Abstract
Oxygen is critically important to metazoan life, and the EGL-9/PHD enzymes are key regulators of hypoxia (low oxygen) response. When oxygen levels are high, the EGL-9/PHD proteins hydroxylate hypoxia-inducible factor (HIF) transcription factors. Once hydroxylated, HIFα subunits bind to von Hippel-Lindau (VHL) E3 ligases and are degraded. Prior genetic analyses in Caenorhabditis elegans had shown that EGL-9 also acted through a vhl-1-independent pathway to inhibit HIF-1 transcriptional activity. Here, we characterize this novel EGL-9 function. We employ an array of complementary methods to inhibit EGL-9 hydroxylase activity in vivo. These include hypoxia, hydroxylase inhibitors, mutation of the proline in HIF-1 that is normally modified by EGL-9, and mutation of the EGL-9 catalytic core. Remarkably, we find that each of these treatments or mutations eliminates oxygen-dependent degradation of HIF-1 protein, but none of them abolishes EGL-9-mediated repression of HIF-1 transcriptional activity. Further, analyses of new egl-9 alleles reveal that the evolutionarily conserved EGL-9 MYND zinc finger domain does not have a major role in HIF-1 regulation. We conclude that C. elegans EGL-9 is a bifunctional protein. In addition to its well-established role as the oxygen sensor that regulates HIF-1 protein levels, EGL-9 inhibits HIF-1 transcriptional activity via a pathway that has little or no requirement for hydroxylase activity or for the EGL-9 MYND domain.
CELLS and tissues are often deprived of oxygen during normal development and during disease. Examples include animals that encounter hypoxic soil or aqueous microenvironments, mammalian tissues that receive insufficient oxygen when the cardiovascular system is taxed or disabled, and cells at the center of a poorly vascularized tumor. Most metazoans rely on aerobic respiration as a primary source of energy, and adaptation to hypoxia is of central importance. The hypoxia-inducible factor (HIF) transcription complexes have been termed master regulators of hypoxia response, because they regulate most hypoxia-induced changes in gene expression in animals as diverse as humans and the nematode Caenorhabditis elegans (Kaelin and Ratcliffe 2008). In mammals, these HIF targets include genes that regulate growth, energy metabolism, cellular differentiation, apoptosis, inflammation, and angiogenesis (Siddiq et al. 2007; Rankin and Giaccia 2008; Weidemann and Johnson 2008).
The EGL-9/PHD proteins act as cellular oxygen sensors, and they are at the core of HIF regulatory networks. When oxygen levels are sufficiently high, PHD/EGL-9 proteins hydroxylate conserved proline residues in the HIFα subunits. Once hydroxylated, HIFα proteins bind to the von Hippel-Lindau tumor suppressor protein (VHL) (Bruick and McKnight 2001; Ivan et al. 2001; Jaakkola et al. 2001; Min et al. 2002). VHL targets HIFα for polyubiquitination and proteasomal degradation (Maxwell et al. 1999; Ohh et al. 2000).
The nematode C. elegans has provided important insights into hypoxia signaling. The egl-9 gene was first identified in genetic screens for mutations that disrupted egg laying (Trent et al. 1983) and for mutations that conferred resistance to the bacterial pathogen Pseudomonas aeruginosa (Darby et al. 1999). Subsequent studies identified C. elegans EGL-9 as the oxygen-sensitive enzyme that controlled oxygen-dependent degradation of HIF-1, and EGL-9 was shown to be orthologous to mammalian PHD1, PHD2, and PHD3 (Epstein et al. 2001). C. elegans that carry a deletion in hif-1 are not able to survive development in hypoxia (Jiang et al. 2001; Padilla et al. 2002). hif-1 and egl-9 have been shown to have roles in other important processes, including heat acclimation, neural development, behavioral responses to oxygen or carbon dioxide, cyanide resistance, and aging (Gallagher and Manoil 2001; Jiang et al. 2001; Treinin et al. 2003; Bretscher et al. 2008; Chang and Bargmann 2008; Pocock and Hobert 2008; Chen et al. 2009; Mehta et al. 2009; Miller and Roth 2009; Zhang et al. 2009).
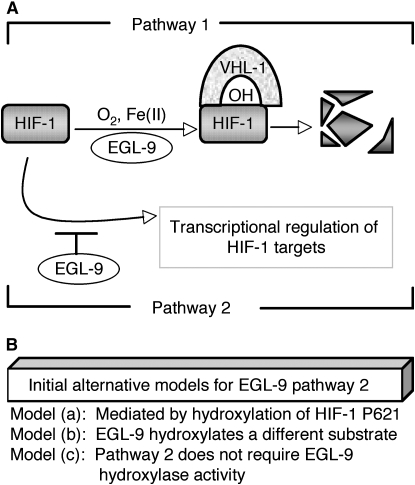
Genetic analyses in C. elegans have shown that EGL-9 regulates HIF-1 via two distinct pathways: oxygen-dependent degradation of HIF-1 and an uncharacterized vhl-1-independent pathway in which EGL-9 represses HIF-1 transcriptional activity (illustrated in Figure 1A). In previous studies, we had discovered that the mRNA transcripts for HIF-1 target genes were expressed at much higher levels in egl-9 mutants, compared to vhl-1 mutants (Shen et al. 2006). Other studies had suggested that mammalian PHD proteins might also regulate HIF activity in some VHL-independent contexts (Ozer et al. 2005; To and Huang 2005). These findings supported the intriguing hypothesis that EGL-9/PHD proteins had VHL-independent roles that might not involve HIF hydroxylation.
