Abstract
Nosema ceranae, a microsporidian parasite originally described from Apis cerana, has been found to infect Apis melllifera and is highly pathogenic to its new host. In the present study, data on the ultrastructure of N. ceranae, presence of N. ceranae-specific nucleic acid in host tissues, and phylogenetic relationships with other microsporidia species are described. The ultrastructural features indicate that N. ceranae possesses all of the characteristics of the genus Nosema. Spores of N. ceranae measured approximately 4.4 × 2.2 μm on fresh smears. The number of coils of the polar filament inside spores was 18--21. PCR signals specific for N. ceranae were detected not only in the primary infection site, the midgut, but also in the tissues of hypopharyngeal glands, salivary glands, Malpighian tubules, and fat body. The detection rate and intensity of PCR signals in the fat body were relatively low compared to other examined tissues. Maximum parsimony analysis of the small subunit rRNA gene sequences showed that N. ceranae appeared to be more closely related to the wasp parasite, N. vespula than to N. apis, a parasite infecting the same host.
Keywords: Spore-forming microorganism, disease of digestive tract, tissue specificity of infection, phylogenetic relationship
Since their first recognition as pathogens in silkworms by Nagëli (1857), microsporidia have been identified as the sources of many infectious diseases in vertebrates and invertebrates, including human, fishes, and insects (Canning et al. 1986; Franzen and Muller 2001; Sprague and Vavra 1977; Wasson and Peper 2000; Wittner and Weiss 1999). Of 143 genera and over 1,200 microsporidian species described (Wittner and Weiss 1999), insects are frequently found to be their primary host. Most of the entomopathogenic microsporidia occur in the genus Nosema, which has more than 150 described species and infects nearly all taxonomic orders of insect, especially the orders of Lepidoptera and Hymenoptera (Becnel and Andreadis 1999; Sprague 1978).
Nosemosis is a serious disease of adult honey bees, caused by Nosema species. Honey bee colonies are frequently infected, and all colony members, including adult worker bees, drones, and queens, can be infected. Nosema infection occurs mostly through ingestion of spores with food or water. The physical and chemical conditions of the midgut trigger the germination of spores and the vegetative stage of Nosema begins to grow and multiply inside midgut cells. Bailey and Ball (1999) showed that 30--50 millions of spores could be found inside a bee's midgut within two weeks after initial infection. Eventually the spores pass out of the bee in its feces, providing new sources of the infection through cleaning and feeding activities in the colonies.
Nosema infections have significant negative impacts on honey bees, causing dysentery, shortened life spans of honey bees, supersedure of infected queens, and decrease in colony size (Hassanein, 1953; Rinderer and Sylvester, 1978; Malone et al., 1995). Nosema ceranae and Nosema apis are two species of Nosema that are reported to infect the European honey bee, Apis mellifera. For years, nosemosis of the European honey bee was exclusively attributed to Nosema apis. Nosema ceranae, a species originally found in the Asian honey bee, Apis cerana (Fries et al., 1996), is now a common infection of European honey bees and is highly pathogenic to its new host (Cox-Foster et al.,2007; Fries et al. 2006; Higes et al. 2006; 2007; Huang et al., 2007; Klee et al 2007). Chen et al. (2007) demonstrated that N. ceranae was transferred from A. cerana to A. mellifera at least a decade ago and is now replacing N. apis as the predominant microsporidian infection in A. mellifera of the U. S. populations. Although widespread infection by N. ceranae in the U.S. population of A. mellifera has been identified, many biological features of this parasite in the host A. mellifera remain to be elucidated. To remedy this deficiency, we describe key morphological features of N. ceranae based on light and electron microscopy, and we use PCR to determine the presence of N. ceranae-specific nucleic acid in different tissues of the infected hosts. By comparing the sequences of SSUrRNA genes of different microsporidia, we construct a phylogenetic tree to determine the genetic relationship of N. ceranae with other species of microsporidia infecting insects.
