Abstract
The [URE3] and [PSI+] prions are the infections amyloid forms of the Saccharomyces cerevisiae proteins Ure2p and Sup35p, respectively. Randomizing the order of the amino acids in the Ure2 and Sup35 prion domains while retaining amino acid composition does not block prion formation, indicating that amino acid composition, not primary sequence, is the predominant feature driving [URE3] and [PSI+] formation. Here we show that Ure2p promiscuously interacts with various compositionally similar proteins to influence [URE3] levels. Overexpression of scrambled Ure2p prion domains efficiently increases de novo formation of wild-type [URE3] in vivo. In vitro, amyloid aggregates of the scrambled prion domains efficiently seed wild-type Ure2p amyloid formation, suggesting that the wild-type and scrambled prion domains can directly interact to seed prion formation. To test whether interactions between Ure2p and naturally occurring yeast proteins could similarly affect [URE3] formation, we identified yeast proteins with domains that are compositionally similar to the Ure2p prion domain. Remarkably, all but one of these domains were also able to efficiently increase [URE3] formation. These results suggest that a wide variety of proteins could potentially affect [URE3] formation.
AMYLOID fibril formation is associated with numerous human diseases, including Alzheimer's disease, type II diabetes, and the transmissible spongiform encephalopathies. Yeast prions provide a powerful model system for examining amyloid fibril formation in vivo. [URE3] and [PSI+] are the prion forms of the Saccharomyces cerevisiae proteins Ure2p and Sup35p, respectively (Wickner 1994). In both cases, prion formation is thought to result from conversion of the native protein into an inactive amyloid form (Glover et al. 1997; King et al. 1997; Taylor et al. 1999). Both proteins contain an N-terminal glutamine/asparagine (Q/N)-rich prion-forming domain (PFD) and a C-terminal functional domain (Ter-Avanesyan et al. 1993; Ter-Avanesyan et al. 1994; Masison and Wickner 1995; Liebman and Derkatch 1999; Maddelein and Wickner 1999). Sup35p contains an additional highly charged middle domain (M) that is not required either for prion formation or for normal protein function, but stabilizes [PSI+] aggregates (Liu et al. 2002).
Amyloid fibril formation is thought to occur through a seeded polymerization mechanism. In vitro, amyloid fibril formation from native proteins is generally characterized by a significant lag time, thought to result from the slow rate of formation of amyloid nuclei; addition of a small amount of preformed amyloid aggregates (seeds) eliminates the lag time, resulting in rapid polymerization (Glover et al. 1997; Taylor et al. 1999; Serio et al. 2000).
Despite considerable study, the mechanism by which amyloid seeds initially form is unclear. At least some of the amyloid proteins involved in human disease can interact with unrelated amyloidogenic proteins, resulting in cross-seeding and modulation of toxicity. Injecting mice with amyloid-like fibrils formed by a variety of short synthetic peptides promotes amyloid formation by amyloid protein A, a protein whose deposition is found in systemic AA amyloidosis (Johan et al. 1998). In yeast, [PSI+] and [PIN+], the prion form of the protein Rnq1p (Sondheimer and Lindquist 2000; Derkatch et al. 2001), both promote the aggregation of and increase toxicity of expanded polyglutamine tracts, like those seen in Huntington's disease (Osherovich and Weissman 2001; Meriin et al. 2002; Derkatch et al. 2004; Gokhale et al. 2005; Duennwald et al. 2006); however, in Drosophila, [PSI+] aggregates reduce polyglutamine toxicity (Li et al. 2007). Thus, interactions between heterologous amyloidogenic proteins can influence amyloid formation both positively and negatively in vivo.
A variety of interactions have been observed among the yeast prions. Under normal cellular conditions, efficient formation, but not maintenance, of [PSI+] requires the presence of [PIN+] (Derkatch et al. 2000). Overexpression of various Q/N-rich proteins can effectively substitute for [PIN+], allowing [PSI+] formation in cells lacking [PIN+] (Derkatch et al. 2001; Osherovich and Weissman 2001). In vitro and in vivo evidence suggest that the ability of [PIN+] to facilitate [PSI+] formation is the result of a direct interaction between Rnq1p aggregates and Sup35p (Derkatch et al. 2004; Bardill and True 2009; Choe et al. 2009). [PIN+] also increases the frequency of [URE3] formation, while [PSI+] inhibits [URE3] formation (Bradley et al. 2002; Schwimmer and Masison 2002).
It is unclear whether the ability of Ure2p, Sup35p, and Rnq1p to cross-react is an intrinsic feature of all similar amyloidogenic proteins, or whether it has specifically evolved to regulate prion formation. There is debate as to whether yeast prion formation is a beneficial phenomenon, allowing for regulation of the activity of the prion protein (True and Lindquist 2000; True et al. 2004), or a deleterious event analogous to human amyloid disease (Nakayashiki et al. 2005). Either way, it is likely that interactions between the yeast prion proteins have specifically evolved, either to minimize the detrimental effects of amyloid formation or to regulate beneficial amyloid formation.
For both Ure2p and Sup35p, the amino acid composition of the PFD is the predominant feature that drives prion formation. Scrambled versions of Ure2p and Sup35p (in which the order of the amino acids in the PFD was randomized while maintaining amino acid composition) are able to form prions when expressed in yeast as the sole copy Ure2p or Sup35p (Ross et al. 2004, 2005). To examine whether amino acid composition can similarly drive interactions between heterologous proteins, we tested whether the scrambled PFDs can interact with their wild-type counterparts to stimulate prion formation. When overexpressed, scrambled Ure2 PFDs promoted de novo prion formation by wild-type Ure2p, suggesting that the Ure2p PFD can promiscuously interact with compositionally similar PFDs during prion formation. When we searched the yeast proteome for proteins with regions of high compositional similarity to Ure2p, four of the top five proteins were able to efficiently stimulate [URE3] formation. However, there were limits to this promiscuity; overexpression of wild-type or scrambled Sup35 PFDs did not increase [URE3] levels. We propose that this ability to promiscuously interact may have evolved as a mechanism to regulate Ure2p activity and/or prion formation.
MATERIALS AND METHODS
Strains and media:
Standard yeast media were as previously described (Sherman 1991). Galactose/raffinose dropout medium contained 2% galactose and 1% raffinose. In all experiments, yeast were grown at 30°.
[URE3] induction by scrambled PFDs:
Strain YER135 (MATa ura2 leu2 his3 trp1 URE2∷HIS3 [PIN+], Σ1278b background) was transformed with either pH 317 (Edskes and Wickner 2000), a 2-μm, LEU2 plasmid carrying the GAL1 promoter, or with the previously described derivatives of pH 317 in which the inducing PFD was inserted under control of the GAL1 promoter (Ross et al. 2004). Strains were grown for 3 days in galactose/raffinose dropout medium lacking leucine. Serial 10-fold dilutions were plated on SD +ureidosuccinate (USA) +Trp to select for [URE3] cells. Colonies were counted after 5 days and the plates were photographed after 7 days. Frequencies of USA+ colony formation were determined as the mean of at least three independent experiments.
Stability, dominance, curability, and cytoduction of [URE3]:
To test for stability of the USA+ phenotype, USA+ colonies were resuspended in water in a 96-well microtiter plate and spotted onto YPAD plates. After 48 hr, the cells from the YPAD plates were resuspended in water in a 96-well microtiter plate and spotted onto SD +USA +Leu +Trp to test for maintenance of the USA+ phenotype (this method is similar to replica plating, but transfers a more reproducible, lower density of cells).
To test for dominance of the USA+ phenotype, USA+ colonies were spotted from the microtiter plates onto YPAD plates spread with a lawn of YER214 (MATα ura2 leu2 his3 ade2 URE2∷HIS3). These plates were grown for 24 hr and replica plated to SD +Ura +Leu to select for diploids. The SD +Ura +Leu plates were grown for 48 hr. Diploids were resuspended in water in a 96-well microtiter plate and spotted onto SD +USA +Leu to test diploids for [URE3].