Figure 1.—
EGL-9 functions and models tested in this study. (A) EGL-9 regulates HIF-1 by two pathways, and they are illustrated here. First, EGL-9 controls oxygen-dependent degradation of HIF-1 (labeled pathway 1). EGL-9 hydroxlates HIF-1 on a conserved proline residue (P621), and this enables binding of HIF-1 to the VHL-1 E3 ligase. HIF-1 is then degraded. Molecular oxygen, Fe(II), and 2-oxoglutarate are required for the hydroxylation reaction. EGL-9 also suppresses expression of HIF-1 targets by a second pathway that does not require VHL-1 (labeled pathway 2 here). (B) Initial alternative models for the VHL-1-independent functions of EGL-9 (pathway 2). Each model predicts a different combination of experimental outcomes. Model a postulates that pathway 2 (like pathway 1) requires hydroxylation of HIF-1 proline 621. Model b is that EGL-9 hydroxylates a different target to inhibit HIF-1 transcriptional activity. This model predicts that all EGL-9 functions would be abrogated by mutations or treatments that eliminated EGL-9 hydroxylase activity. Model c is that EGL-9 represses HIF-1-mediated transcription by a mechanism that does not require EGL-9 hydroxylase activity.
In this study, we investigate the vhl-1-independent mechanism by which C. elegans EGL-9 represses HIF-1 activity. We find that while hydroxylation of HIF-1 at proline residue 621 by EGL-9 is required for HIF-1 destabilization, it is not essential for the vhl-1-independent functions of EGL-9. Further, we show that the two EGL-9 pathways have differing sensitivities to mutations or pharmacological treatments that impair hydroxylase activity. Collectively, these data show that EGL-9 represses HIF-1 transcriptional activity via a pathway that has little or no requirement for EGL-9 hydroxylase activity.
MATERIALS AND METHODS
Alleles and worm culture:
C. elegans were grown at 20° using standard methods (Brenner 1974). The loss-of-function alleles, transgenes, and strains described in this study are listed in supporting information (Table S1, Table S2, and Table S3). All new mutations and integration events were outcrossed to wild-type animals at least four times.
Constructs and worm transformation:
The Pegl-9∷egl-9∷tag expression construct includes 1.6 kb of egl-9 5′ regulatory sequence, genomic sequence for the first three egl-9 exons and the remaining exons from the cDNA for the predominant egl-9 mRNA isoform (egl-9a, illustrated in Figure S1). The egl-9 coding sequences are fused in frame to green fluorescent protein (GFP). Further details of plasmid construction are in supplemental methods. To create the Pegl-9∷egl-9(H487A)∷tag construct, the egl-9 codon for histidine 487 was changed to encode alanine.
The Phif-1∷hif-1∷tag construct contains 5.2 kb of hif-1 5′ regulatory sequence, the genomic sequence for the first exon and first intron of hif-1, cDNA sequence for hif-1 exons 2–9, and an epitope tag. To create the Phif-1∷hif-1(P621G)∷tag construct, the codon for proline 621 was modified to encode glycine. The hif-1 transgenes are further characterized in Figure S2 and in Zhang et al. (2009).
The egl-9 constructs were introduced to the strain ZG305 [egl-9(sa307); unc-119(ed3)], and the hif-1 constructs were introduced to ZG228 [hif-1(ia04), unc-119(ed3)], by microparticle bombardment with the unc-119 rescue plasmid (pPD#MMO16b) as a cotransformation marker (Praitis et al. 2001). The resulting transgenic strains were each backcrossed at least four times.
MOS1 mediated mutagenesis:
We generated the egl-9 loss-of-function alleles ia58, ia60, and ia61 in a screen for Mos1 transposon-mediated mutations that dramatically increased the expression of the Pnhr-57∷GFP reporter (Shen et al. 2006). The methods for Mos1 mobilization have been described previously (Granger et al. 2004). We screened ∼164,600 genomes, as diagrammed in Figure S3.
Protein blots:
Protein blots were probed with monoclonal antibodies recognizing the following epitopes: GFP (antibody from Roche at 1:1000 dilution); HA (antibody from Cell Signaling Technology clone 6E2 at 1:1000 dilution); myc (mouse ascites, clone 9E10, from the Developmental Studies Hybridoma Bank at 1:1000 dilution), or AHA-1 (Jiang et al. 2001) (1:100 dilution). The secondary antibody (goat anti-mouse IgG+IgM from Bio-Rad) was diluted 1:2000. Further information about experimental and statistical analyses are in supplemental methods (File S1).
Real-time PCR:
We used Trizol reagent (Invitrogen) to isolate total RNA from developmentally synchronized populations of L4 or young adult stage worms. Total RNA from each sample was treated by RNase free DNase (Promega) and reverse transcribed to complementary DNA using Oligo(dT18) primers and AffinityScript reverse transcriptase (Stratagene). Quantitative RT–PCR was performed using the iQ SYBR GREEN supermix (Bio-Rad) real-time PCR system, and each reaction included cDNA from 50–100ng total RNA. The primers for K10H10.2 and inf-1 have been published previously (Shen et al. 2006). inf-1 is not regulated by hypoxia and was used as an input control (Shen et al. 2005, 2006). At least three biological replicates were analyzed for each experiment, and each PCR reaction was performed in duplicate. The standard curve method was used to analyze the expression levels. Two-sample paired t-tests were used to assess statistical significance of differences.
RESULTS
Prior studies had shown that egl-9 inhibited HIF-1 by two genetic pathways: (1) VHL-1-mediated oxygen-dependent degradation (Epstein et al. 2001) and (2) repression of HIF-1 transcriptional activity via a pathway that did not require vhl-1 (Shen et al. 2006) (illustrated in Figure 1A). While oxygen-dependent degradation is well characterized, the mechanisms by which EGL-9 inhibits HIF-1 activity are not understood. In the studies described here, we have conducted a series of experiments to distinguish between alternative models for how EGL-9 represses expression of HIF-1 targets independent of VHL-1 (pathway 2 in Figure 1A). The simplest model was that EGL-9-mediated hydroxylation of HIF-1 at proline 621 resulted in both degradation of HIF-1 protein and inhibition of HIF-1 transcriptional activity (model a in Figure 1B). An alternative model was that EGL-9 hydroxylated another region of HIF-1 or a different protein to repress HIF-1-mediated transcription (model b in Figure 1B). Finally, we considered the possibility that EGL-9 inhibited HIF-1 transcriptional activity via a novel mechanism that did not require EGL-9 prolyl hydroxylase activity (model c in Figure 1B). The experiments described herein are designed to distinguish between these three models.