Materials and Methods
Honey Bee Sample Collection
Honey bees were collected from colonies maintained in Beltsville, MD. The abdomens of ten honey bees from each colony were ground up in 2 ml of sterile distilled water. One drop of the homogenate was examined by a light microscope for presence of Nosema spores. When spores of Nosema were observed under the microscope, the remaining portion of the homogenized abdomens was used for DNA extraction and PCR assays to determine the species status of Nosema infection. Once the Nosema infection of bee colonies was identified, additional adult bees were collected from those heavily infected colonies and stored at -20 °C for subsequent morphological and molecular analyses.
Purification of Nosema Spores
To obtain purified Nosema spores, the alimentary tracts of honey bees from Nosema-infected colonies were removed individually by grasping the sting with forceps and gently pulling the alimentary tract away from the abdomen. The midgut was separated from the hindgut and 25 pieces of midgut were crushed in 2 ml of sterile distilled water and filtered through a Corning Netwell (Corning Incorporated Life Science, Lowell, MA) insert (24 mm diameter, 74 μm mesh size) to remove tissue debris. The filtered suspension was centrifuged at 1,500 × g for 5 min and the supernatant was discarded. The pellet was resuspended in 1 ml of sterile water and overlayed very gently on a discontinuous 25%, 50%, 75% and 100% of Percoll (Sigma-Aldrich, St. Louis, MO) gradient from top to the bottom and centrifuged twice at 8,000 × g for 20 minutes at 4 °C using a Beckman rotor (SW 28) in a Beckman L8-70M ultracentrifuge to collect spores having the same size, shape, and density. The supernatant was discarded and the spore pellet was resuspended in distilled sterile water and collected by centrifugation. After a final centrifugation at 8,000 × g for 10 minutes at 4 °C, the spore pellet was resuspended in distilled sterile water and stored at 4 °C until used. Spore sizes were measured under an Eclipse TE 300 light microscope (Nikon, Melville, NY) and photographed with a Nikon Digital Camera (DXM 1200).
Light and Electron Microscopy
Midguts of adult bees from a Nosema-infected colony were dissected out as described above and midgut tissue was fixed for 2 h at room temperature by immersion in 3% (v/v) glutaraldehyde in 0.05 M sodium cacodylate buffer, pH 7.0. After overnight incubation in a refrigerator at 4°C, tissues were washed in a sodium cacodylate buffer rinse, 6 times over 1 h, post fixed in 2% (w/v) osmium tetroxide in 0.05 M sodium cacodylate buffer, pH 7.0, for 2 h, dehydrated in ethanol and imbedded in Spurr's low-viscosity embedding resin. One-micron thick sections were cut on a Reichert/AO Ultracut microtome (Leica, Inc., Deerfield, IL) with a Diatome (Hatfield, PA) diamond knife. The sections for light microscopy were mounted onto slides, stained with 0.5% (w/v) toluidine blue and photographed with the same system for spores. Sections for electron microscopy were mounted onto 200-mesh Ni grids, stained with 4% (w/v) uranyl acetate and 3% (w/v) lead citrate, and viewed in a H-7000 Hitachi (Tokyo, Janpan) microscope at 75kV.
Tissue Dissection
Fifteen adult worker bees collected from Nosema-infected colonies were used for tissue dissection. Tissues of alimentary canal, Malpighian tubules, muscle, fat body, hypopharyngeal gland, and salivary gland were carefully separated under a Zeiss SV11 Stereomicroscope (Thornwood, NY) and photographed with a Zeiss AxioCam digital camera. All tissues were rinsed once with 1 X PBS and twice with nuclease-free water to prevent possible contamination and then subjected to subsequent molecular analysis to determine the presence of N. ceranae-specific nucleic acid in tissues.
DNA isolation, PCR amplification, and DNA sequencing
Tissues were individually ground HOW? in liquid nitrogen and genomic DNA was extracted using the DNAzol DNA purification kit (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. Two pairs of primers specific for N. apis (N-apis-F: 5′-ccattgccggataagagagt-3′; N-apis-R: 5′-cacgcattgctgcatcattgac-3′) and N. ceranae (N-ceranae-F: 5′-cggataaaagagtccgttacc-3′; N- ceranae-R: 5′-tgagcagggttctagggat-3′) were used in the study as described previously (Chen et al., 2007). The specificity of amplification was confirmed by cloning the purified PCR fragments from 1.5% low-melting-point agarose gel using Wizard PCR Prep DNA Purification System (Promega, Madison, WI) in pCR 2.1 vector (Invitrogen, Carlsbad, CA), and sequencing the PCR fragments from both directions using M13-forward and M13-reverse primers. The sequence data was analyzed using the BLAST server at the National Center for Biotechnology Information.