To test curability, USA+ cells were resuspended in water in a 96-well microtiter plate and spotted onto YPAD plates and YPAD plus 5 mm guanidine. After 48 hr, the cells from both plates were resuspended in water in a 96-well microtiter plate respotted onto the same medium (YPAD or YPAD plus 5 mm guanidine HCl). After another 48 hr, cells were resuspended in water in a 96-well microtiter plate and spotted onto SD +USA +Leu +Trp to test for [URE3].
YER216 (MATα kar1 ura2 ade2 leu2 his3 ρ0) was used as a cytoduction recipient. kar1 reduces the efficiency of karyogamy during mating (Conde and Fink 1976), allowing for cell fusion and transfer of cytoplasmic material, without nuclear fusion. Donor (ρ+) and recipient (ρ0) cells were mixed in water and spotted onto YPAD. After incubation for 8 hr at 30°, cells were streaked onto medium selecting for recipient cells. Cytoductants were identified as ρ+ cells with the recipient's nuclear genotype (Ridley et al. 1984).
[PSI+] generation:
Yeast strains 780-1D/pJ533 (Song et al. 2005; from Dan Masison, National Institutes of Health) expressing wild-type SUP35, and versions of 780-1D/pJ533 modified to express each of the scrambled versions of SUP35 (YER259, 289, 290, 292, and 293) were previously described (Ross et al. 2005). Strains were transformed with either pKT24 (from Kim Taylor, NABI, Rockville, MD), a 2-μm, TRP1 plasmid carrying the GAL1 promoter, or with a derivative of pKT24 in which the inducing PFD was inserted under control of the GAL1 promoter (Ross et al. 2005). Strains were grown for 3 days in galactose/raffinose dropout medium lacking tryptophan. Serial 10-fold dilutions were spotted onto SC −ade medium to select for [PSI+] cells and grown for 5 days.
Western blot analysis:
The PFDs of URE2; URE2-21–25; SUP35; and SUP35-21, -24–27, as well as the fragments from SAP30, GPR1, GIS1, PDC2, and YLR278C, were amplified by PCR from the respective inducing plasmids using primer EDR68 paired with EDR1057-1068, respectively (see supporting information, Table S1 for oligonucleotides). These PCR products were then reamplified with EDR1055 and 1056. Together, these PCR reactions inserted a GGSGGSY spacer, hemagglutinin (HA2) tag and stop codon at the carboxyl terminus of each PFD. PCR products were digested with BamHI and XhoI and inserted into BamHI/XhoI cut pH 317 (Edskes and Wickner 2000). Ligation products were transformed into Escherichia coli and analyzed by DNA sequencing.
The resulting LEU2 plasmids were transformed into YER135. Cells were grown overnight in galactose/raffinose dropout medium lacking leucine, then diluted to OD600 = 0.1 and grown to OD600 = 0.4–0.6. Cells from 10 ml of culture were collected by centrifugation and washed once with and resuspended in 25 mm Tris phosphate supplemented with 2 mm phenylmethanesulphonylfluoride (PMSF). Cells were lysed by vortexing with glass beads (10 × 15 sec). Protein concentrations were determined by Bradford assay (Sigma). Five micrograms of protein was separated electrophoretically on SDS/12% PAGE gels and detected by Western blot. Mouse monoclonal anti-HA antibody (Covance; HA.11) was used as the primary antibody and Alexa Fluor IR800 goat anti-mouse (Rockland) was used as the secondary antibody.
[URE3] loss:
Four independent [URE3] isolates were tested. The genotype of YER2 (Figure 4A) is MATα ura2 leu2, Σ1278b background. YER223–225 (Figure 4, B, C, and D, respectively) are [URE3] isolates of YER135. Each [URE3] isolate was transformed with the same plasmids used for the [URE3] induction experiments. Strains were grown for 4 days in galactose/raffinose dropout medium lacking leucine. Cultures were maintained in log phase by monitoring the OD600 and diluting 10- to 100-fold when the OD reached 0.2–0.6. At 24-hr intervals, cells were plated for single colonies on YPAD. Single colonies were resuspended in water in a 96-well microtiter plate and spotted onto SD +USA +Trp +Leu to test for loss of [URE3]. For each strain/plasmid combination at each time point, a minimum of 20 colonies were tested. The fraction of cells that maintained the ability to grow on SD +USA +Trp +Leu is reported. Confidence intervals were calculated using the adjusted Wald method (Agresti and Coull 1998).
Figure 4.—
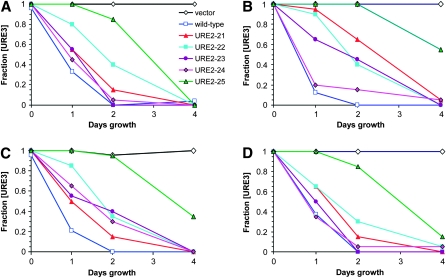
Scrambled PFDs destabilize wild-type [URE3]. Plasmids containing the GAL1 promoter (vector) or expressing wild-type or scrambled Ure2 PFD from the GAL1 promoter were introduced into four different [URE3] strains expressing wild-type URE2 from the URE2 genomic locus. Cells were grown in galactose/raffinose dropout medium for varying lengths of time and then tested for loss of [URE3]. Prion loss in yeast strain YER2 (A), YER223 (B), YER224 (C), and YER225 (D).
Colocalization studies:
To cherry tag the wild-type and scrambled Ure2 PFDs, the PFDs were amplified by PCR from the respective inducing plasmids using primer EDR68 paired with EDR1057–1061, respectively (see Table S1 for oligonucleotides). The mCherry (Shaner et al. 2004) ORF was amplified with EDR1187 and 1189. The product of the mCherry reaction was combined with each of the PFD PCRs and reamplified with EDR68 and 1189. PCR products were digested with BamHI and XhoI and inserted into BamHI/XhoI cut pH 317 under control of the GAL1 promoter. Ligation products were transformed into E. coli and analyzed by DNA sequencing.
pH 327, a CEN plasmid expressing Ure2-GFP from the URE2 promoter (Edskes et al. 1999) was transformed into a [URE3] isolate of YER135. Each of the plasmids expressing cherry-tagged PFDs was transformed into this strain. Cells were grown for 12 hr in galactose/raffinose dropout medium lacking leucine and tryptophan. Cells were visualized by confocal microscopy.
Protein expression and purification:
Plasmid pER94 expresses His6-tagged full-length Ure2p (Ross et al. 2004). Ure2p was expressed in E. coli BL21 in 2× YT medium (1.6% Bacto yeast extract, 1% Bacto tryptone, and 0.5% sodium chloride, pH 7.0) containing 0.1 mg/ml ampicillin at 37° for 4 hr after induction with 1 mm isopropyl β-d-thiogalactosidase at an A600 of ∼1.0. After harvesting, cells were resuspended and sonicated in native lysis buffer (50 mm Tris-HCl, 300 mm NaCl, 5% glycerol, 0.02% NaN3, 10 mm imidazole), containing protease inhibitors (complete EDTA-free, Roche Applied Science). Insoluble material was removed by centrifugation (25 min at 15,000 × g).
Full-length Ure2p was recovered using an 11-ml Ni2+ HiTrap Chelating HP column (GE Heathcare). Protein was bound to the column in 50 mm Tris-HCl, 300 mm NaCl, 5% glycerol, 0.02% NaN3, 10 mm imidazole. Elution buffer was 50 mm Tris-HCl, 300 mm NaCl, 5% glycerol, 0.02% NaN3, 500 mm imidazole. The column was washed in three steps with a mixture of binding and elution buffer of 5, 10, and 30% elution buffer, respectively. Protein was eluted in 100% elution buffer. Purified protein was diluted to 60 μm in 50 mm Tris-HCl, 0.2 m NaCl) and frozen at −70°.
