Abstract
Colloidal quantum dots (QDs) have a large fraction of their atoms arrayed on their surfaces and are capped with bifunctional ligands, which make their photoluminescence highly sensitive to potential charge transfer to, or from, the surrounding environment. In this report, we used peptides as bridges between CdSe-ZnS QDs and metal complexes to promote charge transfer between the metal complexes and QDs. We found that quenching of the QD emission is highly dependent on the relative position of the oxidation levels of QDs and metal complex used; it also traces the number of metal complexes brought in close proximity of the nanocrystal surface. In addition, partial bleaching of the absorption was measured for the QD-metal complex assemblies. These proximity driven interactions were further used to construct sensing assemblies to detect proteolytic enzyme activity.
Keywords: quantum dot, metal complex, electrochemistry, charge transfer, self-assembly, peptide, biosensing, enzyme
Introduction
Semiconductor nanocrystals or quantum dots (QDs) with sizes smaller than the Bohr exciton radius of the constituent bulk material have often been described as “artificial atoms,” due to the appearance of discrete energy states in both the conduction and valence bands. This is a direct result of carrier (electron and hole) confinement within the physical dimensions of the nanocrystals 1-4. They are, however, far more complex than discrete atoms and are affected by a variety of parameters including defects in the crystal structure and the interface with the surrounding medium. Colloidal QDs, in particular, have a large fraction of their atoms arrayed on their surfaces and are always capped with bifunctional ligands, which provide surface passivation and promote compatibility with the surrounding medium (e.g., dispersion in solutions and polymer matrices)2, 5, 6. These ligands along with the surrounding matrix affect the overall optical and electronic properties of the nanocrystals, due to the presence of defect surface states, often attributed to dangling bonds and incomplete passivation1, 2, 7, 8. QDs are also very sensitive to the presence of additional charges (electrons or holes) either on their surfaces or in the surrounding environment, which can alter both the nanocrystal photoluminescence (PL) and absorption alike 9-12.
As a result, bringing redox-active complexes in close proximity to the QDs may promote transfer of external electrons (and holes) to either (i) the QD core conduction (valence) band or (ii) the QD surface states 9, 12. The presence of additional charges can lead to quenching of the QD PL due to an efficient Auger recombination 8, 13. The rate of PL loss depends on the location of the added charge, with a complete quenching measured for charge(s) residing inside the QD core, due to strong spatial overlap between charge(s) and exciton, while partial quenching is measured for charge(s) residing on the nanocrystal surface (due to weaker overlap with the exciton) 8, 9, 12, 13. These unique characteristics have generated intense interest in these materials, motivated by the potential use in developing devices, such as lasers, photovoltaic cells and light emitting diodes (LEDs) 1, 4, 12, 14-18.
Since the development of hydrophilic QDs and the advent of successful techniques to interface them with biological systems 19-21, there has been a strong desire to exploit the interactions of QDs with redox-active complexes for monitoring specific biological events 11, 22, 23. A few preliminary studies have reported the coupling of QDs and redox-active complexes and their use to identify specific biological processes. For example, in one study Niemeyer’s group reported that photoexcitation of core-only CdS QDs conjugated with cytochrome P450 produced free radicals that activated the P450 enzyme and catalyzed the oxygenation of fatty acids 24. In another study, Clarke and co-workers reported that QD-dopamine conjugates could label cells in redox-sensitive patterns 25. They reported that under reducing conditions the QD fluorescence is detected only in the cell periphery and lysosomes, whereas under more oxidizing conditions the QD emission is limited to the perinuclear region. They used these findings to suggest that phototoxicity results from the creation of singlet oxygen and this can be reduced with antioxidants25. Charge-transfer between QDs and proximal metal complexes has also been exploited as a potential sensing mechanism. For instance, CdSe core-only QDs self-assembled with maltose binding protein, site-specifically labeled with a ruthenium complex (Ru), were used to detect the sugar maltose from variations in the QD PL in the presence of maltose 26. The authors attributed the loss in QD PL to changes in the QD-Ru complex separation distance, caused by alteration in the protein conformation following interactions with maltose 26. In a similar conjugate configuration, a fatty acid binding protein was labeled with this Ru complex and attached to CdSe-ZnS core-shell QDs, but in this case changes in QD PL upon ligand binding were ascribed to changes in the solvation conditions 27. Overall, only a partial understanding of these systems exists and there is a strong need for additional studies where rational design of QD-redox assemblies with control over architecture and redox levels can provide insight into the underlying mechanisms.
In this report, we evaluate the effects of three different redox-active compounds with distinct oxidation potentials, namely ruthenium phenanthroline (Ru-phen), ruthenium bypyridine phenanthroline (Ru-bpy-phen), and ferrocene (Fc) metal complexes on the spectroscopic properties of hydrophilic CdSe-ZnS QDs. Peptides engineered with polyhistidine tracts were site-specifically labeled with these compounds and self-assembled onto the surface of CdSe-ZnS QDs functionalized with either charged or neutral surface-capping ligands (Figure 1). The QD-peptide-redox-complex conjugates were characterized using optical absorption along with steady-state and time-resolved fluorescence measurements. In addition, the free QDs, free metal-complexes and metal-complex-labeled peptides were characterized using cyclic voltammetry. We found that Ru-phen complexes induced a significant loss in the QD PL, with quenching rates dependent on the ruthenium complex-to-QD ratio; it also changed the absorption properties of the nanocrystals. In contrast, Fc and Ru-bpy-phen complexes induced little to no quenching of QD PL and had no effects on the absorption spectra. We attribute these results to differences in the match between the oxidation levels of the metal complex and QDs, with the Ru-phen-complex providing a more favorable energy configuration (thus higher charge-transfer and concomitant PL quenching of the nanocrystals) than Fc and Ru-bpy-phen complexes. We further demonstrate the utility of these interactions by applying QD-peptide-metal complex conjugates as substrates for the specific detection of proteolytic enzyme activity in biological assays.