Effects of the HIF-1(P621G) mutation:
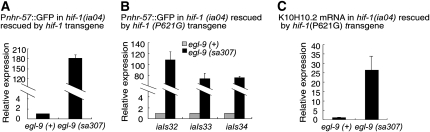
EGL-9 hydroxylates proline 621 of HIF-1 (Epstein et al. 2001). This is the only known EGL-9 target in the HIF-1 protein, and HIF-1 transgenes that carry the P621G mutation are not degraded through the EGL-9/VHL-1 pathway (Zhang et al. 2009) (Figure S2C). To determine whether the HIF-1 P621G mutation eliminated all regulation of HIF-1 by EGL-9, we examined the effects of egl-9 mutations in animals expressing HIF-1(P621G). In these experiments, the endogenous hif-1 gene was knocked out, and hif-1 function was restored by an integrated transgene expressing either epitope-tagged wild-type HIF-1 or stabilized HIF-1(P621G). To assess the effects of the mutations on HIF-1 activity, we assayed the expression of two HIF-1 target genes: K10H10.2 and the Pnhr-57∷GFP reporter (Shen et al. 2006). In control experiments, we confirmed that in animals expressing the wild-type HIF-1 transgene, a strong loss-of-function mutation in egl-9 resulted in dramatic overexpression of Pnhr-57∷GFP (Figure 2A). We then assayed the expression of HIF-1 targets in three independently isolated lines expressing HIF-1(P621G), and we found that the egl-9(sa307) mutation caused markedly higher expression of Pnhr-57∷GFP in these animals (Figure 2B). K10H10.2 mRNA levels were also significantly increased by the egl-9(sa307) loss-of-function mutation in animals expressing the stabilized HIF-1 (Figure 2C). These data demonstrated that the HIF-1(P621G) mutation did not abolish all regulation of HIF-1 by EGL-9, and this effectively disproved “model a” as described in Figure 1B.
Figure 2.—
The HIF-1(P621G) mutation does not prevent egl-9-mediated repression of HIF-1 activity. (A) In control experiments, the transgene encoding wild-type hif-1, rescued expression of Pnhr-57∷GFP in hif-1(ia04) mutant animals, as measured by protein blots. The egl-9(sa307) loss-of-function mutation dramatically increased expression of the Pnhr-57∷GFP reporter. +, the wild-type allele. (B and C) To determine whether the HIF-1(P621G) mutation abrogated all regulation by egl-9, the expression of two HIF-1 targets, Pnhr-57∷GFP and K10H10.2, were compared in egl-9(+) and egl-9(sa307) animals. These experiments were conducted in hif-1(ia04) mutants rescued by the hif-1(P621G)∷tag transgene. (B) The HIF-1(P621G) mutation did not prevent repression of Pnhr-57∷GFP by egl-9. Pnhr-57∷GFP expression was assayed by protein blots. This result was consistent across three independently isolated hif-1(P621G)∷tag transgenic lines (iaIs32, iaIs33, and iaIs34) (P < 0.01 in each case). (C) The hif-1(P621G) iaIs32 mutation does not abolish regulation of K10H10.2 expression by egl-9. Quantitative RT–PCR experiments established a significant difference in K10H10.2 mRNA levels between egl-9(+) and egl-9(sa307) (P < 0.05).
Hypoxia or iron chelator treatments that inhibit oxygen-dependent degradation of HIF-1 do not eliminate all EGL-9 functions:
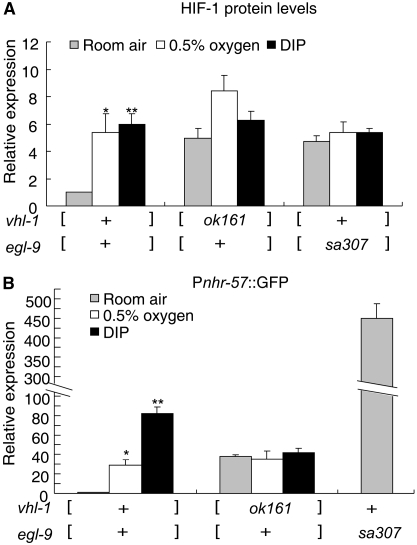
Having established that EGL-9 had functions other than hydroxylation of HIF-1 proline 621, it was important to determine whether those other functions required EGL-9 hydroxylase activity. We reasoned that if EGL-9 hydroxylated other substrates to repress HIF-1 activity (model b in Figure 1B), then all EGL-9 functions would be repressed by treatments that inhibited EGL-9 hydroxylase activity. The HIF-1 hydroxylation reaction requires oxygen, 2-oxoglutarate, and Fe+2 (Kaelin and Ratcliffe 2008). Oxygen deprivation or the iron chelator 2, 2′-dipyridyl (DIP) inhibits this reaction (Bishop et al. 2004). In control experiments, we confirmed that HIF-1 protein was stabilized by hypoxia (0.5% oxygen) or DIP treatments (Figure 3A). We next asked whether inhibitors of EGL-9 hydroxylation activity could fully phenocopy a strong loss-of-function mutation in egl-9. We found that hypoxia, DIP, or a deletion mutation in vhl-1 increased Pnhr-57∷GFP to similar levels (Figure 3B). Although the hypoxia and DIP treatments prevented oxygen-dependent degradation of HIF-1, they did not increase expression of Pnhr-57∷GFP to the levels caused by strong loss-of-function mutations in egl-9 (compare the last bar in Figure 3B to the other conditions). Hypoxia or DIP had no effect on Pnhr-57∷GFP levels in vhl-1 mutant animals. Taken together, these data demonstrated that the vhl-1-independent functions of EGL-9 were relatively insensitive to inhibitors of hydroxylase activity.