Plasmids pER107, pER108, and pER111 expressing His6-tagged versions of the Ure2-21, -22, and -25 PFDs, respectively, are previously described (Ross et al. 2004). PFDs were expressed in the same manner as full-length Ure2p and purified as previously described (Baxa et al. 2003). Purified PFDs were dialyzed into water to initiate fibril formation.
In vitro fibril formation assay:
Amyloid fibril formation was monitored at 37° by thioflavin T (ThT, Calbiochem) fluorescence using a Victor3 microplate reader (Perkin Elmer, Waltham, MA) with excitation and emission wavelengths of 460 and 490 nm, respectively. ThT solution was preincubated at 37° for 1 hr. Soluble full-length Ure2p was added to a concentration of 50 μm, with 1 mm ThT. High concentration seeding reactions (Figure 6A) contained 4% seed relative to full-length protein (mol/mol). Plates were sealed with sealing tape and shaken slowly with a 5-mm circular motion for 5 sec every min with emission measurements taken every 5 min. For unseeded reactions, fluorescence of a ThT blank was subtracted; for seeded reactions, the fluorescence signal from reactions containing only ThT and seed was subtracted. Low concentration seeding reactions (Figure 6B) contained 0.4% (mol/mol) seed relative to full-length Ure2p. Additionally, for low concentration seeding reactions, a 5-mm glass bead was added to each well of the microtiter plate and between scans, plates were shaken continuously at 230 rpm at 37°. Each sample was prepared in triplicate.
Figure 6.—
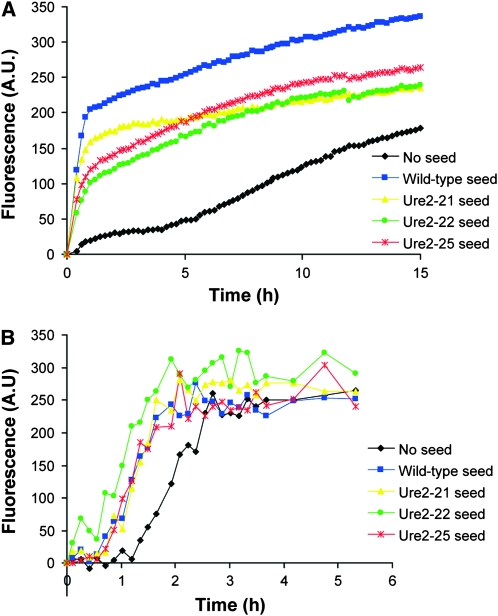
Scrambled PFD aggregates seed wild-type Ure2p amyloid formation in vitro. Polymerization of Ure2p in a microplate was monitored by thioflavin T fluorescence with and without addition of preformed wild-type URE2 or scrambled PFD seed. (A) Four percent seed relative to full-length protein (mol/mol) was added and plates were shaken for 5 sec every min with emission measurements taken every 5 min. (B) Seed [0.4% (mol/mol)] relative to full-length Ure2p was added. To accelerate amyloid fibril formation, plates were shaken continuously at 230 rpm at 37° except during reading of fluorescence, resulting in a shorter lag time for amyloid formation.
Plasmids expressing fragments of yeast proteins:
The yeast proteome was scanned using a 45-amino-acid window size, scoring every window on the basis of the Euclidian distance of its amino acid composition from that of the Ure2 PFD. For the top five scoring window, the window size was expanded to 89 amino acids in three steps. The 89-amino-acid region of maximum Q/N content that contained the original 45-amino-acid fragment was selected. If adjusting the boundaries while maintaining the 89-amino-acid length could eliminate a proline residue without eliminating more than one Q/N, such adjustments were made. If possible without changing proline content, and without changing Q/N content by more than one, the boundaries were further adjusted to bring the net charge closer to ±1.
Selected regions were amplified by PCR (see Table S1 for oligonucleotides) from yeast strain YER135, installing a stop codon at the end of the selected region. PCR products were digested with BamHI and XhoI and inserted into BamHI/XhoI cut pH 317 (Edskes and Wickner 2000). Ligation products were transformed into E. coli and analyzed by DNA sequencing. The resulting LEU2, 2-μm plasmids expressing fragments from SAP30, PDC2, GIS1, GPR1, and YLR278C were named pER334, 336, 338, 340, and 342, respectively.
RESULTS
[URE3] induction by scrambled PFDs:
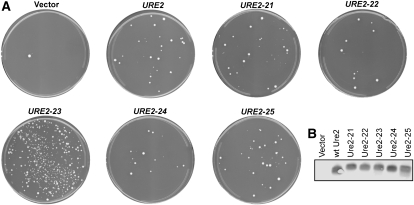
The ability to utilize USA can be used to monitor [URE3] formation. In the presence of a good nitrogen source, Ure2p prevents uptake of USA, an intermediate in uracil biosynthesis. Loss of Ure2p function, either due to prion formation or a mutation in the URE2 gene, allows cells to take up USA. Transient overexpression of the Ure2p PFD in [ure-o] cells, cells lacking the [URE3] prion, significantly increases the appearance of USA+ colonies (Wickner et al. 1995). Surprisingly, overexpression of each of the scrambled Ure2 PFDs also efficiently increased the frequency of USA+ colony formation (Table 1, Figure 1).
TABLE 1.
Induction of wild-type [URE3] by heterologous prion domains
| Overexpressed prion domaina | USA+ colonies/ 106 cellsb | USA+ stabilityc | USA+ dominanced | Cytoductione | |
|---|---|---|---|---|---|
| Isolate A | Isolate B | ||||
| Vector | 1.3 ± 0.3 | 7/20 | 4/7 | 20/20 | 19/20 |
| Wild typef | 23 ± 3 | 12/20 | 8/12 | 18/20 | 20/20 |
| URE2-21 | 17 ± 3 | 12/20 | 7/12 | 20/20 | 20/20 |
| URE2-22 | 10 ± 5 | 18/20 | 17/18 | 20/20 | 20/20 |
| URE2-23 | 130 ± 12 | 14/20 | 7/14 | 17/20 | 20/20 |
| URE2-24 | 18 ± 1 | 15/20 | 7/15 | 14/20 | 18/20 |
| URE2-25 | 27 ± 6 | 19/20 | 18/19 | 20/20 | 18/20 |
| SUP35 | <1.0 | 5/20 | 1/5 | ND | ND |
| SUP35-21 | <1.0 | 6/20 | 4/6 | ND | ND |
| SUP35-24 | <1.0 | 5/20 | 2/5 | ND | ND |
| SUP35-25 | <1.0 | 7/20 | 3/7 | ND | ND |
| SUP35-26 | <1.0 | 4/20 | 0/4 | ND | ND |
|
SUP35-27 |
<1.0 |
6/20 |
5/6 |
ND |
ND |
Yeast strain YER135 was transformed with either a plasmid containing the GAL1 promoter (pH 317, vector) or pH 317 modified to express the indicated prion domain from the GAL1 promoter.
Yeast were grown in galactose/raffinose medium for 3 days and plated onto USA medium to select for prion-containing cells. Data are the number of USA+ colonies per 106 cells and represent the mean of three independent experiments. Standard errors are indicated.
The fraction of USA+ colonies that remained USA+ after 48 hr of growth on YPAD. ND, not determined.
The fraction of stable USA+ colonies whose USA+ phenotype was dominant when mated with [ure-o] cells carrying a chromosomal copy of wild-type URE2.
Two stable dominant USA+ isolates were used as cytoduction donors. The [ure-o] recipient strain carried a wild-type chromosomal copy of URE2. The numbers indicated the fraction of cytoductants that were USA+.
Efficient induction by the wild-type prion domains has previously been reported (Masison and Wickner 1995), but is included here as a positive control.
