monodentate mode of iminodiacetate is retained. Usual method for the synthesis of [Zn(ida)2]2− unit starts from the zinc salt and excess iminodiacetate ligand [21]. However, in the present report, the structures of polymeric units [Zn(Hida)Cl2]− and [Zn(Hida)2] are both constructed by hydrogen iminodiacetato ligand that is unusual with the protonation of imine group.
At pH 7.0, a monomeric phenanthroline iminodiacetato zinc complex [Zn(ida)(phen)(H2O)]·2H2O (4) was isolated by the reaction of zinc chloride with iminodiacetic acid in the presence of 1,10-phenathroline (phen) in an aqueous solution, which could be also obtained by the addition of phen with the solution of 2 in 78% yield. As a “terminal” chelating ligand, 1,10-phen can often reduce structural dimensionality and lead to the formation of discrete metal complexes in the carboxylate system [34]. The reactions proceed as shown in Scheme 1. At lower pH value of 2.0, the bridging monodentate coordination mode of the iminodiacetate ligand has been observed in tetracoordinated zinc complexes 1 and 3. When pH value was further increased to 4.0 ~ 7.0, hexacoordinated zinc complexes 2 and 4 were formed, the ida ligand displays tetradentate and tridentate coordination modes respectively. The result shows that the structure geometry is controlled and modulated by the pH value in the reaction [35, 36].
3.2. Crystal structures
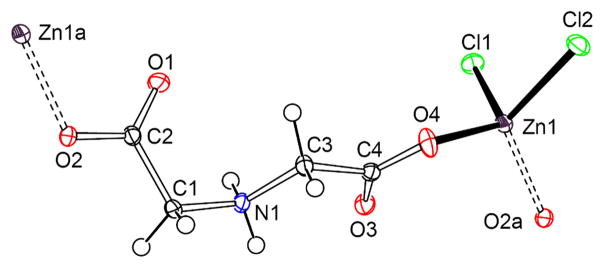
Single-crystal X-ray diffraction reveals that the asymmetric unit of 1 contains one Zn(II) atom, one iminodiacetate and two chloride ion. Fig. 1 shows a remarkable feature of polymeric configuration with respect to the bridging monodentate bridge of 1. Each zinc atom is four-coordinate by two oxygen atoms from ida ligand and two chloride ions in an approximately tetrahedral geometry. The iminodiacetate ion coordinates to zinc atom as a bridging monodentate ligand via the two terminal carboxy groups into one dimensional chain structure (Fig. S1), while the imino nitrogen atom remains free and protonated. The bridging monodentate coordination mode of the iminodiacetate ion in 1 has also been found in diiminodiacetato manganese complex in two dimensional structure [Mn(Hida)2(H2O)2] [37, 38].
Fig. 1.
ORTEP plot of the anion structure for (NH4)n[Zn(Hida)Cl2]n (1) at the 30% probability level.
The Zn(1)–O distances [1.989(1), 1.966(1) Å] in 1 are shorter than the other iminodiacetato zinc complexes reported as shown in Table 2, this may be related to the low coordination number with less hindrance. Furthermore, the coordination of the other oxygen atoms in carboxy groups is very weak [Zn(1)–O(1a) 2.971(3), Zn(1)–O(3) 3.102(3) Å, symmetry code: a−1 + x, y, −1 + z], which is consistent with the tetrahedral structure. Protonation in iminodiacetato ligand of 1 is unique, which is different from the other Zn-ida species [22–25]. The iminodiacetate ion in 1 uses its protonated imine group forming inter-molecular hydrogen bonding with the deprotonated terminal carboxy groups [N (1)···O(3b) 2.774(2) A, N–H…O 151.2°, Symmetry code: b 2−x, y, 2−z]. This, in turn, contributes to the stabilization of [Zn(Hida)Cl2]nn− core.
Table 2.
Comparisons of Zn O and Zn N bond distances (Å) in iminodiacetato zinc complexes (adenine = Hade, imidazole = im)
| CN | Complexes | Zn–N | Zn–O (β-carboxy) | Ref. |
|---|---|---|---|---|
| 4 | (NH4)n[Zn(Hida)Cl2]n(1) | – | 1.989(1), 1.966(1) | This work |
| [Zn(Hida)]n.4H2O(3) | – | 1.963(2), 1.973(2) | ||
| 6 | [Zn(ida)(H2O)2]n (2) | 2.114(2) | 2.010(2), 2.092(2), 2.251(2) | |
| [Zn(ida)(phen)(H2O)]·2H2O (4) | 2.132(2), 2.091(2), 2.153(2) | 2.187(2), 2.070(2) | ||
| K2[Zn(ida)2]·3H2O | 2.16(2) | 2.12(2), 2.03(2) | 23 | |
| La[Zn(Hida)(ida)2]·½H2O | 2.137(3) | 2.092(3), 2.126(3) | 24 | |
| [Zn(ida)(Hade)(H2O)2] | 2.167(2) | 2.112(2), 2.125(2) | 25 | |
| [Zn(ida)(CH2im2)]·H2O | 2.080(3)–2.177(3) | 2.106(2)–2.209(3) | 26 |
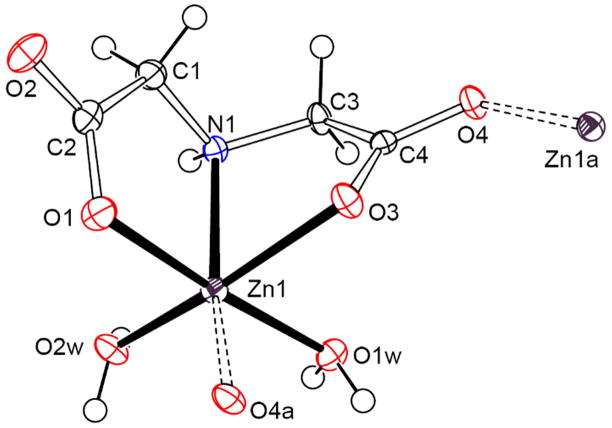
Polymeric species [Zn(ida)(H2O)2]n in 2 is joined by the carboxy group of iminodiacetate as shown in Fig. 2. The iminodiacetate ligand coordinates in a tridentate fashion to zinc atom through the nitrogen atom and the oxygen atoms of carboxy groups, while one of the uncoordinated oxygen atoms of carboxy group further connects with the neighboring zinc atom in the other unit, forming a zigzag chain structure (Fig. S2). The coordination of the zinc atom exists in a distorted octahedron. The principal dimensions of Zn–Ocarboxy distances [2.010(2), 2.092(2) and 2.251(2) Å] are longer than the tetracoordinate zinc complex 1, while the bond distance of bridged carboxy group [Zn(1)–O(3) 2.251(2) Å] is the longest.
