Abstract
The intraflagellar transport machinery is required for the assembly of cilia. It has been investigated by biochemical, genetic, and computational methods that have identified at least 21 proteins that assemble into two subcomplexes. It has been hypothesized that complex A is required for retrograde transport. Temperature-sensitive mutations in FLA15 and FLA17 show defects in retrograde intraflagellar transport (IFT) in Chlamydomonas. We show that IFT144 and IFT139, two complex A proteins, are encoded by FLA15 and FLA17, respectively. The fla15 allele is a missense mutation in a conserved cysteine and the fla17 allele is an in-frame deletion of three exons. The flagellar assembly defect of each mutant is rescued by the respective transgenes. In fla15 and fla17 mutants, bulges form in the distal one-third of the flagella at the permissive temperature and this phenotype is also rescued by the transgenes. These bulges contain the complex B component IFT74/72, but not α-tubulin or p28, a component of an inner dynein arm, which suggests specificity with respect to the proteins that accumulate in these bulges. IFT144 and IFT139 are likely to interact with each other and other proteins on the basis of three distinct genetic tests: (1) Double mutants display synthetic flagellar assembly defects at the permissive temperature, (2) heterozygous diploid strains exhibit second-site noncomplemention, and (3) transgenes confer two-copy suppression. Since these tests show different levels of phenotypic sensitivity, we propose they illustrate different gradations of gene interaction between complex A proteins themselves and with a complex B protein (IFT172).
CILIA and flagella are microtubule-based organelles that are found on most mammalian cells. They provide motility to cells and participate in many sensory processes. Defects in or loss of cilia/flagella cause a variety of human diseases that include polycystic kidney disease, retinal degeneration, infertility, obesity, respiratory defects, left–right axis determination, and polydactyly (Fliegauf et al. 2007). Mouse mutants demonstrate that cilia are essential for viability, neural tube closure, and bone development (Eggenschwiler and Anderson 2007; Fliegauf et al. 2007). Cilia and flagella are also present in protists, algae, moss, and some fungi.
The assembly and maintenance of cilia and flagella require intraflagellar transport (IFT) (Kozminski et al. 1995). IFT involves the movement of 100- to 200-nm-long protein particles from the basal body located in the cell body to the tip of the flagella using the heterotrimeric kinesin-2 (anterograde movement) (Kozminski et al. 1995) and movement back to the cell body (retrograde movement) using the cytoplasmic dynein complex (Pazour et al. 1999; Porter et al. 1999). IFT particles change their direction of movement as well as their size, speed, and frequency at the ends of the flagella as they switch from anterograde to retrograde movement (Iomini et al. 2001). Biochemical isolation of IFT particles reveals that they are composed of at least 16 proteins and that these particles can be dissociated into two complexes in vitro by changing the salt concentration (Cole et al. 1998; Piperno et al. 1998). Recent genetic and bioinformatics analysis adds at least 7 more proteins to the IFT particle (Follit et al. 2009) (Table 1). Complex A contains at least 6 polypeptides and complex B contains at least 17 polypeptides. Analysis of mutations in Chlamydomonas, C. elegans, zebrafish, and mice demonstrates a requirement of IFT proteins and their motors for ciliary/flagellar assembly (Eggenschwiler and Anderson 2007).
TABLE 1.
Proteins and gene names for the intraflagellar transport particles in Chlamydomonas, C. elegans, and mouse
| Protein | Motif | Chlamydomonas gene | C. elegans gene | Mouse gene | References to worm and mouse genes |
|---|---|---|---|---|---|
| Complex A | |||||
| IFT144 | WD | FLA15 | |||
| IFT140 | WD | — | che-11 | Qin et al. (2001) | |
| IFT139 | TRP | FLA17 | dyf-2 | THM1 | Efimenko et al. (2006); Tran et al. (2008) |
| IFT122 | WD | — | IFTA-1 | Blacque et al. (2006) | |
| IFT121 | WD | — | daf-10 | Bell et al. (2006) | |
| IFT43 | — | ||||
| Complex B | |||||
| IFT172 | WD | FLA11 | osm-1 | Wimple | Huangfu et al. (2003); Pedersen et al. (2005); Bell et al. (2006) |
| IFT88 | TRP | IFT88 | osm-5 | Tg737/Polaris | Pazour et al. (2000); Qin et al. (2001) |
| IFT81 | Coil | — | ift-81 | CDV1 | Kobayashi et al. (2007) |
| IFT80 | WD | — | che-2 | Wdr56 | Fujiwara et al. (1999) |
| IFT74/72 | Coil | — | ift-74 | Cmg1 | Kobayashi et al. (2007) |
| IFT57/55 | Coil | — | che-13 | Hippi | Haycraft et al. (2003) |
| IFT54 | Microtubule binding domain MIP-T3 | — | dyf-11 | Traf3IP1 | Kunitomo and Iino (2008); Li et al. (2008); Omori et al. (2008); Follit et al. (2009) |
| IFT52 | ABC type | BLD1 | osm-6 | Ngd2 | Brazelton et al. (2001); Bell et al. (2006) |
| IFT46 | IFT46 | dyf-6 | Bell et al. (2006); Hou et al. (2007) | ||
| IFT27 | G protein | — | Not present | Rabl4 | |
| IFT25 | Hsp20 | — | Not present | HSP16.1 | Follit et al. (2009) |
| IFT22 | G protein | — | IFTA-2 | Rabl5 | Schafer et al. (2006) |
| IFT20 | Coil | — | Follit et al. (2006) | ||
| FAP22 | Cluamp related protein | — | dyf-3 | Cluamp1 | Murayama et al. (2005); Follit et al. (2009) |
| DYF13 |
— |
dyf-13 |
Ttc26 |
Blacque et al. (2005) |
|
—, no mutant found to date in Chlamydomonas.
A collection of temperature-sensitive mutant strains that fail to assemble flagella at the restrictive temperature of 32° was isolated in Chlamydomonas (Huang et al. 1977; Adams et al. 1982; Piperno et al. 1998; Iomini et al. 2001). Analysis of the flagella at 21° permits the measurement of the velocity and frequency of IFT particles in the mutant strains. This analysis suggested that assembly has four phases: recruitment to the basal body, anterograde movement (phases I and II), retrograde movement, and return to the cytoplasm (phases III and IV) (Iomini et al. 2001). Different mutants were classified as defective in these four phases. However, because different alleles of FLA8 were classified as defective in different phases (Iomini et al. 2001; Miller et al. 2005), we combined mutants with IFT defects into just two classes. The first group (phases I and II) includes mutant strains that show decreased anterograde velocities, a decreased ratio of anterograde to retrograde particles, and an accumulation of complex A proteins at the basal body. This group includes mutations in the FLA8 and FLA10 genes, which encode the two motor subunits of kinesin-2 (Walther et al. 1994; Miller et al. 2005), as well as mutations in three unknown genes (FLA18, FLA27, and FLA28). The second group includes mutant strains that show the reciprocal phenotype (phases III and IV); these phenotypes include decreased retrograde velocities, an increased ratio of anterograde to retrograde particles, and an accumulation of complex B proteins in the flagella. With the exception of the FLA11 gene, which encodes IFT172, a component of complex B (Pedersen et al. 2005), the gene products in this class are unknown (FLA2, FLA15, FLA16, FLA17, and FLA24). One might predict that mutations in this group would map to genes that encode complex A or retrograde motor subunits. Interestingly, IFT particles isolated from fla11, fla15, fla16, and fla17-1 flagella show depletion of complex A polypeptides (Piperno et al. 1998; Iomini et al. 2001). The inclusion of IFT172 in this class is explained by the observations that IFT172 plays a role in remodeling the IFT particles at the flagellar tip to transition from anterograde to retrograde movement (Pedersen et al. 2005). The remaining mutant strains do not show obvious defects in velocities, ratios, or accumulation at 21° and may reflect a less severe phenotype at the permissive temperature or a non-IFT role for these genes.