Figure 1.—
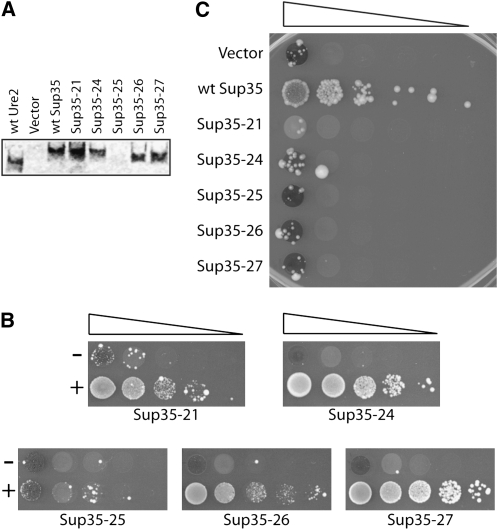
Induction of USA+ colonies by scrambled Ure2p PFDs. (A) Yeast strain YER135 expressing wild-type URE2 was transformed with either a plasmid containing the GAL1 promoter (vector) or expressing the indicated PFD from the GAL1 promoter. Yeast were grown in galactose/raffinose medium for 3 days. Cells (5 × 105) were then plated onto USA medium to select for prion-containing cells. Colonies were counted after 5 days and photographed after 7. (B) Western blots of wild-type and scrambled Ure2 PFDs. Plasmids containing the GAL1 promoter (vector) or HA-tagged PFDs expressed from the GAL1 promoter were introduced into yeast strain YER135. Cells were grown in galactose medium and harvested in log phase. Cell lysates were analyzed by Western blot.
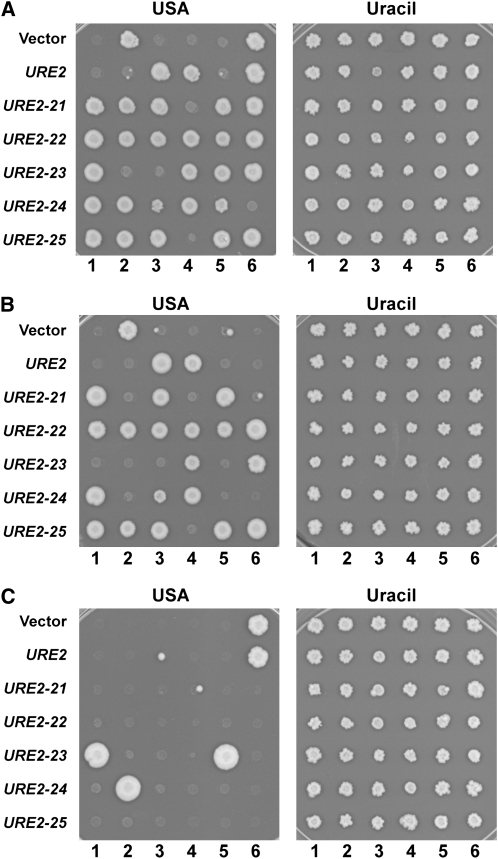
Because the USA+ phenotype results from loss of Ure2p activity, it can result either from a loss-of-function mutation in the URE2 gene or from [URE3] formation. To confirm that the increase in USA+ colony formation was a result of prion formation, we tested for each USA+ colony whether the USA+ phenotype was stable, dominant, curable, and transmissible. For a significant fraction of the USA+ colonies induced by each of the scrambled PFDs, the USA+ phenotype was stable (Table 1; Figure 2A) and dominant (Table 1 and Figure 2B). Additionally, for all stable dominant USA+ colonies, the USA+ phenotype was curable by low concentrations of guanidine HCl (Figure 2C), which cures [URE3] and [PSI+] (Tuite et al. 1981; Wickner et al. 1995) by inhibiting the chaperone Hsp104p (Ferreira et al. 2001; Jung and Masison 2001; Jung et al. 2002). Finally, in all cases the USA+ phenotype was efficiently transmitted by cytoduction, a technique that allows for transmission of cytoplasmic elements such as prions, but not of chromosomal elements (Conde and Fink 1976). Together, these data demonstrate that the increase in USA+ colony formation upon scrambled PFD overexpression is due to an increase in [URE3]-containing cells.
Figure 2.—
Scrambled Ure2 PFDs induce stable, dominant, and curable USA+ colonies. (A) Stability of the USA+ phenotype. USA+ colonies isolated upon overexpression of the wild-type Ure2 PFD, scrambled PFDs (URE2-21–25) or without overexpression (vector) were spotted onto YPAD and grown for 48 hr. Cells were then transferred to medium with uracil (right) or USA (left) to test for the ability to utilize USA. Shown are representative results, with six independent isolates inducing each PFD. (B) Dominance of the USA+ phenotype. USA+ colonies were spotted onto a lawn of [ure-o] cells of the opposite mating type and grown for 24 hr. Cells were replica plated to select for diploids and grown for 48 hr. Diploids were spotted onto medium with uracil (right) or USA (left) to test for the ability to utilize USA. Shown are representative results, from the same USA+ isolates as in A. (C) Curability of the USA+ phenotype. USA+ colonies were spotted onto YPAD supplemented with 5 mm guanidine HCl and grown for 48 hr. Cells were then spotted at low density onto fresh YPAD medium supplemented with 5 mm guanidine HCl and grown for an additional 48 hr. Cells were then transferred to medium with uracil (right) or USA (left) to test for the ability to utilize USA. Shown are representative results, from the same USA+ isolates as in A.
Limits of prion promiscuity:
The wild-type and scrambled versions of Ure2p have identical amino acid compositions. We therefore tested whether less closely related prion proteins could similarly stimulate wild-type [URE3] formation. Overexpression of both wild-type and scrambled Sup35 PFDs actually appeared to slightly suppress USA+ colony formation by wild-type Ure2p (Table 1). This suggests that although Ure2p may interact with wild-type and scrambled Sup35 PFDs, such interactions are not able to productively nucleate [URE3] formation. That the wild-type Sup35 PFD could not induce [URE3] formation was not surprising, as [PSI+] is known to destabilize [URE3] aggregates (Schwimmer and Masison 2002). However, the scrambled Sup35 PFDs, although compositionally identical, have no primary sequence identity with the wild-type Sup35 PFD. Therefore, these results highlight the critical role that amino acid composition plays in the induction of [URE3] by heterologous PFDs.
To confirm that differences in ability to induce [URE3] formation were not due to differences in efficiency of overexpression, expression of each of the wild-type and scrambled Sup35 and Ure2 PFDs was analyzed by Western blot. Each PFD was HA2-tagged and inserted into a plasmid under control of the GAL1 promoter. Each of the HA-tagged PFDs stimulated [URE3] formation with similar efficiency to its untagged counterpart (data not shown). The expression levels of all of the PFDs except SUP35-25 were comparable to the wild-type URE2 PFD (Figures 1B and 3A), demonstrating that the failure of the wild-type and scrambled Sup35 PFDs to induce [URE3] was not a result of inefficient overexpression.
Figure 3.—
[PSI+] formation is not induced by scrambled Sup35p PFDs. (A) Western blots of wild-type and scrambled Sup35 PFDs. PFDs were HA tagged and detected as in Figure 1B. (B) Induction of prion formation by scrambled Sup35s by homologous PFDs. Strains expressing the indicated full-length scrambled SUP35 as the sole copy of SUP35 were transformed with either a plasmid containing the GAL1 promoter (−) or expressing the same scrambled Sup35 PFD from the GAL1 promoter (+). Strains were grown in galactose/raffinose dropout medium, and then serial 10-fold dilutions were spotted onto SC −ade medium to select for [PSI+] cells. (C) Induction of wild-type [PSI+] formation by each of the scrambled PFDs.
Furthermore, each of the scrambled Sup35 PFDs was able to induce [PSI+] prion formation in strains expressing the full-length version of the same scrambled SUP35 (Figure 3B), although consistent with its poor expression (Figure 3A), prion formation by SUP35-25 was induced with the lowest efficiency. [PSI+] was detected by monitoring nonsense suppression of the mutant ade2-1 allele (Cox 1965). ade2-1 mutants are unable to grow without adenine; however, in cells containing the weak nonsense suppressor tRNA SUQ5 (SUP16), [PSI+] allows for growth of ade2-1 mutants in the absence of adenine. These results further confirm that their failure to induce [URE3] was not a result of inefficient overexpression.