Fig. 2.
ORTEP plot of the molecular structure for [Zn(ida)(H2O)2]n (2) at the 30% probability level.
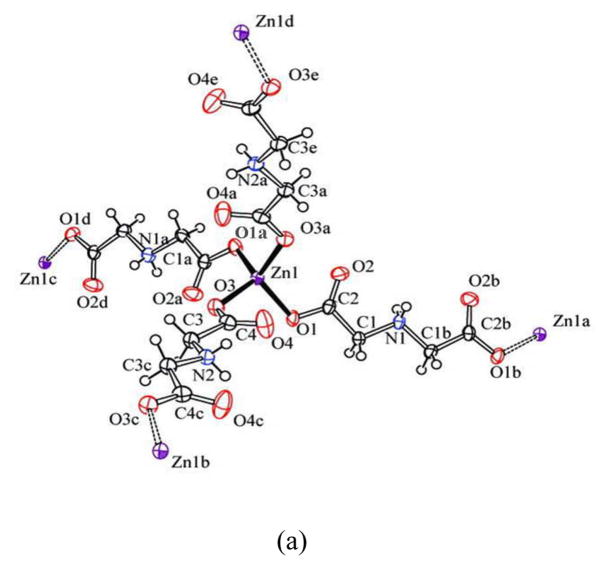
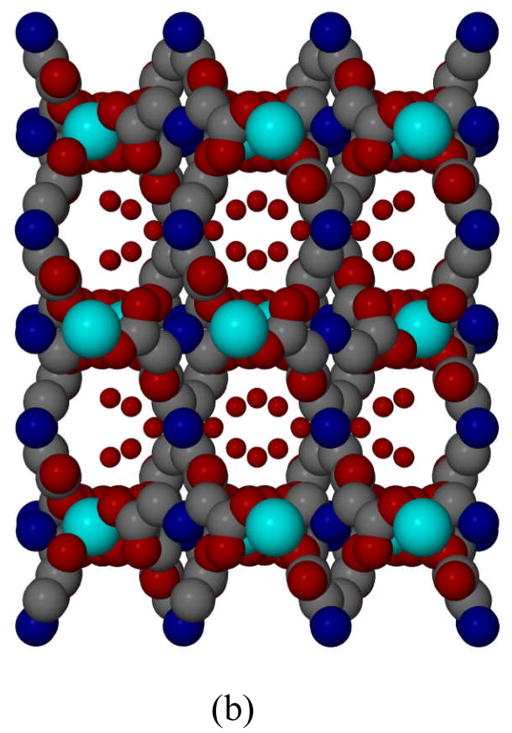
Unlike the preparations of 1 and 2, when zinc nitrate was used to react with iminodiacetic acid instead of zinc chloride, a new 3-D interpenetrating diamondoid network 3 is produced. The ZnII ions of the product exist only in one kind of coordination environment. Each unit of 3 contains one Zn atom, two ida ligand and four water molecules. As illustrated in Fig. 3a, Zn atom is tetracoordinated by four carboxy oxygen atoms [Zn(1) O(1) 1.973(2), Zn(1) O(1a) 1.973(2), Zn(1)–O(3) 1.963(2), Zn(1) O(3a) 1.963(2) Å], which are from different bridging ida ligands to complete a distorted tetrahedral coordination geometry. Compared with the structure of 1, the similar uncoordination of N donor atoms is observed. This is due to the protonation of imine group in ida ligand. However, the Zn(1) ion in 3 is connected to the four adjacent ZnII ions through the hydrogen iminodiacetate (Hida) bridges. This leads to a robust 3-D polymeric network with inter-penetrating diamondoid architecture shown in Fig. 3b. The size of the channel is 11.57 Å that comparable to the other compounds [39]. The diameter of these channels is defined by the O–O distance of the opposite ida ligands. Moreover, the space filling model clearly shows the open space still exists within the network of 3 after the interpenetration, which is occupied by the free water molecules. The water molecules were filled in the void space left after the interpenetration by N–H … O hydrogen bond interaction between aqua molecules and protonated imine group of ida ligand. [N(1)…O(1w) 2.756(4) Å, N–H…O 161(11)°]
Fig. 3.
(a) ORTEP drawing of [Zn(Hida)2]n·4nH2O (3), drawn with displacement ellipsoids at the 50% probability level. Free water molecules are omitted; (b) Schematic representation of 3-D diamondoid architecture by tetrahedral ZnII bridge in complex 3 viewed down the a axis (Zn: light blue, O: red, N: blue, C: gray, partial H atoms omitted for clarity).
