Abstract
The purpose of this work is to test several density functional models (namely, OPBE, O3LYP, OPW91, BPW91, OB98, BPBE, B971, OLYP, PBE1PBE, and B3LYP) to determine their accuracy and speed for computing 13Cα chemical shifts in proteins. The test is applied to 10 NMR-derived conformations of the 76-residue α/β protein ubiquitin (protein data bank id 1D3Z). With each functional, the 13Cα shielding was computed for 760 amino acid residues by using a combination of approaches that includes, but is not limited to, treating each amino acid X in the sequence as a terminally blocked tripeptide with the sequence Ac-GXG-NMe in the conformation of the regularized experimental protein structure. As computation of the 13Cα chemical shifts, not their shielding, is the main goal of this work, a computation of the 13Cα shielding of the reference, namely, tetramethylsilane, is investigated here and an effective and a computed tetramethylsilane shielding value for each of the functionals is provided. Despite observed small differences among all functionals tested, the results indicate that four of them, namely, OPBE, OPW91, OB98, and OLYP, provide the most accurate functionals with which to reproduce observed 13Cα chemical shifts of proteins in solution, and are among the faster ones. This study also provides evidence for the applicability of these functionals to proteins of any size or class, and for the validation of our previous results and conclusions, obtained from calculations with the slower B3LYP functional.
Keywords: computational chemistry, density functional theory, DFT, chemical shifts, protein structure determination, protein structure validation
Introduction
It is well known that the backbone and side-chain conformations of a residue are influenced by interactions with the rest of the protein but, once these conformations are established by these interactions, the 13Cα shielding of each residue depends, mainly, on its backbone1–3 and its side-chain4–9 conformation, with no significant influence of either the amino acid sequence6,8–10 or the position of the given residue in the sequence.10 These properties, together with the fact that 13Cα is ubiquitous in proteins, make this nucleus an attractive candidate for computation of theoretical chemical shifts at the quantum chemical level of theory to determine, refine, and validate protein structures.10–12
For an accurate computation of observed chemical shifts in proteins, it is very important to determine the influence of factors that affect 13Cα-shielding. For example, we have recently investigated the sensitivity of the shielding/deshielding of 13Cα nuclei to changes in the protonation/deprotonation of distant ionizable groups.12 Once these factors have been identified and properly considered, a more important test is to determine the accuracy and speed of the computation of the 13Cα-shielding which are limited by the ability of the density functional model adopted.
Density-functional theory (DFT)13 has become the choice for first-principles quantum chemical calculations of the electronic structure and properties of many molecular and solid systems. There is, however, a large number of DFT methods in the literature because the exact exchange-correlation functional is unknown, making it essential to pursue more and more accurate and reliable approximate functionals, a process which, on the other hand, depends on the applications. Selection of the most appropriate DFT method for a particular application becomes one of the main problems of DFT methods. To the best of our knowledge, tests of the most appropriate DFT method with which to compute chemical shifts have been carried out only for small, and mainly rigid, molecules but not for proteins which exist as an ensemble of conformations in solution. The reason for this is that the factors affecting the shielding in proteins depend strongly on the nuclei to be studied (1Hα, 1HN, 13C′, 17O, 15N, 13Cα, etc.). Thus, among all of them, the 13Cα chemical shifts emerges as one of the most accessible for computational purposes because of the properties mentioned earlier.1–10 These properties enable us to develop a method to compute 13Cα chemical shifts in proteins of any class and size rapidly and efficiently. The selection of the most appropriate density functional, however, is at the core of this method. The main goal of the present work is to determine the most reliable DFT method with which to compute the 13Cα chemical shifts, not the shielding, and the transferability of the results to study proteins of any size and class, accurately and rapidly.
Thus far, we have been using the B3LYP functional in all our previous studies9–12 because there was evidence14 indicating that this functional is one of the most accurate with which to compute 13C chemical shifts. Lately, however, several new DFT functionals have been developed and, hence, new evidence15 has been presented indicating that some of the recently developed functionals, such as OPBE and OPW91 perform significantly better than B3LYP. Recently, Wu et al.15 explored the ability of 21 functionals to reproduce the observed 15N, 13C, 17O, and 19F chemical shifts from 23 small molecules, and carried out a detailed comparison with existing results from other laboratories. Wu et al.15 concluded that, “…OPBE or OPW91 is a good DFT functional currently available for the prediction of NMR data…” In proteins, however, the observed 13Cα chemical shifts represent the contributions from several conformers that coexist in solution and, hence, a discussion of the ability of different functionals to reproduce the observed 13Cα chemical shifts in solution must consider such dispersion in the conformations of the molecule explicitly. To take this effect into account, a set of 11 conformations of the 76-residue protein ubiquitin, solved at high accuracy, were chosen, namely, 10 of them are NMR-derived16 protein data bank (PDB code 1D3Z), shown in Figure 1a, and one is X-ray-derived17 at 1.8 Å resolution (PDB code 1UBQ). The 10 density functionals tested here are OPBE, O3LYP, OPW91, BPW91, OB98, BPBE, B971, OLYP, PBE1PBE, and B3LYP. These 10, out of 21 studied by Wu et al.,15 are those for which the mean absolute deviation (MAD) between computed and observed 13C shielding was lower than 5.2 ppm (as for B3LYP). In other words, all functionals displaying a MAD better than that of B3LYP were chosen to be tested.