To test whether the ability to interact with compositionally identical PFDs was unique to Ure2p, we tested whether overexpression of heterologous PFDs could similarly increase [PSI+] colony formation. None of the scrambled Sup35 PFDs stimulated Ade+ colony formation in strains expressing wild-type SUP35 (Figure 3C). Similarly, overexpression of wild-type and scrambled Ure2 PFDs failed to increase Ade+ colony formation (data not shown).
[URE3] destabilization by scrambled PFDs:
The observed ability of the scrambled PFDs to increase [URE3] populations could be a result of either direct cross-seeding or an indirect mechanism such as titration of an inhibitor of [URE3] formation. Overexpression of the Ure2p PFD fused to GFP results in loss of [URE3] (Edskes et al. 1999). It was proposed that these fusion proteins bind to prion fibrils and “poison” their growth (Edskes et al. 1999) or that overexpression results in aggregates that are too big to propagate (Crapeau et al. 2009). We hypothesized that if the scrambled PFDs can interact with wild-type Ure2p, they might similarly be able to destabilize [URE3] prions.
A plasmid overexpressing either wild-type or scrambled Ure2p PFDs from the GAL1 promoter was introduced into four strains carrying different [URE3] prion variants, each expressing a chromosomal copy of wild-type URE2. Each of these four [URE3] variants was isolated in a different manner—by de novo [URE3] formation (YER2; Figure 4A), by overexpression of the wild-type Ure2 PFD (YER223; Figure 4B), by overexpression of the Ure2-21 PFD (YER224; Figure 4C), and by overexpression of the Ure2-22 PFD (YER225; Figure 4D). Cells were grown in galactose medium to induce expression from the GAL1 promoter; loss of the ability to utilize USA was used to monitor prion loss. Each of the scrambled PFDs destabilized wild-type [URE3] in all strains tested (Figure 4). The scrambled PFDs varied in their efficiencies of [URE3] destabilization, and none were as efficient as the wild-type PFD, although in many cases the differences were not statistically significant (see Table S2 for raw data and confidence intervals). Curing by the scrambled PFDs was only modestly affected by the manner in which the [URE3] prion was originally isolated; although both the Ure2-21 and Ure2-22 PFDs were most efficient at curing prions that had originally been formed by their overexpression (strains YER224 and 225, respectively), these differences were not statistically significant. Similar results were seen for prions induced by each of the other scrambled PFDs (data not shown). These results are consistent with a direct physical interaction between wild-type and scrambled Ure2p.
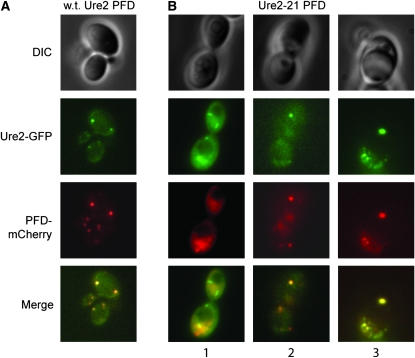
Scrambled Ure2p PFDs colocalize with wild-type Ure2p aggregates:
Ure2-GFP fusions form foci in [URE3] cells, but not in [ure-o] cells (Edskes et al. 1999). We expressed Ure2-GFP fusions in a [URE3] cell, and then transiently overexpressed mCherry (Shaner et al. 2004) tagged wild-type or scrambled Ure2 PFD. As expected, the wild-type PFD consistently colocalized with Ure2-GFP foci (Figure 5A). The results for the cherry-tagged scrambled Ure2 PFDs were more variable. In many cells containing wild-type Ure2-GFP foci, the scrambled Ure2 PFDs appeared diffuse, or in foci that were distinct from the GFP foci [Figure 5B shows one such example for Ure2-21; similar results were seen for the other scrambled PFDs (data not shown)]. However, in a subset of cells, the scrambled PFDs were colocalized with the wild-type Ure2-GFP foci (Figure 5B). Although the frequency of this colocalization varied among the prion variants, in all cases some colocalization was observed (data not shown). This is consistent with a direct interaction between wild-type Ure2p and scrambled Ure2 PFDs, but suggests that this interaction is transient and/or less efficient than the interaction between Ure2 and the wild-type Ure2 PFD.
Figure 5.—
Colocalization of PFDs with wild-type [URE3] aggregates. [URE3] cells expressing Ure2-GFP were transformed with plasmids expressing the mCherry labeled PFD of wild-type Ure2 (A) or Ure2-21 (B). For the Ure2-21 PFD, cells with (columns 2 and 3) and without (column 1) colocalization are shown. DIC, differential interference contrast.
Scrambled Ure2p PFDs directly seed Ure2p amyloid formation:
The rate limiting step for in vitro amyloid formation is the initial formation of amyloid nuclei. Once nuclei are formed, polymerization proceeds rapidly. Therefore, in vitro Ure2p amyloid formation is characterized by a significant lag phase, followed by rapid polymerization; addition of a small amount of preformed Ure2p amyloid fibrils eliminates the lag phase (Taylor et al. 1999; Figure 6). To determine whether the increase in wild-type [URE3] formation upon overexpression of the scrambled Ure2p PFDs is a result of direct cross seeding, we examined in vitro whether amyloid fibrils of the scrambled PFDs could similarly seed amyloid formation by wild-type Ure2p. Three of the scrambled Ure2p PFDs were purified under denaturing conditions and dialyzed into water to initiate amyloid formation. A small amount of these fibers was added to purified full-length Ure2p. At 4% concentration relative to the full-length Ure2p (mol/mol), each scrambled PFD was able to completely eliminate the lag time for Ure2p amyloid formation (Figure 6A). At much lower concentrations (0.4%), each scrambled PFD was able to shorten the lag time with efficiencies comparable to that of wild-type URE2 seed (Figure 6B). These data are consistent with direct seeding of wild-type Ure2p amyloid formation by the scrambled PFDs.
Induction of [URE3] formation by fragments of yeast proteins:
The yeast proteome contains ∼100 proteins with regions of similar Q/N content to Ure2p (Michelitsch and Weissman 2000). We performed a search to identify the yeast proteins with regions that are most compositionally similar to that of the Ure2p PFD. Because ∼40–50 amino acids is the minimum length required for efficient [URE3] induction (Ross et al. 2005), we scanned the yeast proteome using a 45-amino-acid window size, scoring each window on the basis of the Euclidean distance of its amino acid composition from that of the Ure2p PFD (amino acids 1–89).