Phylogenetic analysis
The 21 species of microsporidian SSUrRNA sequences with the highest BLAST similarity score against the complete sequence of the N. ceranae SSUrRNA were retrieved from GenBank database The hosts of microsporidian species used for phylogenetic analysis were all insects from the Orders Hymenoptera, Lepidoptera, and Coleoptera. Trachipleistophora hominis infecting Homo sapiens was used as an outgroup to root the phylogenetic tree. Sequences were aligned using MegAlign (DNASTAR Lasergene software program, Madison, WI) and sequences that could not be aligned unambiguously at both 3′- and 5′-ends were truncated. The percentage identity and divergence of sequences between equivalent microsporidian SSUrRNA was generated by the MegAlign. Aligned sequences of 20 microsporidia species and the outgroup were imported into the phylogenetic analysis program PAUP 4.03 (Sinauer Associates, Sunderland, MA). Maximum Parsimony under a heuristic search with random stepwise addition and TBR branch swapping was used to construct the phylogenetic trees. Phylogenies were assessed by a 1,000 bootstrap replication.
Results
Nosema ceranae infection was found in adult bees of A. mellifera collected in Beltsville, MD. When the abdomens of infected bees were crushed in water, a large numbers of mature spores were released, although most infected bees did not exhibit overt behavior and morphological signs of infection. The samples examined in this study were exclusively N. ceranae-positive: none of the PCR reactions using N. apis specific primers yielded any product (data not shown).
Light microscopy revealed that fresh N. ceranae spores were oval or rod shaped, varied in size with a length 3.9--5.3 μm (mean ± S.E. = 4.4 ± 0.41 μm) and a width 2.0--2.5 μm ((mean ± S.E. = 2.2 ± 0.09 μm) (N = 50) (Fig. 1). Observation of Nosema-infected midgut tissue showed that mature spores not only accumulate in midgut epithelial cells, but also are released into the gut lumen (Fig. 2, 3).
Fig. 1.
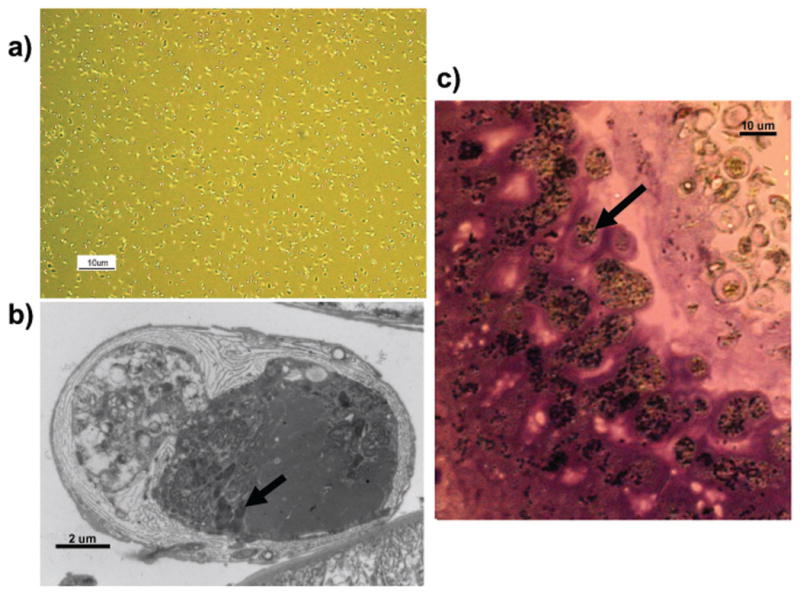
Nosema ceranae spores. Light micrograph of oval to rod-shaped spores of Nosema ceranae after Percoll purification. Scale bar: 10 μM.