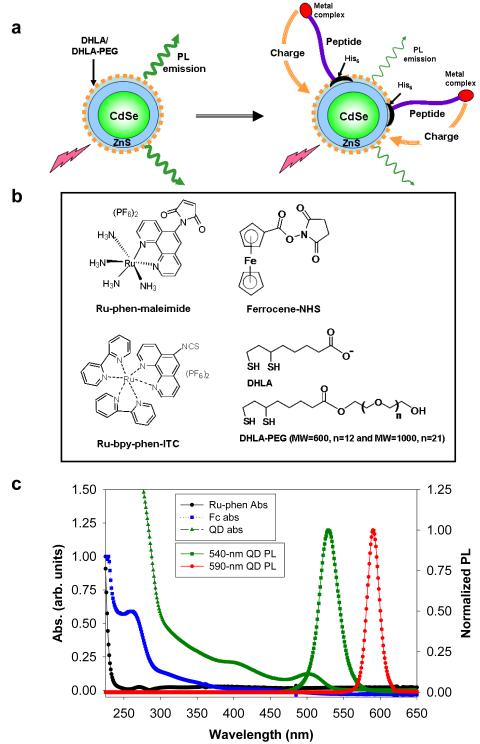
Figure 1. Schematic, chemical structure and optical spectra.
(a) Schematic representation of the charge transfer induced quenching of the QD PL. His-tagged peptides are labeled with redox-active metal complexes and self-assembled on CdSe-ZnS QDs. Due to proximity and redox interactions, electrons can be transferred to the QD resulting in quenching of its PL. The peptide has alpha helical central section, a His tract on one terminal and a reactive group on the other end. (b) Chemical structures of ruthenium-phenanthroline maleimide (Ru-phen), ferrocene-NHS (Fc), ruthenium bypyridine phenanthroline isothiocyanate (Ru-bpy-phen-ITC), DHLA and DHLA-PEG ligands. (c) Absorption of 540-nm emitting QDs and Ru-phen/Fc-labeled peptides along with emission of 540-nm and 590-nm emitting QDs.
Results
Quantum Dots, Ligands, and Conjugate Self-Assembly
We used CdSe-ZnS QDs made hydrophilic via cap exchange of the native trioctyl phosphine and trioctyl phosphine oxide (TOP/TOPO) mixture with either dihydrolipoic acid (negatively-charged) or polyethylene glycol-terminated dihydrolipoic acid (neutral) ligands (DHLA-QDs or DHLA-PEG-QDs, respectively, see Figure 1); 21, 28 PEG of molecular weight 600 was used unless otherwise noted. To bring QDs in close proximity with the metal complexes we used peptide bridges in combination with metal-affinity driven self-assembly to form QD-metal complex conjugates. We have previously shown that self-assembly of polyhistidine-terminated peptides onto DHLA- and DHLA-PEG-capped CdSe-ZnS QDs alike was effective and rapid, as discussed in reference 29. This high-affinity conjugation strategy is simple and allows control over conjugate valence (i.e. number of proteins/peptides per QD) 6, 29, 30. A generic peptide sequence expressing a unique terminal cysteine residue was labeled with the Ru-phen-maleimide complex, while the Ru-bpy-phen-isothiocyanate or the Fc-succinimidyl ester were used to label the single terminal primary amine on a glutathione-modified peptide. These will be referred to as Ru-phen, Ru-bpy-phen- and Fc-labeled peptides unless otherwise noted. Both peptides expressed hexa-histidine (His6) tags, at the opposite end of the metal complex labels, to promote self-assembly on the nanocrystals. Additional details on the peptide sequences, their site-specific labeling along with QD immobilization on electrodes and conjugate self-assembly are described in the experimental section.
Cyclic Voltammetry and Determination of the Formal Potentials
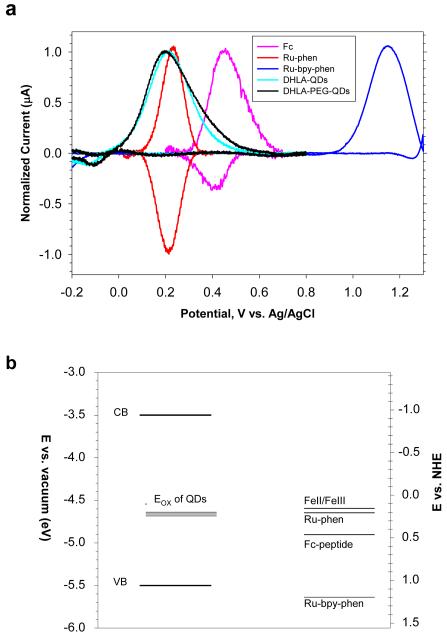
We began by evaluating the redox levels of the QDs and metal complexes used. Figure 2 shows the background-corrected cyclic voltammograms (CVs) collected from single layers of unconjugated QDs, Ru-phen-labeled, Ru-bpy-phen-labeled and Fc-labeled peptides immobilized on ITO electrodes and immersed in phosphate buffered saline (PBS) solution; raw data are provided in Figure S2 (Supporting Information). Both free Ru-phen and Ru-phen-peptide exhibited reversible CV curves with a formal potential centered at 0.23V vs. Ag/AgCl reference, indicating that after coupling the peptide did not change the intrinsic redox properties of the Ru-phen complex. In comparison, samples of Fc-peptides showed a formal potential centered at 0.440V, which is shifted by ~0.15V relative to that measured for free Fc-complex (0.325V, see Table S-II, Supporting Information), whereas Ru-bpy-phen-peptide showed a formal potential centered at 1.16 V (which is slightly shifted from the one measured for the free complex, 1.32 V, see Table S-II, Supporting Information). Both Fc- and Ru-bpy-phen-peptide samples showed a strong current peak in the forward (oxidation) scan but a rather weak peak in the reverse (reduction) scan, indicating that the CV curves are not fully reversible as was the case of Ru-phen. Data in Figure 2 also show that for all QD samples tested, the CV’s exhibited a single irreversible broad peak centered between 0.2 and 0.24 V; the exact location of this peak seems to slightly vary depending on the size and the type of ligand used (charged DHLA or neutral DHLA-PEG). A list of anodic peak potentials recorded for the QDs, and formal potentials for Fc-, Ru-bpy-phen- and Ru-phen-complex samples along with the formal potentials of some additional ruthenium and ferrocene complexes is provided in Tables S-I,II (Supporting Information). In addition, we found that for QD samples the current vs. voltage profiles could be both enhanced and regenerated by maintaining the voltage of the working electrode at -0.2 V (versus Ag/AgCl reference) for ~5 minutes. We attribute the measured current peak to oxidation of the QDs (as done for metal complexes), i.e., transfer of an electron from an occupied electronic state in the nanocrystals to the Fermi level of the ITO electrode. The oxidation potentials show little variation from one sample to another for the set of water soluble QDs used in this study. The CV curves were limited to applied voltages ≥ -0.5 V as experiments were carried out using buffered aqueous media (rich in counterion); it was thus not possible to sweep the potential to much lower values. A comparison between our data and previous results, reported by Nann and co-workers, suggests that the QD oxidation state corresponds to surface defect states (which can be constituted by atomic vacancies or oxygen adsorption)31. Positions of the lowest (highest) energy level in the QD conduction (valence) band, extracted from previously reported measurements on CdSe nanocrystals emitting at ~590 nm are displayed in comparison (Figure 2)31, 32. We should also emphasize that our experiments were carried out in buffer solutions where electrolytic conduction is rather high compared to organic electrolytic solutions. The CV data for the QDs sample show rather small currents. These observations combined indicate that Ru-phen-complex with its lower oxidation potential could promote charge transfer from the ruthenium center to the QD when assembled in QD-peptide-Ru-phen conjugates. In contrast, ferrocene and Ru-bpy-phen present higher oxidation potentials (than Ru-phen) compared to the nanocrystals, and offer a less favorable configuration for charge-transfer in these conjugates.
