Abstract
Maintenance and expression of mitochondrial DNA (mtDNA) are essential for the cell and the organism. In humans, several mutations in the adenine nucleotide translocase gene ANT1 are associated with multiple mtDNA deletions and autosomal dominant forms of progressive external ophthalmoplegia (adPEO). The mechanisms underlying the mtDNA instability are still obscure. A current hypothesis proposes that these pathogenic mutations primarily uncouple the mitochondrial inner membrane, which secondarily causes mtDNA instability. Here we show that the three adPEO-associated mutations equivalent to A114P, L98P, and V289M introduced into the Podospora anserina ANT1 ortholog dominantly cause severe growth defects, decreased reactive oxygen species production (ROS), decreased mitochondrial inner membrane potential (Δψ), and accumulation of large-scale mtDNA deletions leading to premature death. Interestingly, we show that, at least for the adPEO-type M106P and A121P mutant alleles, the associated mtDNA instability cannot be attributed only to a reduced membrane potential or to an increased ROS level since it can be suppressed without restoration of the Δψ or modification of the ROS production. Suppression of mtDNA instability due to the M106P and A121P mutations was obtained by an allele of the rmp1 gene involved in nucleo-mitochondrial cross- talk and also by an allele of the AS1 gene encoding a cytosolic ribosomal protein. In contrast, the mtDNA instability caused by the S296M mutation was not suppressed by these alleles.
THE maintenance and expression of mitochondrial DNA (mtDNA) depend on many nuclear-encoded gene products. Recent studies have shown that defects in this maintenance can have devastating consequences for the cell and the organism. In humans, these defects are an important cause of neurological diseases including autosomal dominant (or recessive) progressive external ophthalmoplegia (adPEO) (Chinnery 2003; Copeland 2008). These disorders are characterized by multiple large-scale deletions of mtDNA. Three different genes that can cause PEO with multiple mtDNA deletions have been identified: the mtDNA polymerase (POLG), the heart/muscle isoform of the adenine nucleotide translocator (ANT1), and the mitochondrial DNA helicase, Twinkle.
The adenine nucleotide translocator (ANT), also known as the ADP/ATP mitochondrial translocator, is the most abundant protein in the inner mitochondrial membrane (Riccio et al. 1975; Nury et al. 2006; Klingenberg 2008). It exports ATP produced by mitochondrial oxidative phosphorylation toward the cytosol to meet the energy requirements of the cell; in exchange, it transports ADP into the mitochondrial matrix to fuel the conversion of ADP to ATP by the F1FO-ATP synthase. In humans, four isoforms of the ANT protein exist, and they are differently expressed in a tissue-specific manner (Stepien et al. 1992; Palmieri 2004; Dolce et al. 2005). The human ANT1 isoform is predominantly expressed in skeletal and cardiac muscle, and specific ANT1 mutations are associated with adPEO characterized by mtDNA instability (Kaukonen et al. 1999, 2000; Napoli et al. 2001; Komaki et al. 2002; Siciliano et al. 2003). In mice, Ant1 knockout induces mitochondrial myopathy (Graham et al. 1997), increased H2O2 production, and mtDNA damage and inhibits oxidative phosphorylation (Esposito et al. 1999). Some of these mutations were introduced in the AAC2 gene of Saccharomyces cerevisiae that encodes the major ADP/ATP mitochondrial translocator isoform in this organism. Numerous and sometimes contradictory effects have been reported depending in particular on the yeast laboratory strains examined (Kaukonen et al. 2000; Chen 2002, 2004; Fontanesi et al. 2004; Palmieri et al. 2005; Wang et al. 2008b).
In an attempt to better understand how these mutations affect mitochondrial DNA stability and their functional consequences on mitochondrial metabolism, we decided to introduce them in the unique ADP/ATP translocator gene of Podospora anserina, PaAnt. Like S. cerevisiae, the filamentous fungus P. anserina is an excellent system for genetic and molecular analyses. In contrast to S. cerevisiae, it is a strict multicellular aerobe that can display heteroplasmic states in which intact and rearranged mitochondrial genomes coexist. In this organism, life span is a reflection of mtDNA stability, and death is always associated with large mtDNA rearrangements. “Natural death” or aging is accompanied by large-scale reorganizations of the mtDNA whereas a nuclear-controlled premature death syndrome is accompanied by the accumulation of site-specific mtDNA deletions (Belcour et al. 1999; Silar et al. 2001 for reviews). P. anserina therefore occupies an interesting position among model systems for studying the cellular consequences of mutations in the ADP/ATP translocase gene.
We show here that the mutations M106P, A121P, and S296M, equivalent to the L98P, A114P (familial), and V289M (sporadic) human mutations, severely impair the vegetative and sexual development of the fungus and are responsible for decreased ROS production and for decreased inner membrane potential (Δψ). The severity of the phenotypes differs according to the mutation. The three mutations show mtDNA instability, which leads to premature death. All these mutated traits are dominant. Interestingly, the mtDNA instability associated with the M106P and A121P mutations depends on the rmp1 gene. This gene exists under two naturally occurring alleles, rmp1-1 and rmp1-2, which control mtDNA integrity in some genetic contexts (Belcour et al. 1991; Contamine et al. 1996, 2004). When associated with the rmp1-1 allele, the M106P and A121P mutations lead to rapid mtDNA instability whereas, in the presence of the rmp1-2 allele, mtDNA instability is suppressed, and life span is considerably increased. Surprisingly, suppression is not accompanied by a restoration of the Δψ or a modification in the ROS level, demonstrating that these parameters are not sufficient to explain the M106P and A121P mtDNA instability. Mitochondrial DNA instability due to the M106P and A121P mutations is also suppressed by a mutation in the AS1 gene encoding a ribosomal protein. The suppressor effects are not observed for the S296M mutation.
MATERIALS AND METHODS
P. anserina strains, growth conditions, life-span measurements, and transformation experiments:
The genetics and biological properties of P. anserina have been described and reviewed (Esser 1974). Strains used in this study were derived from the s wild-type strain (Rizet 1952). The aox∷hygro (Δaox) strain is inactivated for the endogenous aox gene (Lorin et al. 2001). The wild-type strain carrying an ectopic copy of the rmp1-1 allele associated with a hygromycin-resistance cassette was described previously (Contamine et al. 2004). The AS1-4 mutation was selected as an informational antisuppressor. Cultures were grown on standard minimal synthetic (M2) medium (Esser 1974) at 27°. When necessary, hygromycin 100 μg/ml (Boehringer-Mannheim), phleomycin 10 μg/ml (Boehringer-Mannheim), or nourseothricin 50 μg/ml (Werner BioAgents) were added to the medium. Medium for germination contains ground cornmeal (50 g/liter), agar (12.5 g/liter), and ammonium acetate (6 g/liter).
Life spans were measured on M2 medium in race tubes at 27° in the dark for three to five subcultures derived from 5 to 10 independent spores of a given strain. The life span of a strain is defined as the mean time (given with standard errors) of the growth of parallel cultures between the inoculation and the death of the culture. Protoplast preparation and transformation experiments were conducted as described previously (Bergès and Barreau 1989; El-Khoury et al. 2008).
Site-directed mutagenesis and gene replacement:
The PaAnt+ gene was cloned in pUC19 and then digested with SmaI to give a 4.2- kb SmaI restriction fragment, which was then introduced into the plasmid pAPI508, conferring resistance to nourseothricin to give pAPI-PaAnt+ (El-Khoury et al. 2008), and into pBCHygro, conferring resistance to hygromycin (Silar 1995) to give pBC-PaAnt+. pAPI-PaAnt+ was then used as a template for mutagenesis of the PaAnt+ gene by the QuikChange site-directed mutagenesis kit (Stratagene). The list of base changes and corresponding modified primers used to generate them is given in the supporting information, Table S1. After mutagenesis, the different constructs were sequenced to verify the presence of the correct base change. The resulting plasmids were used to replace the endogenous PaAnt+ as described previously (El-Khoury et al. 2008). The pBC-PaAnt+ plasmid was used to introduce by transformation an ectopic PaAnt+ copy into the wild- type strain to obtain the PaAnt+ (PaAnt+) strain.