Fig.2-3. Cross section of the midgut showing spores. 2. Spores accumulated in midgut lumen. 3 Epithelial cells of the midgut infected with Nosema ceranae. Arrows indicate infected epithelial cells with tightly packed parasites.
Ultrastructural studies showed that different developmental stages, including meronts, sporonts, sporoblasts, and mature spores are found in the midgut epithelial cells. Meronts, the earliest developmental stage, had two nuclei in diplokarytic arrangement and were bound by a plasma membrane in direct contact with host cytoplasm (Fig. 4). Sporonts were elongated and oval in shape with dense cytoplasm and no discernible internal structures (Fig. 5). Sporoblasts were generally smaller than sporonts with a more clearly defined cell wall and two nuclei (Fig. 4). Electron micrographs of longitudinal sections of mature spores showed that spore wall consisted of a dense exospore, 48-53 nm thick, and a lucent layer endospore and that the sporoplasm was enclosed by a plasma membrane (Fig. 6). The anchoring disc was located in the anterior pole of the spore. The lamellate polaroplast occupied the anterior part of the spore adjacent to the anchoring disc (Fig. 6). A vacuole was located in the posterior end of the spore and not prominent. Two nuclei in diplokaryotic arrangement were closely apposed in the central region of the spore between the polarplast and the posterior vacuole and the polar filaments were arranged in 18--21 isofilar coils in two rows (Fig. 7). When a spore had an extruded polar tube, the posterior vacuole swelled and became very prominent inside the spore (Fig. 8).
Fig. 4,5.
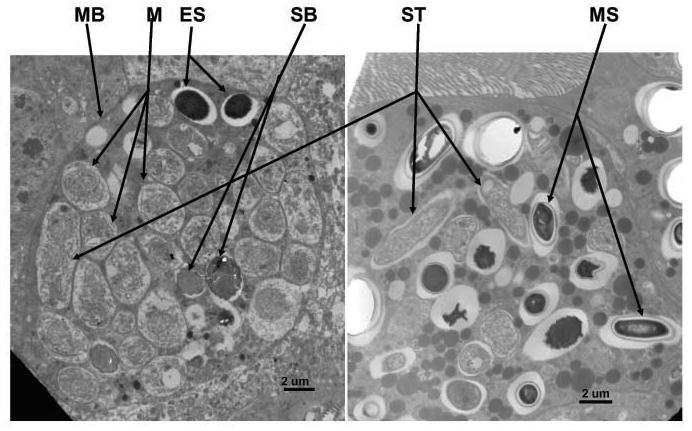
Epithelial cells infected with different developmental stages of Nosema ceranae. The developmental stages include meront (M), sporont (ST), sporoblast (SB), and mature spore (MS). MB = Membrane of the infected host cell. ES = Empty shell of the hatched spore.
Fig. 6--8.
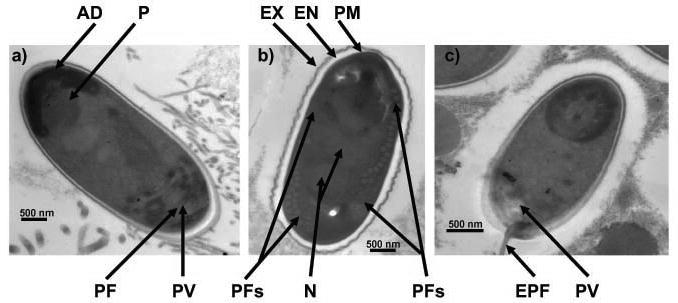
Electron-micrographs of longitudinal sections of spores of Nosema ceranae. 6. Micrograph showing anchoring disk (AD), polaroplast (P), posterior vacuole (PV), polar filament (PF). 7. Micrograph showing endospore (EN), exospore (EX), plasmamembrane (PM), nucleus (N), 20--22 isofilar coils of the polar filament (PFs). 8. A spore with an extruded polar filament (EPF). Note the more conspicuous PV.
The PCR assays revealed that N. ceranae-specific nucleic acid was detected in 100 % of the alimentary canals, Malpighian tubules, and hypopharyngeal glands, in 87% of the salivary glands, and in 20% of the fat bodies (N = 15). No N. ceranae-specific PCR signal was detected in the muscle tissue examined (Lane 5, Fig. 9).