Figure 2. Cyclic voltammetry.
(a) Superimposed and background-corrected linear scan cyclic-voltammograms collected from immobilized 590-nm emitting QDs capped with DHLA or DHLA-PEG, Ru-phen-, Ru-bpy-phen- and Fc-labeled peptides. (b) Positions of the energy levels corresponding to the QD oxidation peak (Eox) and the Fc-, Ru-bpy-phen- and Ru-phen-labeled peptide oxidation potentials along with that of FeII/FeIII compound. Representative positions of CdSe QD conduction band (CB) and valence band (VB) were extracted from references 31 and 32; the location of both bands slightly vary with the nanocrystal size as a result of quantum confinement effects.
Quenching of the Quantum Dot Photoluminescence
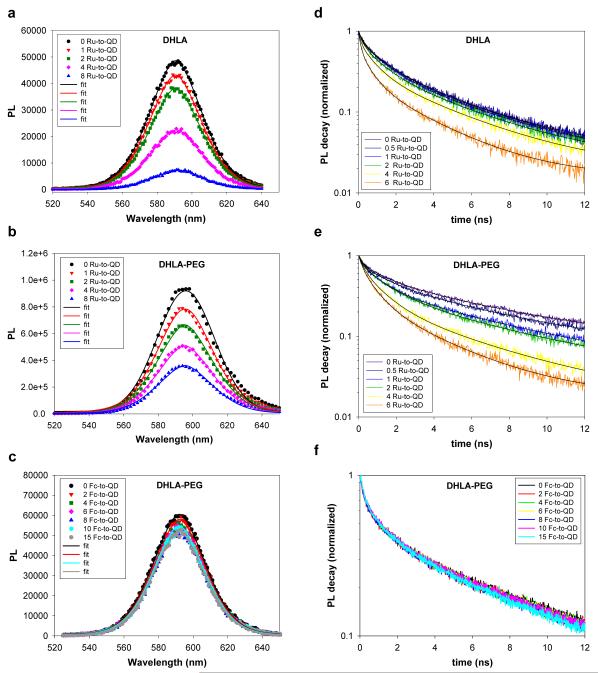
We next examined the effects of varying the metal complex oxidation potential (using Ru-phen-, Fc- or Ru-bpy-phen-labeled peptides) on the QD photoemission using both steady-state and time-resolved fluorescence experiments. Effects of varying the number of metal complexes arrayed around a single QD (afforded by our centro-symmetric conjugates) were also examined. Given the difference between the oxidation levels of the metal complexes (see Figure 2) our experiments were designed to: 1) allow a side-by-side comparison of the effects of mismatch in the oxidation potentials on the exciton recombination and PL of the QDs; and 2) investigate the effects of QD surface charges on the interactions with proximal metal complexes through the use of QDs capped with either negatively charged DHLA or neutral DHLA-PEG ligands. Figures 3a-c show the typical progression of the PL spectra versus an increasing average number of Ru-phen-peptides per QD, collected using either DHLA- or DHLA-PEG-capped nanocrystals, together with those from solutions of DHLA-PEG-QDs self-assembled with Fc-peptides. Figures 3d-f show the corresponding time-resolved PL decays. Data clearly indicate that assembling an increasing number of Ru-labeled peptides on the nanocrystals produced a significant and progressive loss of PL coupled with shortening of the exciton lifetime, for both DHLA- and DHLA-PEG-QDs. In comparison, conjugation of QDs (capped with either ligand) to Fc-labeled or Ru-bpy-phen-labeled peptides produced a very small loss in the PL signal (< 20% loss) and essentially no change in the exciton lifetime (Figure 3 and Figure S1). No loss in QD PL intensity and no change in the QD exciton lifetime were measured for nanocrystals self-assembled with unlabeled peptides in all cases (data not shown).
Figure 3. Steady-state and time resolved fluorescence.
PL spectra collected from solutions of QD-peptide-metal-complex conjugates at increasing metal-to-QD ratio, n: (a) 590-nm DHLA-QDs with Ru-phen-labeled-peptide, (b) 590-nm DHLA-PEG-QDs with Ru-phen-labeled-peptide, and (c) 590-nm DHLA-PEG-QDs with Fc-labeled peptide. (d-f) Time-resolved PL decay corresponding to the ensemble data shown in a-c. Ru-phen is referred to as Ru in the legends.