To delete the PaAnt+ gene, targeting fragments of 1.3 kb (5′) and 1.1 kb (3′) flanking the PaAnt gene were prepared by PCR on genomic DNA using the primer pairs E-5′-UTR (5′-TCGAATTGGAGTTGGGAAAG-3′) and S-5′-UTR (5′-GTCAGCCACAGCAAGATGAA-3′) and N-3′-UTR (5′-GGATCAGAGGCTTCATCGTC-3′) and S-3′-UTR (5′-TGACAACCCAGGTTCTTTGA-3′), respectively. The two fragments bear EcoRI–SpeI and NotI–SpeI restriction sites and were introduced sequentially into the corresponding sites of the polylinker of plasmid pPable that contains a phleomycin- resistance cassette (Coppin and Debuchy 2000). The resulting vector, pPable-ΔPaAnt+, was digested by SpeI, and ∼10 μg of linear plasmid was used to transform the PaAnt+ (PaAnt+) strain. The deletion of the endogenous PaAnt+ gene was verified by PCR and Southern blot.
Mitochondrial DNA analysis:
The mtDNA of the different growing or dying cultures was extracted by minipreparation (Lecellier and Silar 1994). The probes used in this study were EcoIV, EcoI, and 2604 ( described previously by Begel et al. 1999); they cover ∼80% of the mitochondrial genome.
Western blot:
Mitochondria were isolated as previously described (Sellem et al. 2007). Thirty micrograms of mitochondrial protein were fractionated by SDS–PAGE and transferred to a nitrocellulose membrane. Immunochemistry was performed with an anti-Aac2 monoclonal antibody generated against the Aac2 protein of S. cerevisiae. This antibody, kindly donated by E. Kunji, was raised against the Aac2 conserved motif SYPLDTVRRRMMMT (Bamber et al. 2007). In addition, blots were reprobed with an anti-βATPase antibody (a gift from J. Velours) as a standardization control. The bound antibodies were detected using an enhanced chemiluminescence detection system (Pierce Supersignal West picochemiluminescent substrate).
Quantitative RT–PCR:
Total RNA was extracted using the RNeasy plant mini kit (Qiagen) from cultures grown for 48 hr on cellophane disks overlying M2 medium. The mycelium was broken with glass beads in a Fastprep apparatus (40 sec, intensity 6.5). For quantitative RT–PCR (qRT–PCR), cDNA was synthesized with SuperScript II reverse trancriptase (Invitrogen) from 2 μg of RNA, using a T15 primer. Subsequent quantitative real-time PCRs were performed in a Lightcycler (Roche), using the LightCycler FastStart DNAMaster SYBR Green I kit (Roche). At least three independent experiments were performed on one to three different RNA preparations for each strain. Primers QAOX4-F (5′-TGATCTCGCCACGAATTACA-3′) and QAOX4-R (5′-TATAGGTGTGGACCGCTTCC-3′) were used for the aox gene.
Mitochondrial morphology:
The wild type, PaAntM106P, PaAntA121P, and PaAntS296M strains were crossed with a strain that carries the pAPI213 transgene consisting of the mitochondrial targeting sequence of the Neurospora crassa atp9 gene cloned in frame with the enhanced green fluorescent protein (EGFP) sequence and a hygromycin- resistance cassette (Sellem et al. 2007) Wild-type and mutant strains expressing this transgene were obtained. They were grown on thin synthetic solid medium for 12–24 hr. The area of medium containing the mycelium was cut out and transferred onto a microscopic slide. Fluorescence was observed with a Zeiss Axioplan 2 upright microscope equipped with a photometrics CoolSnap HQ (Roper Scientific) camera and Metamorph software.
Respiration analysis:
The rate of respiration was estimated in vivo in an oxytherm chamber with a Clark-type O2 electrode (Hansatech), using mycelia grown on cellophane disks overlying M2 medium for 48 hr or using protoplast suspensions (3.107 protoplasts/ml in 0.6 m sucrose). Cyanide (KCN, 1 mm) was added to inhibit the cytochrome pathway.
Flow cytometry, ROS, and mitochondrial membrane potential (Δψ) measurements:
ROS elimination was measured by the production of dichlorofluorescin (DCF) resulting from the oxidation of the diacetate form (H2DCF-DA). For each strain, 107 protoplasts were incubated in H2DCF-DA (80 μm in 0.4 m sucrose/50 mm phosphate buffer, pH 6), and measurements of the fluorescent DCF were performed after 90 min. The cytometric measurements were performed in a Partec PAS-III flow cytofluometer. Mitochondrial membrane potential (Δψ) was measured by incubating protoplasts (106/ml) in 0.1 μm of 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)] (Sigma Aldrich) for 30 min at 25°. Protoplasts were centrifuged to remove the excess fluorochrome and resuspended in fresh medium before analysis. Control samples were prepared with 17 μm final carbamoyl cyanide m-chlorophenylhydrazone (mClCCP), an ionophore that dissipates the mitochondrial membrane potential. This drug was added to the protoplasts 15 min before DiOC6(3). Fluorescence of the DiOC6(3) was measured in a Partec PAS-III flow cytofluometer.
Nonyl-acridine orange, MitoFluor Green, and MitoTracker Green were not suitable for the quantification of mitochondria because we observed that their accumulation in the mitochondria is dependent on the membrane potential in P. anserina (data not shown); therefore, the fluorescence values of DiOC6(3) were normalized using the quantity of mitochondrial protein determined by the Bradford method (Bio-Rad).
RESULTS
The genome of P. anserina encodes a unique PaAnt gene whose deletion is lethal:
Taking advantage of the recently sequenced genome of P. anserina (Espagne et al. 2008), we identified a unique gene (PaAnt) encoding a homolog of the human ANT1 protein. The 315-amino-acid protein deduced from the PaAnt sequence shares 71.3%, 47.3%, and 48.3% identity with Aac2, ADT1, and hANT1 proteins from S. cerevisiae, Bos taurus, and Homo Sapiens, respectively (Figure S1). The PaAnt gene was cloned and modified as described in materials and methods. It was first deleted in a strain bearing an ectopic copy (PaAnt+-Hygro) of the wild-type gene. The primary transformants PaAnt∷Phleo (PaAnt+-Hygro) were crossed with a wild-type strain. The analysis of >50 asci clearly revealed that all the homocaryotic spores resistant to phleomycin and sensitive to hygromycin were unable to germinate, indicating that the PaAnt∷Phleo genotype is viable only when the ectopic transgene is present. This allows us to conclude that the inactivation of the PaAnt gene is lethal for P. anserina and to confirm that no functionally redundant gene is present in the genome.
PaAnt alleles equivalent to hANT1 mutants responsible for adPEO cause pleiotropic somatic and sexual defects:
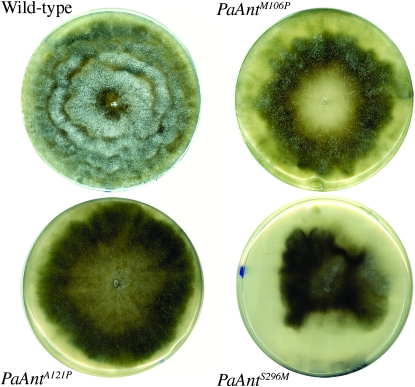
To mimic the L98P, A114P, and V289M human mutations responsible for adPEO, the corresponding Met106, Ala121, and Ser296 codons of the P. anserina ANT protein were changed to proline, proline and methionine codons, respectively (Figure S1). The three mutant alleles PaAntM106P, PaAntA121P, and PaAnt S296M were introduced site specifically in place of the endogenous wild-type gene as previously described (El-Khoury et al. 2008). The recipient strain used for the transformation-mediated gene replacement carried an ectopic wild-type allele. The primary transformants were crossed with wild type. Monocaryotic spores bearing a mutated allele (determined by resistance to nourseothricin) without the ectopic PaAnt+ allele (determined by the hygromycin sensitivity) were obtained for the three mutations, revealing that none of them is lethal. Thus, we obtained PaAntM106P, PaAntA121P, and PaAnt S296M strains in which the mutated allele is subject to the normal regulatory control of the PaAnt gene. The three mutant strains displayed drastic phenotypic alterations. They showed a delayed and reduced rate of germination, a slow vegetative growth rate (Table 1A), a heterogeneous mycelial aspect with very pigmented sectors devoid of aerial hyphea (Figure 1), female sterility, and reduced male fertility (data not shown).