The monomeric complex 4 consists of neutral unit [Zn(ida)(phen)(H2O)] and two water molecules. The zinc ion is located at the center of a distorted octahedral coordination geometry with a typical fac–NO+O (apical) coordination mode by ida ligand. It consists of the imine N atom and two carboxy oxygen atoms [O(1) and O(3)] from the coordinated ida anion and two nitrogen atoms [N(2), N(3)] from phenanthroline and one oxygen atom (O1w) from water molecules in apical position (Fig. 4a). Due to the trans-influence, the axial Zn–O distances [Zn(1)–O(1) 2.187(2), Zn(1)–O(1w) 2.176(2) Å] are much longer than those of the other metal-coordinated atoms [Zn(1)–O(3) 2.070(2), Zn(1) N(1) 2.132(2), Zn(1) N(2) 2.153(2), Zn(1) N(3) 2.091(2) Å], which complete the distorted octahedral coordination. The bond angles O(1w)–Zn(1)–O(1) 173.57(7)°, N(3)–Zn(1)–N(1) 172.90(8)° and O(3)–Zn(1)–N(2) 175.75(8)° (Table S1) also show obvious deviation from the ideal angle of 180°.
Fig. 4.
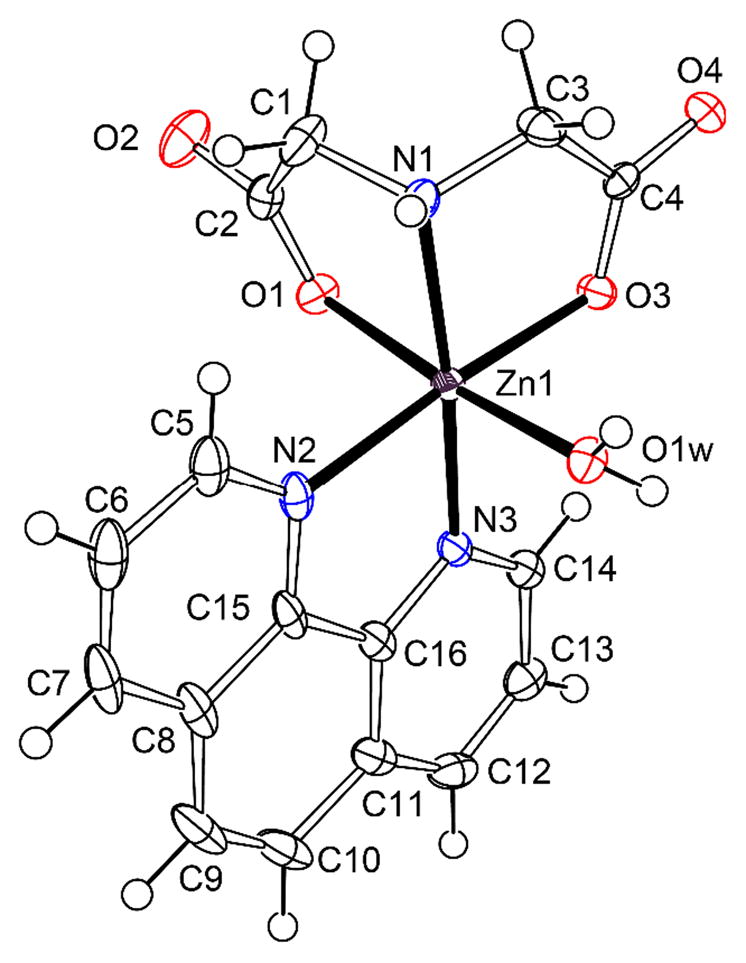
(a) ORTEP plot of the molecular structure for [Zn(ida)(phen)(H2O)]·2H2O (4) at the 30% probability level, Solvent water molecules are omitted for charity; (b) Tetrametric water cluster gathering four neighboring units of complex 4 by hydrogen-bond interaction viewed down the b axis.
Two water molecules [O(2w), O(3w)] in asymmetric unit are connected to the other two centrosymmetrically related water molecules [(O(2wa), O(3wa)], forming a novel cyclic nonplanar tetrameric water cluster (Fig. 4b). The hydrogen bonding distances between O(2w)…O(3w) and O(2w)… O(3wa) are 2.777(6) Å and 2.901(7) Å respectively, compared to that of 2.759 Å in ice with Ih symmetry [40], or 2.85 Å in liquid water [41]. O(2w) in 4 only acts as a double hydrogen-bond donor to O(3w) and O(3wa), while O(3w) not only acts as a double hydrogen-bond acceptor, but also acts as a double hydrogen-bond donor to the nearby free carboxy oxygen atoms. This assembles four [Zn(ida)(phen)(H2O)] units around each tetrameric water cluster. Furthermore, intermolecular hydrogen bonds [O(1w)…H–O(3c), 2.759(3); O(1w)…H–O(1d) 2.806(3); Å] are formed between neighboring molecules. The overall structure is further stabilized by the intermolecular π-π interactions between the two phen entities, in which the centroid centroid distance of the stacked ring is 3.431(2) and 3.581(2) Å respectively. Therefore, these multiple interactions play an important role in the structure and the formation of an extended 3D supramolecular assembly (see Fig. S3). This is associated with the fluorescent property of 4 in solid state, which exhibits a strong peak at 393 nm when excited at 331 nm, which is attributed to π–π* transition of the coordinated phen ligand (Fig. S4).