Steady-state fluorescence experiments on control samples consisting of QDs mixed with free Ru-phen-maleimide complex showed different behaviors depending on whether DHLA- or DHLA-PEG-QDs were used (see Figure S3, Supporting Information). Mixing neutral DHLA-PEG-QDs with free Ru-phen-complex resulted in a PL loss much smaller than that measured for QDs conjugated to the corresponding Ru-phen-peptidyl complex. In comparison, PL losses were essentially identical for solutions of charged DHLA-QDs mixed with either free Ru-phen or Ru-phen-peptide complexes. Mixing free Fc-complex or Ru-bpy-phen-complex with either type of QDs had no effect on the PL (data not shown). We attribute the rather pronounced quenching measured for solutions prepared using DHLA-QDs and free Ru-phen to electrostatic attractions between the charged carboxyl groups on the QDs and the Ru-phen-complex. DHLA-capped QDs present multiple carboxyl groups (negatively charged) while each Ru-phen-complex has a net +2 charge, which can promote strong electrostatic coupling in solution. Indeed, we found that addition of 0.5 M NaCl to a solution of DHLA-QDs and Ru-phen-complex prevented conjugate formation, confirming the electrostatic nature of the observed binding (data not shown). We should also emphasize that no aggregation was observed for any of the solutions studied (made with nanocrystals capped with either set of surface ligands). Based on this, we conclude that the interactions between Ru-phen-complex and nanocrystals are specifically due to conjugation with the bridging peptides for the neutral DHLA-PEG-QDs. In contrast, the interactions in solutions of Ru-phen-peptides and DHLA-QDs may be caused by a combination of peptide-His6-QD binding and direct electrostatic attractions. Even though the interactions for DHLA-QD samples may occur without the presence of the peptide, they are clearly proximity-driven (i.e., due to assembly of QD-redox-complex). Consequently, the subsequent data for samples using both sets of surface ligands are analyzed in the context of a potential difference in the mechanism of charge transfer between the two sets of nanocrystals.
Figure 4a,b shows plots of the quenching efficiency η versus the average ratio of Ru-phen-complex per QD, n, derived from steady-state fluorescence using:
| (1) |
and from time-resolved fluorescence data using the expression:
| (2) |
where PLn and PL0, respectively, designate the PL intensity measured for solutions of QD conjugates with and without an increasing number of Ru-phen-complexes; kn and k0 are the corresponding decay rates33. Because the interaction of a QD with an individual proximal metal complex is independent from all other complexes arrayed around its surface (independent charge-transfer channels), the dependence of the quenching efficiencies vs. ratio n is fit using:
| (3) |
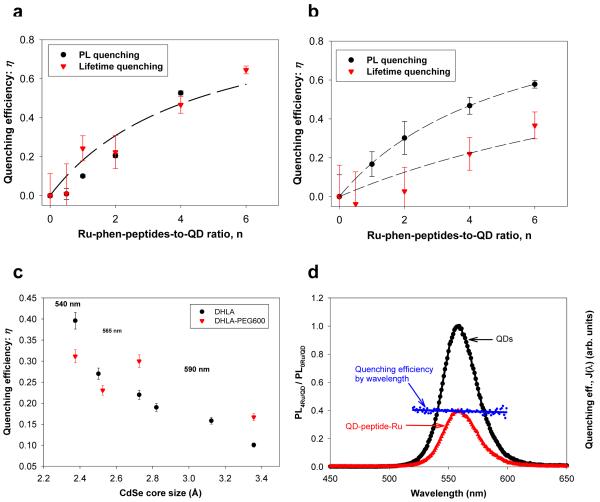
The dependence of η vs. n is characteristic of a homogeneous quenching of QD emission, and was demonstrated for non-radiative energy transfer (driven by Förster dipole-dipole interactions) between a central QD and n dyes arrayed around its surface 34. K accounts for effects of the center-to-center separation distance, r, and is a constant for the self-assembled conjugates discussed here (r is the same for all complexes within a conjugate)34-37. The data show that the efficiencies measured using steady-state fluorescence are comparable to those derived from changes in the lifetime decays for charged DHLA-QDs. However, the values measured using time-resolved fluorescence are consistently smaller than those extracted from steady-state measurements for DHLA-PEG-QDs (further discussed below). To complement the above findings, we tested the effects of core size, ZnS-overcoating and potential spectral overlap on the rate of QD PL quenching.
Figure 4. Quenching efficiencies.
Comparison of the quenching efficiencies derived from steady-state PL and time-resolved excited-state lifetime data for: (a) 590-nm DHLA-QDs and (b) 590-nm DHLA-PEG-QDs samples shown in Figure 3. (c) Quenching efficiency versus CdSe core sizes for DHLA- and DHLA-PEG-capped QDs normalized to 1 Ru-phen-peptide per QD. (d) PL of a QD solution without (black) and with Ru-phen-peptide conjugates (red), together with a superposition of the rate of quenching and spectral overlap, J(λ)~ε(λ)×λ4 versus wavelength (as detailed in the text).
Dependence of the Photoluminescence Quenching on the QD Core Size
We tested the effects of varying the CdSe core size on the PL quenching efficiencies of proximal Ru-phen-complexes. Figure 4c shows a plot of the PL quenching efficiency measured for a set of QDs with increasing core size capped with either DHLA or DHLA-PEG and assembled with Ru-phen-labeled peptides; efficiencies were extracted from steady-state fluorescence experiments and are all normalized to a Ru-phen-complex-to-QD ratio of 1 (n = 1). Data show that rate of PL loss progressively decreases with increasing core radius for both types of surface ligands, even though there is more scattering in the experimental values for QDs capped with neutral PEG ligands. This indicates that the charge transfer and quenching of the PL are more pronounced for nanocrystals with smaller core size, regardless of the type of surface ligands used.
Effects of ZnS-overcoating
To explore the potential effects of the ZnS-overcoating shell on the QD PL quenching due to proximal Ru-phen-complexes, we compared the relative PL loss of samples made with CdSe (core only) QDs side-by-side with those prepared using CdSe-ZnS nanocrystals (both sets are DHLA-capped). Overall, the relative PL quenching efficiency measured for CdSe QDs were slightly higher than those measured for core-shell nanocrystals (see Figure S3, Supporting Information). This result somewhat agrees with prior observations using QDs mixed with molecular scale hole scavengers and dispersed in organic solutions 38. We should also mention that because the PL emission from hydrophilic core-only QDs is very weak (low PL yield), the measured intensities are subject to rather low signal-to-noise ratios.