TABLE 1.
Growth rate and life span of strains carrying different PaAnt mutant alleles
| Strain | Growth rate (mm/day ± SE) | Life span (days ± SE) |
|---|---|---|
| A | ||
| PaAnt+, rmp1-2 mat+ | 6.5 ± 0.2 | 18.2 ± 1.2 |
| PaAnt+, rmp1-1 mat− | 6.7 ± 0.1 | 17.7 ± 0.8 |
| PaAntM106P, rmp1-2 mat+ | 4 ± 0.2 | >180 |
| PaAntM106P, rmp1-1 mat− | 3.6 ± 0.4 | 9.4 ± 2.1 |
| PaAntA121P, rmp1-2 mat+ | 5.2 ± 0.1 | >180 |
| PaAntA121P, rmp1-1 mat− | 4.9 ± 0.4 | 9.4 ± 1.1 |
| PaAntS296M, rmp1-2 mat+ | 4.2 ± 0.4 | 5.6 ± 1 |
| PaAntS296M, rmp1-1 mat− | 4.5 ± 0.7 | 5.8 ± 0.7 |
| B | ||
| PaAntM106P, rmp1-2 mat+ (rmp1-1) | ND | 9.2 ± 1.4 |
| PaAntA121P, rmp1-2 mat+ (rmp1-1) | ND | 9 ± 2 |
| C | ||
| PaAntM106P (PaAnt+) rmp1-2 mat+ | 4.7 ± 0.6 | >180 |
| PaAntM106P (PaAnt+) rmp1-1 mat− | 4.7 ± 0.2 | 11.3 ± 0.9 |
| PaAntA121P (PaAnt+) rmp1-2 mat+ | 5.3 ± 0.1 | >180 |
| PaAntA121P (PaAnt+) rmp1-1 mat− | 4.5 ± 0.1 | 11.4 ± 0.8 |
| PaAntS296M (PaAnt+) rmp1-2 mat+ | 4 ± 0.5 | 5.1 ± 1.3 |
|
PaAntS296M (PaAnt+) rmp1-1 mat− |
3.8 ± 0.2 |
5.2 ± 1.2 |
(A) Characteristics of the homoallelic strains carrying the wild-type or a mutant allele of PaAnt associated with rmp1-1 mat− or rmp1-2 mat+. (B) Characteristics of the PaAntM106P rmp1-2 mat+ and PaAntA121P rmp1-2 mat+ strains carrying an ectopic copy of the rmp1-1 allele. (C) Characteristics of the heteroallelic strains carrying a mutant allele of PaAnt and an ectopic copy of the wild-type allele associated with rmp1-1 mat− or rmp1-2 mat+.
Figure 1.—
Mycelium aspect of the wild type and the three PaAnt mutants. Petri plates of M2 medium were inoculated with an explant of each strain and incubated for 6 days at 27°. The wild type exhibits dense, aerial mycelium whereas the three mutants exhibit a heterogeneous mycelium with highly pigmented sectors devoid of aerial hyphae. The vegetative growth of the PaAntS296M strain stopped prematurely. The strains presented are all rmp1-2 mat+; the equivalent rmp1-1 mat− strains display similar phenotypes.
The three PaAnt mutations cause a strong decrease of mtDNA stability and life span, which are suppressed for M106P and A121P but not S296M by the rmp1-2 allele:
The three hANT1 mutations leading to PEO disease are associated with multiple deletions of the mitochondrial genome in skeletal muscle. In P. anserina, life span is an indicator of mtDNA stability. Life span and death-associated mtDNA instability were therefore analyzed in the three strains PaAntM106P, PaAntA121P, and PaAntS296M. Surprisingly, as shown in Table 1A, except for the PaAntS296M mutant that showed a premature death phenotype whatever the mating type (∼5– 6 days compared to 18 days for the wild-type strain), the life span of the PaAntM106P and PaAntA121P mutants was severely reduced (∼2 fold) or greatly increased (>10- fold) according to the mating type, mat− or mat+, respectively. The mat− cultures displayed a reduction in life span of ∼2-fold whereas the mat+ cultures displayed an increase in life span of >10-fold (>180 days). A gene, rmp1, exists as two naturally occurring alleles, rmp1-1 and rmp1-2. This gene is tightly linked to the mat locus (mat− and mat+, respectively) and is involved in the timing of death in a certain genetic context (Contamine et al. 1996). We asked whether, in the PaAntM106P and PaAntA121P strains, the differences in life span between the two mat loci revealed an interaction between the PaAnt gene and the mat locus or between the PaAnt gene and the rmp1 gene. To address this question, we took advantage of a strain bearing an ectopic copy of the dominant rmp1-1 gene (Contamine et al. 2004) to introduce it by a genetic cross into the PaAntM106P and PaAntA121P rmp1-2 mat+ strains. Interestingly, as shown in Table 1B, the introduction of rmp1-1 into either PaAntM106P or PaAntA121P rmp1-2 mat+ strains led to a premature death phenotype similar to that observed in PaAntM106P and PaAntA121P rmp1-1 mat− strains. This demonstrates that the timing of death in PaAntM106P and PaAntA121P mutants depends on the presence of the rmp1-1 or the rmp1-2 allele and not on the mating type.
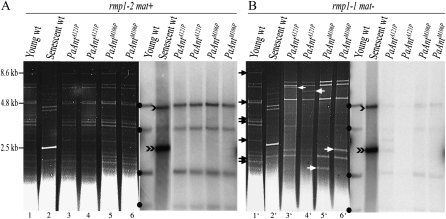
In P. anserina, growth arrest of the wild-type strain by aging is correlated with the accumulation of short mtDNA sequences (senDNAs) as amplified circular multimeric molecules. Large amounts of senDNAα are systematically observed in senescent wild-type cultures (see Belcour et al. 1999 for review). In marked contrast to this situation, in the dying PaAntM106P rmp1-1 and PaAntA121P rmp1-1 cultures (Figure 2B), as in the dying PaAntS296M rmp1-1 and rmp1-2 cultures (Figure 3), death is not associated with the accumulation of senDNAα. The mtDNA profile of independent PaAntM106P rmp1-1 and PaAntA121P rmp1-1 cultures revealed large-scale rearrangements and the loss of numerous fragments covering ∼60% of the wild-type chromosome (Figure 2B and Figure S2). These data were confirmed by the use of different probes covering ∼80% of the mitochondrial genome. Remarkably, some of these fragments were systematically lost in independent cultures, indicating that the deletions cover a common region of the mitochondrial genome. However, different additional fragments presumed to result from circularization of the deleted chromosomes were recovered in the different dying cultures, indicating that these deletions are not site specific and present different boundaries. In contrast, the analysis of the mtDNA content of independent dying PaAntS296M rmp1-1 or rmp1-2 cultures revealed multiple rearrangements different from one culture to another and affecting different parts of the mitochondrial genome (Figure 3). One interesting outcome of the mtDNA analysis (shown in Figure 2A) is that, whereas the PaAntM106P and PaAntA121P mutations are responsible for mtDNA instability in the presence of the rmp1-1 allele as observed in adPEO patients, this instability is suppressed in the presence of the rmp1-2 allele: the mtDNA of the long-lived PaAntM106P rmp1-2 mat+ and PaAntA121P rmp1-2 mat+ mutants remained stable for >180 days.