Fig. 9.
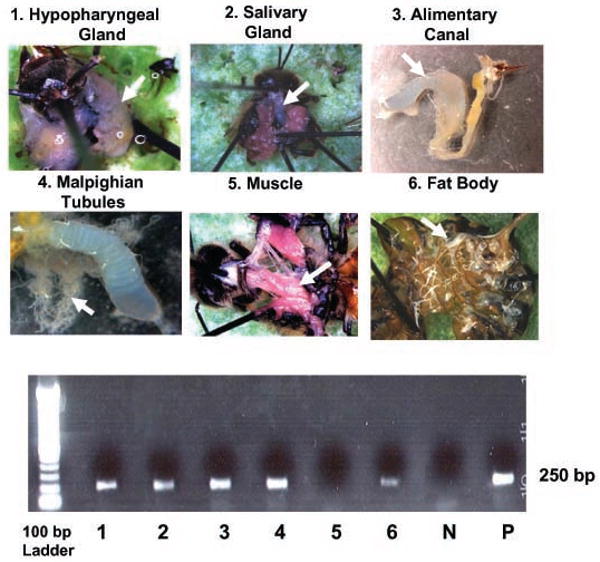
Detection of Nosema ceranae by PCR amplification of nucleic acids from different tissues. DNA was extracted from tissues and examined for the presence of N. ceranae-signal by PCR method and electrophoresis. The gel numbers 1--6 indicate the hypopharyngeal gland, salivary gland, alimentary canal, Malpighian tubules, muscle, and fat body, respectively; N indicates negative control and letter P indicates positive control. The size of PCR fragments is indicated on the right of the gel.
The complete DNA sequences of the rRNA gene is 4,475 bp. The G+C content of the SSUrRNA cistron at positions 1--1,259 was 36.46%. The ITS region consisted of a 39-bp sequence and was located between nucleotides 1260--1298. The DNA sequence of LSUrRNA, located at the 3′-end between nucleotide 1299-3828, contained 2,530 bp and was 32.86% G+C.
The percent of SSUrRNA sequence identity revealed that N. ceranae shared the highest degree of sequence identity (97.5%) with N. vespula and was the most distantly related to N. plutellae with 19.3% sequence divergence among all microsporidia included in this study. Our phylogenetic tree of 20 microsporidian taxa contains two distinct clades (Fig. 10). One clade includes Vairimorpha impecfecta and some species of the “true”Nosema group, a group of lepidopteran Nosema species closely related to Nosema bombycis (Baker et al., 1994). Nosema ceranae, along with several non-lepidoteran Nosema species and true Nosema species forms another clade (Fig. 10). Within this latter clade, N. ceranae is most closely related to N. vespula with 80% bootstrap support, and was distantly related to N. apis.
Fig. 10.
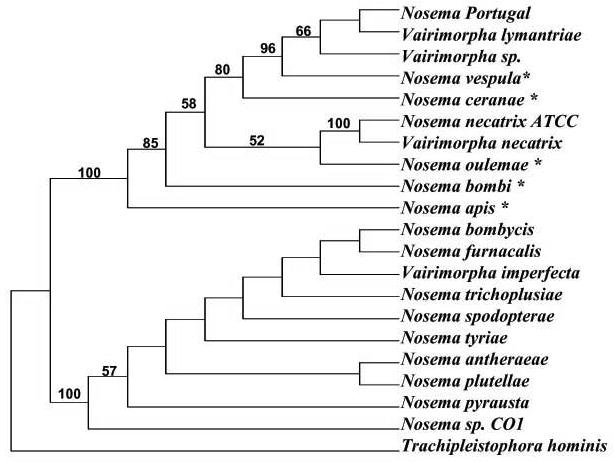
Phylogenetic tree of microsporidia. Phylogenetic tree of microsporidia infecting insects based on the sequences of the small subunit rRNA gene and constructed by Maximum Parsimony analysis under a heuristic search. Trachipleistophora hominis infecting Homo sapiens was used as an outgroup. The non-lepidopteran Nosema species are indicated by an asterisk. The reliability of the tree topology is shown by the bootstrap values located on the tree branches.