Distinction between Charge Transfer- and FRET-induced Quenching
We compare the PL behavior of the present assemblies to that observed using QD-protein-dye and QD-peptide-dye conjugates, where PL quenching results from Förster resonance energy transfer (FRET) and directly depends on J(λ), the spectral overlap between QD PL and dye absorption 34-37. We have shown that because the PL spectrum of a macroscopic QD sample is a superposition of several narrow single QD PL spectra (of different sizes), there is an inhomogeneous donor-acceptor spectral overlap across the PL spectrum of a homogeneous QD sample: J(λ)~ε(λ)×λ4 (with ε(λ) being the acceptor extinction coefficient spectrum). This produces an “inhomogeneous” wavelength-dependent rate of FRET between individual QDs and proximal dyes, and can result in either a blue shift of the QD PL spectrum measured for dye acceptors having an absorption that overlaps with the blue region of the QD emission, or a red shift for dyes with absorption coinciding with the red portion of the QD PL 36. Ru-phen-complex with its weak absorption (ε = 5000 M-1cm-1 at 490 nm, far from the QD emissions) is not expected to produce efficient FRET interactions with the QDs. We thus examined the wavelength-dependent quenching of QD PL as an additional tool to distinguish the interactions between QDs and metal complexes from those due to FRET in QD-dye pairs. Figure 4d shows the PL spectra of a typical QD sample before and after assembly with the peptide-Ru-phen-complex, together with a plot of the experimental wavelength-dependent overlap J(λ). There is no deformation in the QD PL spectrum after quenching due to interactions with the Ru-phen-complex. This shows that the quenching efficiency is essentially constant throughout the full QD emission spectrum, and that wavelength dependence of the quenching mechanism is negligible. These observations are consistent with the expected nature of the interactions and confirm that the measured quenching for each QD-peptide-Ru-phen-complex sample cannot be attributed to Förster dipole-dipole coupling as was the case with QD-dye assemblies 34, 37.
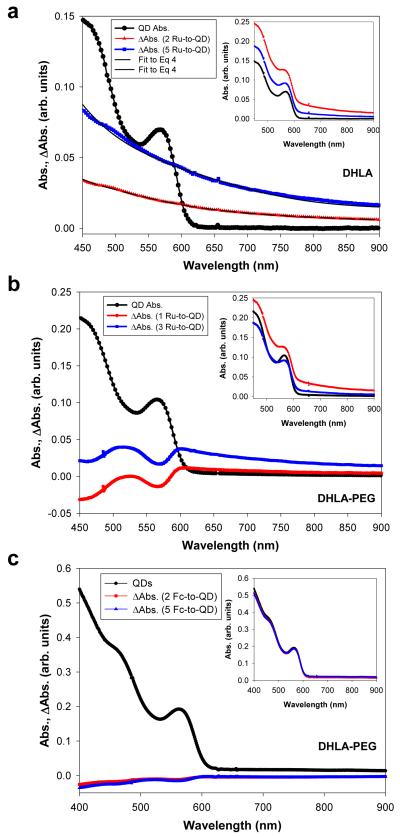
Changes in the QD absorption spectra
Additional insights into the mechanism of redox interactions can be gained from examining changes in the QD absorption spectra following coupling with metal complexes. When assembled with Ru-phen-complex, changes in the QD absorption spectra showed two distinct behaviors, depending on whether QDs were capped with DHLA or DHLA-PEG ligands (Figure 5a,b). Partial bleaching of the first and second absorption (exciton) peaks together with an additional long red tail contribution to the absorption spectrum were measured for DHLA-PEG-QDs. In comparison, only a pronounced background contribution to the absorption spectra was measured for DHLA-capped QDs. The amplitude of the absorption tail increases progressively with increasing number of Ru-phen-complex per QD-conjugate, and the absorption increment (extracted from the broad red tail in Figure 5a) could be fitted to the expression 9:
| (4) |
where E and Eg, respectively, designate the excitation and bandgap energies; λ and λg are the corresponding wavelengths. A broad red tail in ΔAbs with a power dependence on the energy difference as in Eq. 4 has been attributed to electrons populating the QD surface states 9. We should emphasize that no aggregation of the QD-peptide-Ru-phen was observed, indicating that the absorption red tail cannot be attributed to light scattering by potential small aggregates. In comparison, there are essentially no changes in the absorption features of the nanocrystals assembled with Fc-labeled peptides and unlabeled peptides (Figure 5c and Figure S4), regardless of whether DHLA-PEG or DHLA ligands were used; only data for DHLA-PEG-QDs are shown in Figure 5c. Also, no changes in the QD absorption spectra were observed for QDs mixed with Ru-bpy-phen-peptide (data not shown).
Figure 5. Effects of redox coupling on the QD absorption.
Plots of the QD absorption together with differential absorption (ΔAbs) spectra for: DHLA-QDs (a), DHLA-PEG-QDs self-assembled with increasing average number of either Ru-phen-peptides per QD (b), or Fc-labeled peptides (c); 590-nm emitting QDs were used. Absorption spectra are shown in the insets.
Proteolytic Assays
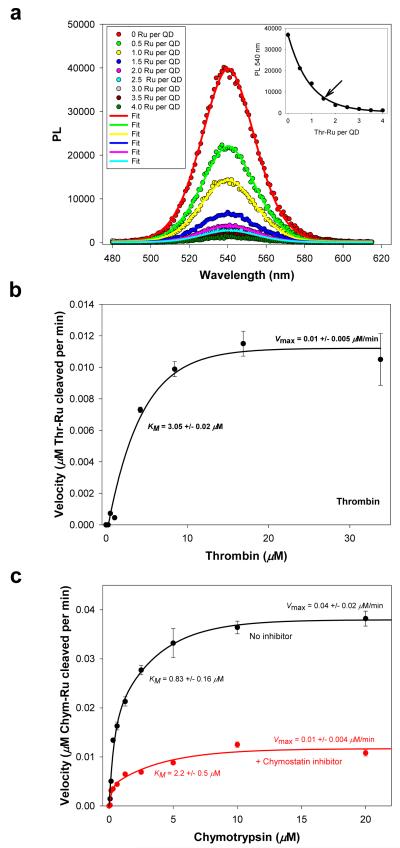
We exploited the proximity-dependent charge-transfer interactions (and the induced PL quenching) to implement the use of QD-peptide-Ru-phen conjugates as specific biosensing assemblies to monitor the activity of proteolytic enzymes. We previously demonstrated the use of self-assembled QD-peptide-dye conjugates as substrates for the FRET-based monitoring of proteolytic activity 37. Enzyme-driven cleavage of the peptides altered the QD-dye FRET signature, and allowed quantitative monitoring of the enzyme activity 37. Since Ru-phen-quenching of the QDs is proximity-driven and the magnitude of the quenching is also dependent on the ratio of Ru-phen-complex to QD (similar to what was observed for the FRET sensor), we reasoned that proteolytic cleavage of the QD-peptide-Ru-phen should also alter the separation distance and concomitant quenching, thus allowing the monitoring of enzymatic activity (see schematic in Figure S5, Supporting Information). Peptides with sequences specifically recognized by the peptidases thrombin (cleaves Arg-Gly) or chymotrypsin (cleaves C-terminal to aromatic or residues with large side-chains) were labeled with Ru-phen-maleimide, and referred to as Thr-Ru and Chym-Ru peptides, respectively.