Figure 2.—
Effects of PaAntM106P and PaAntA121P mutant alleles on mtDNA. (A) rmp1-2 mat+ strains. (B) rmp1-1 mat− strains. (Left, A and B) The ethidium-bromide-stained HaeIII restriction patterns of mtDNA extracted from different cultures. (Right, A and B) The corresponding Southern blots probed with a fragment of the P. anserina mitochondrial genome including the α-region. This probe reveals four fragments indicated by solid dots on intact mtDNA molecules. In senescent wild-type cultures, it reveals a fragment of 2.5 kb (indicated by a black double arrowhead) corresponding to senDNAα and a fragment of 5 kb (black arrowhead) potentially corresponding to a dimer of senDNAα. (A) Lane 1: young wild-type rmp1-2 mat+ culture; lane 2: senescent wild-type rmp1-2 mat+ culture; lanes 3 and 4: independent PaAntA121P rmp1-2 mat+ cultures after 180 days of growth; lanes 5 and 6: independent PaAntM106P rmp1-2 mat+ cultures after 180 days of growth. (B) Lane 1′: young wild-type rmp1-1 mat− culture; lane 2′: senescent wild-type rmp1-1 mat− culture; lanes 3′ and 4′: independent dying PaAntA121P rmp1-1 mat− cultures; lanes 5′ and 6′: independent dying PaAntM106P rmp1-1 mat− cultures. Black arrows indicate fragments present in the wild-type chromosome that are absent or underrepresented in the mtDNA from PaAntM106P rmp1-1 mat− and PaAntA121P rmp1-1 mat− cultures. White arrows indicate additional fragments presumed to correspond to deletion junctions.
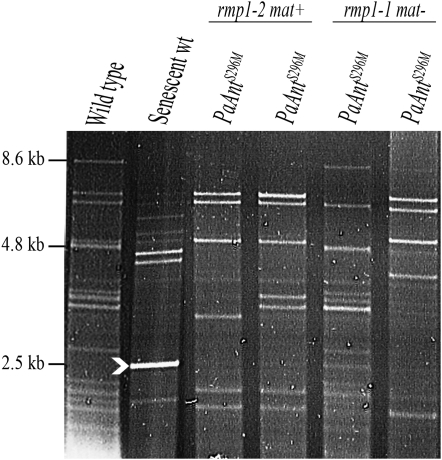
Figure 3.—
Effects of the PaAntS296M mutant allele on mtDNA. Ethidium- bromide- stained HaeIII restriction patterns of mtDNA extracted from different dying PaAntS296M cultures. The white arrowhead indicates the position of senDNAα.
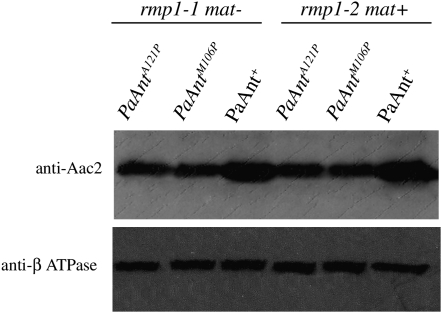
Western blot analyses were performed to determine whether a reduced amount of mutant protein could explain the phenotypic alterations of the mutants and the differences of mtDNA instability between the PaAntM106P and PaAntA121P strains harboring the rmp1-1 or the rmp1-2 allele. Only PaAntM106P and PaAntA121P mutants were studied because we were not able to prepare sufficient mitochondria from the PaAntS296M mutant due to its premature death phenotype. As shown on Figure 4, the amount of the mutant protein is reduced (∼50% of wild type) in both the PaAntM106P and the PaAntA121P strain. However, no difference was observed in either rmp1-1 or rmp1-2 strains, indicating that the mtDNA instability is not correlated to a reduced protein content.
Figure 4.—
Western blot analysis of the PaANT protein. Mitochondrial protein extracts (30 μg) from mitochondria extracted from the wild-type, PaAntM106P, and PaAntA121P strains (rmp1-1 mat− and rmp1-2 mat+) were separated on SDS–PAGE and probed with a yeast Aac2 monoclonal antibody. The blot was reprobed with an antibody directed against the β-subunit of ATPase as loading control.
Mutation in a cytosolic ribosomal protein suppresses the premature death phenotype conferred by the mutations M106P and A121P but not S296M:
In S. cerevisiae, it was recently shown that the aac2A128P mutation leads to the formation of degenerative microcolonies whose frequency increases with age. This degenerative cell death is partially suppressed by reduced cytosolic protein synthesis (Wang et al. 2008a). In P. anserina, mutations that affect the translation apparatus drastically modify life span and mtDNA stability (Silar et al. 2001). However, the nature of the relationships between protein synthesis, aging, and associated mtDNA instability are unclear. The most striking effect observed is that of the AS1-4 mutation identified as affecting the fidelity of translation (Coppin-Raynal 1981). This mutation is located in a cytosolic ribosomal protein (Dequard-Chablat and Sellem 1994) that, in association with the rmp1-1 or the rmp1-2 alleles, drastically shortens or increases life span (Belcour et al. 1991; Contamine et al. 1996). Interestingly, like the PaAntM106P and PaAntA121P strains, dying AS1-4 cultures accumulate large mitochondrial deletions, but these are characterized by precise boundaries (Belcour et al. 1991; Sainsard-Chanet et al. 1998; Figure S2). To test whether the AS1-4 mutation is able to suppress the premature death phenotype due to the PaAnt mutations, it was associated with the three PaAntM106P, PaAntA121P, and PaAntS296M mutations by crosses. Remarkably, the premature death phenotype and the mtDNA instability of PaAntM106P rmp1-1 and PaAntA121P rmp1-1 were suppressed in the presence of the AS1-4 mutation, and a synthetic phenotype was obtained (life span >60 days) (Table 2). In contrast, the premature death of PaAntS296M rmp1-1 and rmp1-2 was not suppressed by AS1-4. Furthermore, whereas the life span of the AS1-4 rmp1-2 strain was greatly increased, that of the double mutant PaAntS296M AS1-4 rmp1-2 was very short, and the mtDNA pattern at the time of death corresponded to multiple rearrangements as in the PaAntS296M single mutant, indicating that PaAntS296M is epistatic to the AS1-4 mutation. In the same way, the double mutants PaAntM106P AS1-4 and PaAntA121P AS1-4 shared the same mycelial aspect, female sterility, and growth rate as the single PaAntM106P and PaAntA121P mutants (Table 2), indicating that, although AS1-4 suppresses the mtDNA instability, the translocase mutations are epistatic to the AS1-4 mutation for the somatic and sexual defects.
TABLE 2.
Suppression of the short-lived phenotype of PaAntM106P rmp1-1 and PaAntA121P rmp1-1 by the AS1-4 mutation
| Strain | Growth rate(mm/day ± SE) | Life span (days ± SE) |
|---|---|---|
| PaAnt+, AS1-4, rmp1-2 mat+ | 5.6 ± 0.2 | >100 |
| PaAnt+, AS1-4, rmp1-1 mat− | 5.5 ± 0.1 | ∼4 |
| PaAntM106P, AS1-4, rmp1-2 mat+ | 4.5 ± 0.2 | >60 |
| PaAntM106P, AS1-4, rmp1-1 mat− | 3.7 ± 0.4 | >60 |
| PaAntA121P, AS1-4, rmp1-2 mat+ | 4.4 ± 0.1 | >60 |
| PaAntA121P, AS1-4, rmp1-1 mat− | 4.9 ± 0.1 | >60 |
| PaAntS296M, AS1-4, rmp1-2 mat+ | 4.2 ± 0.2 | 5.5 ± 0.8 |
|
PaAntS296M, AS1-4, rmp1-1 mat− |
3.9 ± 0.3 |
6 ± 0.5 |
The mutant PaAnt alleles lead to morphological alterations of mitochondria and reduction in respiratory activity, ROS production, and inner membrane potential:
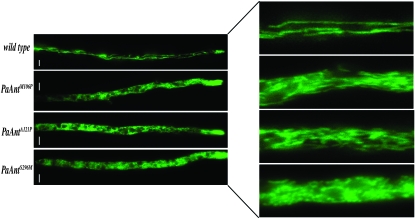
The effect of the different PaAnt mutations on mitochondrial structure and morphology was examined using a reporter system composed of the EGFP protein to which a mitochondrial-targeting presequence was added (Sellem et al. 2007). The construct was integrated after transformation into a wild-type strain and transferred into the three mutants through crosses. Whereas the wild-type strain contained long filamentous mitochondria, the mitochondrial structure of the mutants was considerably modified and fragmented (Figure 5). Interestingly, the mutants showed a similar mitochondrial phenotype regardless of the rmp1 allele, life span, and mtDNA stability. The strongest phenotype was observed in PaAntS296M, in which the majority of mitochondria seemed to be organized in aggregates while, in PaAntM106P and PaAntA121P, they appeared as small fragmented filaments. These results indicate that the three PaAnt mutations affect mitochondrial structure and morphology.