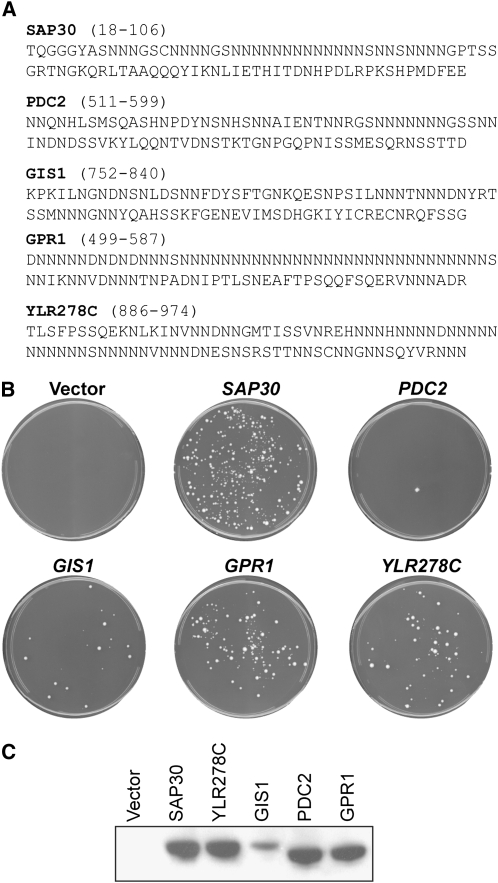
The fragments closest in amino acid composition to the Ure2p PFD were from SAP30, PDC2, GIS1, GPR1, and YLR278C (Table 2). Although 40–50 amino acids of the Ure2p PFD are sufficient for prion induction, in general longer PFD fragments induce more efficiently (Ross et al. 2005). Therefore, for each 45-amino-acid fragment identified by the search, an 89-amino-acid segment containing the 45-amino-acid fragment was selected for testing (Figure 7A). The 89-amino-acid fragment was chosen to (1) maximize Q/N content, (2) minimize proline content, as prolines have been shown to disrupt β-sheet formation, and (3) when possible, have the fragments net charge be ±1, as these net charges have been shown to be ideal for amyloid formation by small peptides (Lopez De La Paz et al. 2002). These fragments were inserted into plasmids under control of the GAL1 promoter and used for induction experiments. Of the five fragments, only that of PDC2 failed to efficiently induce USA+ colony formation (Table 2; Figure 7B). The failure of PDC2 was not due to poor expression; all of the fragments except that of GIS1 showed comparable expression levels (Figure 7C). Remarkably, three of the fragments, from SAP30, GPR1, and YLR278C, increased USA+ colony formation by greater than 30-fold; by contrast, overexpression of the wild-type PFD under the same conditions only increased USA+ colony formation by ∼20-fold. For many of the USA+ colonies induced by overexpression of the SAP30, GIS1, GPR1, and YLR278C fragments, the USA+ phenotype was stable and dominant (Table 2), and all stable dominant USA+ colonies were curable by guanidine HCl (data not shown). Additionally, for all dominant, stable USA+ colonies tested (at least two for each induction), the USA+ phenotype was transmissible by cytoduction (data not shown). Together, these data demonstrate that the fragments from SAP30, GIS1, GPR1, and YLR278C are each able to increase the frequency of [URE3].
TABLE 2.
[URE3] induction by yeast protein fragments
| Gene | Proposed function | Region identified | Fragment tested | USA+ colonies/ 106 cells ± SEM | USA+ stability | USA+ dominance |
|---|---|---|---|---|---|---|
| vector | None | 1.3 ± 0.6 | 9/20 | 7/9 | ||
| URE2 | 1–89 | 24 ± 3 | 16/20 | 11/16 | ||
| SAP30 | Histone deacetylase | 40–84 | 18–106 | 76 ± 9 | 19/20 | 18/19 |
| PDC2 | Transcription regulator | 535–579 | 511–599 | 1.0 ± 0.4 | 13/20 | 10/13 |
| GIS1 | Histone demethylase | 775–819 | 752–840 | 18 ± 3 | 12/20 | 8/12 |
| GPR1 | G-protein coupled receptor | 540–584 | 499–587 | 55 ± 13 | 14/20 | 9/14 |
|
YLR278C |
Unknown; nuclear |
886–930 |
886–974 |
44 ± 12 |
19/20 |
14/19 |
The yeast proteome was searched, using a 45-amino-acid window size, to identify the proteins that contain regions that are most compositionally similar to the Ure2p prion domain. The five proteins containing the highest scoring regions and the proposed molecular functions of the proteins are indicated. For each protein, an 89-amino-acid fragment was chosen that contained the identified region. This fragment was inserted into plasmid pH 317 under control of the GAL1 promoter and transformed into yeast strain YER135. USA+ colony formation, USA+ stability, and USA+ dominance were tested as in Table 1.
Figure 7.—
Induction of [URE3] by compositionally similar domains. (A) Amino acid sequences of the tested domains. The locations of the segments within the respective protein sequences is indicated. (B) Yeast strain YER135 expressing wild-type URE2 was transformed with either a plasmid containing the GAL1 promoter (vector) or expressing the indicated PFD from the GAL1 promoter. USA selection was conducted as in Figure 1A. (C) Western blots of the compositionally similar fragments. PFDs were HA tagged and detected as in Figure 1B.
DISCUSSION
Because the events that initiate amyloid formation in human disease are not well understood, identifying the factors that contribute to amyloid formation is critical for understanding amyloid disease. Experiments from a variety of systems indicate that amyloidogenic proteins can cross-react to seed amyloid formation. However, this cross-seeding is generally highly inefficient; for example, although Rnq1p aggregates can cross-seed Sup35p amyloid formation, Rnq1p aggregates are at least 50-fold less efficient than Sup35p aggregates at seeding aggregation of soluble Sup35p in vitro (Derkatch et al. 2004). The significant finding here is that heterologous PFDs can promote [URE3] formation at efficiencies rivaling that of the homologous PFD.
The increase in [URE3]-containing cells upon overexpression of scrambled PFDs could result from either direct cross-seeding or an indirect mechanism. For example, PFD overexpression could upregulate expression of prion-promoting proteins. Microarray analysis indicates that [URE3] formation, even in the presence of PFD overexpression, does not cause any significant gene expression changes beyond those attributable to nitrogen derepression (Ross and Wickner 2004), arguing against a transcriptional change; nevertheless, such changes could occur post-transcriptionally or could be below the threshold of detection by microarray. PFD overexpression could also titrate away an inhibitor of prion formation, thereby promoting prion formation by the full-length protein. However, the ability of the scrambled PFDs to seed wild-type Ure2p amyloid formation in vitro in the absence of any additional proteins, the colocalization of scrambled PFDs with wild-type [URE3] aggregates, and the ability of scrambled PFDs to destabilize existing [URE3] aggregates all argue against these indirect mechanisms and in favor of a direct cross-seeding mechanism.
Structural studies of amyloid fibrils of the yeast prion proteins provide some insight into how amyloid formation can be seeded by heterologous PFDs. NMR studies indicate that Sup35p, Ure2p, and Rnq1p amyloid fibrils, as well as those of scrambled versions of Ure2p and Sup35p, are composed of in-register parallel β-sheets (Shewmaker et al. 2006, 2008; Baxa et al. 2007; Wickner et al. 2008), although alternative models for both Ure2p and Sup35p amyloid fibrils have been proposed (Bousset et al. 2002; Krishnan and Lindquist 2005). The PFDs of Sup35p, Ure2p, and Rnq1p are all Q/N-rich, and stacking of Q/N residues to form polar zippers has been proposed to stabilize amyloid fibrils (Perutz et al. 2002; Tsai et al. 2005). Therefore, Q/N residues may allow for interactions between Sup35p, Ure2p, and Rnq1p, although it is interesting that [PIN+] can also stimulate amyloid formation in vivo by the non-Q/N-rich Het-s PFD from Podospora anserina (Taneja et al. 2007). Thus, the ends of growing amyloid fibrils may act as an imperfect template for heterologous prion proteins, allowing the heterologous prion protein to add to the fiber end (Wickner et al. 2008).
This idea explains both the ability of [PIN+] to seed amyloid formation by Sup35p and other amyloidogenic proteins (Derkatch et al. 2004), and the relative inefficiency of this cross-seeding. In vivo, even in the presence of [PIN+], significant [PSI+] prion formation requires overexpression of the Sup35p PFD. The rarity with which [PIN+] seeds [PSI+] formation presumably reflects the fact that fibers of Rnq1p are imperfect templates for Sup35p amyloid formation.
Therefore, it was surprising that scrambled versions of Ure2p could seed wild-type Ure2p amyloid formation almost as efficiently as wild-type Ure2p self-seeds. The seeding of [URE3] formation by scrambled PFDs was significantly more efficient than any previously observed cross-seeding among yeast prions, both in vitro and in vivo, reflecting the uniquely promiscuous nature of Ure2p. Furthermore, these results are in sharp contrast to mammalian prion systems, in which changes of a few amino acids can create a species barrier, preventing prion transmission (reviewed in Collinge and Clarke 2007).