We first characterized the quenching efficiencies of the substrates by self-assembling an increasing number of Thr-Ru peptides onto DHLA-PEG1000-QDs emitting at 540 nm, while Chym-Ru peptides were assembled onto 590-nm DHLA-PEG600-QDs (Figure S4, Supporting Information); two QD sizes and two peptide sequences were used. The PL quenching versus number of labeled-peptides per QD were fitted to an expression similar to Eq. 3 (PLn ~ PL0/[1-η]) and used to provide calibration curves (see inset in Figure 6a). Data show higher quenching efficiency at low Thr-peptide-to-QD ratios (n<3) for conjugates using the smaller 540-nm emitting QDs. This contrasts with the need for ~8 peptides to achieve a similar quenching efficiency when using the larger 590-nm emitting QDs (Figure S5), even though the Chym-peptide sequence used was shorter than the Thr-peptide. The more pronounced quenching measured for the QD-Thr-Ru conjugates may reflect a more effective charge-induced PL quenching efficiency for the smaller 540-nm emitting nanocrystals, as shown in Figure 4c. We should stress that the quenching data for the 590-nm QD-Chym-Ru conjugates are consistent with what was observed for the same QDs assembled with generic Ru-peptides (compare Figure 3 and Figure S4, Supporting Information); both peptides have similar lengths.
Figure 6. Proteolytic assay.
(a) Quenching of QDs following self-assembly with an increasing Thr-Ru-peptide-to-QD ratio; 540-nm emitting QDs were used. Gaussian fits of some of the PL spectra are also shown. Inset shows the PL at the peak value vs. Thr-Ru number, which served as a calibration curve. Arrow marks the ratio used as proteolytic substrate. (b) Velocity vs. thrombin concentration. (c) Velocity vs. chymotrypsin concentration in the absence and in the presence of a chymostatin inhibitor. The changes in kinetics parameters (higher KM and lower Vmax) are characteristic of a mixed inhibition process; data analysis within a mixed inhibition model provides a value for the inhibitor dissociation constant Ki of ~ 225 μM for Chymostatin37.
For the proteolytic assay, we chose an average of ~1.5 Thr-Ru-peptide per QD conjugate and ~4 Chym-Ru-peptide per QD conjugate as substrates. These ratios allow substantial initial PL loss for both assays (~80% loss for thrombin substrates and ~70% loss for chymotrypsin substrates), and potentially a large change in PL recovery following proteolysis (i.e., large dynamic range of QD PL recovery) 37. For each assay, the QD-Ru-peptide substrates were exposed to increasing concentrations of enzyme (excess enzyme conditions) in buffer for ~10 minute incubations, followed by collection of the QD PL. The increases in QD PL resulting from proteolysis were converted into enzymatic velocity by correlating quenching efficiencies to the number of intact QD-Ru-peptide conjugates per QD (using the standard/calibration curves shown in Figure 6a and Figure S5). In addition, using QD-Chym-peptide-Ru conjugates we assayed the proteolytic activity of chymotrypsin in the presence of a known inhibitor, chymostatin. The velocity data shown in Figure 6b-c were fit using standard Michaelis-Menten equations (see Supporting Information). Values derived for the Michaelis constant KM, maximum velocity Vmax, turnover number kcat, and kcat/KM ratio are shown in Table I. Importantly, these values are overall comparable to those extracted from FRET measurements using QD-peptide-dye assemblies 37. FRET-based sensing requires substantial spectral overlap between QD PL and dye absorption, which can limit its effectiveness to specific donor-acceptor pairs. In contrast, sensing based on charge transfer could be applied to a series of QDs emitting throughout the optical spectrum. As shown above, the effectiveness of interactions with a given proximal metal complex having a favorable oxidation potential can span a broad range of QD size/emission.
Table I. Enzyme Kinetic Parameters.
| Enzyme | Km | Vmax | kcat1 | kcat/Km |
|---|---|---|---|---|
| Thrombin | 3.05 ± 0.02 μM | 0.01 ± 0.9 μM/min | 0.03 min-1 | 1×106 M-1 min-1 |
| Chymotrypsin | 0.83 ± 0.16 μM | 0.04 ± 0.02 μM/min | 0.04 min-1 | 5×106 M-1 min-1 |
| Chymotrypsin + inhibitor | 2.2 ± 0.5 μM | 0.01 ± 0.004 μM/min | 0.01 min-1 | 1×106 M-1 min-1 |
kcat = Vmax/Stotal
Discussion
There are four main findings that can be extracted from the above set of data. 1-Bringing ruthenium phenanthroline (Ru-phen) complexes in close proximity to the QDs using a peptide bridge alters the spectroscopic properties of the nanocrystals. 2-The measured loss in PL directly traces the number of Ru-phen-complexes attached to a QD, and is verified by both steady-state and time-resolved fluorescence experiments (Figures 3 and 4). 3-Effects of Ru-phen-complex on the QD absorption manifest in a pronounced background contribution to the spectra for both sets of ligands, but additional partial bleaching of the first and second excitonic peaks are observed only for DHLA-PEG-QDs. 4-Due to absence of any effective spectral overlap between the QDs and Ru-phen-complex, the observed PL loss cannot be interpreted within the framework of Förster dipole-dipole coupling (Figures 1c and 4d).