3.3. NMR Spectroscopy
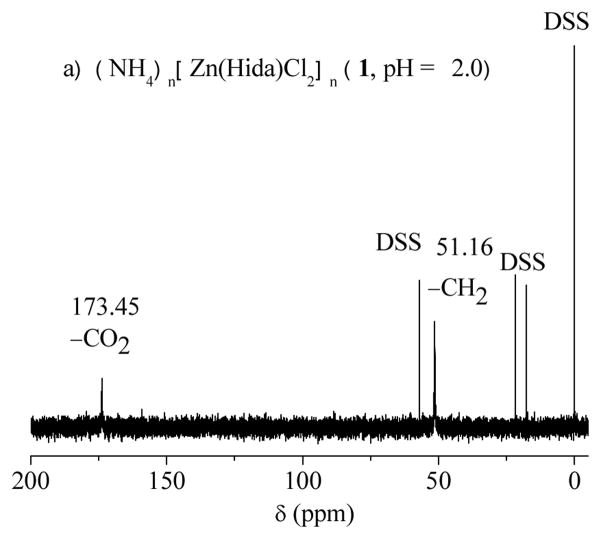
1 and 3 are soluble in water, while 2 is slightly soluble. Although a large diversity of metal coordination polymers has been reported, water-soluble examples remain very scant [42]. In order to obtain more structural information for complexes 1–3 in aqueous solution, solution 13C NMR spectra of 1–3 in D2O were shown in Fig. 5, and the chemical shifts are listed in Table 4. The experiment is further compared with the solution 13C NMR spectra of the free iminodiacetate ligand at various pH values adjusted by ammonium hydroxide (Fig. S5). The later demonstrates that the resonances of iminodiacetate changed with pH value. In the pH range of 1.0 – 7.0, the 13C resonances of iminodiacetate shifts downfield with rising pH value. However, the downfield shifts of the 13C resonances at pH 7.0 are almost the same as that at pH 5.0.
Fig. 5.
13C NMR spectra of the complexes in aqueous solution. a) (NH4)n[Zn(Hida)Cl2]n (1) (pH = 2.0); b) [Zn(Hida)]n.4nH2O (3), pH = 2.0; c) [Zn(ida)(H2O)2]n (2) (pH = 7.0).
Table 4.
13C NMR spectra data (δ, ppm) a of the free iminodiacetateligand and coordination polymers 1–3 in D2O.
| Free idab (pH = 2.0) | 1b (pH = 2.0) | 3b (pH = 2.0) | 2b (pH = 7.0) | Free idab (pH = 7.0) | |
|---|---|---|---|---|---|
| (CO2) | 173.11 | 173.45 (0.34) | 173.84 (0.73) | 181.48 (7.39) | 174.09 |
| -CH2 | 51.05 | 51.16 (0.11) | 51.48 (0.43) | 55.79 (4.02) | 51.77 |
Δδ values in brackets are chemical shifts of 1–3 compared to the corresponding carbons in the free iminodiacetate at the same pH.
13C NMR spectra of 1–3 in D2O solution were taken at autogenous pH, and 13C NMR spectra of the corresponding free iminodiacetate at the same pH values.
In comparison with the corresponding carbons in free iminodiacetic acid at the same pH value (pH = 2.0), the chemical shifts in 1 show a little downfield (173.45 and 51.16 ppm). The related ΔδC values (ΔδC = 0.34, 0.11 ppm) are very small. So the two peaks can be assigned to the resonances of carboxy and methylene carbons for the free iminodiacetate ligand. Moreover, the solution 13C NMR spectrum of 3 at pH 2.0 is very similar to that of 1, which gives only one set of resonances. This is comparable with that of free iminodiacetate ligand at the corresponding pH value of 2.0. The results reveal that both of tetracoordinated zinc species 1 and 3 decompose into free ida ligand in solution. There is a equilibrium between the solid coordination polymer 1 or 3 and the solution zinc ion and free ida ligand. The much stronger ligand of chloride anion in 1 is useful to block the further formation of three dimensional network like 3, which is constructed by the nitrate salt with weak coordination ability.
It is noteworthy that the long time acquisition of 13C NMR spectrum of 2 in aqueous solution (pH 7.0) gives obvious downfield shifts of carbon signals. In particular, it is found that terminal carboxy carbons show large downfield shifts ΔδC of 7.39 ppm compared with the free ida ligand at pH 7.0 [13C NMR δC (D2O) 174.09 (CO2), 51.77 (CH2)]. This is a clear indication of the coordination of ida ligand. The methylene carbons also show observable downfield shift ΔδC of 4.02 ppm. However, only one set of 13C peaks is observed in complex 2, indicating that the two terminal carboxy groups are in one type of coordination mode in solution. On the basis of the NMR experiment, the reasonable conclusion can be drawn: the complex 2 is discrete in aqueous solution. Therefore, the coordination polymer 2 might decompose into trihydrate iminodiacetato zinc complex [Zn(ida)(H2O)3] (5) in aqueous solution, which could be served as a water substituted product of 2 by the bridging oxygen atom of carboxy group in ida ligand. Although an attempt to isolate complex 5 was unsuccessful, a similar compound of trihydrate nitrilotriacetato zinc complex [Zn(Hnta)(H2O)3]·H2O (H3nta = nitrilotriacetic acid) with a free terminal carboxylic acid group has been reported recently [18]. Proposed structure of iminodiacetato zinc complex 5 is also appeared in immobilized metal ion affinity chromatography matrix [3]. Further evidence for the existence of 5 is obtained by the trapping reaction of 1,10-phenanthroline. The trapping reaction results in the formation of [Zn(ida)(phen)(H2O)]·2H2O (4) by the addition of phenanthroline to the aqueous solutions of polymer 2, in which the two water molecules are substituted by phen group, forming a monomeric species (Scheme 2). [Fig. 5 and Scheme 2]
Scheme 2.