Direct interactions occur between components of complex B. IFT81 and IFT74/72 interact to form a scaffold required for IFT complex B assembly (Lucker et al. 2005). IFT57 and IFT20 also interact with each other and kinesin-2 (Baker et al. 2003). While physical interactions are being used to define IFT particle architecture, genetic interactions among loci encoding IFT components should be instructive regarding their function as well. To probe retrograde movement and its function, we have identified the gene products encoded by two retrograde defective mutant strains. They are FLA15 and FLA17 and encode IFT144 and IFT139, respectively. The genetic interactions of these loci provide interesting clues about the assembly of the IFT particles and possible physical interactions in the IFT particles.
MATERIALS AND METHODS
Strains and culture media:
Strains fla15-1, fla16-1, fla17-1, fla17-2, and fla24 (Piperno et al. 1998) and the fla2, fla8-2, fla10-14, and fla11 strains (Adams et al. 1982) were utilized. Each strain was backcrossed to a NIT2 ac17 strain to remove any unlinked modifiers and to introduce selectable markers for diploid strain construction. On the basis of results obtained in this study, fla16 is renamed fla17-3, in agreement with the current nomenclature for Chlamydomonas reinhardtii. Cells were grown as previously described (Holmes and Dutcher 1989). Crosses, maturation of zygotes, and tetrad analysis were carried out as previously described (Dutcher 1995).
Linkage analysis:
To determine linkage between complex A genes and fla15-1, fla16-1, and fla17-1, we designed PCR-based markers for the genes Chlre2_kg.scaffold_2000196 (FAP66), estExt_gwp_1H.C_30331 (FAP60), estExt_fgenesh2_pg.C_290033 (IFT140), estExt_fgenesh2_pg.C_70a0033 (FAP118), and Chlre2_kg.scaffold_ 3000443. Primers were chosen using Primer3 software for the six complex A genes and are shown in Table 2 (Rozen and Skaletsky 2000). DNA was isolated using a Genisol maxiprep kit (ABgene, Epsom, UK). DNA from progeny that pelleted at 32° from tetrads of crosses of fla15, fla16-1, and fla17-1 × CC-1952 was analyzed for segregation of the mutant allele with respect to polymorphic alleles. Similar crosses with fla2 and fla24 were performed.
TABLE 2.
Mapping of the complex A IFT subunits to linkage groups and fla mutants
| IFT gene | Linkage group | Parental:recombinant progeny for mapping to linkage group (map units) | Parental:recombinant progeny dCAP marker to fla mutant (map units) | IFT primers (5′–3′), enzyme |
|---|---|---|---|---|
| IFT43 | VI | Mating-type 36P:2R (2.6) | fla2 38P:0R | AGT GGT GTT TGC AGA ATG GTG TAT TGC CTG AGT GCC TGT CA DdeI |
| fla24 75P:2R (1.3) | ||||
| IFT121 | XI | cDNA19 34P:2R (5.0) | fla5 15P:4R (21) | GAA GGA CCA GGG ACT GGA CT TGG CTT TCC TTC CTC GAT TA AluI |
| IFT122 | I | AGK 25P:22R (46.5) | pf4 44P:3R (6.3) | TCT TCA TGC TGT TGT CCT C ATC CCC ATT CAA CCA AAC CT AluI |
| S18 49P:1R (2.0) | ||||
| IFT 139 | VI | Mating-type 235P:7R (1.4) | fla17-1 216P:0R | GGA GCT CAT GTT CCA CAA GG AAG TGG TAG ATG GCG GTG TC |
| BBS5 75P:9R (5.35) | fla17-2 62P:0R | |||
| fla16-1 43P:0R | ||||
| IFT140 | VIII | STK 31:10 (24) | None | CGT TTG TGG AGG AGG AGG T GGA CAC AGG GTT CAA ATG CT MspI |
| IFT144 | XIV | CCS1 44:2 (4.3) | fla15-1 243P:0R | GGG AGG TGC AGA CCT TGA TA GGA CAC AGG GTT TCA ATG CT AciI |
|
IGPS 36:7 (16.3) |
Mapping with fla5 and pf4 was based on the flagellar phenotypes of the mutants and not PCR-based markers.
Fifty-six progeny from a cross of a 137c-derived strain (NIT2 ac17) × CC-1952 were analyzed to map complex A genes to a linkage group (LG), using 44 markers previously described (Bowers et al. 2003) or new markers described in Table 2.
Sequencing:
DNA was isolated using a Genisol maxiprep kit (ABgene) or as described previously (Johnson and Dutcher 1991). Primers were designed to amplify IFT144 exons as predicted from the JGI web site (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). Sequences of the primers are available in supporting information, Table S1. PCR products from fla15-1 obtained using REDTaq and 1 m betaine (Sigma, St. Louis) at the annealing temperature of 50° were cloned into the pCR4-TOPO vector (TOPO TA cloning kit; Invitrogen, Carlsbad, CA). Plasmid DNA was sequenced by the Nucleic Acid Sequencing Laboratory (Washington University, St. Louis) or in the Department of Genetics using M13 primers. DNA sequences were aligned to the predicted IFT144 gene using SeqMan II (DNASTAR, Madison, WI). A nucleotide substitution T to C was detected in the 823-bp PCR fragment amplified by primers IFT144-14 (Table S1). The nucleotide substitution was confirmed by sequencing an independent PCR product from wild-type and fla15 genomic DNA. A derived cleaved amplified polymorphic sequence (dCAPS) marker that distinguishes the wild-type and mutant alleles in the IFT144-14 PCR was used. MspAI cuts the 137c PCR product into three fragments (380, 299, and 144 bp), while the fla15 fragment is cut into two fragments (443 and 380 bp). This polymorphism segregated with the flagellar phenotype in 59 progeny. This polymorphism was also used for the reversion analysis. True revertants or pseudorevertants of T to G produce three fragments while suppressors or other pseudorevertants will generate only two bands like the mutant strain.
Because of the high level of GC content, genomic DNA amplification and sequencing were difficult for IFT139 in several regions. Consequently, five overlapping primers (available in Table S1) were designed from the predicted cDNA sequence and RT–PCR was performed. RNA was extracted as described (Witman et al. 1972) and single-strand cDNA synthesized using SuperscriptII (Invitrogen) (Li et al. 2004). The RT–PCR fragments were sequenced as described above. IFT139-42 primers (Table S1) showed a size difference between wild type and fla16-1, fla17-1, and fla17-2 DNA.