Figure 3.—
Differential effects of hypoxia or iron chelators on the two EGL9 pathways. (A) In control experiments, hypoxia (0.5% oxygen, 1 hr) or the iron chelator 2, 2′-dipyridyl (DIP, 200 μm, 4 hr) increased HIF-1 protein to levels similar to those caused by loss-of-function mutations in vhl-1 or egl-9. The bar graph shows HIF-1 protein levels relative to that in untreated vhl-1(+) and egl-9(+) animals. In these strains, the only functional copy of hif-1 is the epitope-tagged transgene. The error bars indicate the standard errors from three independent biological replicates. (B) To determine whether hydroxylase inhibitors had similar effects on HIF-1 target genes as a mutation in egl-9, expression of the reporter was assayed by protein blots. Hypoxia or DIP treatment increased expression of the reporter to levels found in vhl-1 loss-of-function mutants, but the treatments did not completely phenocopy the effects of a loss-of-function mutation in egl-9. In the bar graph, Pnhr-57∷GFP levels are shown relative to that in untreated vhl-1(+) and egl-9(+) animals. The vhl-1(ok161) and egl-9(sa307) mutations are strong loss-of-function alleles. +, the wild-type allele. The bars represent average values from three independent replicates, and the error bars reflect standard error. The statistics were comparing the hypoxia or DIP treated data to untreated data with the same genotype. Asterisks represent statistically significant differences between treatment and room air for animals of the same genotype. *P < 0.05; **P < 0.01.
Mutation in the EGL-9 catalytic core has differential effects on the two EGL-9 pathways:
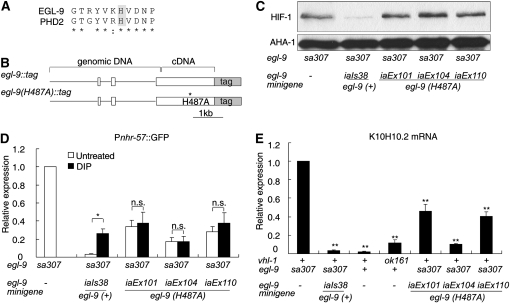
The findings that hypoxia or DIP treatments did not increase expression of Pnhr-57∷GFP in vhl-1 mutants provided support for a model in which EGL-9 had functions that did not require hydroxylase activity (model c in Figure 1B). To test this hypothesis further, we analyzed the consequences of the EGL-9(H487A) mutation. The histidine 487 residue is in the catalytic core of EGL-9. This region of the protein contributes to the Fe(II)-binding pocket and is highly conserved (Figure 4A) (McDonough et al. 2006). Mutation of the analogous residue in the mammalian PHD2 protein (His313) has been shown to eliminate hydroxylase activity (Pan et al. 2007).
Figure 4.—
The egl-9(H487A) mutation does not abolish egl-9-mediated repression of HIF-1 activity. (A) Alignment of human PHD2 and C. elegans EGL-9 at the region around EGL-9 His487. (B) Diagrams of egl-9 minigenes. The minigenes are fusions of genomic and cDNA sequences. Boxes represent coding regions. GFP is fused in frame to egl-9. The H487A mutation disrupts the Fe(II) binding pocket in EGL-9 and impairs EGL-9 catalytic activity. (C) In egl-9(sa307) mutants, HIF-1 destabilization was rescued by the transgene coding for wild-type egl-9(iaIs38), but not by the egl-9(H487A) transgenes (iaEx101, iaEx104, or iaEx110). AHA-1 protein levels have been shown to be unaffected by severe loss-of-function mutations in egl-9 or vhl-1 (Shen et al. 2006), and AHA-1 serves as a loading control. (D and E) Transgenes expressing either wild-type egl-9 or egl-9(H487A) can suppress expression of HIF-1 targets in an egl-9(sa307) background. (D) In strains carrying the wild-type egl-9 transgene, repression of Pnhr-57∷GFP is more effective and is inhibited by DIP. In strains expressing egl-9(H487A), DIP does not have a significant effect. The bar graph shows averages of Pnhr-57∷GFP protein levels from three biological replicates, normalized to expression levels in an egl-9(sa307) mutant. The asterisk indicates DIP causes a statistically significant difference in the expression of the reporter (*P < 0.05) and NS is no significant difference. (E) The wild-type egl-9 transgene and the egl-9(H487) mutant transgene were able to repress K10H10.2 mRNA levels in an egl-9(sa307) mutant. K10H10.2 mRNA levels were also assayed in wild-type N2 and in vhl-1-deficient animals for comparison. The bar graph shows the relative K10H10.2 mRNA levels in each strain, compared to egl-9(sa307) and the error bars reflect standard error. NS, no significant difference. *P < 0.05; **P < 0.01.
To compare the functions of wild-type EGL-9 with EGL-9(H487A), we first constructed and assayed an epitope-tagged wild-type egl-9 minigene, in which genomic sequence including 5′ regulatory sequences, three exons, and two introns were fused to cDNA for the remaining exons in the predominant egl-9 mRNA isoform (Figure 4B). The minigene was able to restore egl-9 function in an egl-9(sa307) mutant, as assayed by destabilization of HIF-1 (compare lanes 1 and 2 in Figure 4C), repression of Pnhr-57∷GFP expression (compare bars 1 and 2 in Figure 4D), or repression of K10H10.2 mRNA levels (compare bars 1 and 2 in Figure 4E). We then introduced the H487A point mutation to the wild-type egl-9 minigene and analyzed the ability of the egl-9(H487A) minigene to rescue egl-9(sa307). As expected, EGL-9(H487A) did not destabilize HIF-1 (lanes 3, 4, and 5 in Figure 4C). The iaIs38, iaEx101, and iaEx110 transgenes all expressed similar levels of EGL-9 wild-type or mutant protein, while expression from iaEx104 was slightly lower (Figure S4).
Having confirmed that EGL-9(H487A) could not rescue oxygen-dependent degradation of HIF-1 in egl-9(sa307) mutants (pathway 1 in Figure 1A), we next asked whether this catalytically deficient EGL-9 was able to repress expression of HIF-1 target genes (pathway 2 in Figure 1A). To address this question, we compared the ability of wild-type egl-9 and egl-9(H487A) minigenes to inhibit expression of Pnhr-57∷GFP in animals that carried a strong loss-of-function mutation in the endogenous egl-9 gene. The wild-type egl-9 minigene (iaIs38) destabilized HIF-1 (see lane 2 of Figure 4C) and repressed Pnhr-57∷GFP (compare first and second bars of Figure 4D). Stabilization of HIF-1 by DIP resulted in increased Pnhr-57∷GFP expression, but not to the levels seen in animals that lack egl-9 function (compare the third bar and the first bar in Figure 4D). In these assays, the HIF-1 reporter gene was similarly regulated in animals containing the wild-type egl-9 locus or in animals expressing egl-9 from the iaIs38 transgene.