Discussion
The transfer of N. ceranae from its described original host, A. cerana, to a possible novel host, A. mellifera, adds a new dimension to the biological and epidemiological aspects of this parasite. Experimental infection of A. mellifera by N. ceranae conducted by Higes et al (2007) clearly showed that this parasite is highly pathogenic to its new host and poses a serious threat to the beekeeping industry
The morphological and molecular characterization of N. ceranae in Asian honey bees was conducted by Fries et al. in 1996. Later, Fries et al. reported (2006) the natural infection N. ceranae in European honey bees. However, many morphological details of spores such as types and sizes of spores in a dense spore purification and the morphology at the different developmental stages of spores in midgut epithelium cells in naturally infected hosts remained to be demonstrated. Our observation with light microscopy showed that spores of N. ceranae from European honey bees are oval shaped and rather uniform in shape. The electron microscopy indicates that N. ceranae contains all of the ultrastructural characteristics of the genus Nosema (Larsson 1986): diplokaryotic nuclei present in all developmental stages; a long flexible polar filament that appears in the mature spores; meronts, the earliest stages in the life cycle of the parasite, which are in direct contact with host cell cytoplasm; mature spores that are bounded by a thickened wall consisting of electron-dense exospore and electron-lucent endospore layers; and the thickness of exospore is 48-52 nm. The number of polar filament coils is an important taxonomic criterion to differentiate different species of Nosema (Burges et al., 1974). The number of coils of polar filament inside N. ceranae spores measured by us was 18--21, overlapping with the range of 20--23 coils reported by Fries et al. (1996), which is much smaller than the more than 30 coils recorded for N. apis (Fries, 1989; Liu, 1984).
Although not all examined tissues showed visible signs of pathological changes, PCR assay followed by sequencing analysis showed that N. ceranae-specific PCR signals are not restricted to the midgut tissue but spread to other tissues, including the Malpighian tubules, hypopharyngeal glands, salivary glands, and fat bodies. The presence of the signal suggests that these tissues may be infected, as was determined microscopically for Nosema bombi in a bumble bee (Fries et al., 2001). However, microscopic studies of N. ceranae in A. mellifera tissues, that would verify the infections, remain to be conducted. The detection of N. ceranae-specific PCR signals in both hypopharyngeal and salivary glands suggests that royal jelly, the secretion of hypopharyngeal and salivary glands of worker bees used to feed the queen and larvae, could be another vehicle for horizontal fecal-oral and food-borne transmission of the parasite in the bee colonies. A weak PCR signal specific for N. ceranae detected in the fat body tissue suggests a low parasite load, arguing that fat body tissue is not a primary target for N. ceranae infection even though fat body tissues are one of the primary sites for microsporidian infection. Infection of fat bodies causes formation of whitish cysts and the infected gut becomes swollen and whitish as a result of impaired fat metabolism in many other insects (Sokolova et al. 2006).
Although honey bee colonies with reduced longevity, decreased population size, higher autumn/winter colony loss, and/or reduced honey production are often reported to be associated with the presence of N. ceranae, the disease signs such as dysentery or crawling behavior or milky white coloration of gut, that are usually associated with N. apis infection, has never been described in N. ceranae infected bees (Fries et al. 2006). The absence of these disease symptoms in N. ceranae infected A. mellifera might reflect the absence or low intensity of N. ceranae specific. PCR signals in the muscles and fat bodies of infected bees, respectively. It is not clear why N. ceranae has different pathological effects on the host, A. mellifera compared to N. apis. Further studies are warranted to ascertain the pathogenesis of both parasites in the A. mellifera.]
The sequences of the rRNA operon have been widely used as a molecular marker for detection of microsporidian infection, differentiation of closely related species, and estimation of phylogenetic relationship among microsporidia. The organization of the rRNA gene of N. ceranae contains one SSUrRNA gene at the 5′ end, one LSUrRNA gene at 3′ end, and an internal transcribed spacer (ITS) located between the SSUrRNA and LSUrRNA genes. Parallel comparison of the rRNA gene sequences of N. ceranae and N. apis showed a sequence identity of 92.7% for SSUrRNA, 91.9 % for LSUrRNA, and 48.5% for ITS. Although N. apis and N. ceranae infect the same host and share similarities in sequences of rRNA gene, phylogenetic analysis based on sequences of SSUrRNA showed that N. apis is not the closest relative of N. ceranae. Within the same clade, N. ceranae appears to be more closely related to N. vespula, a parasite infecting wasps, with 80% bootstrap support. Nosema apis seems to have branched off earlier and is most closely linked to N. bombi, a parasite infecting bumble bees.