Chlamydomonas transformation:
Six micrograms of BAC DNA from I7I9 and 4M13 clones (Dutcher et al. 2002) was introduced into fla17-1 cells, using electroporation (Shimogawara et al. 1998) as modified by Kovar and co-workers (Kovar et al. 2002). DNA from BAC 4M13 was cut with XhoI at 37° for 5 hr prior to transformation. BAC DNA from 12K11 (7 μg) was cut with AatI and StuI and introduced into fla15-1 cells.
Analysis of swimming transformants:
Each rescued transformant (n = 2 for fla17-1 and n = 4 for fla15-1) was crossed to CC-124 to determine if the rescue was extragenic. Thirty tetrads were analyzed for the appearance of the flagellar assembly defect and the presence of the mutant and wild-type allele, using the dCAPS markers in Table S1. All transformants were extragenic. Meiotic progeny from nonparental ditype tetrads that were swimming were retained for further experiments.
SHIRT analysis:
The segregation of heterozygosity in rescued transformants (SHIRT) analysis provides a rapid method to determine which regions of BAC DNA are integrated nonhomologously into the genome and cosegregate with the rescued phenotype (Esparza 2008). It obviates the requirement for subcloning to determine the causative DNA for rescued transformants. It is rare in Chlamydomonas that the entire transforming DNA of a BAC or plasmid integrates into the genome, and independent transformants generally integrate nonidentical segments of the BAC or plasmid DNA (Dutcher and Trabuco 1998; Myster et al. 1999). For this analysis, multiple independent transformants are mated to the polymorphic strain, CC-1952 and meiotic tetrad progeny are recovered. In general, nonparental ditype (NPD) tetrads are identified that have two mutant progeny and two wild-type progeny. A dCAPS marker is made for each BAC gene to be tested in the transformant (Figure 1). Markers predicted by Rymarquis et al. (2005) are tested. If no predictions are available, 3′-UTR sequences are amplified and tested with 3–10 restriction enzymes (Bowers et al. 2003). In these progeny, there are three alleles of each gene. One copy is from the CC-1952 parent, one from the inserted BAC, and one from the mutant parent. Since the BAC library was made from the same strain as the mutant parent, the alleles contributed by the BAC and the mutant parent possess the same polymorphism while the allele contributed by the CC-1952 parent is different. PCR is performed for each marker of the progeny that shows an NPD segregation pattern for the flagellar phenotype and the rescue. The two aflagellate progeny should have only a copy of the parental allele and the two swimming progeny have the BAC and CC-1952 alleles (Figure 1C, see diagram for gene 2), if the gene is responsible for rescue. If the gene from the BAC is not integrated into the genome, then the PCR products will resemble the gene 3 example (Figure 1C). If the gene from the BAC does not rescue, the pattern may resemble gene 1 and not cosegregate (Figure 1C). DNA was isolated from four NPD tetrads for each transformant and PCR was used for various dCAPS markers to determine if the IFT144 or IFT139 genes were present. The primers are available in Table S1.
Figure 1.—
Schematic diagram of segregation of heterozygosity in rescued transformants (SHIRT). SHIRT uses mapping and PCR-based markers to determine which parts of a BAC are responsible (cosegregate) for rescue of a mutant phenotype following transformation. (A) Diagram of three genes present on a BAC that come from the mutant parent (black), the polymorphic mapping strain CC-1952 (red), and the BAC (blue). Primers for PCR are generally made to the 3′-UTR of genes to be tested (arrows). (B) Bands of digested PCR products (dCAP markers) for the three genes from the three sources of DNA. The digested PCR products from the black and blue alleles cannot be distinguished from each other. (C) Three possible outcomes among nonparental ditype (NPD) tetrads in which the mutant phenotype is observed in two of the four progeny (fla and +). The pattern with gene 1 suggests that the BAC DNA is not responsible for rescue. The hybrid (black and blue) band indicates that the band is amplified from both the mutant and the BAC allele. The pattern with gene 2 is consistent with the BAC DNA providing rescue. Further proof requires additional NPD tetrads with this pattern. The pattern with gene 3 suggests that it is not integrated into the genome and therefore not responsible for rescue.
Reversion of fla15-1 and fla17-2:
Cultures of fla15-1 and fla17-2 cells were plated onto Sager and Granick medium at a density of ∼2000 colonies per plate and grown for 4 days at 21°. They were subjected to ultraviolet irradiation (70,000 J) and left in the dark to prevent photoreactivation for 18 hr. Approximately 40 colonies were transferred to 20 ml of Sager and Granick medium in 25 × 150-mm tubes. The 72 tubes were placed at 32° for 3 days. Five milliliters of supernatant was transferred to new tubes at 3 days and this was repeated four times. Swimming cells from the supernatant were plated to isolate individual colonies and one colony from each of the tubes was retained and scored for flagellar assembly. For fla15-1, 56 of the 72 cultures yielded cells that assembled flagella and swam at 32°. DNA was made from each of these 52 strains to examine by PCR with the IFT144-14 primers (GCA TGC ATT CTT CGA TGA CC and GGA GTG TAC TCG TG CCC TA) and digested with MspAI as described for the mutant allele. For fla17-2, 24 of the 72 cultures yielded swimming cells. By tetrad analysis, all were extragenic suppressors, but had no phenotype beyond suppression.
Flagellar isolation and immunoblots:
Cells were prepared as described previously for immunofluorescence (Iomini et al. 2001). The antibodies against IFT74/72, which was previously designated at p71, are described in Iomini et al. (2001, 2004), p28 is described in LeDizet and Piperno (1986), and antibody raised against complex A is described in Iomini et al. (2001).
Immunoblots are as described previously (LeDizet and Piperno 1986; Iomini et al. 2001). Isolated flagella were treated with nonionic detergent to extract them (Piperno et al. 1998).
Diploid construction:
Diploid strains were obtained by screening on minimal medium lacking acetate with 2 mm KNO2 instead of ammonium in the medium. Eight independent diploid strains were picked and heterozygosity of the mating-type locus was assessed by PCR to MID1 and to FUS1 (Miller et al. 2005). The parent strains were ac17 NIT2 and AC17 nit2.
RESULTS
Mapping the retrograde mutant strains:
Each of the temperature-sensitive mutations in Chlamydomonas with decreased retrograde velocities and reduced numbers of retrograde particles was crossed to the polymorphic strain, CC-1952 (Gross et al. 1988) to map the mutant phenotypes with respect to a marker for each of the complex A subunits (IFT144, IFT140, IFT139, IFT122, IFT121, and IFT43), which were developed using sequences kindly provided by D. Cole and H. Qin (personal communication) (Table 2). In addition, if the gene was placed on a chromosome in the JGI assemblies (Merchant et al. 2007), the position was verified by mapping to known markers. If the chromosome was not known, it was determined by crossing to markers on each chromosome (Table 2).