Each of the three egl-9(H487A) transgenic arrays were also able to repress expression of Pnhr-57∷GFP, although not to the level achieved by the wild-type egl-9 transgene (Figure 4D). As expected, DIP had no significant effect when the EGL-9 hydroxylase domain was disabled by the H487A mutation (the solid bars in sets 3, 4, and 5 of Figure 4D). These data demonstrated that while disruption of the EGL-9 iron-binding pocket was sufficient to abolish oxygen-dependent degradation of HIF-1 (Figure 4C), this mutation did not eliminate all EGL-9-mediated inhibition of the HIF-1 reporter (Figure 4D).
These findings supported a model in which EGL-9 had two functions: (i) hydroxylation of HIF-1 in the HIF-1 oxygen-dependent degradation pathway and (ii) repression of HIF-1 transcriptional activity by a pathway that had little or no requirement for EGL-9 hydroxylation activity. To further compare the ability of wild-type and catalytically deficient egl-9 transgenes to repress expression of HIF-1 targets, we analyzed K10H10.2 mRNA by RT–PCR. As shown in Figure 4E, when the wild-type egl-9 transgene was introduced to an egl-9(sa307) mutant, it reduced expression of K10H10.2 >10-fold (compare first and second bars of Figure 4E) to levels similar to those in wild-type animals (third bar in Figure 4E). The egl-9(H487A) transgenes did not destabilize HIF-1 (as shown in Figure 4C), so we expected that if they retained the ability to repress HIF-1 transcriptional activity via vhl-1-independent pathways, then the egl-9(H487A) transgenic animals would express K10H10.2 at levels lower than egl-9(sa307) controls and similar to the levels seen in vhl-1(ok161) mutants. As shown in the last three bars of Figure 4E, K10H10.2 mRNA levels were significantly reduced in each of the three egl-9(H487A) lines, relative to the egl-9(sa307) control. In sum, the data in Figure 4 show that the catalytically deficient EGL-9 protein was unable to destabilize HIF-1, but retained the ability to repress expression of HIF-1 targets.
egl-9 mutations and their effects on HIF-1:
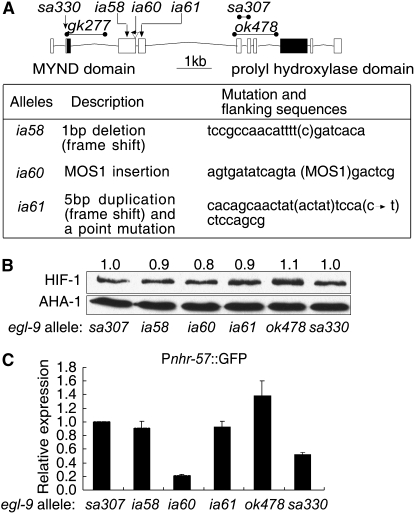
We sought to understand the relationships between EGL-9 functions and EGL-9 structure further by characterizing a series of mutations in the endogenous egl-9 gene. Like its mammalian cognates, EGL-9 includes both an MYND zinc finger domain and a prolyl hydroxylase domain (Figure 5A). The egl-9 locus encodes at least nine mRNA isoforms. We sequenced 19 egl-9 cDNAs and have submitted these sequences to WormBase (Rogers et al. 2008). The structures and relative frequencies of the alternatively spliced egl-9 transcripts are diagrammed in Figure S1. Prior chemical mutagenesis screens had isolated loss-of-function mutations in egl-9 that caused defects in egg laying or resistance to cyanide produced by the bacterial pathogen P. aeruginosa (Darby et al. 1999; Gallagher and Manoil 2001). More recently, the C. elegans gene knockout consortium isolated two egl-9 deletion alleles: gk277 and ok478 (diagrammed in Figure 5A). We isolated three additional loss-of-function mutations in egl-9 in a MOS1 transposon-mediated screen to identify mutations that caused overexpression of Pnhr-57∷GFP. The screen is illustrated in Figure S3, and the new egl-9 mutations are described in Figure 5A.
Figure 5.—
Characterization of egl-9 loss-of-function alleles. (A) Diagram of egl-9 exons and introns and description of new mutations. Boxes represent exons for the predominant egl-9 mRNA isoform, and the exons encoding the MYND or hydroxylase domains are filled. Lines represent deleted sequences in the gk277, sa307, or ok478 alleles; arrows indicate the positions of the ia58 and ia61 mutations; and the position of the ia60 transposon insertion is shown. The alleles isolated in this study are described in the table. (B) Relative effects of egl-9 mutations on HIF-1 protein levels. These animals carry the hif-1(ia04) deletion mutation, and hif-1 function is restored by the hif-1∷tag transgene. The numbers above the lanes reflect HIF-1 levels relative to those detected in egl-9(sa307) mutants, as determined from three replicate experiments. (C) Expression of the Pnhr-57∷GFP reporter in egl-9 mutants, relative to animals homozygous for the egl-9(sa307), a strong loss-of-function egl-9 allele. The bars represent average values relative to those in egl-9(sa307), from three independent replicates, and the error bars reflect standard error.
Initially, we characterized six egl-9 loss-of-function mutations. These included a mutation previously shown to be a strong loss-of-function allele (sa307), the three new alleles from the MOS1-mediated screen (ia58, ia60, and ia61), a deletion allele that removed exons common to all known egl-9 isoforms (ok478), and a mutation that had been shown to confer resistance to P. aeruginosa, but did not cause egg-laying defects (sa330; Darby et al. 1999). For each allele, we characterized its effects on HIF-1 stability and on expression of a HIF-1 target gene, Pnhr-57∷GFP. When comparing the relative effects of these mutations, we used egl-9(sa307) as a reference allele, because prior studies had shown that egl-9(sa307) was a severe mutation that abolished degradation of HIF-1 through the EGL-9/VHL-1 pathway (Epstein et al. 2001; Shen et al. 2006). All six of the egl-9 mutations resulted in HIF-1 overexpression phenotypes very similar to that seen in egl-9(sa307), indicating that each of these alleles severely disabled oxygen-dependent degradation of HIF-1 (Figure 5B).