The comparative analysis of rRNA sequences indicated that ribosomal RNA is conserved and maintains a similar secondary and tertiary structure for all types of organisms (Gutell et al., 1986a, b). While the microsporidian rRNAs contain some of the characteristic features found in the vast majority of the eukaryotic rRNAs, the 16S-like and 23S-like rRNAs of N. ceranae are very unusual. They lack many of the structural elements present in other nuclear-encoded eukaryotic rRNAs, and they are significantly shorter in length. For example the Saccharomyces cerevisiae 16S-like and 23S-like rRNAs are approximately 1800 and 3550 nucleotides in length, the N. ceranae 16S-like and 23S-like rRNAs are 1259 and 2530 nucleotides in length, respectively. To determine how the reduction in size of rRNA contributes to the life cycle of the intracellular parasite in the host, further studies are needed.
As with many other new and emerging pathogens, we are just beginning to scratch the surface of understanding how N. ceranae adopt and establish infection in the new host. Genomic and biochemical characterizations of N. ceranae are currently in progress to study the roles of parasite genetic variability, host physiological conditions, and host immune status in the course of infection and disease.
Acknowledgments
We would like to thank Michele Hamilton, Bart Smith, and Andrew Ulsamer for their excellent technical assistance. The work was supported in part by the 2006 California State Beekeepers' Association (CSBA) research fund. R. Gutell and J. Lee were supported by the National Institutes of Health (GM067317) and the Welch Foundation (F-1427).
Footnotes
Publisher's Disclaimer: Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
References
- Bailey L, Ball BV. Honey bee pathology. 2nd Academic Press; London: 1991. [Google Scholar]
- Baker MD, Vossbrinck CR, Maddox JV, Undeen AH. Phylogenetic relationships among Vairimorpha and Nosema species (Microspora) based on ribosomal RNA sequences. J Invertebr Pathol. 1994;61:100–106. doi: 10.1006/jipa.1994.1077. [DOI] [PubMed] [Google Scholar]
- Becnel JJ, Andreadis TG. Microsporidia in insect. In: Wittner M, Weiss LM, editors. The microsporidia and Microsporidiosis. ASM Press; Washington, DC: 1999. pp. 447–501. [Google Scholar]
- Burges HD, Canning EU, Hulls JK. Ultrastructure of Nosema oryzaephili and the taxonomic value of the polar filament. J Invertebr Pathol. 1974;23:135–139. doi: 10.1016/0022-2011(74)90176-1. [DOI] [PubMed] [Google Scholar]
- Canning EU, Lom J, Dykova I. The Microsporidia of Vertebrates. Academic; New York: 1986. [Google Scholar]
- Chen YP, Evans JD, Smith JB, Pettis JS. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J Invertebr Pathol. 2008;97:186–188. doi: 10.1016/j.jip.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Cox-Foster DL, Conlan S, Holmes E, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, van Engelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- Franzen C, Muller A. Microsporidiosis: human diseases and diagnosis. Microbes Infect. 2001;3:389–400. doi: 10.1016/s1286-4579(01)01395-8. [DOI] [PubMed] [Google Scholar]
- Fries I. Observation on the development and transmission of Nosema apis Z. in the ventriculus of the honey bee. J Api Res. 1989;28:107–117. [Google Scholar]
- Fries I, Feng F, Silva AD, Slemenda SB, Pieniazek NJ. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae) Europ J Protistology. 1996;32:356–365. [Google Scholar]
- Fries I, Granados RR, Morse RA. Intracellular germination of spores Nosema apis Z. Apidologie. 1992;23:61–71. [Google Scholar]