Figure 5.—
Analysis of mitochondrial morphology in wild type, PaAntM106P, PaAntA121P and PaAntS296M strains. (Right) An enlargement of a sector of the filaments. The wild-type strain shows snakelike mitochondria. The PaAntM106P and PaAntA121P mutants show fragmented filamentous mitochondria, and the PaAntS296M mutant shows aggregated mitochondria. The represented strains are rmp1-2 mat+. The rmp1-1 mat− strains display similar phenotypes. Bars, 5 μm.
We then tested the effects of the PaAnt mutations on oxygen consumption, ROS production, and mitochondrial inner membrane potential (Δψ). Only PaAntM106P and PaAntA121P mutants (both the long-lived rmp1-2 mat+ and the short-lived rmp1-1 mat− strains) were studied because we were not able to prepare sufficient protoplasts from the PaAntS296M mutant as previously mentioned. As shown in Table 3, the respiratory activity of the PaAntM106P and PaAntA121P mutants, regardless of the rmp1 allele, life span, and mtDNA stability, was decreased by a factor of 2–3 compared to that of wild type. Furthermore, in contrast to the wild-type strain in which oxygen consumption is very sensitive to cyanide, respiration was ∼50% resistant to cyanide in the two mutants. In P. anserina, resistance to cyanide has previously been reported in several respiratory mutants partially or completely deficient for complexes III or IV and is due to the induction of an alternative oxidase (AOX) (Dufour et al. 2000; Stumpferl et al. 2004; Sellem et al. 2007). Quantitative RT–PCR experiments revealed that the amount of aox transcripts increased ∼7- to 20-fold in the mutants compared to the wild type (Table 3). The induction of the alternative oxidase and the reduction of cellular respiration in the PaAntM106P and PaAntA121P strains strongly suggest that these mutations cause damage to the cytochrome part of the electron transport chain. However, as the strains display similar respiratory properties whatever their mtDNA stability, this damage is probably independent of the mtDNA instability.
TABLE 3.
Mitochondrial characteristics of strains carrying different PaAnt mutant alleles
| Strain | Respiration rate(nmol O2 · min−1) | KCN inhibition level (%) | Relative abundance of AOX transcripts | ROS production level | Membrane potential level (Δψ) | Δψ + mClCCP |
|---|---|---|---|---|---|---|
| PaAnt+, rmp1-2 mat+ | 25.9 ± 3.2 | 83 | 1 | 93 ± 6 | 35 ± 3 | 7.66 ± 0.2 |
| PaAnt+, rmp1-1 mat− | 22.1 ± 4.1 | 88 | 1 | 99 ± 2 | 33 ± 2.8 | 7 ± 0.2 |
| PaAntM106P, rmp1-2 mat+ | 9.8 ± 0.7 | 58 | 7.5 ± 0.9 | 37.6 ± 2 | 16 ± 1.1 | 7.3 ± 0.3 |
| PaAntM106P, rmp1-1 mat− | 9.3 ± 2.2 | 62 | 23 ± 8 | 42.5 ± 3.5 | 14.3 ± 1.2 | 7.4 ± 0.3 |
| PaAntA121P, rmp1-2 mat+ | 12.8 ± 2.1 | 33 | 7 ± 0.8 | 34.9 ± 3 | 16 ± 1.2 | 4 ± 0.3 |
| PaAntA121P, rmp1-1 mat− | 8.2 ± 0.9 | 56 | 9.5 ± 1 | 39 ± 2 | 14.9 ± 1.2 | 4.2 ± 0.2 |
| PaAnt+, Δaox, rmp1-2 mat+ | 24.3 ± 3.3 | 95 | — | 81.5 ± 4 | — | — |
| PaAntM106P, Δaox, rmp1-2 mat+ | 10.2 ± 1.6 | 94 | — | 36.1 ± 2.2 | — | — |
| PaAntA121P, Δaox, rmp1-2 mat+ | 8.7 ± 1.4 | 94 | — | 39 ± 2 | — | — |
| PaAnt+ (PaAnt+), rmp1-2 mat+ | 24.3 ± 0.8 | — | — | 96 ± 3.6 | 34.8 ± 3.1 | 8.9 ± 0.6 |
| PaAntM106P (PaAnt+), rmp1-2 mat+ | 14.3 ± 0.4 | — | — | 40 ± 1.7 | 11.1 ± 1.8 | 4.3 ± 0.7 |
|
PaAntA121P (PaAnt+), rmp1-2 mat+ |
12.3 ± 0.8 |
— |
— |
44 ± 3.3 |
15.1 ± 1 |
5.1 ± 1.9 |
Respiration rate was measured on 3.107 protoplasts for each experiment and expressed as nanomoles min−1 ± SE. The relative abundance of the aox transcripts was measured by real-time qPCR experiments. For each strain, the level of the aox transcripts was normalized using the gpd transcript level, which was used as a standard; the aox mRNA copy number was given a value of 1 in the wild-type strain. ROS and Δψ measurements were carried out on protoplasts in a Partec PAS-III flow cytofluometer. The values correspond to the mean fluorescent intensity of the fluorescent probes H2DCF-DA (80 mm) and DIOC6(3) (0.1 mm) used for ROS and Δψ measurements, respectively. The fluorescence of DIOC6(3) was also measured in the presence of the uncoupling agent carbamoyl cyanide m-chlorophenylhydrazone (mClCCP) to confirm that fluorescence is correlated to Δψ.
We then analyzed the effects of the PaAntM106P and PaAntA121P mutations on ROS production by flow cytofluometry, by incubating protoplasts with H2DCF-DA, and by monitoring the formation of fluorescent DCF. In both PaAntM106P and PaAntA121P mutants, regardless of the rmp1 allele, life span, and mtDNA stability, the intensity of fluorescence was about three fold less than in the wild type, indicating a clear decrease in ROS production in these mutants (Table 3). To investigate if the reduced levels of ROS could be explained by the expression of the alternative oxidase known to act in some situations as an antioxidant enzyme (Maxwell et al. 1999), the PaAntM106P and PaAntA121P mutant strains devoid of the aox gene (obtained after crosses with the Δaox strain) were also analyzed. As shown in Table 3, the ROS production and respiration rate were similar in PaAntM106P and PaAntA121P strains whether the aox gene was expressed or inactivated. This result demonstrates that the reduction of ROS production is not due to the presence of the alternative oxidase but rather to the reduced respiratory activity of the mutants. We also measured the expression levels of the PaSod3 and PaSod1 genes encoding the mitochondrial and cytosolic superoxide dismutases, respectively (Borghouts et al. 2002), by qRT–PCR in the PaAntM106P, PaAntA121P, and wild-type strains. These levels were not significantly different between the genotypes (data not shown), which is consistent with the absence of oxidative stress in the PaAnt mutants studied. This result is important; it indicates that (1) the life span of the strains carrying the adPEO alleles is not correlated to the ROS level and (2) the mitochondrial instability of the PaAnt rmp1-1 mutant strains is not associated with increased mitochondrial oxidative stress.