We next asked how each of these mutations affected expression of a HIF-1 target gene. We found that while the Pnhr-57∷GFP reporter was overexpressed in all of these strains, two of the alleles, ia60 and sa330, caused slightly less severe phenotypes (Figure 5C). The ia60 allele is a MOS1 transposon insertion in exon 3, and the sa330 mutation causes an early translational stop in exon 2 of the predominant egl-9 transcript (Figure 5A) (Darby et al. 1999). The egg-laying defects caused by the egl-9(ia60) or egl-9(sa330) alleles were also less severe, when compared to other egl-9 mutations (Darby et al. 1999 and data not shown). These findings suggest that the proteins encoded by the egl-9ia60 and sa330 alleles cannot destabilize HIF-1, but they retain some ability to repress expression of HIF-1 target genes.
Characterization of egl-9(gk277), an allele that deletes the MYND zinc finger domain:
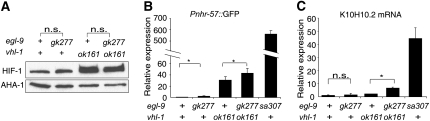
The egl-9(gk277) mutation provided a unique opportunity to investigate the role of the MYND motif in EGL-9 function. This deletion allele removes most of egl-9 exon 2 and intron 2 (Figure 5A). We generated cDNA from egl-9(gk277) animals and determined that in these mutants egl-9 exon 1 was spliced to exon 3, resulting in an in-frame deletion of the evolutionarily conserved MYND zinc finger domain. We determined that HIF-1 protein levels were 1.8-fold higher in egl-9(gk277), relative to animals containing the wild-type egl-9 gene, and the statistical significance of this difference is marginal (lanes 1 and 2 in Figure 6A). By comparison, strong loss-of-function mutations in egl-9 or vhl-1 caused 3- to 4-fold increases in HIF-1 levels. In a vhl-1(ok161) background, the egl-9(gk277) mutation had no effect on HIF-1 protein levels (lanes 3 and 4 in Figure 6A).
Figure 6.—
The egl-9(gk277) mutation removes the MYND zinc finger domain, but has little effect on HIF-1 protein levels or on expression of HIF-1 targets. (A) The egl-9(gk277) mutation does not significantly increase expression of HIF-1 protein in vhl-1(+) or vhl-1(ok161). (B and C) The egl-9(gk277) mutation has a significant effect on the expression of HIF-1 target genes, but these effects are much smaller than the severe egl-9(sa307) mutation. The bar graphs show the expression of the Pnhr-57∷GFP reporter (B) and the endogenous HIF-1 target gene K10H10.2 (C) in each strain, relative to the values for animals carrying wild-type alleles of vhl-1 and egl-9. The bars represent average values from three independent replicates, and the error bars reflect standard error. NS, no significant difference. *P < 0.05.
We next investigated the effects of the gk277 mutation on the expression of two genes regulated by HIF-1. While the Pnhr-57∷GFP reporter and the endogenous K10H10.2 gene were overexpressed in egl-9(sa307) animals by ∼400 and 40-fold, respectively, the gk277 mutation caused only 2- to 3-fold increases in the expression of these HIF-1 targets (Figure 6, B and C) In animals in which HIF-1 was stabilized by the vhl-1(ok161) mutation, the gk277 mutation had relatively small effects on the expression of either gene (third and fourth bars in Figure 6, B and C). Thus, deletion of the MYND domain in egl-9(gk277) animals had little effect on the ability of EGL-9 to inhibit HIF-1 transcriptional activity.
DISCUSSION
EGL-9 is the key regulator of HIF-1 protein stability and HIF-1 activity. In C. elegans and in mammals, HIF induces the expression of EGL-9/PHD, and this establishes a negative feedback loop that attenuates HIF activity (Bishop et al. 2004; Shen et al. 2005; Berra et al. 2006). The catalytic functions of EGL-9/PHD enzymes are of central importance in normal development, homeostasis, and disease states and have been studied intensively (Kaelin and Ratcliffe 2008). In humans, mutations in the PHD2 active site and other mutations that stabilize HIF have been linked to familial erythrocytosis (Percy et al. 2006, 2007; Al-Sheikh et al. 2008; Martini et al. 2008). Efforts are underway to develop small molecule inhibitors of PHD activity for the treatment of anemia and other diseases that may be mitigated by increased HIF protein stability (Hewitson et al. 2004). Here, we have shown that EGL-9 has a second function: it represses HIF-1 activity through a pathway that has little or no requirement for EGL-9 hydroxylase activity.
Hydroxylase-deficient EGL-9 still represses HIF-1 activity:
The evidence that C. elegans EGL-9 regulates HIF-1 stability and HIF-1 activity through two distinct mechanisms is multifold: (i) Loss-of-function mutations in egl-9 or vhl-1 stabilize HIF-1 protein, but mutations in egl-9 cause much higher levels of HIF-1 target gene expression (Shen et al. 2006) (Figure S2B; Figure 3, A and B); (ii) mutation of the proline normally hydroxylated by EGL-9 [in HIF-1(P621G)] releases HIF-1 from oxygen-dependent degradation, but it does not prevent EGL-9-mediated inhibition of HIF-1 target gene expression (Figure S2C; Figure 2, B and C); (iii) treatments that inhibit EGL-9 catalytic activity do not fully phenocopy the egl-9 phenotype, and hypoxia or DIP have little effect on HIF-1 activity in vhl-1 mutants (Bishop et al. 2004) (Figure 3B); and (iv) the H487A mutation in the iron-binding pocket of the EGL-9 catalytic domain stabilizes HIF-1, but EGL-9(H487A) still inhibits transcriptional activity (Figure 4, C, D, and E).