Although Ure2p appears to have a unique ability to efficiently cross-react with compositionally similar domains, there are limits to its promiscuity. That wild-type Sup35 PFD could not seed [URE3] formation was not surprising, as the Ure2p and Sup35p have coevolved, and therefore may have specifically evolved to minimize their interaction. However, all of the scrambled Sup35p PFDs efficiently aggregate, yet none induced [URE3] formation. This suggests that amino acid composition may be critical for determining whether Ure2p can productively interact with heterologous proteins to stimulate [URE3] formation. Although both the Ure2 and Sup35 PFDs are Q/N-rich (48.3 and 45.6%, respectively), they are only 46.9% compositionally identical (Table S3). Interestingly, the Ure2 PFD is far more N rich, while Sup35 is more Q rich, raising the possibility that interactions between asparagines (but not interactions between asparagines and glutamines) may be involved in induction of [URE3] by heterologous domains.
The failure of wild-type [PSI+] formation to be seeded by scrambled versions of Sup35p demonstrates that similar amino acid composition is not sufficient for efficient cross-seeding between heterologous Q/N-rich proteins. Clearly there must be some unique feature of Ure2p (either of the PFD or the C terminus) that allows it to be cross-seeded with such remarkable efficiency. Therefore, further studies on the basis for Ure2p's promiscuity will be critical for determining whether similar heterologous cross-seeding events may be involved in disease-related amyloid formation.
The demonstration that fragments of other yeast proteins were also able to induce [URE3] formation raises the possibility that Ure2p's promiscuity could be physiologically relevant. The identified domains may be less accessible for interaction within the context of their respective full-length proteins. Therefore, further study will be required to determine whether the respective full-length proteins can influence [URE3], either by forming prions themselves or by forming aggregates that are not stably propagated, but are nonetheless able to affect [URE3] formation or stability.
Nevertheless, the ability of all but one of these fragments to induce [URE3] formation indicates that the requirements for heterologous cross-seeding are quite broad. Although these fragments were identified as having similar composition to the Ure2p PFD, the search algorithm was quite simple. The fragments have significant compositional deviations from that of the Ure2 PFD, with 66.2–79.7% compositional identity (Table S3 for detailed amino acids compositions). We did not bias the algorithm in favor of fragments with similar Q/N content to the Ure2 PFD or against fragments that have residues such as prolines that are known to inhibit β-sheet formation. Consequently, these fragments had from 25 to 56 Q/N residues and from one to four prolines; by contrast, the Ure2 PFD has 43 Q/N residues and no prolines. Given the high success rate of such a simple search algorithm, it is likely that at least some of the many other Q/N-rich proteins in yeast would similarly be able to induce [URE3] formation.
Three of the identified domains (GPR1, SAP30, and PDC2) were recently identified in a separate bioinformatics search for potential PFDs (Alberti et al. 2009). Intriguingly, fragments from GPR1 and SAP30 were shown to form SDS-resistant aggregates when overexpressed and visible foci when fused to GFP; however, neither efficiently formed amyloid fibrils in vitro. By contrast, PDC2, which was the only fragment identified by our search that failed to induce [URE3], failed to form both foci and SDS-resistant aggregates upon overexpression (Alberti et al. 2009). These results suggest that PDC2 may fail to induce [URE3] formation simply because it has a lower propensity to aggregate in vivo.
The function, if any, of Ure2p's unique promiscuity is unclear. It has been proposed that yeast prions may serve a beneficial function in cells (True and Lindquist 2000; True et al. 2004), although the failure to identify [URE3] or [PSI+] in any wild yeast strains argues that such beneficial prion formation is at most a rare event (Nakayashiki et al. 2005). Over the past year, the list of yeast prion proteins has rapidly grown and now includes Mot3p (Alberti et al. 2009), Cyc8p (Patel et al. 2009), Swi1p (Du et al. 2008), Mca1p (Nemecek et al. 2009), Rnq1p (Derkatch et al. 2001; Sondheimer and Lindquist 2000), Ure2p (Wickner 1994), and Sup35p (Wickner 1994). Ure2p's promiscuity, combined with the growing list of yeast prions, raises the intriguing possibility that a complex network of prion protein interactions may affect [URE3] formation and propagation.
The theory of beneficial prions states that cells normally exist in a nonprion state but are constantly sampling the prion state through spontaneous prion formation. If the prion state confers a selective advantage, prion-containing cells take over the population. Because [URE3] cells grow slower than cells lacking [URE3] under optimal growth conditions (Wickner 1994), it might be beneficial to suppress prion formation under ideal growth conditions and to select for it during times of cellular stress; a cell that is about to die has little to lose by sampling the prion state. A variety of cellular stresses, many of which disrupt protein folding, have been shown to increase the frequency of prion formation (Chernoff 2007; Tyedmers et al. 2008). Perhaps the promiscuity of the Ure2p PFD has evolved as a mechanism to increase prion formation in response to cellular stress. Because of Ure2p's ability to promiscuously interact, aggregation of other Q/N-rich proteins as a result of cellular stress might increase [URE3] formation.
Alternatively, the promiscuity of the Ure2 PFD may reflect a normal function of the protein. The PFD is intrinsically disordered. For many proteins, regions of intrinsic disorder are used to recognize multiple binding targets, with the disordered domain adopting different structures upon binding to each target (Hansen et al. 2006). Although it is unclear exactly what function the PFD serves, when the PFD is removed, Ure2p is less active and shows reduced ability to bind to Gzf3p, a component of the nitrogen regulation system (Shewmaker et al. 2007). The PFD may have evolved to promiscuously interact with multiple targets. Thus, the ability to self-interact to form prion fibers and the ability of the prion formation to be seeded by heterologous proteins may simply be an unfortunate byproduct of this promiscuity.
Further experiments are needed to determine the function, if any, of Ure2p's ability to interact with other Q/N-rich proteins during prion formation. However, our results clearly show that [URE3] formation can be modulated with remarkable efficiency by a variety of unrelated peptides. This demonstration that heterologous seeding can be quite efficient raises the important question of whether any human amyloidogenic proteins are similarly promiscuous.
Acknowledgments
We thank Reed Wickner for his generous advice and discussion; Karenia Soto for assistance with cloning; Jake and Keith Deluca for help with microscopy; and Frank Shewmaker and members of the Ross lab for materials, discussion, and assistance. This work was supported by Colorado State University and a Basil O'Connor Starter Scholar Research award (5-FY07-104) from the March of Dimes (to E.D.R.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.109322/DC1.