Mitochondrial inner membrane potential was then monitored by flow cytometry using DiOC6(3) dye. The results showed a twofold decrease of the Δψ in the mutant strains compared to that of the wild type in both rmp1 contexts (Table 3 and Figure S3). This indicates that there is no correlation between a reduction of the Δψ and mtDNA instability in the PaAntM106P and PaAntA121P mutants.
All the phenotypic defects caused by the PaAnt mutant alleles behave as dominant traits:
The L98P, A114P, and V289M mutations are dominant in humans. To mimic the diploid organization of mammal somatic tissues and study whether the three alleles also behave as dominants in P. anserina, heteroallelic strains containing a mutant and a wild-type allele were constructed. The PaAnt+(PaAnt+-Hygro) strain bearing a functional ectopic copy of the wild-type allele able to complement the deletion of the PaAnt+ gene was crossed with the PaAntM106P, PaAntA121P, and PaAntS296M strains. The germination, growth rate, and longevity phenotypes of the three heteroallelic PaAntM106P (PaAnt+), PaAntA121P (PaAnt+), and PaAntS296M (PaAnt+) strains were indistinguishable from the corresponding monoalleleic mutant strains (Table 1C). The presence of the wild-type allele did not modify the rmp1 effect in the PaAntM106P and PaAntA121P strains or the premature death phenotype of the PaAntS296M strains irrespective of the rmp1 allele present. As in homoallelic strains, death in PaAntM106P and PaAntA121P but not in PaAntS296M was accompanied by the accumulation of large-scale deletions affecting the same part of the mitochondrial genome without accumulation of senDNAα (data not shown). Importantly, the presence of the wild-type allele does not restore wild-type respiration rate, wild-type ROS production, or wild-type Δψ to the PaAntM106P and PaAntA121P strains (Table 3). Taken together, these data demonstrate that all the phenotypic traits due to the adPEO alleles behave as dominant traits in P. anserina.
DISCUSSION
In this study, we analyzed the properties of strains carrying three adPEO-associated missense mutations introduced into the PaAnt gene of P. anserina. The M106P, A121P, and S296M mutations in P. anserina correspond to the L98P, A114P (familial cases), and V289M (sporadic case) mutations of the hANT1 gene in humans and to the M114P, A128P, and S303M mutations in S. cerevisiae. Alanine 114 and leucine 98 in the hANT1 protein map in highly conserved or conservatively substituted residues throughout eukaryotes, while valine 289 does not. The M114 and S303 residues of the S. cerevisiae Aac2 protein were replaced by their corresponding residues from the wild-type ANT1, resulting in “humanized wild type” AAC2 alleles. These were functional and able to complement oxidative growth defects due to the AAC2 deletion (Fontanesi et al. 2004).
The unique PaAnt gene is an essential gene:
In contrast to humans and S. cerevisiae, in which several functional isoforms of the ADP/ATP translocator are present (Fiore et al. 1998; Dolce et al. 2005), only one gene has been found in P. anserina as in N. crassa (Arends and Sebald 1984) and numerous other filamentous fungi. Considering the central role of ATP/ADP transport across the mitochondrial inner membrane (MIM) in oxidative metabolism and the strict aerobic metabolism of P. anserina, it is not surprising that the PaAnt gene is essential. It is probable that the deletion of the ATP/ADP translocator results in ADP depletion in the mitochondrial matrix, which blocks the F1-FO ATPase and thus respiration. Also the absence of the most abundant inner membrane protein probably affects the structure of the inner membrane, which leads to respiration inhibition. Recently, it was demonstrated that in S. cerevisiae the abundant Aac2 isoform exists in physical interaction with the cytochrome bc1-COX supercomplex and the TIM23 machinery and that, in the absence of Aac2, the assembly of the surpercomplex is altered (Dienhart and Stuart 2008).
Characteristics of the mtDNA instability distinguish between the severity of the three mutant alleles:
In contrast to the ΔPaAnt allele, the three mutant alleles were viable but responsible for several detrimental effects on vegetative and sexual development, independently of the rmp1 allele. These defects were more pronounced in the PaAntS296M mutant than in PaAntM106P and PaAntA121P. A reduced concentration of the mutant ANT protein was observed in the PaAntM106P and PaAntA121P strains. This could be due to a reduced import efficiency (De Marcos Lousa et al. 2002) or to a reduced stability of the mutant proteins. However, it seems unlikely that this reduction is responsible for the defects observed in the mutants since the mutations are dominant and the deletion of PaAnt is recessive (not shown) as is the deletion of AAC2 in S. cerevisiae (Chen 2004). This strongly suggests that the mutant phenotypes are due to an alteration of, and not to a decreased amount of, mutant proteins.
In humans, the three adPEO mutations are linked to the formation of multiple mtDNA deletions (Kaukonen et al. 2000); in S. cervisiae, only aac2A128P and aac2S303M, but not aac2M114P, were reported to cause an increase in abnormal mtDNA genomes (Fontanesi et al. 2004). In P. anserina, the three mutations are linked to the formation of large mitochondrial rearrangements. However, several properties differentiate PaAntM106P and PaAntA121P from the PaAntS296M mutation. First, in the PaAntM106P and PaAntA121P strains, the mitochondrial rearrangements correspond to large-scale mtDNA deletions covering a common region that represents ∼60% of the genome with no identified fixed boundaries, whereas, in the PaAntS296M strain, they correspond to variable rearrangements and deletions involving different regions of the genome. Since in P. anserina, mitochondrial instability leads to death, the timing of the death of a strain is an indicator of its mtDNA instability; it can therefore be concluded that the PaAntS296M allele causes greater mtDNA instability than either the PaAntM106P and the PaAntA121P allele.
A second difference is that the mtDNA instability caused by the PaAntM106Pand the PaAntA121P mutations is observed in the presence of the rmp1-1 allele and suppressed by the rmp1-2 allele. In contrast, the PaAntS296M mutant showed huge mtDNA instability in the presence of either rmp1 allele. Suppression of mtDNA instability by the rmp1-2 allele has previously been reported in strains carrying the AS1-4 allele, which alters a cytosolic ribosomal protein (Dequard-Chablat and Sellem 1994) and causes the accumulation of site-specific mtDNA deletions (Belcour et al. 1991; Contamine et al. 1996). The rmp1-2 allele differs from rmp1-1 by the presence of a nonsense mutation that eliminates the last 19 amino acids of the protein (Contamine et al. 2004). Thus rmp1-1 is probably fully functional, and it could be directly or indirectly involved in the accumulation of large-scale deletions of the mtDNA genome such as those caused by the AS1-4, PaAntM106P, and PaAntA121P mutations, while the rmp1-2 allele would delay the rate of accumulation of these deletions. In contrast, rmp1 does not appear to be involved in the rate of accumulation of other types of mtDNA rearrangements such as those generated during senescence (senDNAs) or by the PaAntS296M mutation. Rmp1 is an essential gene whose function is presently unknown. The encoded protein is localized in mitochondria and/or cytosol compartments depending on the cell type and the developmental stage. Putative homologs have been found only in filamentous fungi to date, but rmp1 is thought to evolve rapidly, rendering the recognition of its homologs in other species very difficult (Contamine et al. 2004).
A third difference between the PaAntM106P/PaAntA121P and the PaAntS296M alleles is the synthetic phenotype obtained when PaAntM106P and PaAntA121P are associated with AS1-4 rmp1-1. The association of PaAntM106P/PaAntA121P but not PaAntS296M with AS1-4 rmp1-1 leads to a spectacular stabilization of the mtDNA. A possible reason for the synthetic phenotype is that the deletions observed in AS1-4 rmp1-1 and those observed in PaAntM106P rmp1-1 and PaAntA121P rmp1-1 are generated by mutually exclusive mechanisms.