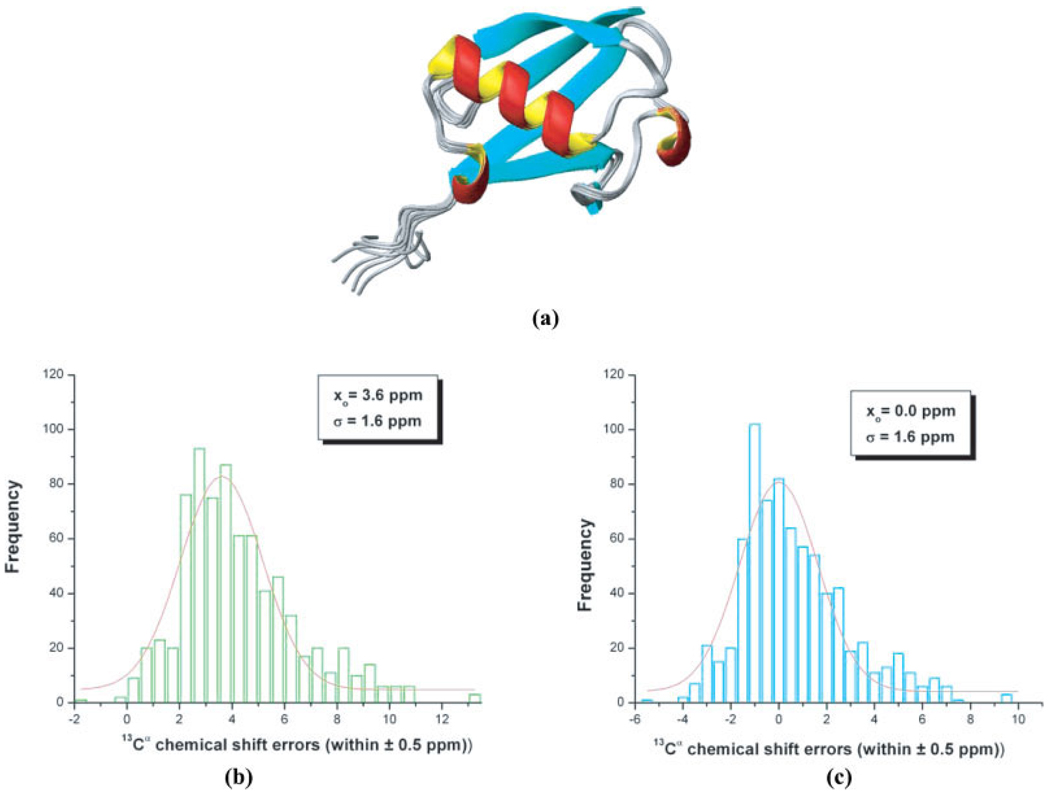
Figure 1.
(a) Ribbon diagram of the superposition of 10 NMR-derived conformations of the protein ubiquitin (PDB id: 1D3Z); (b) bars indicate the frequency of the error distribution, computed assuming a reference TMS value of 188.10 ppm, within a ±0.5 ppm interval between predicted and computed 13Cα chemical shifts (with the OB98 functional) from 10 conformations of ubiquitin (PDB code: 1D3Z) as explained in the Materials and Methods section. The distribution was generated by binning the data between −2 and 13 ppm. The solid red line represents the fitting of the data, only for the range of values provided, by a Gaussian (or Normal) distribution. The values of the mean, xo, and the standard deviation, σ, for the Gaussian (or Normal) distribution are inserted in the panel; (c) same as (b) but assuming an effective TMS value of 184.50 ppm, rather than 188.10 ppm used in (b).
The test proposed here is very CPU time consuming, and this is one of the reasons to limit the number of functionals to be tested. For example, for each of the 10 functionals studied here, 760 DFT calculations with the 6-311+G(2d,p) basis set are carried out, whereas each of the 21 functionals in the analysis of Wu et al.,15 were tested on only 32 small molecules (using the same basis set).
It is worth noting that 13Cα chemical shifts, not absolute shielding, are the primary focus of this work because 13Cα chemical shifts from proteins are determined with high accuracy in NMR experiments and they are the main goal of quantum chemical research. Hence, the ability of the functionals to reproduce the observed 13Cα chemical shifts for each of the 76 residues of protein ubiquitin was evaluated here by a test that includes: the standard deviation (SD) and the correlation coefficient18 (R) between observed and computed conformational-averaged 13Cα chemical shifts; the characteristic mean (xo), and SD (σ) of the Normal (or Gaussian) fit of the frequency of the error distribution; the conformational average root-mean-square-deviation (ca-rsmd)10; and the average-total CPU time, i.e., an average over all 76 residues in the sequence from conformation one of 1D3Z. As a protein in solution exists as an ensemble of conformations, the ca-rmsd was adopted as a scoring function, rather than the individual or the mean, rmsd value, because the ca-rmsd provides a better representation of the physical nature of the observed 13Cα chemical shifts in solution.10
Given that our interest is in the 13Cα chemical shifts, not their shielding, a computation of an accurate value for the shielding of the reference, namely, tetramethylsilane (TMS), is crucial and, hence, these shielding values will be computed here by using two independent methods, and the results of such calculations will be included in the evaluation. Unfortunately, computation of the shielding of TMS is a nontrivial problem because it is a large molecule, the geometry is complicated and account must be taken of vibrational and rotational averaging for a specific temperature19 in calculating the shieldings. Moreover, the shielding of the reference TMS molecule should be calculated with the same computational parameters used for the desired system, in particular, the basis set of each selected exchange-correlation functional and the convergence cutoff must be included.
Materials and Methods
Experimental set of Structures
The experimental set of structures for ubiquitin contains 11 conformations experimentally determined by high-resolution X-ray crystallography17 and NMR methods,16 and their coordinates were obtained from the PDB.20 These proteins are identified by a four-symbol PDB code, namely, 1UBQ for the X-ray-derived conformation and 1D3Z for the 10 NMR-derived conformations.
The 76 observed 13Cα chemical shifts can be found in the Biological Magnetic Resonance Data Bank under accession number 6457. Conversion of the computed TMS-referenced values for the 13Cα chemical shifts to the 3-(trimethylsilyl) propionate sodium salt (TSP) used as the reference during the NMR experiments were carried out by adding 1.82 ppm to the computed TMS values.21
Method Used to Compute the 13Cα Chemical Shifts in Ubiquitin
The computations involve a series of approximations described later. All the experimentally determined conformations were regularized as explained10,11 previously, i.e., all residues were replaced by the standard ECEPP/322 residues in which bond lengths and bond angles are fixed (rigid-body geometry approximation), and hydrogen atoms were added, if necessary.