- Fries I, Martín R, Meana A, García-Palencia P, Higes M. Natural infections of Nosema ceranae in European honey bees. J Api Res. 2006;45:230–233. [Google Scholar]
- Fries I, de Ruijter A, Paxton RJ, da Silva AJ, Slemeda SB, Pieniazek NJ. Molecular characterization of Nosema bombi (Microsporidia: Nosematidae) and a note on its sites of infection in Bombus terrestris (Hymenoptera: Apidae) J Api Res. 2001;40:91–96. [Google Scholar]
- de Graaf DC, Raes H, Sabbe G, de Rycke PH, Jacobs FJ. Early development of Nosema apis (Microspora : Nosematidae) in the midgut epithelium of the honey bee (Apis mellifera) J Invertebr Pathol. 1994;63:74–81. [Google Scholar]
- Gutell RR, Noller HF, Woese CR. Higher order structure in ribosomal RNA. EMBO J. 1986a;5:1111–3. doi: 10.1002/j.1460-2075.1986.tb04330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF. Comparative Anatomy of 16S-like ribosomal RNA. Prog Nuc Acid Res Mol Biol. 1986b;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hassanein MH. The influence of Nosema apis on the larval honeybee. Ann Appl Biol. 1953;38:844–846. [Google Scholar]
- Higes M, Garcίa-Palencίa P, Martίn-Hernάndez R, Meana A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia) J Inverteb Pathol. 2007;94:211–217. doi: 10.1016/j.jip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Higes M, Martín R, Meana A. Nosema ceranae, a new microsporidian parasite in honey bees in Europe. J Invert Pathol. 2006;92:93–95. doi: 10.1016/j.jip.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Huang WF, Jiang JH, Chen YW, Wang CH. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie. 2007;38:30–37. [Google Scholar]
- Klee J, Besana AM, Genersch E, Gisder S, Nanetti A, Tam DQ, Chinh TX, Puerta F, Ruz JM, Kryger P, Message D, Hatjina F, Korpela S, Fries I, Paxton RJ. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J Invertebr Pathol. 2007;96:1–10. doi: 10.1016/j.jip.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Larsson R. Ultrastructure, function, and classification of microsporidia. In: Corliss JD, Patterson DJ, editors. Progress in protistology. Vol. 1. Biopress; Bristol, England: 1986. pp. 325–390. [Google Scholar]
- Liu TP. Ultrastructure of the midgut of the worker honey Apis mellifera heavily infected with Nosema apis. J Invertebr Pathol. 1984;44:103–105. [Google Scholar]
- Malone LA, Giacon HA, Newton MR. Comparison of the responses of some New Zealand and Australian honey bees (Apis mellifera L) to Nosema apis Z. Apidologie. 1995;26:495–502. [Google Scholar]
- Nageli KW. Uber die neue Krankheit der Seidenraupe und verwandte Organismen. Bot Z. 1857;15:760–761. [Google Scholar]
- Rinderer TE, Sylvester HA. Variation in response to Nosema apis, longevity, and hoarding behavior in a free-mating population of the honey bee. Ann Entomol Soc Am. 1978;71:372–374. [Google Scholar]
- Sokolova YY, Kryukova NA, Glupov VV, Fuxa JR. Systenostrema alba Larsson 1988 (Microsporidia, Thelohaniidae) in the Dragonfly Aeshna viridis (Odonata, Aeshnidae) from South Siberia: Morphology and Molecular Characterization. J Euk Microb. 2006;53:49–57. doi: 10.1111/j.1550-7408.2005.00075.x. [DOI] [PubMed] [Google Scholar]
- Sprague V. Characterization and composition of the genus Nosema. Misc Publ Entomol Soc Am. 1978;11:5–16. [Google Scholar]
- Sprague V, Vavra J. Systematics of the microsporidia. In: Bulla LA, Cheng TC, editors. Comparative Pathobiology. Plenum; New York: 1977. [Google Scholar]
- Wasson K, Peper RL. Mammalian microsporidiosis. Vet Pathol. 2000;37:113–128. doi: 10.1354/vp.37-2-113. [DOI] [PubMed] [Google Scholar]
- Wittner M, Weiss LM. The Microsporidia and Microsporidosis. ASM Press; Washington, DC: 1999. p. 553. [Google Scholar]