We conclude that EGL-9 is a bifunctional protein, with some functions that require hydroxylase activity and others that do not. We have considered the formal possibility that the EGL-9(H487A) mutant protein retains some catalytic function. However, it is clear that the mutation in the EGL-9 iron-binding pocket abrogates hydroxylase activity, as assayed by destabilization of HIF-1 (Figure 4C). Further, some EGL-9 functions are insensitive to inhibitors of hydroxylase activity. In EGL-9(H487A) animals or in vhl-1 mutants, DIP treatments do not increase HIF-1 activity (Figures 3B and 4D). We conclude that the two EGL-9 pathways have dramatically different requirements for EGL-9 prolyl hydroxylase activity.
It has been proposed that mammalian PHD proteins may also have functions that do not require hydroxylase activity. PHD2 overexpression was shown to reduce expression of HIF targets in a cell line lacking VHL and in a manner independent of HIF destabilization. Further, PHD2 exerted this inhibitory effect on the HIF-1α transcriptional activation region fused to a heterologous DNA binding domain (To and Huang 2005). These data are consistent with a model in which PHD2 could recruit repressors to the HIF transcriptional complex, independent of its role in VHL-mediated degradation of HIF. In an independent study of hypoxia-treated HeLa cells, PHD2 was shown to be associated with HIF DNA binding sites. PHD2 was also shown to bind the ING4 tumor suppressor, which has been proposed to act as a transcriptional repressor (Ozer et al. 2005). In a third study, expression of a catalytically deficient form of PHD2 was shown to inhibit endothelial cell proliferation (Takeda and Fong 2007). Most recently, PHD2 has been shown to have functions in angiogenesis that do not require hydroxylase activity (Chan et al. 2009). These findings do not take away from the central importance of PHDs as oxygen sensors, but they provide strong evidence that mammalian PHD proteins have additional functions that do not require hydroxylase activity.
Characterization of egl-9 loss-of-function alleles:
Several egl-9 loss-of-function alleles have been isolated in screens for mutations that result in egg-laying deficiencies (Trent et al. 1983), resistance to P. aeruginosa infection (Darby et al. 1999) or overexpression of HIF-1 target genes (this study). In all cases that have been tested, depletion of hif-1 by RNAi or mutation has been shown to suppress egl-9 mutant phenotypes (Shen et al. 2006; Chang and Bargmann 2008; Gort et al. 2008; Pocock and Hobert 2008). We assayed several of these alleles to determine whether they had differential effects on HIF-1 stabilization or VHL-1-independent suppression of HIF-1 activity. With the exception of the gk277 deletion of the MYND domain, all of the mutations impaired both pathways. Two findings are of particular interest. First, there is a correlation between the degree of Pnhr-57∷GFP overexpression and the egl phenotypes caused by the egl-9 mutations. egl-9(sa330) and egl-9(ia60) mutants are not egg-laying defective. Second, the egl-9(sa330) mutation causes a translational stop in exon 2 of the predominant egl-9 transcript (Darby et al. 1999) (Figure 5C), but it does not abolish all egl-9 function. Six of the nine egl-9 mRNA isoforms do not include exon 2 or the MYND domain. It is possible that these other transcripts provide some function in egl-9(sa330) mutants.
The EGL-9 MYND domain:
The MYND zinc finger domain is an evolutionarily conserved feature of PHD proteins. Since the divergence of nematode and mammalian lineages, the PHD gene amplified in mammals. The PHD1, PHD2, and PHD3 genes have diverged and have acquired different tissue distributions, subcellular localization patterns, and functions (Fong and Takeda 2008; Kaelin and Ratcliffe 2008). PHD3 lacks an MYND domain, and, although it has different substrate specificities than PHD1 and PHD2, it can hydroxylate some substrates (Epstein et al. 2001). Deletion of the MYND domain in PHD2 resulted in increased HIF-1 destabilization in HeLa cells (Choi et al. 2005), suggesting that the MYND domain might modulate PHD activity in some contexts. The gk277 mutation presents a unique opportunity to examine the in vivo effects of deleting the MYND domain, and C. elegans egl-9(gk277) mutants had been reported to have defects in neuronal pathfinding (Pocock and Hobert 2008). It was, therefore, of particular interest to determine whether the egl-9(gk277) mutation impaired one or both of the EGL-9 pathways for HIF-1 regulation. In our assays, the egl-9(gk277) mutation did not change total HIF-1 levels and had only marginal effects on the expression of HIF-1 targets.
This study provides a foundation for further experiments to understand the mechanism by which EGL-9 inhibits expression of HIF-1 transcriptional targets. We have established that mutations or treatments that inhibit hydroxylase activity do not eliminate all regulation of HIF-1 by EGL-9. The next important steps will be to identify EGL-9/PHD interaction partners and to characterize their respective roles in regulating HIF-1 stabilization or activity.
Acknowledgments
Mutant strains were obtained from the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health (NIH) National Center for Research Resources. Yuji Kohara and colleagues provided the egl-9 cDNAs. We are grateful to Clark Coffman, Maggie Pruitt, and members of the C. elegans research community for helpful discussions and for comments on this manuscript. This work was supported by NIH award GM078424.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107284/DC1.