For each amino acid residue X in the protein sequence (a) it is assumed that the observed 13Cα chemical shift is a conformational-averaged one [see eq. (2)]; (b) computation of the 13Cα shielding was carried out on a terminally blocked tripeptide with the sequence Ac-GXG-NMe in the conformation of the regularized experimental protein structure10; (c) computation of the 13Cα shielding was carried out with a 6-311+G(2d,p) locally dense basis set, whereas the remaining residues in the tripeptide were treated with a 3-21G basis set10; (d) all ionizable residues were considered neutral during the gas-phase quantum chemical calculations12; (e) no geometry optimization is necessary as such optimization by ab-initio (HF) or DFT methods has only a small effect on the computed chemical shifts4,23; (f) the computed 13Cα shieldings (σsubst,th) were converted to 13Cα chemical shifts (δ) by employing the equation δth = σref – σsubst,th, where the indices denote a theoretical (th) computation, the reference substance (ref), and the substance of interest (subst), i.e., the 13Cα shielding of a given amino acid residue X. Initially, as a value for the reference, the observed shielding value of TMS in the gas phase,24 namely, 188.1 ppm, was adopted.
Computation of the Conformationally Averaged rmsd
Computation of this scoring function relies on the following assumption: a protein in solution exists as an ensemble of conformations. As a consequence, we can assume that the observed chemical shifts for a given amino acid μ can be interpreted as a conformational average over different rotational states represented by a discrete number of different conformations, all of which satisfied the NMR constraints, such as NOEs, vicinal coupling constants, Residual Dipolar Coupling constants, etc., from which the conformations were derived.10 Thus, we can compute the following quantity: where is the computed chemical shift for amino acid μ in conformation i out of Ω protein conformations (Ω = 10 in this work), and λi is the weight (Boltzmann) factor for conformation i, with the condition . It is not feasible, however, to determine λi at the quantum chemical level, with the existing computational resources and, hence, some additional approximation must be adopted. In this work, we assume that, under conditions of fast conformational averaging, the following equality holds λi = 1/Ω, i.e., all weight factors contribute equally. Hence, for each amino acid μ, we define an error function:
| (1) |
with
| (2) |
where 1 ≤ μ ≤ N, with N being the number of observed 13Cα chemical shifts (N = 76 for ubiquitin). Then, the conformationally averaged rmsd (ca-rmsd) is defined as10:
| (3) |
and
| (4) |
For a single structure, as an X-ray-derived one, Ω = 1, and hence,
| (5) |
Computation of the Averaged-CPU Time
The averaged computational time for a given functional, listed in Table 1, was computed as an average over all 76 residues of conformation one out of 10 of 1D3Z:
| (6) |
where Tμ represent the total CPU time (in seconds) for residue μ, as reported by the output file of the Gaussian 03 suite of programs. 25 All DFT calculations were carried out in a system with 64 1.15 GHz EV7 processors with 256 Gbytes of shared memory located at the Pittsburgh Supercomputing Center.
Table 1.
Effective TMS Value for the 10 Density Functionals.
| TMSb (ppm) |
||
|---|---|---|
| Densityafunctional | Effective | Computed |
| OPBE | 190.82 | 188.68 |
| O3LYP | 186.61 | 185.29 |
| OPW91 | 190.30 | 188.29 |
| BPW91 | 182.25 | 182.96 |
| OB98 | 184.50 | 184.79 |
| BPBE | 182.79 | 183.38 |
| B971 | 185.29 | 185.53 |
| OLYP | 185.56 | 184.76 |
| PBE1PBE | 187.98 | 187.32 |
| B3LYP | 181.90 | 182.47 |
List of functionals for which the 13Cα shielding for TMS was computed. Those functionals showing a difference between computed and effective TMS lower than 0.3 ppm are highlighted in bold face.
The values from the columns effective and computed TMS were obtained as described in “Materials and Methods” section.
Determination of Two Sets of TMS Values
Two independent methods were used to compute the shielding values of TMS.
Determining an Effective TMS Shielding Value
By adopting the observed TMS value of 188.124 ppm as a reference, it is possible to find the characteristic mean (xo) and SD (σ) of the Normal (or Gaussian) fit of the frequency of the error distribution, computed by eq. (1), for each of the 10 functionals. For all functionals, the characteristic mean value (xo) appears displaced from the ideal value of 0.0 by a positive, or negative, amount. This is illustrated in Figure 1b for OB98 where a value of xo = 3.6 ppm is shown. This indicates that, for any of the 10 functionals tested here, a straightforward use of the observed TMS shielding value (188.1 ppm) is not an appropriate reference if no further corrections are introduced. Hence, for each functional it is feasible to find an “effective” TMS shielding value for which the Normal (or Gaussian) fit shows a zero displacement, i.e., an effective TMS value that gives a value of xo = 0.0. In other words, by subtracting 3.6 ppm (see the value of xo in the panel of Fig. 1b) from 188.1 ppm and recomputing the Gaussian fit with an effective TMS value of 184.5 ppm gives xo = 0.0 (see the panel of Fig. 1c). The effective values, computed with this procedure are listed in Table 1, for all functionals.
Computing a TMS Shielding Value
For each selected functional, the TMS shielding value was computed with the 6-311+G(2d,p) basis set by first carrying out an extremely tight geometry optimization26,27 of the fully staggered conformation in Td symmetry. Table 1 shows the results of the computed TMS shieldings (as opposed to the effective TMS shieldings) for each of the 10 functionals. All calculations were performed with the Gaussian 03 suite of programs.25
There is good agreement between both the computed and effective sets of values listed in Table 1, and a detailed analysis concerning the observed differences is given later in the results and discussion section.