IFT43 is tightly linked to the FLA2 locus, but no changes in the sequence of the gene were found between the mutant and the parental strain (data not shown). IFT122 and IFT121, the homolog of Caenorhabditis elegans daf-10, map to linkage group I near the PF4 locus and to linkage group XI near the FLA5 locus, respectively. They are both separable from these loci (Table 2). IFT140 maps to linkage group VIII, but no flagellar mutants map to this region. Complete linkage was observed between fla15-1 and IFT144 (n = 243) and between fla16 (n = 43), fla17-1 (n = 216), and fla17-2 (n = 62) with IFT139. The respective genes were sequenced from fla15-1 and fla17-1 and compared to sequences in the JGI database (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html).
Temperature-sensitive mutation in IFT144:
The fla15-1 allele has a T to C change that changes a C to an R at amino acid 1283 (EF592033). This cysteine is conserved in IFT144 homologs that range from Giardia to Drosophila to humans (Figure 2B). To further ascertain that this change is responsible for the temperature-sensitive phenotype of the fla15-1 allele, we screened for revertants of the temperature-sensitive flagellar assembly defect after mutagenesis of cells with ultraviolet light (see materials and methods). Fifty-six independent strains that assemble functional flagella at 32° were isolated. Tetrad analysis of 8 strains indicated that the events were likely to be intragenic as they were tightly linked to fla15-1; the flagellar assembly defect was not recovered in at least 35 tetrads for each of the eight strains. A dCAPS marker was developed that allowed detection of the mutant vs. the wild-type or a pseudorevertant allele in PCR products. Forty-six of the revertants were examined by PCR and the C was changed to either a T or a G (see materials and methods). Eight of these strains were sequenced and all were true revertants of C to T rather than to the pseudorevertant G. These reversion studies support the identification of the FLA15 gene.
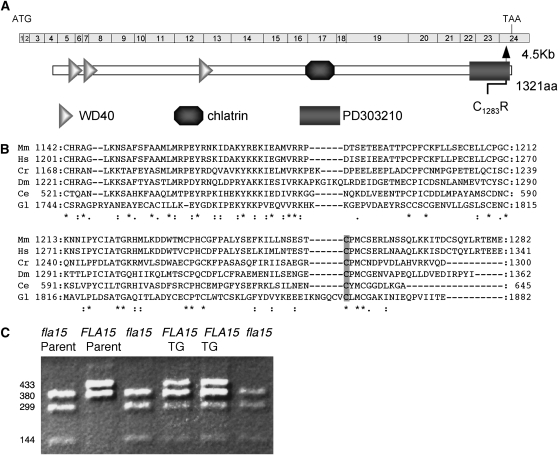
Figure 2.—
Identification of IFT144 as the gene product of the FLA15 locus. (A) Diagram of the structure of the gene. IFT144 is a WD repeat protein (Cole 2003). It also has a clathrin domain, which has been implicated in vesicle transport in the testis (Oyhenart et al. 2003), as well as an uncharacterized domain (PD303210). (B) The region of IFT144 from mouse (Mm), human (Hs), Chlamydomonas (Cr), Drosophila (Dm), C. elegans (Ce), and Giardia (Gl) that contains the cysteine that is mutated in fla15-1. This cysteine is invariant (shaded). (C) SHIRT analysis of the transgene. PCR products from a nonparental ditype are shown. The parents (fla15 + rescue; CC-1952) are in lanes 1 and 2. The four progeny are in lanes 3–6. Lanes 3 and 6 are from cells aflagellate at 32° and the band is consistent with the mutant alleles. Lanes 4 and 5 are from flagellated cells and the bands are consistent with the wild-type FLA15 allele from the CC-1952 parent and the BAC contributed transgene.
The fla15-1 strain was transformed with an ∼80-kb BAC (12K11) that contains the IFT144 gene and four independent transformants were obtained. Each contained the wild-type and mutant IFT144 gene as monitored by PCR and the dCAPS marker (data not shown) and the suppression segregated independently from the fla15-1 mutation. The IFT144 transgene segregated in eight tetrads with the rescue of the flagellar assembly defect, using the SHIRT analysis method (Figure 2C; see materials and methods). Although at least two additional predicted genes are present on the rescuing BAC (data not shown), these multiple lines of evidence strongly suggest that IFT144 is the correct gene.
Temperature-sensitive mutation in IFT139:
The IFT139 gene was sequenced from the fla17-1 allele and contains a deletion that precisely removes three predicted exons (Figure 3A, highlighted exons). The fla17-1 allele and the fla16-1 allele contain the same deletion by PCR analysis (Figure 3B) as well as by sequencing of the fla17-2 allele. Given that FLA16 and FLA17 were unlinked in a previous analysis (Iomini et al. 2001), it is likely that a stock-keeping error occurred at some time in the past before the strains were deposited at the Chlamydomonas Genetics Center and the original fla16 allele is lost. We refer to this locus as FLA17 and the extant fla16-1 allele is referred to as fla17-3. The sequence in the three deleted exons is highlighted in bold in Figure 3C, and it deletes a TPR repeat.
Figure 3.—
Identification of IFT139 as the gene product of the FLA17 locus. (A) Diagram of the structure of the gene. IFT139 contain multiple TRP domains that are thought to mediate short-term protein interactions and two half A TPR (HAT) domains. Shaded exons represent the three exons deleted in the fla17 alleles. (B) PCR amplification of IFT139 with primers in exons 16 and 20 (5′-ATC CGC GAG ACG CCT CTG TAC and CTG TGC GCC GCC GCG GGC GT). The wild-type product is ∼700 bp while the mutant product of fla17-1 and fla17-3 (also known as fla16-1) is ∼325 bp. (C) Alignment of protein around and including the deletion in human, mouse, Chlamydomonas, and C. elegans. The deleted region is shown in boldface type. (D) Exon 10 in the IF139 gene shows alternative splicing. In black are the shared introns before and after exon 10. Exonic sequence that is present in both splicing variants is shown in red. In green is the exon that is present in only one splice variant. The four splice donor and acceptor sites are underlined and in uppercase. The 58 amino acids encoded by the longer splice variant are shown in blue and the 28 amino acids encoded by the short splice variant are shown in purple.
The exon structure presented in Figure 3A does not agree with the GenBank sequence from Cole and co-workers (EF592032) for exon 10 (Figure 3C). We sequenced four independent RT–PCR reactions from wild-type cells. One of them agreed with the GenBank entry (EF592032) to produce an exon that encodes 58 amino acids (Figure 3C, blue protein sequence) and three of them used internal splice sites to produce an exon with only 28 amino acids (Figure 3C, purple protein sequence). Thus, the IFT139 gene shows alternative splicing in this exon.
The fla17-1 strain was transformed with the BACs 17I9 and 4M13, which contain the IFT139 gene, and two independent transformants were obtained. Each one contained the IFT139 gene and the transgene segregated independently from the fla17-1 mutation in 35 tetrads. By SHIRT analysis, in one of the transformants the only gene integrated into the genome from the BAC is the IFT139 gene (data not shown). Intragenic revertants of the fla17-1 allele were sought using the same methodology described for the fla15-1 allele, and only unlinked suppressors were identified (n = 24) as might be expected for a deletion allele.