Possible intermediate species and reaction in aqueous solutions of coordination polymer 2
3.4. Thermogravimetric analyses
In order to understand the coordination polymers more fully in terms of thermal stability, we examined 1–3 using TGA. The experiment was performed on samples consisting of single crystals of 1 – 3 under a N2 atmosphere with heating rate of 10 °C min−1 (Fig. S6). The TGA profile of 1 shows a weight loss of 31.1% that is attributed to the loss of one NH3 and two HCl molecules during 30–349 °C (calcd 31.5%), and then decomposition of the organic ligand occurs from 349 to 687 °C, leading to formation of zinc oxide as the residue (observed 27.1%, calcd 28.3%). Complex 2 loss two coordinated water molecules (observed 15.4%, calcd 15.5%) between 95 and 135°C, and sharp weight loss of 55.4% (calcd 56.3%) in the temperature range of 382–619°C, corresponds approximately to the loss of one ida ligand and the final residue is zinc oxide (calcd 35.0%). For complex 3, the first weight loss corresponding to the release of four lattice water molecules from room temperature to 234 °C (observed 19.3%, calcd 17.9%). From 234 to 871°C, the anhydrous compound decomposes with stepwise loss of the organic portion, leading to the collapse of the 3-D framework. However, it is difficult to determine these weight losses due to the dissociation of the organic fractions. The remaining weight should be attributed to the formation of zinc oxide observed 20.1%, calcd 20.2%). Therefore, this fact suggests that the framework of 3 may serve as potential porous materials.
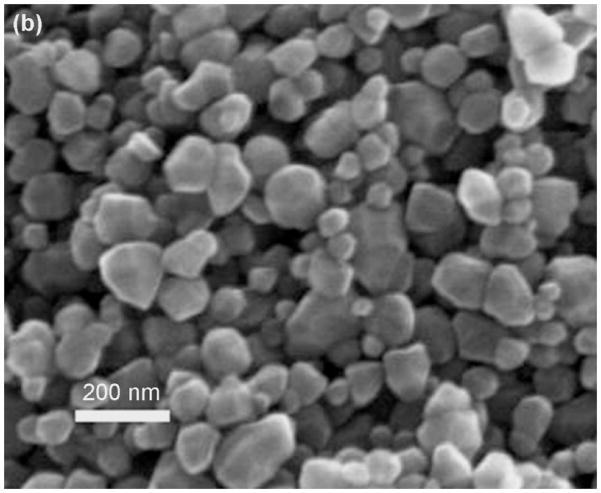
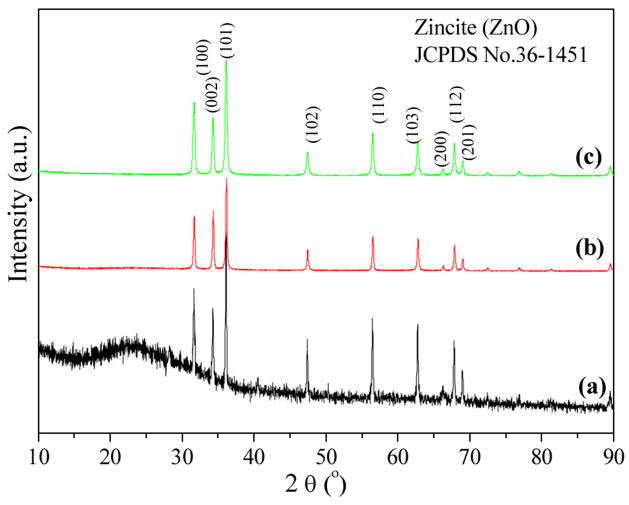
Thermal decomposition of coordination polymers 1 –3 at 500 °C for 4h in a muffle furnace results in the formation of crystals of zinc oxide, whose diameters in ranging from 25 nm ~ 11 μm respectively. Figure 6a shows the product with the shape of a regular hexagonal pyramid from 1. The mean diameter and length of ZnO crystal are about 2.6 and 4.6 μm respectively, and the aspect (length/width) ratio is about 1.8:1. The micrographs in Fig. 6b and Fig. S8 show the irregular granular crystallites of ZnO from 2 and 3, which have diameters in range of 25 – 202 nm. The XRD patterns of these nanoparticles’ crystallinity are illustrated in Fig. 7. All diffraction peaks can be indexed as the zincite phase of ZnO (JCPDS No. 36-1451), which indicates the complexes have been thermally decomposed into ZnO. The EDS measurement indicates that only zinc and oxygen were detected in ZnO nanocrystal samples (Fig. S7).
Fig. 6.

SEM images of (a) (NH4)n[Zn(Hida)Cl2]n (1) calcined at 500°C for 4h; (b) [Zn(ida)(H2O)2]n (2) calcined at 500°C for 4h; (c) [Zn(Hida)]n.4H2O (3) calcined at 500°C for 4h.
Fig. 7.
The powder XRD patterns of (a) (NH4)n[Zn(Hida)Cl2]n (1) calcined at 500°C for 4 h, (b) [Zn(ida)(H2O)2]n (2) calcined at 500°C for 4 h, (c) [Zn(Hida)]n.4nH2O (3) calcined at 500°C for 4 h.