About the Selected Density Functionals
We have used 10 different exchange-correlation functionals, denoted as OPBE, O3LYP, OPW91, BPW91, OB98, BPBE, B971, OLYP, PBE1PBE, and B3LYP.28–37
For these density functional methods, the correlation functional is taken from either Lee-Yang-Parr (LYP),28,29 Perdew-Wang (PW91),30–34 Perdew-Burke-Ernerhof (PBE),35,36 or Becke’s revisions of the B9737,38 functional, namely, B98,38 whereas the exchange functional is from either Becke’s three-parameters HF/DFT hybrid exchange functional (B3),39 Becke’s pure DFT exchange functional (B),40 or Handy and Cohen’s OPTX modification of Becke’s exchange functional (O)41 and its corresponding three-parameter HF/DFT hybrid exchange functional form (O3).42 B97143 is a hybrid HF/DFT functional proposed by Handy, Tozer and coworkers as a modification of the B97 functional.37,38 PBE1PBE (also known as PBE0) is a hybrid HF/DFT functional based on the generalized gradient approximation (GGA) which combines PBE36,37 exchange-correlation with exact exchange using a one-parameter equation.37
Results and Discussion
Analysis of the Effective and Computed TMS Shielding Values
The determination of a proper TMS value for each functional is crucial for an accurate computation of the 13Cα chemical shifts because, among other reasons, it will enable us to minimize the presence of systematic errors which might bias the chemical shift-based analysis. From this point of view, the effective TMS value provides the most accurate approach to solve the problem because it will not require arbitrary further adjustments. On the other hand, because the 13Cα shielding of any amino acid residue is computed by a using a series of approximations, as described in the Materials and Methods section, the effective TMS value adopted might, or might not, be associated with any physically reasonable TMS geometry. Table 1 shows a comparison for each of the computed and effective TMS values (as described in the Materials and Methods section). From Table 1, it can be seen that, for the first three functionals, namely, OPBE, O3LYP, and OPW91, the difference between “effective” and “computed” TMS values is greater than 1.3 ppm while, for the remaining seven, is lower than, or equal to, 0.8 ppm. For two of them, namely, OB98 and B971, the differences are lower than 0.3 ppm. Table 2 shows the TMS optimized geometry, from which the TMS shielding values listed in Table 1 were obtained, for each of the 10 functionals and how these geometry values compare with the molecular geometry derived from electron diffraction data by Campanelli et al.44 (footnote b of Table 2). From both Table 1 and Table 2, we conclude that, for most of the functionals, but in particular for OB98 and B971, the “effective” TMS shielding value can be associated with an experimentally observed TMS geometry, i.e., with a structure determined from gas-phase electron diffraction data.
Table 2.
Geometry of TMSa
| Parameters of the calculated TMS geometryb |
|||
|---|---|---|---|
| Density functional | Si–C (Å) | C–H (Å) | Si–C–H (degrees) |
| OPBE | 1.874 | 1.092 | 111.19 |
| O3LYP | 1.890 | 1.097 | 111.15 |
| OPW91 | 1.894 | 1.100 | 111.19 |
| BPW91 | 1.890 | 1.100 | 111.22 |
| OB98 | 1.894 | 1.100 | 111.19 |
| BPBE | 1.886 | 1.096 | 111.18 |
| B971 | 1.896 | 1.097 | 111.24 |
| OLYP | 1.879 | 1.094 | 111.12 |
| PBE1PBE | 1.886 | 1.093 | 111.23 |
| B3LYP | 1.874 | 1.093 | 111.18 |
Obtained from the TMS optimized geometry, as described in the “Materials and Methods” section.
The experimental data for the TMS molecular geometry from an electron diffraction study44 is: 1.877 ± 0.004 Å, for the Si–C bond length; 1.110 ± 0.003 Å for the C–H bond length; and 111.0° ± 0.2°, for the Si–C–H bond angle. As noted by Campanelli et al.44 these experimental data are consistent with a model of Td symmetry.
Transferability of the Results
Once the residue conformations are established by their interactions with the rest of the protein, the 13Cα shielding of each residue depends, mainly, on its backbone and its side-chain conformation, with no significant influence of either the amino acid sequence or the position of the given residue in the sequence. This observation, discussed in the Introduction section, is crucial for the current methodology. In fact, it allows us to parallelize the 13Cα shielding calculations in proteins, making it feasible. Furthermore, it also means that a given set of highly accurately determined amino acid residue conformations, representing the accessible conformational space for all the 20 naturally occurring amino acids and showing a good distribution of side-chain conformations, will constitute a reasonable ensemble with which to carry out tests for the current methodology. In other words, the results of such tests will not depend on whether such ensembles of residue conformations belong to a single or to many proteins and, hence, their results should be transferable to proteins of any class or size.
For this purpose, the ubiquitin protein was chosen. It contains 18 out of the 20 naturally occurring amino acid residues, i.e., without Cys and Trp, with their residue backbone torsional angles populating the α-helical, β-sheet, turn and extended regions of the Ramachandran map for a given amino acid residue. These residues display a significant variability of side-chain conformations among the different conformers of 1D3Z (see the rmsd’s in Fig. 2a). Thus, we expect that all conclusions obtained here, regarding the ability of different functionals to reproduce the observed 13Cα chemical shifts of proteins in solution, to be transferable to proteins of any size or class.