We attribute these results to charge transfer between Ru-phen-complexes and primarily the surface energy states of the QD, independently of the QD photo-excitation. This introduces an additional non-radiative pathway for the QD exciton recombination, and reduces its photoemission. These effects are much more pronounced in the presence of Ru-phen-complexes than for either Fc- or Ru-bpy-phen-complexes, due to a more favorable match between the oxidation level(s) of the QDs and Ru-phen-complex, as anticipated by the Marcus theory (briefly discussed in the Supporting Information). The dependence of the PL quenching efficiency on QD size shown in Figure 4c supports the proposed interaction mechanism, where the more pronounced quenching for the smaller size may be attributed to a combination of slightly higher density of surface states (larger surface-to-volume ratio) and better spatial overlap between charge carriers and surface states. A more pronounced QD PL quenching in the presence of Ru-phen-complexes thus results for smaller size nanocrystals. In comparison, Fc and Ru-bpy-phen with their higher formal oxidation potentials (Figure 2) do not promote effective coupling and charge transfer to the proximal QDs. The surface states at the origin of the oxidation peak observed for the QD samples and used to interpret our findings are different from deep trap states often used to explain the very broad emission measured for poor quality samples. These surface states reduce the measured PL quantum yields compared to an ideal sample for which the yield should be close to unity.
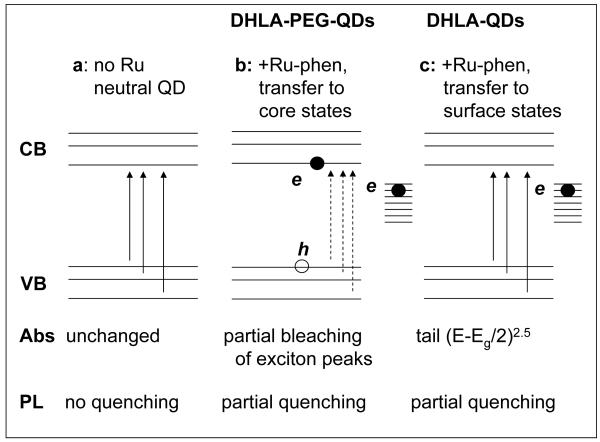
The Ru-phen-complex interactions with QDs, however, do not seem to be identical for assemblies using QDs capped with either neutral DHLA-PEG or negatively charged DHLA. There is a difference between the time-resolved PL and absorption changes collected for the two sets of QDs shown in Figures 3-5. In a simplified model sketched in Figure 7a-c, we attribute this somewhat unexpected difference between the two sets of QDs (DHLA and DHLA-PEG surface) to additional effects of electrostatic interactions on the nature of charge transfer from Ru-phen-complex to QD. Assembly of Ru-phen with DHLA-QDs causes charge transfer primarily to the QD surface states (corresponding to the observed oxidation peak at ~0.2-0.23V vs. Ag/AgCl). This produces a partial and rather homogeneous quenching of the QD population, which is equally reflected in the efficiencies extracted from steady-state and time-resolved measurements. This also manifests in a strong background contribution to the absorption spectra. The presence of electrons populating the surface states is also supported by the fact that the differential absorption follows a (E-Eg/2)2.5 profile (Eq. 4) 9.
Figure 7. Schematic representation of the proposed model for charge-transfer.
(a) Absorption and PL spectra are unaffected for neutral QDs. (b) With DHLA-PEG-QDs a combined transfer to surface states as well as core states leads to additional partial bleaching of the exciton absorption features along with quenching of the QD PL. (c) With DHLA-QDs charge transfer to QD surface states dominate the interactions and lead to partially quenched PL and a broad absorption tail.
DHLA-PEG-capped QDs on the other hand do not present a density of surface charges. Interestingly, even though their oxidation potential is similar to DHLA-QDs, these QDs exhibit a different behavior when assembled with Ru-phen complexes. In addition to the dominant charge transfer to surface states as above, which produces partial homogenous quenching of the PL, there is a small contribution derived from a transfer to the conduction or valence bands, susceptible to cause a complete quenching of the QD PL. While PL intensities measured using steady-state fluorescence are sensitive to both types of quenching, time-resolved measurements are only sensitive to changes in the PL decay rates of emitting nanocrystals. In the time-resolved PL data, the fraction of fully quenched QDs essentially does not contribute to the measured signal, which results in lower measured efficiencies, compared to ensemble PL. As a consequence, we attribute the difference between time-resolved and steady-state quenching efficiencies for DHLA-PEG-QDs to the likely presence of small fraction of completely quenched QDs due to charge transfer directly to conduction or valence bands (primarily to the 1S and possibly to higher states). The possibility of realizing charge transfer to core states results in partial bleaching of the first and potentially second absorption peaks, a property that is reasonably well reflected in the differential absorption spectra shown in Figure 5b, in addition to the apparition of a red tail corresponding to charge transfer to surface states as above. Furthermore, the observed quenching efficiency and the changes in the absorption features are clearly dependent on the ratio of Ru-phen-to-QD in both sets of conjugates. Even though we observe the signature of partial charge transfer to core states for these QD assemblies, we should remark that this scenario is rather unexpected for these systems given the relative position of the oxidation potential of the various components with respect to the CB of the QDs. Further investigations using for example additional redox compounds with varying redox levels and QDs with other functional ligands are needed to understand its underlying mechanisms.
Charge transfer from redox complexes to QDs and its effect on the QD PL and absorption properties are dependent on the relative positions of their oxidation energies/levels. The Ru-phen oxidation level stands slightly above that of the QD, so that electron transfer from Ru-phen to this level is energetically favorable. In comparison, Fc and Ru-bpy-phen, with their higher formal oxidation potentials, i.e., lower energy levels (Figure 2), do not promote effective coupling and charge transfer to the proximal QDs. Consistently with the respective positions of these energy levels, we observe the spectroscopic signature of charge transfer (for PL and absorption) from Ru-phen to QDs, but not from Fc- or Ru-bpy-phen-complexes. We further tested whether the QD spectroscopic changes indeed corresponded to charge transfer using a much simpler atomic redox system with favorable redox properties, as a control. The energy level corresponding to the Fe(III)/Fe(II) redox couple is higher than that of Ru-phen and stands at approximately 250 meV above the QD redox level. If interacting with the QD, its effects on the QD PL and absorption should be similar to those observed for Ru-phen. Indeed, mixing Fe(II) ions with DHLA-QDs causes a progressive QD PL quenching along with red tail in the QD absorption spectrum (Figure S7), similar to what was observed with Ru-phen complexes at comparable concentrations; coupling between DHLA-QDs and Fe(II) is driven by electrostatic interactions. This confirms that the interactions between Ru-phen and QDs are promoted by favorable redox interactions (or coupling).