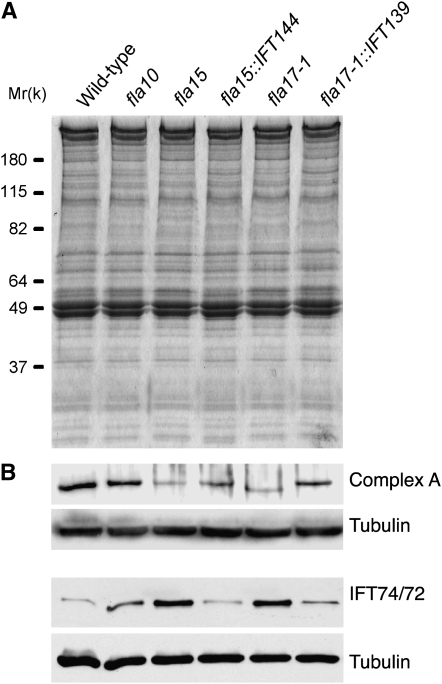
As further evidence that FLA17 encodes IFT139, we examined flagellar extracts from wild-type, mutant, and transgenic strains at 21° by immunoblot, using an antibody to IFT139 (Piperno et al. 1998). A deletion of exons 17, 18, and 19 from FLA17 predicts an ∼13-kDa decrease in the molecular weight of IFT139. In agreement with previously published electrophoretograms of 17S complexes (Piperno et al. 1998), we found that the polypeptide recognized by the anti-IFT139 antibody in fla17-1 flagella was smaller than the polypeptides detected by the same antibody in flagella from wild-type or other strains. In the fla17-1∷IFT139 transgenic strain, only the wild-type polypeptide is detected (Figure 4A), which suggests that the wild-type band is incorporated more effectively. Immunoblots also show a reduction of IFT139 in flagellar extracts from fla17 and fla15 (Figure 4A) (Piperno et al. 1998). The smaller polypeptide is also detected in isolated cell bodies (data not shown). This evidence supports the molecular data that FLA17 encodes IFT139.
Figure 4.—
Immunoblots for IFT components with flagellar extracts of wild-type and mutant strains grown at 21°. (A) Coomassie blue-stained gels of wild type, fla15, fla17, and rescued strains fla15-1∷IFT144 and fla17-1∷IFT139. (B) Immunoblots of the region around 145–130 kDa reacted with antibodies to complex A, IFT74/72, and α-tubulin. The complex A antiserum has been shown previously to recognize the IFT139 polypeptide (Iomini et al. 2001) and IFT74/72 is described in Iomini et al. (2004).
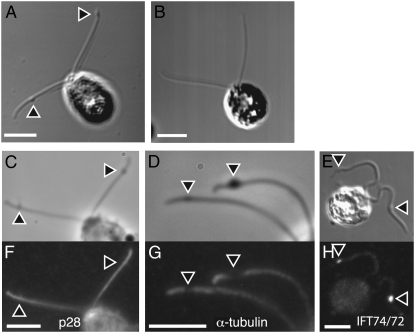
Accumulation of flagellar bulges:
In fla15 and fla17 mutants, bulges form in the distal one-third of the flagella (Piperno et al. 1998). The flagella of a fla15 strain have bulges (arrowhead in Figure 5A) and this phenotype is rescued by the transgene (Figure 5B). By immunofluorescence, these bulges contain the complex B component IFT74/72 (Figure 5, E and H) (Iomini et al. 2001, 2004), but not α-tubulin (Figure 5, D and G) or p28, a component of the inner arm (Figure 5, C and F).
Figure 5.—
Retrograde mutant bulges contain some but not all flagellar proteins. (A and B) Phase contrast of bulges (arrowheads) on fla15-1 cells (A) and their absence in the rescued fla15-1∷IFT144 strain (B). In the mutant strain, >80% of the cells have bulges (n = 200) and no bulges were observed in the rescued strains. (C–E) Phase images of cells with bulges. (F–H) Immunofluorescence of cells with bulges. (F) p28 antibody. (G) IFT74/72 antibody. (H) α-Tubulin antibody. IFT74/72 is present in the bulges, but α-tubulin and p28 are not.
Synthetic interactions of fla15 and fla17:
Synthetic phenotypes have been used to build networks of interactions in many cellular processes (Ooi et al. 2006). Many double-mutant combinations of hypomorphic alleles for genes that act in the same pathway show synthetic phenotypes, but mutants in two independent pathways can also underlie synthetic phenotypes. We have investigated if temperature-sensitive mutations in flagellar assembly show synthetic phenotypes in double-mutant combinations. Haploid strains with any combination of fla11, fla15, and fla17 alleles lack flagella at 21° and 32°, but are not aflagellate in combination with mutant alleles in the subunits of the heterotrimeric kinesin-2 (fla10-14 and fla8-2) at 21°. Double mutants of fla2 and fla24 with fla15 or fla17 were also aflagellate at both temperatures (Table 3). Since there are no known temperature-sensitive complex B mutations other than IFT172, we could not examine this class of double-mutant combinations. These genetic interactions support the biochemical phenotypes that the complex A genes act together for assembly and with IFT172 in complex B.
TABLE 3.
Synthetic phenotypes of double mutants of anterograde and retrograde defective mutants at 21°
| Locus | Defect | Protein | fla15-1 | fla17-1 |
|---|---|---|---|---|
| fla2-1 | R | Unknown | − | − |
| fla8-2 | A | Kinesin-2 motor subunit | + | + |
| fla10-14 | A | Kinesin-2 motor subunit | + | + |
| fla11-1 | R | IFT172 | − | − |
| fla17-3a | R | IFT139 | − | NA |
|
fla24-1 |
R |
Unknown |
− |
− |
R, retrograde defect based on previous results (Iomini et al. 2001); A, anterograde defect based on previous results (Iomini et al. 2001); −, aflagellate cells (>97% of cells have no flagella); +, flagellate cells (>80% of cells have flagella); NA, not applicable.
Formerly fla16-1.
We also examined the ability of these mutations to complement one another in doubly heterozygous diploid strains. The phenomenon of second-site noncomplementation, also known as dominant enhancers (Hawley and Gilliland 2006), is defined as the lack of complementation between two recessive mutations in two different loci. It can arise via a number of mechanisms, which include the creation of a poisonous interaction complex or the reduction in the dose of a complex formed by the two mutant gene products. Diploid strains heterozygous for fla2, fla11-1, fla15-1, fla17-1, or fla24-1 alleles with the wild-type allele are able to swim at 21° and 32°; all of these mutations are recessive to the wild-type allele. However, diploid strains that are heterozygous for all pairwise combinations of the fla11-1, fla15-1, and fla17-1 mutations are unable to assemble flagella at 21° or 32° (Table 4). Surprisingly, the double heterozygous diploids show a more extreme phenotype than the homozygous diploid strains, which suggests that the heterozygous diploid has multiple defects in the pathway. For example, homozygous fla17-1 diploid strains have functional flagella at 21° and lack flagella only at 32°, while the double heterozygous diploid strain (FLA15/fla15-1; FLA17/fla17-1, for example) fails to assemble flagella at either temperature. Double heterozygous diploids with mutations in the heterotrimeric kinesin-2 subunits assemble flagella at both temperatures, which is consistent with the haploid strains. The double heterozygous strain with the fla24 mutation and either fla15 or fla17 becomes aflagellate at 32° but remains flagellated with the fla11 mutant (Table 4). Although mutations in FLA2 show retrograde defects, it fails to show this type of genetic interaction. This mutant allele may be less severe or the loss has fewer effects on complex A particles.