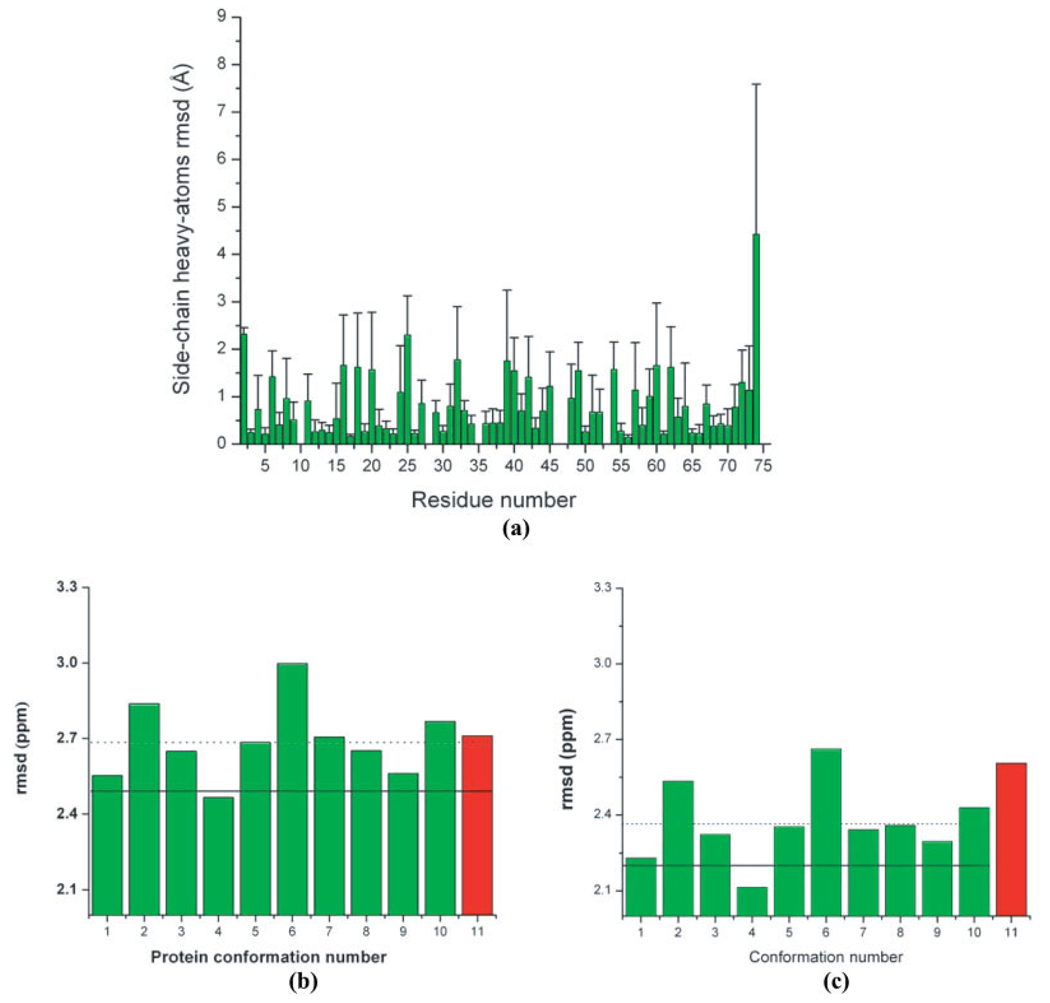
Figure 2.
(a) Bars indicate the per-residue all-heavy atoms side-chain rmsd (Å) between the 10 models of 1D3Z, i.e., by adopting model 1 as reference; (b) Green bars indicate the rmsd (ppm) between observed and computed 13Cα chemical shifts as described in the Materials and Methods section for each of the 10 conformations of ubiquitin (PDB code: 1D3Z). The red vertical bar indicates the rmsd (ppm) computed for the X-ray-derived structure of ubiquitin (PDB code: 1UBQ). The horizontal dotted line represents the mean value for the rmsd computed from the 10 NMR conformations of ubiquitin. The solid horizontal line indicates the ca-rmsd value computed from the 10 conformations of 1D3Z (as explained in the Materials and Methods section). All these results were obtained by using the B3LYP functional10; (c) as (b) but with the OB98 functional.
Statistical Analysis Among the Selected Functionals
A comparison between different parameters adopted to decide on the best functional is shown in Table 3.
Table 3.
Statistical Analysis for Application of 10 Density Functionals to Ubiquitin.
| DensityaFunctional | Rb | SDc (ppm) | Ca-rmsdd (ppm) | CPU Timee (s) |
|---|---|---|---|---|
| OPBE | 0.902 | 2.04 (1.72) | 2.12 | 3475.97 |
| O3LYP | 0.905 | 2.01 (1.69) | 2.16 | 6587.23 |
| OPW91 | 0.903 | 2.03 (1.69) | 2.12 | 3488.24 |
| BPW91 | 0.908 | 1.99 (1.60) | 2.30 | 3624.62 |
| OB98 | 0.908 | 1.98 (1.62) | 2.19 | 3559.61 |
| BPBE | 0.907 | 1.99 (1.59) | 2.30 | 3605.91 |
| B971 | 0.905 | 2.02 (1.80) | 2.22 | 6531.22 |
| OLYP | 0.906 | 2.00 (1.60) | 2.18 | 3668.72 |
| PBE1PBE | 0.902 | 2.04 (1.84) | 2.21 | 6767.88 |
| B3LYP | 0.905 | 2.01 (1.70) | 2.34 | 6686.93 |
This column contains the list of the 10 functionals tested in this work. The best result for each functional (in terms of these parameters) is high-lighted in bold face in Columns 2–4.
The correlation coefficient, R, (or Pearson coefficient) between the observed and the conformational-averaged 13Cα chemical shifts, computed with eq. (2), for each of the functionals listed in Column 1.
Standard deviation of the computed conformational-averaged 13Cα chemical shifts from a linear regression. The standard deviation (σ) of the Normal (or Gaussian) curve that fits the frequency of the error distribution computed by using an effective TMS value as given by Table 2 (see “Materials and Methods” section) is shown in parentheses.
Ca-rmsd was computed by using eq. (3), as described in the “Materials and Methods” section. For each functional, the ca-rmsd was computed by using the effective TMS value listed in Table 1.
Averaged-CPU time computed by using eq. (5) from conformation 1 out of 10 conformations of 1D3Z. The CPU times that, on average, require ~1 h are set in bold face.
The frequency distribution of the errors for each functional, computed with eq. (1), can be modeled by a Normal (or Gaussian) function with a characteristic mean (xo = 0) and SD (σ), and the results for σ are shown in Table 3. Instead of using the type of analysis used for small molecules,14,15 in which the maximum and minimum errors between computed and observed 13Cα chemical shift are frequently reported, we prefer to discuss the frequency of the error distribution. In other words, the frequencies of the error distribution for ubiquitin, rather than the maximum or minimum errors are the most important quantities with which to assess the quality of the functionals. For example, Figure 1c shows that the highest errors are ~10 ppm; their frequency, however, is very low. In fact, due to the Gaussian distribution of the frequency of the errors, 99.7% of these errors lie within 3σ, i.e., with a Δα ≤ 4.8 ppm for calculations carried out with the OB98 functional. This result also highlights the relevance of the SD (σ) in the assessment of the functionals. From Table 3, some dispersion of σ values, namely, 1.59 ≤ σ ≤ 1.84 can also be seen. This range of σ values is within the range of the SD (0.90 ppm ≤ σ ≤ 2.25 ppm) observed by Wang and Jardetzky 45 for 13Cα chemical shifts (from a database containing more than 6000 amino acid residues in the α-helix, β-sheet, and statistical-coil conformations).