An explanation for the formation of different distinctive shape and size of ZnO crystallites under the same calcination conditions is related to the different network structures of the coordination polymers 1 – 3, and also the existence of chloride anion. Compared with ZnO nanparticles from one dimensional coordination polymers 1 and 2, ZnO from 3 with smaller particle size might be attributed to the 3D polymeric network of 3, which collapses slower and inhibits the growth of ZnO particles. This nucleation and growth of nanocrystalline materials was affected by the underlying molecular chemistry of the precursors [43, 44].
3.5. Photoluminescence (PL) spectra
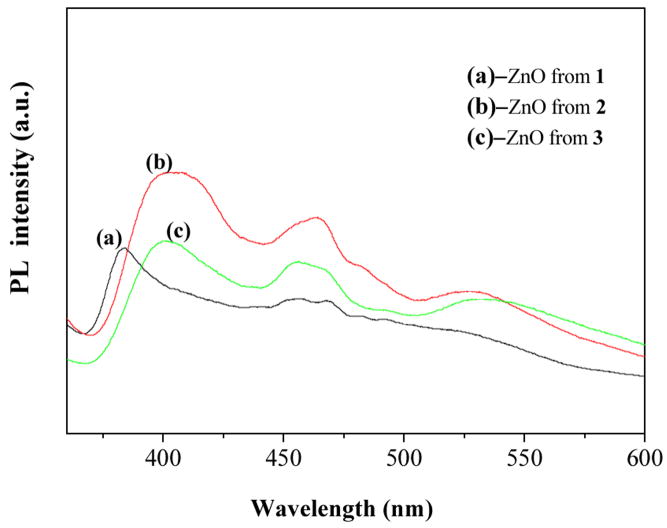
To investigate the optical properties of ZnO from the thermal decomposition of 1 – 3, PL spectra of ZnO nanocrystals using a Xe lamp as the excitation source at room temperature are shown in Fig. 8. There are three emitting bands, including a strong UV bandgap emission at round 386 nm (3.21eV), a shoulder blue band at 452 nm (2.74eV) and 462 nm (2.68 eV), and a green band emission at 532 nm (2.33eV). The strong UV emission is attributed to the near-band-edge emission arising from the excitonic recombition [45, 46]. The red shift can be observed due to the large band tail of ZnO nanoparticles [47, 48]. The green emission of ZnO is usually ascribed to the single ionized oxygen vacancy in ZnO and originates from the radiative recombination of a photogenerated hole when an oxygen vacancy traps an electron [46, 49]. The explanation for the blue shoulder emission peaks has been reported for ZnO films and particles before, yet the exact mechanism is still not clear [50, 51]. Because the ratio of UV/visible emission is a key criterion to evaluate crystalline quality of ZnO [52, 53], both sample (b) and sample (c) exhibit two relative strong UV emissions compared with sample (a), which implies that they have relative good crystal quality and it is consistent with results of XRD patterns.
Fig. 8.
Room temperature PL spectra of the ZnO nanoparticles from the decomposition of coordination polymers 1 – 3.
4. Conclusion
In conclusion, we have successfully isolated and characterized four new zinccomplexes with iminodiacetate ligand. Complexes 1 and 2 show 1-D chain like structures, while complex 3 reveals a 3-D polymeric network with a diamondiod structure. Complex 4 has mononuclear [Zn(phen)(ida)] structure with the cyclic nonplanar tetrameric water cluster, which plays an important role in stabilizing the structure in 4. It is of interest that coordination polymers 1, 2 and 3 are obtained depending on controlling pH values and the metal-to-ligand molar ratio in aqueous solution. The detailed solution studies by NMR analysis show that coordination polymers 1 and 3 with good solubility and decompose into free ida ligand in solution, while coordination polymer 2 might decompose into discrete species like 5. We have also demonstrated complexes 1–3 can serve as single molecular precursors for the preparation of zinc oxide. The same idea is applicable to the synthesis of a wide range of nanomaterials with a coordination polymer as a precursor.
Supplementary Material
Table 3.
Hydrogen bonds (Å and °) in 4
| D–H…A | D–H | H…A | D…A | (DHA) |
|---|---|---|---|---|
| O(2w)—H(2w1)…O(3wa) | 0.87(1) | 2.20(7) | 2.901(7) | 138(8) |
| O(2w)—H(2w2)…O(3w) | 0.88(1) | 1.94(3) | 2.777(6) | 161(8) |
| O(3w)—H(3w1)…O(2b) | 0.86(1) | 2.34(7) | 2.809(3) | 115(7) |
| O(3w)—H(3w2)…O(4) | 0.86(1) | 1.94(3) | 2.757(3) | 160(6) |
| O(1w)—H(1w1)…O(3c) | 0.85(1) | 1.91(1) | 2.759(3) | 173(3) |
| O(1w)—H(1w2)…O(1d) | 0.85(1) | 1.96(1) | 2.806(3) | 173(4) |
Symmetry codes: a, −x, −y, −z − 1; b, 1 − x, −y, −z; c, −x, 1 −y, −z; d, x − 1, y, z.
Acknowledgments
This work is supported by the Ministry of Science and Technology (2005CB221408) and the authors thank the Cramer research group and the NIH support of its instruments (GM65440).
Appendix A. Supplementary material
CCDC 694048, 694049, 724739 and 694050 contain the supplementary crystallographic data for 1 – 4. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html, or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223-336-033; or e-mail: deposit@ccdc.cam.ac.uk. Supplementary data of Photoluminescence spectra, UV-Vis diffuse reflection spectra, IR and 1H NMR spectra; TGA; SEM images and EDS results in the online version, at doi:10.****/j.jssc.2009.