Taken together, these results clearly show that the S296M mutation and the M106P/A121P mutations lead to different functional consequences. The predicted localization of the three PaAnt mutations derived from the crystal structure of the bovine ANT1 protein places the residues M106 and A121 in close proximity in the cytosolic gating region (Wang et al. 2008b), suggesting that the two mutations might induce similar structural changes. The S296M mutation is localized in a different region and thus may affect the translocator in a different and more severe manner, which may explain the particular properties of this mutation. These properties could also result from the sequence divergence between humans and P. anserina at this position (V289 in humans), which might suggest a specific role for this residue in P. anserina.
mtDNA instability is not systematically correlated with an increased ROS production or a reduced Δψ:
Several mechanisms have been suggested to explain how the adPEO alleles of hANT1 could lead to mtDNA instability: nucleotide imbalance causing defects in dATP biosynthesis and mtDNA replication (Kaukonen et al. 2000; Fontanesi et al. 2004); mtDNA damage by overproduction of ROS (Esposito et al. 1999; Palmieri et al. 2005); and the low Δψ model in which the pathogenic mutations uncouple the mitochondrial inner membrane primarily affecting mitochondrial biogenesis, which would have secondary effects on mtDNA stability (Chen 2002, 2004; Wang et al. 2008b). It was not possible to measure nucleotide exchange properties of the mutant strains because mitochondria isolated from these strains were of low quality, but it is presumed that a reduced availability of matrix ADP would inhibit the ATP synthase, causing hyperpolarization of the MIM and an increased production of ROS. This was demonstrated in tissues of Ant1-knockout mice (Esposito et al. 1999). In S. cerevisiae, the aac2A128P and aac2M114P alleles, corresponding to the PaAntA121P and PaAntM106P alleles, respectively, have been shown to uncouple the mitochondrial respiration and decrease the Δψ (Wang et al. 2008b). A more extensive study of aac2A128P revealed that this allele is responsible for mitochondrial degeneration and degenerative cell death, probably due to the low Δψ. Interestingly, reduced cytosolic protein synthesis suppressed both membrane depolarization and mitochondrial degeneration, revealing a strong link between these two parameters (Wang et al. 2008a).
In P. anserina, the pathogenic mutations are responsible for defects in growth and sexual development, reduction of respiration rate, strong decline of the Δψ and of ROS production regardless of the rmp1 allele, and mtDNA instability (and premature death) only in presence of the rmp1-1 allele. The rmp1-2 allele suppresses mtDNA instability (and therefore premature death) but not the other defects and is not accompanied by reestablishment of a wild-type Δψ. These results strongly suggest that, as already suggested for yeast (Wang et al. 2008a, b), the primary consequence of the adPEO-associated mutations is not the mtDNA instability but rather the damages to the electron transport chain which lead to growth and sexual development defects. These results also suggest that the decline of the Δψ alone is not sufficient to cause mtDNA instability and that other functions are implicated. The rmp1-1 allele would have such a function, which would be lost for the rmp1-2 allele that encodes a truncated protein.
All the traits associated with the adPEO alleles are dominant:
Finally, it is of interest to note that all the phenotypes associated with PaAntM106P and PaAntA121P (and PaAntS296M when tested) are mediated by a dominant mechanism in P. anserina as in humans. These include slow growth rate, mitochondrial morphology modifications, mtDNA instability, timing of death, reduced respiration rate, reduced ROS production, and reduced Δψ. The dominant nature of the three PaAntM106P, PaAntA121P, and PaAntS296M alleles strongly suggests that they correspond to a gain of function and that this function is deleterious for the cell. In S. cerevisiae, the three equivalent mutations behave as dominant for mtDNA instability and respiratory rate but recessive for oxidative growth (Fontanesi et al. 2004). In fact, it seems that at least for the equivalent A114P and L98P mutations, the dominant/recessive character of the mtDNA instability depends on the genetic balance between the mutated and the wild-type alleles (Wang et al. 2008b).
Taken together, the results presented in this work validate P. anserina as a model organism for studying the pathogenecity of different human mutations and underscore the importance of using a variety of model systems to dissect human mitochondrial disorders. They also show that, in P. anserina, the mtDNA instability associated with mitochondrial adenine translocator mutations is not due to increased ROS levels or to reduced Δψ alone. Thus, any proposed therapy increasing Δψ or limiting ROS production should be considered carefully. Finally, it is important to realize the central role of the genetic background in developing the pathogenic effects of a mutation.
Acknowledgments
We are very grateful to Carole Sellem for her constant interest in this work. We thank Sandrine Bach-Berkowicz for the construction of the vectors carrying the PaAnt mutant alleles and Edmund R. S. Kunji for his generous gift of the Aac2 monoclonal antibody. We thank Guy Lauquin and Véronique Trézéguet for useful discussions and critical comments, Vincent Rincheval for his help in the flow cytometry analysis, Chris Herbert and Linda Sperling for critical reading of the manuscript, and Delphine Petit for technical help. This work was supported by grants from the Association Française contre les Myopathies and the European Community's Sixth Framework Programme (LSHM-CT-200-512020).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107813/DC1.
References
- Arends, H., and W. Sebald, 1984. Nucleotide sequence of the cloned mRNA and gene of the ADP/ATP carrier from Neurospora crassa. EMBO J. 3 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamber, L., D. Slotboom and E. R. S. Kunji, 2007. Yeast mitochondrial ADP/ATP carriers are monomeric in detergents as demonstrated by differential affinity purification. J. Mol. Biol. 371 388–395. [DOI] [PubMed] [Google Scholar]
- Begel, O., J. Boulay, B. Albert, E. Dufour and A. Sainsard-Chanet, 1999. Mitochondrial group II introns, cytochrome c oxidase, and senescence in Podospora anserina. Mol. Cell. Biol. 19 4093–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcour, L., O. Begel and M. Picard, 1991. A site-specific deletion in mitochondrial DNA of Podospora is under the control of nuclear genes. Proc. Natl. Acad. Sci. USA 88 3579–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcour, L., A. Sainsard-Chanet, C. Jamet-Vierny and M. Picard, 1999. Stability of the mitochondrial genome of Podospora anserina and its genetic control, pp. 209–228 in Mitochondrial Diseases, edited by P. Lestienne. Springer-Verlag, Berlin/New York.