TABLE 4.
Phenotype of double-heterozygous strains at 21° and 32°
 |
||||
|---|---|---|---|---|
| Dominance test: FLA+ | fla11-1 | fla15-1 | fla17-1 | |
| fla2 | + | + | + | + |
| fla8-2 | + | + | + | + |
| fla10-14 | + | + | + | + |
| fla11-1 | + | +21° | − | − |
| −32° | ||||
| fla17-1 | + | − | − | +21° |
| −32° | ||||
| fla24-1 | + | + | +21° | +21° |
| −32° |
−32° |
|||
−, aflagellate cells (>97% of cells have no flagella); +, flagellate cells (>90% of the cells have flagella, n = 200).
Two-copy suppression:
The presence of multiple copies of a gene or overexpression of a gene can suppress the phenotype of mutations in a noncognate gene in Saccharomyces cerevisiae and Schizosaccharomyces pombe. This observation has been termed multicopy suppression and has been used extensively to find genes that act in the same pathway (Forsburg 2001). Dosage suppressors may occur because the protein at the increased dosage helps to stabilize the mutant protein or the increased dosage protein may act downstream to bypass the requirement for the mutant protein. We find that two copies of the wild-type IFT144 gene (FLA15) can rescue the fla17-1 mutant phenotype at 32° (Table 5). Meiotic crosses and allele-specific PCR verified the genotypes; the presence of two copies and rescue were observed in six independent progeny (Figure 2D). Two copies of the IFT144 gene partially rescue the phenotype of fla11-1 cells (Table 5), and the genotypes were verified for three independent meiotic progeny. Two copies of the IFT144 gene do not rescue the mutant phenotypes of the fla2, fla8, fla10, or fla24 strains (Table 5). We also find that two copies of the wild-type IFT139 gene (FLA17) rescue the fla15-1 mutant phenotype. The presence of two copies as well as the mutant fla15-1 allele was verified in eight independent progeny. A single wild-type copy of IFT139 or IFT144 rescues the aflagellate phenotype of the fla15; fla17 double mutant at 21°, but not at 32° (Table 5). This demonstrates that the rescue requires two wild-type copies of the noncognate gene. Two copies of the IFT139 gene do not rescue the fla2, fla8, fla10, fla11, or fla24 mutant phenotypes (Table 5).
TABLE 5.
Two-copy suppression: flagellar number at 32°
| FLA15 transgene | FLA17 transgene | No transgene | |
|---|---|---|---|
| fla2 | 100:0:0 | 100:0:0 | 100:0:0 |
| fla8-1 | 100:0:0 | 100:0:0 | 100:0:0 |
| fla10-14 | 100:0:0 | 100:0:0 | 100:0:0 |
| fla11-1 | 38:1:61 | 92:0:8 | 89:0:11 |
| fla15-1 | 7:1:92 | 9:2:89 | 100:0:0 |
| fla17-1 | 5:1:94 | 6:0:94 | 100:0:0 |
| fla24 | 100:0:0 | 100:0:0 | 100:0:0 |
| fla15, fla17, 21° | 11:2:87 | 8:3:89 | 100:0:0 |
|
fla15, fla17, 32° |
100:0:0 |
100:0:0 |
100:0:0 |
Numbers represent percentage of cells with zero, one, or two flagella (n = 250 cells) in logarithmically growing culture (∼1 × 106/ml) shifted to 32° for 8 hr.
DISCUSSION
IFT is required to assemble cilia and flagella. Mutations in complex B genes have been identified in a variety of organisms (Table 1). Nonconditional mutations in Chlamydomonas IFT172, IFT88, and IFT52 result in aflagellate cells, while mutations in IFT46 result in cells with short, paralyzed flagella. A reduction in the level of IFT27 protein by RNA interference also causes aflagellate cells as well as defects in cytokinesis (Qin et al. 2007). These mutants demonstrate that anterograde IFT is essential for ciliary/flagellar assembly. Mutations in the genes that encode complex A proteins in Chlamydomonas have not been previously identified (Table 1). However, mutations in the motor subunit DYNC2H1 of cytoplasmic dynein 2 complex (previously called cytoplasmic dynein 1b) (Pfister et al. 2005) in Chlamydomonas lead to cessation of the retrograde IFT movement and result in cells with short (1–2 μm) flagella stumps that contain even shorter axonemal microtubules (Pazour et al. 1999; Porter et al. 1999). The dynein light chain LC8 is encoded by the FLA14 locus and the fla14 mutant strain assembles short flagella filled with unassembled flagellar precursors (Pazour et al. 1998). A dynein light intermediate chain, DYNC2LI1, has been isolated and is also required for retrograde transport (Hou et al. 2004). We report that IFT144 and IFT139 are encoded by FLA15 and FLA17, respectively. Unlike the motor mutants, the complex A mutants are aflagellated, which suggests a slightly different requirement for the retrograde motors and complex A.
Complex B, which has been studied extensively by biochemical techniques, has a set of core proteins (IFT88, -81, -74/72, -52, -46, and -27) that do not require the other complex B components (IFT172, -80, -57, and -20) to remain associated (Lucker et al. 2005). In fact, fla11 (IFT172) mutant strains have phenotypes that more closely resemble the retrograde mutants (Pedersen et al. 2005). However, these relationships are not maintained through evolution in other organisms. IFT88 in C. elegans is encoded by the osm-5 gene and acts as if it were a peripheral component rather than a core component of the B complex (Haycraft et al. 2003). In Physcomitrella patens, a moss with motile sperm, several of the core complex B IFT genes in Chlamydomonas (IFT81, -72/74, and -20) cannot be found in the genome by bioinformatics tools (Rensing et al. 2008).
Our genetic interaction studies suggest that IFT144 and IFT139 interact intimately and IFT144 may interact with IFT172 (Figure 6). All pairwise combinations of these three mutations show a synthetic aflagellate phenotype and also have an aflagellate phenotype in doubly heterozygous diploid strains. The FLA15 transgene (IFT144) suppresses the phenotypes of fla15 and fla17 strains and partially suppresses the phenotype of the fla11 strain. In contrast, the FLA17 transgene (IFT139) suppresses the phenotypes of the fla15 and fla17 strains, but not the phenotype of the fla11 strain. Because there are no null alleles for these genes, we cannot ask if these interactions occur by a bypass mechanism. However, it seems likely that the suppression could result from stabilization of the mutant gene product in the presence of an excess of the noncognate transgene product together with the endogenous copy of the gene. This hypothesis is supported by the observations that the double mutant (fla15; fla17) is not rescued by either of the transgenes when there is only one copy of the wild-type gene. We suggest that IFT144 may interact with both IFT139 and IFT172 while IFT139 is less likely to interact with IFT172 (Figure 6). This model might also suggest that there is more than one copy of IFT144 per IFT particle. Biochemical experiments will be needed in the future to address these hypotheses.