Regarding a comparison between observed and computed 13Cα chemical shifts, Table 3 shows that there are five functionals, namely, OPBE, O3LYP, OPW91, OB98, and OLYP, which behave very similarly in their ability to reproduce the observed 13Cα chemical shifts, in terms of the ca-rmsd, accurately for ubiquitin in solution. In summary, we observe that, although OB98 appears to be slightly better than any other functional in terms of R and SD, other functionals such as OPBE, O3LYP, OPW91, and OLYP are slightly better in terms of ca-rmsd.
The assessment was intended to find not only the most accurate functional with which to reproduce the observed 13Cα chemical shifts in solution but also the faster ones, in terms of CPU time, because the CPU time could severely limit the applicability for proteins. In fact, this is crucial for applications such as determination, validation, and refinement for which the 13Cα chemical shifts must be computed for several dozens of conformations of, usually, molecules containing more than 60–70 residues. The averaged-CPU time listed in Table 3 (computed as indicated in the Materials and Method section) can be used to classify all 10 functionals into two groups: first, those for which the averaged computational time of the 13Cα chemical shifts for all 76 residues, for a given conformation of the protein ubiquitin, would take ~1 hour (highlighted in bold face in Table 3), such as OPBE, OPW91, BPW91, OB98, BPBE, and OLYP, and second, those for which the same calculation takes twice the time (~2 hours), as for hybrid DFT/HF exchange functional O3LYP, B971, PBE1PBE, and B3LYP. The significant CPU time difference among functionals clearly indicates the need to consider the timing during the final evaluation.
A Comparison Regarding Results Obtained with OB98 and B3LYP
The main goal of this work is to determine the most accurate, and rapid, DFT functional model with which to compute 13Cα chemical shifts in proteins. Once this new functional has been selected, it is necessary to determine whether the results obtained earlier9–12,23 with B3LYP have to be revisited. In order to answer such an important question, we carried out a series of analyses involving only two functionals, namely, OB98 and B3LYP. This comparison is very important because B3LYP has been used extensively during the last few years for determination and validation of protein structures in our laboratory.10–12,23 On the other hand, OB98 was arbitrarily chosen among a group of four functionals, namely, OPBE, OPW91, OB98, and OLYP, that show very similar accuracy and CPU time according to the results shown in Table 3.
A comparison of the rmsd distribution for each of the 10 NMR-derived (1D3Z) conformations (green bars) and the X-ray-derived structure, solved at 1.8 Å resolution (red bar) by using B3LYP,10 is shown in Figure 2b; a similar analysis with OB98 is shown in Figure 2c. The dashed and solid black horizontal lines represent the corresponding averaged ca-rmsd and the 〈rmsd〉, computed by using eq. (3) and eq. (4), respectively, for the 10 NMR-derived conformations. Despite the differences seen in the heights of the bars in Figures 2b and 2c, which are not relevant for the purpose of the current analysis, the results obtained with OB98 (Fig. 2c) follow the same trend observed with the calculations carried out previously with the B3LYP functional (Fig. 2b),10 except for the relative values of rmsd between conformations seven and eight. Even more important, the use of either B3LYP or OB98 leads to the same conclusion: the NMR-derived set of conformations appears to be a better representation of the observed 13Cα chemical shifts in solution than the X-ray structure. Regarding this comparison, it is interesting to note that the gap (0.4 ppm) between the rmsd value for the X-ray (2.6 ppm) and the ca-rmsd (2.2 ppm), computed, with OB98 is twice the corresponding one (0.2 ppm) computed with the B3LYP functional.10 As the 10 NMR-derived conformations and the X-ray-derived structure are very similar among themselves, i.e., with an averaged backbone all-heavy atoms rmsd of 0.97 ± 0.41 Å, this result suggests that OB98 is more sensitive than B3LYP to discriminate subtle differences among the NMR-derived conformations and the X-ray structure.
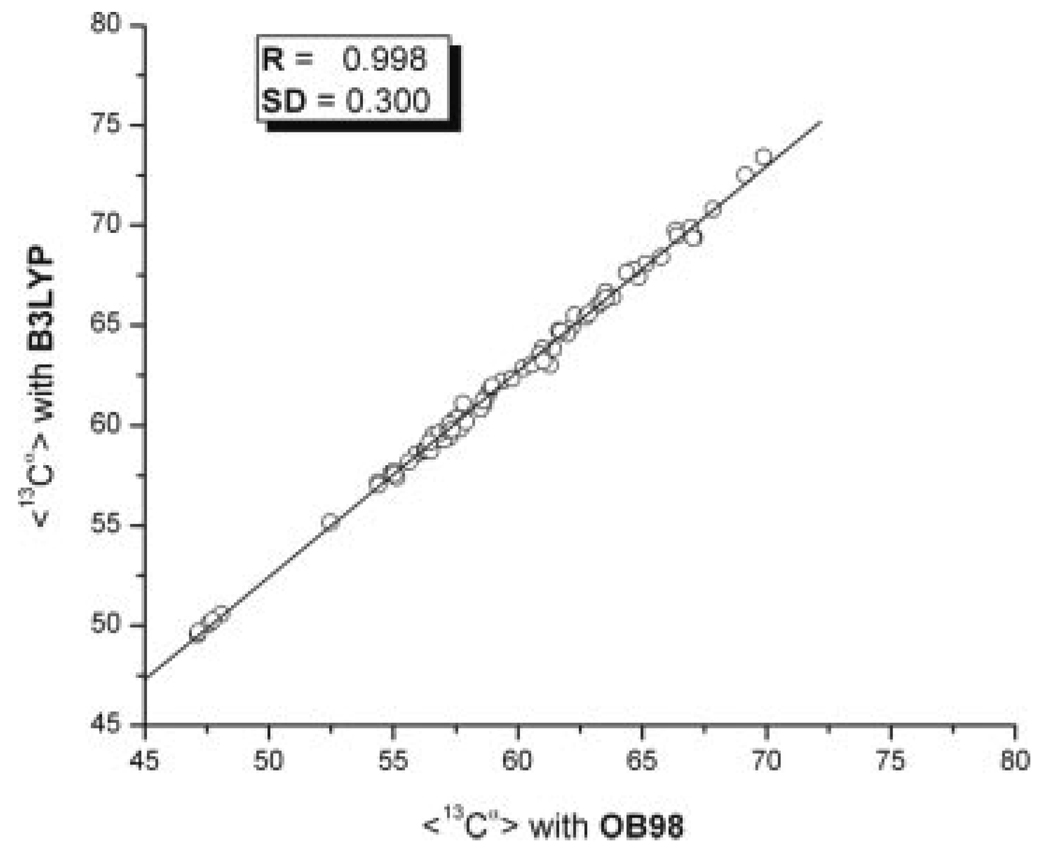
Finally, Figure 3 shows the correlation existing between the averaged 13Cα chemical shifts values, computed for the 10 conformations of 1D3Z computed with the eq. (2), for the OB98 and B3LYP functionals. The excellent correlation coefficient obtained (R = 0.998) and the low SD (0.3) provides evidence that the results and conclusion obtained by using B3LYP do not need to be revisited if the OB98 functional is adopted.