References
- 1.Porath J, Carlsson J, Olsson I, Belfrage G. Nature. 1975;258:598. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 2.Chaga GS. J Biochem Biophys Methods. 2001;49:313. doi: 10.1016/s0165-022x(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 3.(a) Gaberc-Porekar V, Menart V. J Biochem Biophys Methods. 2001;49:335. doi: 10.1016/s0165-022x(01)00207-x. [DOI] [PubMed] [Google Scholar]; (b) Hochuli E, Doebeli H, Schacher A. J Chromatogr. 1987;411:177. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 4.Oleinikova M, Muraviev D, Valiente M. Anal Chem. 1999;71:4866. doi: 10.1021/ac9902345. [DOI] [PubMed] [Google Scholar]
- 5.Lin LC, Juang RS. Chem Engineering J. 2007;132:205. [Google Scholar]
- 6.Tikhonov NA, Zagorodni AA. Sepa Sci Technol. 1998;33:633. [Google Scholar]
- 7.Hochuli E, Doebeli H, Schacher A. J Chromatogr. 1987;411:177. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 8.Koike T, Kajitani S, Nakamura I, Kimura E, Shiro M. J Am Chem Soc. 1995;117:1210. [Google Scholar]
- 9.Tanase T, Yun JW, Lippard SJ. Inorg Chem. 1995;34:4220. [Google Scholar]
- 10.Abufarang A, Vahrenkamp H. Inorg Chem. 1995;34:2207. [Google Scholar]
- 11.Kirn SI, Happel CM, Hrubanova S, Weyhernuller T, Klein C, Metzler-Nolte N. J Chem Soc, Dalton Trans. 2004:1201. doi: 10.1039/b313634e. [DOI] [PubMed] [Google Scholar]
- 12.Ng CH, Kong KC, Von ST, Pauline B, Jensen P, Eswary T, Hamada H, Chikira M. J Chem Soc, Dalton Trans. 2008:447. doi: 10.1039/b709269e. [DOI] [PubMed] [Google Scholar]
- 13.Kunkely H, Volger A. J Chem Soc, Chem Commun. 1990:1204. [Google Scholar]
- 14.Johnson AL, Hollingsworth N, Kociok-kohn G, Molloy KC. Inorg Chem. 2008;47:12040. doi: 10.1021/ic801591d. [DOI] [PubMed] [Google Scholar]
- 15.Rebilly JN, Gardner PW, Darling GR, Bacsa J, Rosseinsky MJ. Inorg Chem. 2008;47:9390. doi: 10.1021/ic801097w. [DOI] [PubMed] [Google Scholar]
- 16.Wu MC, Lee CS. Inorg Chem. 2006;45:9634. doi: 10.1021/ic061531c. [DOI] [PubMed] [Google Scholar]
- 17.Yin M, Gu Y, Kuskovsky IL, Andelman T, Zhu Y, Neumark GF, O’Brien S. J Am Chem Soc. 2004;126:6206. doi: 10.1021/ja031696+. [DOI] [PubMed] [Google Scholar]
- 18.Thakuria H, Borah BM, Das G. Eur J Inorg Chem. 2007:524. [Google Scholar]
- 19.(a) Ren YP, Long LS, Mao BW, Yuan YZ, Huang RB, Zheng LS. Angew Chem, Int Ed. 2003;42:532. doi: 10.1002/anie.200390153. [DOI] [PubMed] [Google Scholar]; (b) Mukhopadhyay S, Chatterjee PB, Mandal D, Mostafa G, Caneschi A, Slageren JV, Weakley TJR, Chaudhury M. Inorg Chem. 2004;43:3413. doi: 10.1021/ic035422z. [DOI] [PubMed] [Google Scholar]; (c) Baggio R, Garland MT, Perec M, Vega D. Inorg Chem. 1996;35:2396. doi: 10.1021/ic951078x. [DOI] [PubMed] [Google Scholar]; (d) Drew MGB, Rice DA, Timewell CW. J Chem Soc, Dalton Trans. 1975:144. [Google Scholar]; (e) Delaunay J, Kappenstein C, Hugel RP. J Chem Res. 1978;48:801. [Google Scholar]
- 20.(a) Policar C, Lambert F, Cesario M, Morgenstern-Badarau I. Eur J Inorg Chem. 1999:2201. [Google Scholar]; (b) Levstein PR, Calvo R. Inorg Chem. 1990;29:1581. [Google Scholar]; (c) Rueff J-M, Masciocchi N, Rabu P, Sironi A, Skoulios A. Eur J Inorg Chem. 2001:2843. [Google Scholar]; (d) Jin S, Wang D, Chen W. Inorg Chem Commun. 2007;10:685. [Google Scholar]
- 21.Rabenstein DL, Blakney G. Inorg Chem. 1973;12:128. [Google Scholar]
- 22.Ramanujam VV, Selvarajan VM. J Indian Chem Soc. 1981;58:125. [Google Scholar]
- 23.Vasil’ev VP, Romanova LM, Egorushkina NA. Zh Neorg Khim. 1983;28:1944. [Google Scholar]
- 24.Anderegg G. Inorg Chim Acta. 1991;180:69. [Google Scholar]
- 25.(a) Sinkha UC, Kramarenko FG, Polynova TN, Porai-Koshits MA, Mitrofanova ND. Zh Strukt Kim. 1975;16:144. [Google Scholar]; (b) Kramarenko FG, Polynova TN, Porai-Koshits MA, Chalyi VP, Mitrofanova ND. Koord Khim. 1975;1:1423. [Google Scholar]