Figure 6.—
Diagram of possible interactions in the IFT particle. (A) IFT complexes A and B with kinesin-2 (in green) and cytoplasmic dynein (in red). (B) Synthetic phenotypes suggest interactions between all of the mutants with retrograde defects. (C) Noncomplementation phenotypes show more limited interactions. The fla2 mutant no longer interacts. (D) Two-copy suppression shows interactions of IFT139 and IFT144 while IFT172 shows a weak interaction with IFT144.
The analysis of double mutants suggests that this genetic test is the most permissive for finding mutant phenotypes (Figure 6). Synthetic phenotypes are observed in all combinations of mutants with retrograde defects to produce an aflagellate phenotype at 21°, but not with the anterograde motor mutants. This analysis suggests that FLA24 and FLA2 may identify genes not identified by biochemical isolation of IFT particles. Second-site noncomplementation is observed in all combinations of fla11, fla15, and fla17 alleles at 21° and 32°. Aflagellate phenotypes are observed in heterozygous diploid strains for fla24 with either fla15 or fla17, but not with fla11. No phenotype is observed in diploid strains with the fla2 allele or with the anterograde motor mutants. The lack of phenotype in heterozygotes with fla24 and fla11 suggests that there is a more direct interaction of FLA24 with complex A components (IFT144 and IFT130) than with IFT172 from complex B.
FLA15 is a homolog of dyf-2 in C. elegans (Efimenko et al. 2006). Efimenko and co-workers suggested that this WD (tryptophan aspartic acid) repeat protein, which is an ortholog of the human WDR19 protein (Lin et al. 2003), associates with complex B proteins, but interferes with function of complex A proteins. WDR19 has been associated with prostrate cancer as well (Lin et al. 2008) and localizes to cilia (Efimenko et al. 2006). The Chlamydomonas biochemical and phenotypic data reported previously (Piperno et al. 1998; Iomini et al. 2001) show that FLA15 is a component of the A complex. Our genetic data suggest that IFT144 interacts with IFT172, which may mediate between the two complexes.
The Sonic hedgehog (Shh) pathway is uniquely impaired in mice carrying the aln null mutation in the Ttc21b gene that encodes THM1, the mouse ortholog of IFT139 (Tran et al. 2008). Mutant aln embryos show an inappropriate activation of the Shh pathway, which contrasts with anterograde ciliary mutant mice that show a disruption in the activation of the Shh pathway (Eggenschwiler and Anderson 2007). This set of phenotypes suggests that retrograde IFT may modulate Shh signaling. Ciliary signaling is needed for mating in Chlamydomonas (Lux and Dutcher 1991; Piperno et al. 1996; Pan and Snell 2002; Wang et al. 2006). We attempted to ask if retrograde IFT is needed for mating, but both fla15 and fla17 cells retain some retrograde IFT movement after 4 hr at the restrictive temperature. We were unable to find a time in which IFT stopped and the cells retained flagella (data not shown). The role of retrograde IFT may be to recycle components or to help in retention of various proteins. Screens for new mutants that give synthetic phenotypes in a fla15 or fla17 haploid strain or fail to complement in heterozygous diploid strains may be useful to ask about the role IFT in the signaling for mating and to find new regulators and possibly novel pathways.
Acknowledgments
This work was made possible by the kindness of Doug Cole (University of Idaho). He provided the amino acid sequences for the complex A IFT proteins. We thank Kat Lyle, Camille Starback, and Brett Havranek for help with various experiments; Laura Kyro for help with Figure 6; and Alison Albee for critical reading of the manuscript. This work was supported by a grant to S.K.D. from the National Institutes of Health (NIH) (GM-03842). J.M.E. was supported by a fellowship from the Ford Foundation and an NIH supplement. Camille Starback was supported by the Washington University Summer Scholars Program.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101915/DC1.
References
- Adams, G. M., B. Huang and D. J. Luck, 1982. Temperature-sensitive, assembly-defective flagella mutants of Chlamydomonas reinhardtii. Genetics 100 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S. A., K. Freeman, K. Luby-Phelps, G. J. Pazour and J. C. Besharse, 2003. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J. Biol. Chem. 278 34211–34218. [DOI] [PubMed] [Google Scholar]
- Bell, L. R., S. Stone, J. Yochem, J. E. Shaw and R. K. Herman, 2006. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics 106 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque, O. E., E. A. Perens, K. A. Boroevich, P. N. Inglis, C. Li et al., 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15 935–941. [DOI] [PubMed] [Google Scholar]
- Blacque, O. E., C. Li, P. N. Inglis, M. A. Esmail, G. Ou et al., 2006. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell 17 5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, A. K., J. A. Keller and S. K. Dutcher, 2003. Molecular markers for rapidly identifying candidate genes in Chlamydomonas reinhardtii. ery1 and ery2 encode chloroplast ribosomal proteins. Genetics 164 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton, W. J., C. D. Amundsen, C. D. Silflow and P. A. Lefebvre, 2001. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11 1591–1594. [DOI] [PubMed] [Google Scholar]
- Cole, D. G., D. R. Diener, A. L. Himelblau, P. L. Beech, J. C. Fuster et al., 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D. G., 2003. The intraflagellar transport machinery in Chlamydomonas reinhardtii. Traffic 4 435–442. [DOI] [PubMed] [Google Scholar]
- Dutcher, S. K., 1995. Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 47 531–540. [DOI] [PubMed] [Google Scholar]
- Dutcher, S. K., and E. C. Trabuco, 1998. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9 1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S. K., N. S. Morrissette, A. M. Preble, C. Rackley and J. Stanga, 2002. Epsilon-tubulin is an essential component of the centriole. Mol. Biol. Cell 13 3859–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko, E., O. E. Blacque, G. Ou, C. J. Haycraft, B. K. Yoder et al., 2006. Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol. Biol. Cell 17 4801–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler, J. T., and K. V. Anderson, 2007. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23 345–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza, J. M., 2008. Epsilon tubulin mutants affect basal body integrity, disrupt katanin localization, and increase microtubule stability. Ph.D. Thesis, Washington University, St. Louis.