Figure 3.
Correlation between average, 〈13Cα〉, chemical shifts computed from the 10 conformations of ubiquitin (PDB code: 1D3Z) by using eq. (2) with B3LYP versus the same value computed with the OB98 functional. The line represents the linear regression. Values for the correlation coefficient (R) and the standard deviation from the linear regression SD are inserted in the panel.
Conclusions
Our test with 10 NMR-derived conformations (1D3Z) of the protein ubiquitin, solved at high-accuracy, focuses on the ability of the 10 selected exchange-correlation functionals to reproduce the observed 13Cα chemical shifts, not their shielding. The calculations were always carried out with the 6-311+G(2d,p) basis set by using the Gaussian 03 suite of programs,25 and some approximations to make such calculations feasible for proteins were adopted here. The main result of this test indicated that four of these functionals, namely OPBE, OPW91, OB98, and OLYP behave similarly in their ability to reproduce the observed 13Cα chemical shifts in proteins accurately, and faster than other functionals.
This conclusion is in line with the analysis of Wu et al.15 of the agreement between observed and computed 13C chemical shifts from small molecules, indicating that OLYP and OB98 are, together with OPBE and OPW91, among the best 21 DFT functionals analyzed. Their results illustrate that the OLYP and OB98 functionals show very similar minimum (0.0 and −0.1 ppm, respectively) and maximum errors (6.0 and 6.3 ppm, respectively) between observed and computed chemical shifts to those from OPBE and OPW91 (see Table 4 of Wu et al.15).There is a small difference15 in terms of the MAD with respect to the observed chemical shifts, namely, 2.0 for OPBE and OPW91 and 3.0 and 2.9 for OLYP and OB98, respectively. It is worth noting that, from the analysis of Wu and coworkers,15 O3LYP shows comparable accuracy with that of OPBE and OPW91, namely, with a minimum and maximum error of 0.3 ppm and 7.1 ppm, respectively, and a MAD value of 2.4. However, Wu and coworkers15 do not include the CPU time in their test, although it is crucial for treatment of larger molecules such as proteins. In other words, although O3LYP also shows comparable accuracy with that of OLYP and OB98 in our test (see Table 3), O3LYP is significantly more CPU time-consuming than OLYP and OB98 and, hence, it is not recommended for calculating 13Cα chemical shifts of proteins. This result should not be surprising because GAA functionals such as the OPBE, OPW91, OLYP, and OB98 functionals are faster, by construction, than hybrid functionals such as O3LYP, because coupled-perturbed equations are actually uncoupled in GGA functionals.
In summary, we have been able to show that (a) a test involving several conformations for a protein, not small molecules, is feasible; (b) computation of the 13Cα chemical shifts by using one of the four functionals, namely, OPBE, OPW91, OB98, and OLYP leads to more accurate results than any other functional tested here on 10 conformations of ubiquitin, without requiring a significant increase in computational cost, as with O3LYP; (c) the above conclusions, based on a consideration of ubiquitin structures, should be transferable to the computation of the 13Cα chemical shifts for protein structures of any size or class; (d) the results, and therein, the conclusions derived from the use of B3LYP should not be revisited if any of these four functionals were adopted; (e) our results, in full agreement with those from an analysis15 of small molecules, indicate that B3LYP does not represent the best approximation for computing 13Cα chemical shifts in proteins; this conclusion is also in line with that of Hoe et al.46 who pointed out that BLYP should be considered out-dated for computational chemistry and recommended OLYP instead; and (f) The GGA functionals such as PBE, PW91, LYP, and B98, using Handy’s optimized exchange functional (OPTX),41 consistently provide the best performance for both magnetic shieldings and chemical shifts, suggesting the importance of a good exchange functional to obtain a good prediction of NMR observables.15
In conclusion, the results of this work indicate that accurate and fast quantum chemistry calculations of the 13Cα chemical shifts in proteins can be obtained by adopting any of the four functionals recommended here, namely, OPBE, OPW91, OB98, or OLYP, in combination with the corresponding TMS shielding value listed in Table 1.
Acknowledgments
The research was conducted using the resources of a Beowulf-type cluster located at the Baker Laboratory of Chemistry and Chemical Biology, Cornell University; and the National Science Foundation Terascale Computing System at the Pittsburgh Supercomputer Center.