- 26.Xu HB, Zhao YH, Su ZM, Li GH, Ma Y, Shao KZ, Zhu DX, Zhang HJ, Yue SM. Chem Lett. 2004;33:446. [Google Scholar]
- 27.Morel AC, Choquesillo-Lazarte D, Alarcon-Payer C, Gonzalez-Perez JM, Castineiras A, Niclos-Gutierrez J. Inorg Chem Commun. 2003;6:1354. [Google Scholar]
- 28.Jin SW, Wang DQ, Chen WZ. Inorg Chem Commun. 2007;10:685. [Google Scholar]
- 29.Selvakumar B, Rajendiran V, Maheswari PU, Stoeckli-Evans H, Palaniandavar M. J Inorg Biochem. 2006;100:316. doi: 10.1016/j.jinorgbio.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Oxford Diffraction. CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd; Abingdon, United Kingdom: 2005. [Google Scholar]
- 31.Farrugia LJ. J Appl Cryst. 1999;32:837. [Google Scholar]
- 32.Sheldrick GM. Programs for Crystal Structure Analysis. University of Gottingen; Germany: 1999. SHELX-97. [Google Scholar]
- 33.(a) Holliday BJ, Mirkin CA. Angew Chem Int Ed. 2001;40:2023. [PubMed] [Google Scholar]; (b) Roesky HW, Andruh M. Coord Chem Rev. 2003;236:91. [Google Scholar]; (c) Bu XH, Xie YB. [Google Scholar]; (d) Li JR, Zhang RH. Inorg Chem. 2003;42:7422. doi: 10.1021/ic034454j. [DOI] [PubMed] [Google Scholar]; (e) Zhou YF, Lou BY, Yuan DQ, Xu YQ, Jinag FL, Hong MC. Inorg Chim Acta. 2005;358:3057. [Google Scholar]; (f) Wang JJ, Gou L, Hu HM, Han ZX, Li DS, Xue GL, Lin M. QZ Shi Cryst Growth Des. 2007;7:1514. [Google Scholar]
- 34.(a) Ye BH, Tong ML, Chen XM. Coord Chem Rev. 2005;249:545. [Google Scholar]; (b) He X, Li YN, Zhang Y, Zhang LP, Xu JN, Wang Y. Inorg Chem Commun. 2005;8:983. [Google Scholar]; (c) Li XJ, Sun DF, Cao R, Sun YQ, Wang YQ, Bi WH, Gao SY, Hong MC. Inorg Chem Commun. 2003;6:908. [Google Scholar]
- 35.Wang RH, Han L, Jiang FL, Zhou YF, Yuan DQ, Hong MC. Cryst Growth Des. 2005;5:129. [Google Scholar]
- 36.Go YB, Wang XQ, Anokhina EV, Jacobson AJ. Inorg Chem. 2005;44:8265. doi: 10.1021/ic050644d. [DOI] [PubMed] [Google Scholar]
- 37.Yong GP, Jiang C, Wang ZY. Z Anorg Allg Chem. 2003;629:1898. [Google Scholar]
- 38.Zhao QH, Li HF, Fang RB. Trans Metal Chem. 2003;28:220. [Google Scholar]
- 39.Kremer C, Morales P, Torres J. Inorg Chem Commun. 2008;11:862. [Google Scholar]
- 40.Eisenberg D, Kauzmann W. The structure and Properties of Water. Oxford University Press; Oxford: 1969. [Google Scholar]
- 41.Lu J, Yu JH, Chen XY, Cheng P, Zhang X, Xu JQ. Inorg Chem. 2005;44:5978. doi: 10.1021/ic0507621. [DOI] [PubMed] [Google Scholar]
- 42.Lidrissi C, Romerosa A, Saoud M, Serrano-Ruiz M, Gonsalvi L, Peruzzini M. Angew Chem, Int Ed. 2005;44:25. doi: 10.1002/anie.200462790. [DOI] [PubMed] [Google Scholar]
- 43.Rautio J, Per P, Honkamo J, Jantunen H. Microchem J. 2009;91:272. [Google Scholar]
- 44.Cho KS, Hong JI, Chung CI. Polymer Engineering and Science. 2004;44:1702. [Google Scholar]
- 45.Liang J, Liu J, Xie Q, Bai S, Yu W, Qian Y. J Phys Chem B. 2005;109:9463. doi: 10.1021/jp050485j. [DOI] [PubMed] [Google Scholar]
- 46.Vanheusden K, Warren WL, Ceager CH, Tallant DR, Voigt JA. J Appl Phys. 1996;79:7983. [Google Scholar]
- 47.Xu XL, Lau SP, Chen JS, Sun Z, Tay BK, Chai JW. Mater Sci Semicond Process. 2001;4:617. [Google Scholar]
- 48.Dijken AV, Meulenkamp EA, Vanmaekelbergh D, Meijerink A. J Lumin. 2000;87–89:454. [Google Scholar]
- 49.Wu XL, Siu GG, Fu CL, Ong HC. Appl Phys Lett. 2001;78:2285. [Google Scholar]
- 50.Dai L, Chen XL, Wang WJ, Zhou T, Hu BQ, Phys J. Condents Matter. 2003;15:2221. [Google Scholar]
- 51.Li F, Jiang Y, Hu L, Liu LY, Li Z, Huang XT. Jouranl of Alloys and Compounds. 2009;474:531. [Google Scholar]
- 52.Wang M, Zhang Lidw. Materials Letter. 2009;63:301. [Google Scholar]
- 53.Xing YJ, Xi ZH, Xue ZQ, Zhang XD, Song JH, Wang RM. Apply Phys Lett. 2003;83:1689. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.