- Berges, T., and C. Barreau, 1989. Heat shock at an elevated temperature improves transformation efficiency of protoplasts from Podospora anserina. J. Gen. Microbiol. 139 601–604. [DOI] [PubMed] [Google Scholar]
- Borghouts, C., C. Q. Scheckhuber, A. Werner and H. D. Osiewacz, 2002. Respiration, copper availability and SOD activity in P. anserina strains with different lifespan. Biogerontology 3 143–153. [DOI] [PubMed] [Google Scholar]
- Chen, X. J., 2002. Induction of an unregulated channel by mutations in adenine nucleotide translocase suggests an explanation for human ophthalmoplegia. Hum. Mol. Genet. 11 1835–1843. [DOI] [PubMed] [Google Scholar]
- Chen, X. J., 2004. Sal1p, a calcium-dependent carrier protein that suppresses an essential cellular function associated with the Aac2 isoform of ADP/ATP translocase in Saccharomyces cerevisiae. Genetics 167 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery, F. P., 2003. Searching for nuclear-mitochondrial genes. Trends Genet. 19 60–62. [DOI] [PubMed] [Google Scholar]
- Contamine, V., G. Lecellier, L. Belcour and M. Picard, 1996. Premature death in Podospora anserina: sporadic accumulation of the deleted mitochondrial genome, translational parameters and innocuity of the mating types. Genetics 144 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine, V. R., D. Zickler and M. Picard, 2004. The Podospora rmp1 gene implicated in nucleus-mitochondria cross-talk encodes an essential protein whose subcellular location is developmentally regulated. Genetics 166 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, W. S., 2008. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 59 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin, E., and R. Debuchy, 2000. Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics 155 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin-Raynal, E., 1981. Ribosomal suppressors and antisuppressors in Podospora anserina: altered susceptibility to paromomycin and relationships between genetic and phenotypic suppression. Biochem. Genet. 19 729–740. [DOI] [PubMed] [Google Scholar]
- Cummings, D. J., K. L. McNally, J. M. Domenico and E. T. Matsuura, 1990. The complete DNA sequence of the mitochondrial genome of Podospora anserina. Curr. Genet. 17 375–402. [DOI] [PubMed] [Google Scholar]
- De Marcos Lousa, C., V. Trezeguet, A.C. Dianoux, G. Brandolin and G.J. Lauquin, 2002. The human mitochondrial ADP/ATP carriers: kinetic properties and biogenesis of wild-type and mutant proteins in the yeast S. cerevisiae. Biochemistry 41 14412–14420. [DOI] [PubMed] [Google Scholar]
- Dequard-Chablat, M., and C. H. Sellem, 1994. The S12 ribosomal protein of Podospora anserina belongs to the S19 bacterial family and controls the mitochondrial genome integrity through cytoplasmic translation. J. Biol. Chem. 269 14951–14956. [PubMed] [Google Scholar]
- Dienhart, M. K., and R. A. Stuart, 2008. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell. 19 3934–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolce, V., P. Scarcia, D. Iacopetta and F. Palmieri, 2005. A fourth ADP/ATP carrier isoform in man: identification, bacterial expression, functional characterization and tissue distribution. FEBS Lett. 579 633–637. [DOI] [PubMed] [Google Scholar]
- Dufour, E., J. Boulay, V. Rincheval and A. Sainsard-Chanet, 2000. A causal link between respiration and senescence in Podospora anserina. Proc. Natl. Acad. Sci. USA 97 4138–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury, R., C. H. Sellem, E. Coppin, A. Boivin, M. F. Maas et al., 2008. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53 249–258. [DOI] [PubMed] [Google Scholar]
- Espagne, E., O. Lespinet, F. Malagnac, C. Da Silva, O. Jaillon et al., 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9 R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, L. A., S. Melov, A. Panov, B. A. Cottrell and D. C. Wallace, 1999. Mitochondrial disease in mouse results in increased oxidative stress. Proc. Natl. Acad. Sci. USA 96 4820–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser, K., 1974. Podospora anserina, pp. 531–551 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Fiore, C., V. Trezeguet, A. Le Saux, P. Roux, C. Schwimmer et al., 1998. The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie 80 137–150. [DOI] [PubMed] [Google Scholar]
- Fontanesi, F., L. Palmieri, P. Scarcia, T. Lodi, C. Donnini et al., 2004. Mutations in AAC2, equivalent to human adPEO-associated ANT1 mutations, lead to defective oxidative phosphorylation in Saccharomyces cerevisiae and affect mitochondrial DNA stability. Hum. Mol. Genet. 13 923–934. [DOI] [PubMed] [Google Scholar]
- Graham, B. H., K. G. Waymire, B. Cottrell, I. A. Trounce, G. R. MacGregor et al., 1997. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 16 226–234. [DOI] [PubMed] [Google Scholar]
- Kaukonen, J., M. Zeviani, G. P. Comi, M. G. Piscaglia, L. Peltonen et al., 1999. A third locus predisposing to multiple deletions of mtDNA in autosomal dominant progressive external ophthalmoplegia. Am. J. Hum. Genet. 65 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen, J., J. K. Juselius, V. Tiranti, A. Kyttälä, M. Zeviani et al., 2000. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289 782–785. [DOI] [PubMed] [Google Scholar]
- Klingenberg, M., 2008. The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778 1978–2021. [DOI] [PubMed] [Google Scholar]
- Komaki, H., T. Fukazawa, H. Houzen, K. Yoshida, I. Nonaka et al., 2002. A novel D104G mutation in the adenine nucleotide translocator 1 gene in autosomal dominant progressive external ophthalmoplegia patients with mitochondrial DNA with multiple deletions. Ann. Neurol. 51 645–648. [DOI] [PubMed] [Google Scholar]
- Lecellier, G., and P. Silar, 1994. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 25 122–123. [DOI] [PubMed] [Google Scholar]
- Lorin, S., E. Dufour, J. Boulay, O. Begel, S. Marsy et al., 2001. Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol. Microbiol. 42 1259–1267. [DOI] [PubMed] [Google Scholar]
- Maxwell, D. P., Y. Wang and L. McIntosh, 1999. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli, L., A. Bordoni, M. Zeviani, G. M. Hadjigeorgiou, M. Sciacco et al., 2001. A novel missense adenine nucleotide translocator-1 gene mutation in a Greek adPEO family. Neurology 57 2295–2298. [DOI] [PubMed] [Google Scholar]
- Nury, H., C. Dahout-Gonzalez, V. Trezeguet, G. J. M. Lauquin, G. Brandolin et al., 2006. Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu. Rev. Biochem. 75 713–741. [DOI] [PubMed] [Google Scholar]
- Palmieri, F., 2004. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447 689–709. [DOI] [PubMed] [Google Scholar]
- Palmieri, L., S. Alberio, I. Pisano, T. Lodi, M. Meznaric-Petrusa et al., 2005. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum. Mol. Genet. 14 3079–3088. [DOI] [PubMed] [Google Scholar]
- Riccio, P., H. Aquila and M. Klingenberg, 1975. Solubilization of the carboxy-atractylate binding protein from mitochondria. FEBS Lett. 56 192–232. [PubMed] [Google Scholar]
- Rizet, G., 1952. Les phénomènes de barrage chez Podospora anserina. I. Analyse génétique des barrages entre souches S et s. Rev. Cytol. Biol. Vég. 13 51–92. [Google Scholar]
- Sainsard-Chanet, A., O. Begel and Y. d'Aubenton-Carafa, 1998. Two co-existing mechanisms account for the large-scale deletions of mitochondrial DNA in Podospora anserina that involve the 5′ border of a group-II intron. Curr. Genet. 34 326–335. [DOI] [PubMed] [Google Scholar]
- Sellem, C. H., S. Marsy, A. Boivin, C. Lemaire and A. Sainsard-Chanet, 2007. A mutation in the gene encoding cytochrome c1 leads to a decreased ROS content and to a long-lived phenotype in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 44 648–658. [DOI] [PubMed] [Google Scholar]
- Siciliano, G., A. Tessa, S. Petrini, M. Mancuso, C. Bruno et al., 2003. Autosomal dominant external ophthalmoplegia and bipolar affective disorder associated with a mutation in the ANT1 gene. Neuromuscul. Disord. 13 162–165. [DOI] [PubMed] [Google Scholar]
- Silar, P., 1995. Two new easy to use vectors for transformation. Fungal Genet. Newsl. 42 73. [Google Scholar]
- Silar, P., H. Lalucque and C. Vierny, 2001. Cell degeneration in the model system Podospora anserina. Biogerontology 2 1–17. [DOI] [PubMed] [Google Scholar]
- Stepien, G., A. Torroni, A. B. Chung, J. A. Hodge and D. C. Wallace, 1992. Differential expression of adenine nucleotide translocator isoforms in mammalian tissues and during muscle cell differentiation. J. Biol. Chem. 267 14592–14597. [PubMed] [Google Scholar]
- Stumpferl, S. W., O. Stephan and H. D. Osiewacz, 2004. Impact of a disruption of a pathway delivering copper to mitochondria on Podospora anserina metabolism and life span. Eukaryot. Cell 3 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., X. Zuo, B. Kucejova and X. J. Chen, 2008. a Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nat. Cell. Biol. 10 1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., K. Salinas, X. Zuo, B. Kucejova and X. J. Chen, 2008. b Dominant membrane uncoupling by mutant adenine nucleotide translocase in mitochondrial diseases. Hum. Mol. Genet. 17 4036–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]