Contract/grant sponsor: National Institutes of Health; contract/grant numbers: GM-14312, GM-24893
Contract/grant sponsor: National Science Foundation; contract/grant numbers: MCB05-41633
Contract/grant sponsor: CONICET, FONCyT-ANPCyT; contract/grant numbers: PAV 22642/22672
Contract/grant sponsor: Universidad Nacional de San Luis; contract/grant numbers: P-328501
References
- 1.Spera S, Bax A. J Am Chem Soc. 1991;113:5490. [Google Scholar]
- 2.deDios AC, Pearson JG, Oldfield E. Science. 1993;260:1491. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- 3.Kuszewski J, Qin JA, Gronenborn AM, Clore GM. J Magn Reson Ser B. 1995;106:92. doi: 10.1006/jmrb.1995.1017. [DOI] [PubMed] [Google Scholar]
- 4.Pearson JG, Le H, Sanders LK, Godbout N, Havlin RH, Oldfield E. J Am Chem Soc. 1997;119:11941. [Google Scholar]
- 5.Havlin RH, Le H, Laws DD, deDios AC, Oldfield E. J Am Chem Soc. 1997;119:11951. [Google Scholar]
- 6.Iwadate M, Asakura T, Williamson MP. J Biomol NMR. 1999;13:199. doi: 10.1023/a:1008376710086. [DOI] [PubMed] [Google Scholar]
- 7.Xu XP, Case DA. J Biomol NMR. 2001;21:312. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ. J Am Chem Soc. 2001;123:2970. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 9.Villegas ME, Vila JA, Scheraga HA. J Biomol NMR. 2007;37:137. doi: 10.1007/s10858-006-9118-6. [DOI] [PubMed] [Google Scholar]
- 10.Vila JA, Villegas ME, Baldoni HA, Scheraga HA. J Biomol NMR. 2007;38:221. doi: 10.1007/s10858-007-9162-x. [DOI] [PubMed] [Google Scholar]
- 11.Vila JA, Ripoll DR, Scheraga HA. J Phys Chem B. 2007;111:6577. doi: 10.1021/jp0683871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vila JA, Scheraga HA. Proteins: Struct Funct Bioinform. 2008;71:641. doi: 10.1002/prot.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parr RG, Yang W. Density Functional Theory of Atoms and Molecules. New York: Oxford University Press; 1989. [Google Scholar]
- 14.Cheeseman JR, Trucks GW, Feith TA, Frisch MJ. J Comp Chem. 1996;104:5497. [Google Scholar]
- 15.(a) Zhang Y, Wu A, Xu X, Yan Y. Chem Phys Letters. 2006;421:383. [Google Scholar]; (b) Wu A, Zhang Y, Xu X, Yan Y. J Comp Chem. 2007;28:243. doi: 10.1002/jcc.20641. [DOI] [PubMed] [Google Scholar]
- 16.Cornilescu G, Marquardt JL, Ottiger M, Bax A. J Am Chem Soc. 1998;120:6836. [Google Scholar]
- 17.Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 18.Press HW, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in Fortran 77. The Art of Scientific Computing. 2nd ed. Cambridge, UK: Cambridge University Press; 1992. Chapter 14; p. 630. [Google Scholar]
- 19.Jameson CJ, de Dios AC. In: Nuclear Magnetic Shieldings and Molecular Structure, NATO ASI Series C. 286. Tossell JA, editor. Dordrech: Kluwer; 1993. p. 95. [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J Biomol NMR. 1995;6:135. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 22.Némethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. J Phys Chem. 1992;96:6472. [Google Scholar]
- 23.Vila JA, Ripoll DR, Baldoni HA, Scheraga HA. J Biomol NMR. 2002;24:245. doi: 10.1023/a:1021633403715. [DOI] [PubMed] [Google Scholar]
- 24.Jameson AK, Jameson CJ. J Chem Phys Lett. 1997;134:461. [Google Scholar]
- 25.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision E. 01. Wallingford, CT: Gaussian, Inc; 2004. [Google Scholar]
- 26.Csaszar P, Pulay P. J Mol Struct (Theochem) 1984;114:31. [Google Scholar]
- 27.Farkas Ö, Schlegel HB. J Chem Phys. 1999;111:10806. [Google Scholar]
- 28.Lee C, Yang W, Parr RG. Phys Rev B. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 29.Miehlich B, Savin A, Stoll H, Preuss H. Chem Phys Lett. 1989;157:200. [Google Scholar]
- 30.Burke K, Perdew JP, Wang Y. In: In Electronic Density Functional Theory: Recent Progress and New Directions. Dobson JF, Vignale G, Das MP, editors. New York: Plenum; 1998. p. 81. [Google Scholar]
- 31.Perdew JP. In: In Electronic Structure of Solids 91. Ziesche P, Eschrig H, editors. Berlin: Akademie Verlag; 1991. p. 11. [Google Scholar]
- 32.Perdew JP, Chevary JA, Vosko SH, Jackson KA, Pederson MR, Singh DJ, Fiolhais C. Phys Rev B. 1992;46:6671. doi: 10.1103/physrevb.46.6671. [DOI] [PubMed] [Google Scholar]
- 33.Perdew JP, Chevary JA, Vosko SH, Jackson KA, Pederson MR, Singh DJ, Fiolhais C. Phys Rev B. 1993;48:4978. doi: 10.1103/physrevb.48.4978.2. [DOI] [PubMed] [Google Scholar]
- 34.Perdew JP, Burke K, Wang Y. Phys Rev B. 1996;54:16533. doi: 10.1103/physrevb.54.16533. [DOI] [PubMed] [Google Scholar]
- 35.Perdew JP, Burke K, Ernzerhof M. Phys Rev Lett. 1996;77:3865. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 36.Perdew JP, Burke K, Ernzerhof M. Phys Rev Lett. 1997;78:1396. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 37.Becke AD. J Chem Phys. 1997;107:8554. [Google Scholar]
- 38.Schmider HL, Becke AD. J Chem Phys. 1998;108:9624. [Google Scholar]
- 39.Becke AD. J Chem Phys. 1993;98:5648. [Google Scholar]
- 40.Becke AD. Phys Rev A. 1988;38:3098. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 41.Handy NC, Cohen A. J. Mol Phys. 2001;99:403. [Google Scholar]
- 42.Cohen AJ, Handy NC. Mol Phys. 2001;99:607. [Google Scholar]
- 43.Hamprecht FA, Cohen AJ, Tozer DJ, Handy NC. J Chem Phys. 1998;109:6264. [Google Scholar]
- 44.Campanelli AR, Ramondo F, Domenicano A, Hargittai I. Struct Chem. 2000;11:155. [Google Scholar]
- 45.Wang Y, Jardetzky O. Protein Sci. 2002;11:852. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoe WM, Cohen AJ, Handy N. Chem Phys Lett. 2001;341:319. [Google Scholar]