References
- Al-Sheikh, M., K. Moradkhani, M. Lopez, H. Wajcman and C. Prehu, 2008. Disturbance in the HIF-1alpha pathway associated with erythrocytosis: further evidences brought by frameshift and nonsense mutations in the prolyl hydroxylase domain protein 2 (PHD2) gene. Blood Cells Mol. Dis. 40 160–165. [DOI] [PubMed] [Google Scholar]
- Berra, E., A. Ginouves and J. Pouyssegur, 2006. The hypoxia-inducible-factor hydroxylases bring fresh air into hypoxia signalling. EMBO Rep. 7 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, T., K. W. Lau, A. C. Epstein, S. K. Kim, M. Jiang et al., 2004. Genetic analysis of pathways regulated by the von Hippel-Lindau tumor suppressor in Caenorhabditis elegans. PLoS Biol. 2 e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, A. J., K. E. Busch and M. de Bono, 2008. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105 8044–8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick, R. K., and S. L. McKnight, 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294 1337–1340. [DOI] [PubMed] [Google Scholar]
- Chan, D. A., T. L. Kawahara, P. D. Sutphin, H. Y. Chang, J. T. Chi et al., 2009. Tumor vasculature is regulated by PHD2-mediated angiogenesis and bone marrow-derived cell recruitment. Cancer Cell 15 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A. J., and C. I. Bargmann, 2008. Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105 7321–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., E. L. Thomas and P. Kapahi, 2009. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in C. elegans. PLoS Genet. 5 e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K. O., T. Lee, N. Lee, J. H. Kim, E. G. Yang et al., 2005. Inhibition of the catalytic activity of hypoxia-inducible factor-1alpha-prolyl-hydroxylase 2 by a MYND-type zinc finger. Mol. Pharmacol. 68 1803–1809. [DOI] [PubMed] [Google Scholar]
- Darby, C., C. L. Cosma, J. H. Thomas and C. Manoil, 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96 15202–15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke et al., 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107 43–54. [DOI] [PubMed] [Google Scholar]
- Fong, G. H., and K. Takeda, 2008. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 15 635–641. [DOI] [PubMed] [Google Scholar]
- Gallagher, L. A., and C. Manoil, 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183 6207–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gort, E. H., G. van Haaften, I. Verlaan, A. J. Groot, R. H. Plasterk et al., 2008. The TWIST1 oncogene is a direct target of hypoxia-inducible factor-2alpha. Oncogene 27 1501–1510. [DOI] [PubMed] [Google Scholar]
- Granger, L., E. Martin and L. Segalat, 2004. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res. 32 e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson, K. S., L. A. McNeill and C. J. Schofield, 2004. Modulating the hypoxia-inducible factor signaling pathway: applications from cardiovascular disease to cancer. Curr. Pharm. Des. 10 821–833. [DOI] [PubMed] [Google Scholar]
- Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando et al., 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292 464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert et al., 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292 468–472. [DOI] [PubMed] [Google Scholar]
- Jiang, H., R. Guo and J. A. Powell-Coffman, 2001. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. USA 98 7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin, Jr., W. G., and P. J. Ratcliffe, 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30 393–402. [DOI] [PubMed] [Google Scholar]
- Martini, M., L. Teofili, T. Cenci, F. Giona, L. Torti et al., 2008. A novel heterozygous HIF2AM535I mutation reinforces the role of oxygen sensing pathway disturbances in the pathogenesis of familial erythrocytosis. Haematologica 93 1068–1071. [DOI] [PubMed] [Google Scholar]
- Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux et al., 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399 271–275. [DOI] [PubMed] [Google Scholar]
- McDonough, M. A., V. Li, E. Flashman, R. Chowdhury, C. Mohr et al., 2006. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc. Natl. Acad. Sci. USA 103 9814–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, R., K. A. Steinkraus, G. L. Sutphin, F. J. Ramos, L. S. Shamieh et al., 2009. Proteasomal Regulation of the Hypoxic Response Modulates Aging in C. elegans. Science 324 1196–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D. L., and M. B. Roth, 2009. C. elegans are protected from lethal hypoxia by an embryonic diapause. Curr. Biol. 19 1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, J. H., H. Yang, M. Ivan, F. Gertler, W. G. Kaelin, Jr. et al., 2002. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science 296 1886–1889. [DOI] [PubMed] [Google Scholar]
- Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim et al., 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell. Biol. 2 423–427. [DOI] [PubMed] [Google Scholar]
- Ozer, A., L. C. Wu and R. K. Bruick, 2005. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF). Proc. Natl. Acad. Sci. USA 102 7481–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla, P. A., T. G. Nystul, R. A. Zager, A. C. Johnson and M. B. Roth, 2002. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol. Biol. Cell 13 1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y., K. D. Mansfield, C. C. Bertozzi, V. Rudenko, D. A. Chan et al., 2007. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol. Cell. Biol. 27 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy, M. J., Q. Zhao, A. Flores, C. Harrison, T. R. Lappin et al., 2006. A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc. Natl. Acad. Sci. USA 103 654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy, M. J., P. W. Furlow, P. A. Beer, T. R. Lappin, M. F. McMullin et al., 2007. A novel erythrocytosis-associated PHD2 mutation suggests the location of a HIF binding groove. Blood 110 2193–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock, R., and O. Hobert, 2008. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat. Neurosci. 11 894–900. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin, E. B., and A. J. Giaccia, 2008. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 15 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, A., I. Antoshechkin, T. Bieri, D. Blasiar, C. Bastiani et al., 2008. WormBase 2007. Nucleic Acids Res. 36 D612–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C., D. Nettleton, M. Jiang, S. K. Kim and J. A. Powell-Coffman, 2005. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280 20580–20588. [DOI] [PubMed] [Google Scholar]
- Shen, C., Z. Shao and J. A. Powell-Coffman, 2006. The Caenorhabditis elegans rhy-1 gene inhibits HIF-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1. Genetics 174 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq, A., L. R. Aminova and R. R. Ratan, 2007. Hypoxia inducible factor prolyl 4-hydroxylase enzymes: center stage in the battle against hypoxia, metabolic compromise and oxidative stress. Neurochem. Res. 32 931–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, K., and G. H. Fong, 2007. Prolyl hydroxylase domain 2 protein suppresses hypoxia-induced endothelial cell proliferation. Hypertension 49 178–184. [DOI] [PubMed] [Google Scholar]
- To, K. K., and L. E. Huang, 2005. Suppression of hypoxia-inducible factor 1alpha (HIF-1alpha) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J. Biol. Chem. 280 38102–38107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treinin, M., J. Shliar, H. Jiang, J. A. Powell-Coffman, Z. Bromberg et al., 2003. HIF-1 is required for heat acclimation in the nematode Caenorhabditis elegans. Physiol. Genomics 14 17–24. [DOI] [PubMed] [Google Scholar]
- Trent, C., N. Tsuing and H. R. Horvitz, 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann, A., and R. S. Johnson, 2008. Biology of HIF-1alpha. Cell Death Differ. 15 621–627. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Z. Shao, Z. Zhai, C. Shen and J. A. Powell-Coffman, 2009. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One 4 e6348. [DOI] [PMC free article] [PubMed] [Google Scholar]