- Fliegauf, M., T. Benzing and H. Omran, 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell. Biol. 8 880–893. [DOI] [PubMed] [Google Scholar]
- Follit, J. A., R. A. Tuft, K. E. Fogarty and G. J. Pazour, 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell 17 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit, J. A., F. Xu, B. T. Keady and G. J. Pazour, 2009. Characterization of mouse IFT complex B. Cell Motil. Cytoskeleton 66 447–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S., 2001. The art and design of genetic screens: yeast. Nat. Rev. Genet. 2 659–668. [DOI] [PubMed] [Google Scholar]
- Fujiwara, M., T. Ishihara and I. Katsura, 1999. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126 4839–4848. [DOI] [PubMed] [Google Scholar]
- Gross, C. H., L. P. Ranum and P. A. Lefebvre, 1988. Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13 503–508. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., and W. D. Gilliland, 2006. Sometimes the result is not the answer: the truths and the lies that come from using the complementation test. Genetics 174 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft, C. J., J. C. Schafer, Q. Zhang, P. D. Taulman and B. K. Yoder, 2003. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 284 251–263. [DOI] [PubMed] [Google Scholar]
- Holmes, J. A., and S. K. Dutcher, 1989. Cellular asymmetry in Chlamydomonas reinhardtii. J. Cell Sci. 94 273–285. [DOI] [PubMed] [Google Scholar]
- Hou, Y., G. J. Pazour and G. B. Witman, 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell 15 4382–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., H. Qin, J. A. Follit, G. J. Pazour, J. L. Rosenbaum et al., 2007. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., M. R. Rifkin and D. J. Luck, 1977. Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. J. Cell Biol. 72 67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., A. Liu, A. S. Rakeman, N. S. Murcia, L. Niswander et al., 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426 83–87. [DOI] [PubMed] [Google Scholar]
- Iomini, C., V. Babaev-Khaimov, M. Sassaroli and G. Piperno, 2001. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini, C., K. Tejada, W. Mo, H. Vaananen and G. Piperno, 2004. Primary cilia of human endothelial cells disassemble under laminar shear stress. J. Cell Biol. 164 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. E., and S. K. Dutcher, 1991. Molecular studies of linkage group XIX of Chlamydomonas reinhardtii: evidence against a basal body location. J. Cell Biol. 113 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., K. Gengyo-Ando, T. Ishihara, I. Katsura and S. Mitani, 2007. IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells 12 593–602. [DOI] [PubMed] [Google Scholar]
- Kovar, J. L., J. Zhang, R. P. Funke and D. P. Weeks, 2002. Molecular analysis of the acetolactate synthase gene of Chlamydomonas reinhardtii and development of a genetically engineered gene as a dominant selectable marker for genetic transformation. Plant J. 29 109–117. [DOI] [PubMed] [Google Scholar]
- Kozminski, K. G., P. L. Beech and J. L. Rosenbaum, 1995. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo, H., and Y. Iino, 2008. Caenorhabditis elegans DYF-11, an orthologue of mammalian Traf3ip1/MIP-T3, is required for sensory cilia formation. Genes Cells 13 13–25. [DOI] [PubMed] [Google Scholar]
- LeDizet, M., and G. Piperno, 1986. Cytoplasmic microtubules containing acetylated alpha-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J. Cell Biol. 103 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., P. N. Inglis, C. C. Leitch, E. Efimenko, N. A. Zaghloul et al., 2008. An essential role for DYF-11/MIP-T3 in assembling functional intraflagellar transport complexes. PLoS Genet. 28 e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. B., J. M. Gerdes, C. J. Haycraft, Y. Fan, T. M. Teslovich et al., 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117 541–552. [DOI] [PubMed] [Google Scholar]
- Lin, B., J. T. White, A. G. Utleg, S. Wang, C. Ferguson et al., 2003. Isolation and characterization of human and mouse WDR19, a novel WD-repeat protein exhibiting androgen-regulated expression in prostate epithelium. Genomics 82 331–342. [DOI] [PubMed] [Google Scholar]
- Lin, B., A. G. Utleg, K. Gravdal, J. T. White, O. J. Halvorsen et al., 2008. WDR19 expression is increased in prostate cancer compared with normal cells, but low-intensity expression in cancers is associated with shorter time to biochemical failures and local recurrence. Clin. Cancer Res. 14 1397–1406. [DOI] [PubMed] [Google Scholar]
- Lucker, B. F., R. H. Behal, H. Qin, L. C. Siron, W. D. Taggart et al., 2005. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 280 27688–27696. [DOI] [PubMed] [Google Scholar]
- Lux, III, F. G., and S. K. Dutcher, 1991. Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics 128 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S. S., S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz et al., 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. S., J. M. Esparza, A. M. Lippa, F. G. Lux, III, D. G. Cole et al., 2005. Mutant kinesin-2 motor subunits increase chromosome loss. Mol. Biol. Cell 16 3810–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama, T., Y. Toh, Y. Ohshima and M. Koga, 2005. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 346 677–687. [DOI] [PubMed] [Google Scholar]
- Myster, S. H., J. A. Knott, K. M. Wysocki, E. O'Toole and M. E. Porter, 1999. Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas. J. Cell Biol. 146 801–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori, Y., C. Zhao, A. Saras, S. Mukhopadhyay, W. Kim et al., 2008. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat. Genet. 10 437–444. [DOI] [PubMed] [Google Scholar]
- Ooi, S. L., X. Pan, B. D. Peyser, P. Ye, P. B. Meluh et al., 2006. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 22 56–63. [DOI] [PubMed] [Google Scholar]
- Oyhenart, J., R. Le Goffic, M. Samson, B. Jégou and N. Raich, 2003. Phtf1 is an integral membrane protein localized in an endoplasmic reticulum domain in maturing male germ cells. Biol. Reprod. 68 1044–1053. [DOI] [PubMed] [Google Scholar]
- Pan, J., and W. J. Snell, 2002. Kinesin-II is required for flagellar sensory transduction during fertilization in Chlamydomonas. Mol. Biol. Cell 13 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., C. G. Wilkerson and G. B. Witman, 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., B. L. Dickert and G. B. Witman, 1999. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., B. L. Dickert, Y. Vucica, E. S. Seeley, J. L. Rosenbaum et al., 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, L. B., M. S. Miller, S. Geimer, J. M. Leitch, J. L. Rosenbaum et al., 2005. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 15 262–266. [DOI] [PubMed] [Google Scholar]
- Pfister, K. K., E. M. Fisher, I. R. Gibbons, T. S. Hays, E. L. Holzbaur et al., 2005. Cytoplasmic dynein nomenclature. J. Cell Biol. 171 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., K. Mead and S. Henderson, 1996. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1(FLA10) to reach the distal part of flagella in Chlamydomonas. J. Cell Biol. 133 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., E. Siuda, S. Henderson, M. Segil, H. Vaananen et al., 1998. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M. E., R. Bower, J. A. Knott, P. Byrd and W. Dentler, 1999. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell 10 693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., J. L. Rosenbaum and M. Barr, 2001. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11 457–461. [DOI] [PubMed] [Google Scholar]
- Qin, H., Z. Wang, D. Diener and J. Rosenbaum, 2007. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, S. A., D. Lang, A. D. Zimmer, A. Terry, A. Salamov et al., 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Rymarquis, L. A., J. M. Handley, M. Thomas and D. B. Stern, 2005. Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 137 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, J. C., M. E. Winkelbauer, C. L. Williams, C. J. Haycraft, R. A. Desmond et al., 2006. IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J. Cell Sci. 119 4088–4100. [DOI] [PubMed] [Google Scholar]
- Shimogawara, K., S. Fujiwara, A. Grossman and H. Usuda, 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P. V., C. J. Haycraft, T. Y. Besschetnova, A. Turbe-Doan, R. W. Stottmann et al., 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 40 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, Z., M. Vashishtha and J. L. Hall, 1994. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., J. Pan and W. J. Snell, 2006. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell 125 549–562. [DOI] [PubMed] [Google Scholar]
- Witman, G. B., K. Carlson, J. Berliner and J. L. Rosenbaum, 1972. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54 507–539. [DOI] [PMC free article] [PubMed] [Google Scholar]