References
- Agresti, A., and B. Coull, 1998. Approximate is better than ‘exact’ for interval estimation of binomial proportions. Am. Stat. 52 119–126. [Google Scholar]
- Alberti, S., R. Halfmann, O. King, A. Kapila and S. Lindquist, 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill, J. P., and H. L. True, 2009. Heterologous prion interactions are altered by mutations in the prion protein rnq1p. J. Mol. Biol. 388 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa, U., K. L. Taylor, J. S. Wall, M. N. Simon, N. Cheng et al., 2003. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J. Biol. Chem. 278 43717–43727. [DOI] [PubMed] [Google Scholar]
- Baxa, U., R. B. Wickner, A. C. Steven, D. E. Anderson, L. N. Marekov et al., 2007. Characterization of beta-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 46 13149–13162. [DOI] [PubMed] [Google Scholar]
- Bousset, L., N. H. Thomson, S. E. Radford and R. Melki, 2002. The yeast prion Ure2p retains its native alpha-helical conformation upon assembly into protein fibrils in vitro. EMBO J. 21 2903–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, M. E., H. K. Edskes, J. Y. Hong, R. B. Wickner and S. W. Liebman, 2002. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 99(Suppl 4): 16392–16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff, Y. O., 2007. Stress and prions: lessons from the yeast model. FEBS Lett. 581 3695–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, Y. J., Y. Ryu, H. J. Kim and Y. J. Seok, 2009. Increased [PSI+] appearance by fusion of Rnq1 with the prion domain of Sup35 in Saccharomyces cerevisiae. Eukaryot. Cell 8 968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge, J., and A. R. Clarke, 2007. A general model of prion strains and their pathogenicity. Science 318 930–936. [DOI] [PubMed] [Google Scholar]
- Conde, J., and G. R. Fink, 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA 73 3651–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, B. S., 1965. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 26 211–232. [DOI] [PubMed] [Google Scholar]
- Crapeau, M., C. Marchal, C. Cullin and L. Maillet, 2009. The cellular concentration of the yeast ure2p prion protein affects its propagation as a prion. Mol. Biol. Cell 20 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, S. V. Masse, S. P. Zadorsky, G. V. Polozkov et al., 2000. Dependence and independence of [PSI(+)] and [PIN(+)]: A two-prion system in yeast? EMBO J. 19 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, J. Y. Hong and S. W. Liebman, 2001. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106 171–182. [DOI] [PubMed] [Google Scholar]
- Derkatch, I. L., S. M. Uptain, T. F. Outeiro, R. Krishnan, S. L. Lindquist et al., 2004. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 101 12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z., K. W. Park, H. Yu, Q. Fan and L. Li, 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald, M. L., S. Jagadish, F. Giorgini, P. J. Muchowski and S. Lindquist, 2006. A network of protein interactions determines polyglutamine toxicity. Proc. Natl. Acad. Sci. USA 103 11051–11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes, H. K., V. T. Gray and R. B. Wickner, 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. USA 96 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes, H. K., and R. B. Wickner, 2000. A protein required for prion generation: [URE3] induction requires the Ras-regulated Mks1 protein. Proc. Natl. Acad. Sci. USA 97 6625–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, P. C., F. Ness, S. R. Edwards, B. S. Cox and M. F. Tuite, 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40 1357–1369. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., A. S. Kowal, E. C. Schirmer, M. M. Patino, J. J. Liu et al., 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89 811–819. [DOI] [PubMed] [Google Scholar]
- Gokhale, K. C., G. P. Newnam, M. Y. Sherman and Y. O. Chernoff, 2005. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J. Biol. Chem. 280 22809–22818. [DOI] [PubMed] [Google Scholar]
- Hansen, J. C., X. Lu, E. D. Ross and R. W. Woody, 2006. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281 1853–1856. [DOI] [PubMed] [Google Scholar]
- Johan, K., G. Westermark, U. Engstrom, A. Gustavsson, P. Hultman et al., 1998. Acceleration of amyloid protein A amyloidosis by amyloid-like synthetic fibrils. Proc. Natl. Acad. Sci. USA 95 2558–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., G. Jones and D. C. Masison, 2002. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion- curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA 99 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., and D. C. Masison, 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43 7–10. [DOI] [PubMed] [Google Scholar]
- King, C. Y., P. Tittmann, H. Gross, R. Gebert, M. Aebi et al., 1997. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 94 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, R., and S. L. Lindquist, 2005. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. B., K. Xu and N. M. Bonini, 2007. Suppression of polyglutamine toxicity by the yeast sup35 prion domain in Drosophila. J. Biol. Chem. 282 37694–37701. [DOI] [PubMed] [Google Scholar]
- Liebman, S. W., and I. L. Derkatch, 1999. The yeast [PSI+] prion: making sense of nonsense. J. Biol. Chem. 274 1181–1184. [DOI] [PubMed] [Google Scholar]
- Liu, J. J., N. Sondheimer and S. L. Lindquist, 2002. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc. Natl. Acad. Sci. USA 99(Suppl 4): 16446–16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez De La Paz, M., K. Goldie, J. Zurdo, E. Lacroix, C. M. Dobson et al., 2002. De novo designed peptide-based amyloid fibrils. Proc. Natl. Acad. Sci. USA 99 16052–16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein, M. L., and R. B. Wickner, 1999. Two prion-inducing regions of Ure2p are nonoverlapping. Mol. Cell. Biol. 19 4516–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison, D. C., and R. B. Wickner, 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270 93–95. [DOI] [PubMed] [Google Scholar]
- Meriin, A. B., X. Zhang, X. He, G. P. Newnam, Y. O. Chernoff et al., 2002. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 157 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch, M. D., and J. S. Weissman, 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 97 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki, T., C. P. Kurtzman, H. K. Edskes and R. B. Wickner, 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. USA 102 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek, J., T. Nakayashiki and R. B. Wickner, 2009. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc. Natl. Acad. Sci. USA 106 1892–1896. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Osherovich, L. Z., and J. S. Weissman, 2001. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 106 183–194. [DOI] [PubMed] [Google Scholar]
- Patel, B. K., J. Gavin-Smyth and S. W. Liebman, 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz, M. F., B. J. Pope, D. Owen, E. E. Wanker and E. Scherzinger, 2002. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta -peptide of amyloid plaques. Proc. Natl. Acad. Sci. USA 99 5596–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, S. P., S. S. Sommer and R. B. Wickner, 1984. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 4 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, E. D., and R. B. Wickner, 2004. Prions of yeast fail to elicit a transcriptional response. Yeast 21 963–972. [DOI] [PubMed] [Google Scholar]
- Ross, E. D., U. Baxa and R. B. Wickner, 2004. Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24 7206–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, E. D., H. K. Edskes, M. J. Terry and R. B. Wickner, 2005. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. USA 102 12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer, C., and D. C. Masison, 2002. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio, T. R., A. G. Cashikar, A. S. Kowal, G. J. Sawicki, J. J. Moslehi et al., 2000. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289 1317–1321. [DOI] [PubMed] [Google Scholar]
- Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer et al., 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194 3–21. [DOI] [PubMed] [Google Scholar]
- Shewmaker, F., R. B. Wickner and R. Tycko, 2006. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc. Natl. Acad. Sci. USA 103 19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker, F., L. Mull, T. Nakayashiki, D. C. Masison and R. B. Wickner, 2007. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker, F., E. D. Ross, R. Tycko and R. B. Wickner, 2008. Amyloids of shuffled prion domains that form prions have a parallel in-register beta-sheet structure. Biochemistry 47 4000–4007. [DOI] [PubMed] [Google Scholar]
- Sondheimer, N., and S. Lindquist, 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5 163–172. [DOI] [PubMed] [Google Scholar]
- Song, Y., Y. X. Wu, G. Jung, Y. Tutar, E. Eisenberg et al., 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja, V., M. L. Maddelein, N. Talarek, S. J. Saupe and S. W. Liebman, 2007. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol. Cell 27 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, K. L., N. Cheng, R. W. Williams, A. C. Steven and R. B. Wickner, 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283 1339–1343. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan, M. D., V. V. Kushnirov, A. R. Dagkesamanskaya, S. A. Didichenko, Y. O. Chernoff et al., 1993. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 7 683–692. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan, M. D., A. R. Dagkesamanskaya, V. V. Kushnirov and V. N. Smirnov, 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, H. L., and S. L. Lindquist, 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407 477–483. [DOI] [PubMed] [Google Scholar]
- True, H. L., I. Berlin and S. L. Lindquist, 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431 184–187. [DOI] [PubMed] [Google Scholar]
- Tsai, H.-H. G., M. Reches, C.-J. Tsai, K. Gunasekaran, E. Gazit et al., 2005. Energy landscape of amyloidogenic peptide oligomerization by parallel-tempering molecular dynamics simulation: significant role of Asn ladder. Proc. Natl. Acad. Sci. USA 102 8174–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M. F., C. R. Mundy and B. S. Cox, 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers, J., M. L. Madariaga and S. Lindquist, 2008. Prion switching in response to environmental stress. PLoS Biol. 6 e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R. B., 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., D. C. Masison and H. K. Edskes, 1995. [PSI] and [URE3] as yeast prions. Yeast 11 1671–1685. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., F. Dyda and R. Tycko, 2008. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc. Natl. Acad. Sci. USA 105 2403–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]