We would like to stress the fact that the quenching of QD emission cannot be induced by the peptide bridge alone. Indeed, assembling QDs with unlabeled peptides did not cause any change in the steady state PL, the exciton lifetime or absorbance properties of the samples. It has been reported that peptides and proteins can facilitate through-bond (or bonded) charge (electron) transfer 39-41, and it is likely that through-bond contribution to charge transfer takes place in our samples. However, effects of redox/oxidation potential matching dominate the rate of charge transfer in these conjugates. Since a through-bond transfer may depend on the peptide sequence and QD-metal complex separation distance, we tested if the observed differences between the Ru-phen and Fc systems could be due to differences in peptide sequences used and/or metal complex location within the conjugate used. We labeled a control sequence of His6-appended-peptide with either Ru-phen-complex at the farthest residue from the His-tract or with Fc-complex at a residue close to the His tract, then monitored the self-assembly of these Ru-phen-labeled and Fc-labeled peptides onto DHLA-PEG-QDs (exact sequence and complex location are provided in Figure S6, Supporting Information). Data clearly show that only Ru-phen-complex produced a pronounced quenching of the QD emission, with a loss that traced the Ru-phen-QD ratio. In contrast, Fc-complex had no measurable effects, even though a closer approach (shorter separation distance) was permitted for QD-peptide-Fc assemblies. This result confirms our conclusion that within our QD-peptide-Ru-phen conjugates, PL quenching is primarily due to charge transfer (induced by redox interactions) from the Ru-phen to the nanocrystals. Nonetheless, it still remains unclear whether or not additional interactions between peptides and ligands contribute to charge transfer and quenching. Future studies will try to elucidate any potential contribution resulting from peptide sequence/length and peptide-ligand/metal complex-ligand interactions to the PL quenching.
Finally we should emphasize that our results differ from those reported by Sykora et al., where photoinduced charge transfer between Ru-polypyridine complexes and QDs were examined 42. In that study the authors used Ru complexes containing several bipyridine groups coupled to additional carboxy and methyl groups (Ru(bpy)3, Ru(bpy)2-methyl-carboxy, and Ru(bpy)-methyl-(carboxy)2) that were also luminescent. Those complexes share some of the basic structures of the Ru-bpy-phen complex used here. In comparison, the ruthenium phenanthroline and ferrocene complexed used here are non-emitting. They reported, for example, that interactions with Ru(bpy)-methyl-(carboxy)2 manifested in carrier exchange between the QD and the excited proximal Ru(bpy) complex, with the transfer of an electron from the excited level of the Ru(bpy) complex to the QD conduction band and a hole from the QD valence band to Ru(bpy). This necessitates excitation of the Ru complex and produces a loss of emission for QD and Ru(bpy) alike. Our experiments using the Ru-bpy-phen complex showed that there are essentially no effects of the metal-complex on the QD emission (Figure S1, Supporting Information); similar behavior was also recorded in the presence of ferrocene. There is no eventuality for carrier exchange in our systems, since the non-emitting metal complexes do not present an electron-hole pair next to an excited QD. There were also several practical differences. Their experiments were performed in organic solution as opposed to the aqueous buffer used here. In their samples the QDs and Ru-complexes are mixed in solution at very high ratios (100:1 Ru-complex:QD), and proximity relied on diffusion encounter and non-specific interactions involving a potential replacement of a few TOP/TOPO ligands by a Ru-complex. Control over the proximity interactions between QD and Ru(bpy) complexes as well as the number of complexes simultaneously interacting with a single QD (i.e., “conjugate valence”) is difficult to achieve using such a system. In comparison peptide bridging between the QD and Ru-phen-complexes allows control over proximity and average conjugate valence.
Conclusion
We have characterized the interactions between CdSe-ZnS QDs and proximal redox-active metal-complexes brought in close proximity via a peptide bridge. We found that charge transfer occurs only when there is a favorable match between the oxidation potentials of the QD and metal complex (CdSe QDs and Ru-phenanthroline in this case), and is reflected in systematic loss in QD photoemission and changes in the QD absorption properties. These changes are dependent upon the number of Ru-phen complexes arrayed around a QD center. In contrast to ruthenium phenanthroline, the data collected using Fc-labeled- or Ru-bpy-phen-labeled peptide showed little to no interactions, due to unfavorable matching between QD and proximal complexes. We also found that the nature of ligands used to promote hydrophilicity of the nanocrystals affect the charge transfer interactions for QD-peptide-Ru-phen conjugates. We attributed the differences between data collected for samples made using neutral DHLA-PEG-QDs and positively-charged DHLA-capped nanocrystals to electrostatic effects. In particular, DHLA-QDs present a homogenous charge density on their surfaces, which in turn induces a dominant channel of electron transfer from Ru-phen to the QD electronic surface states. The interactions with neutral DHLA-PEG-QDs on the other hand results in the coexistence of a major population of partially quenched QDs (with transfer to surface states) together with one of fully quenched QDs (with injected charges to core states). We exploited the proximity-driven interactions and their ability to induce quenching of the QD PL to design specific biosensing assemblies capable of detecting enzymatic activity in solutions.
Beyond ruthenium-polypyridine complexes, there is a variety of small electroactive compounds with a broad range of redox potentials that can be used with QDs 12,43. Choice of an “ideal” redox complex, however, will be dictated by key criteria, such as matching potentials, the ability to function in an aqueous environment, and ultimately incorporation into a reactive probe for site-specific labeling of biological molecules if bioapplications are desired. Although much still remains to be understood, these results add to the growing work suggesting that QDs may be far more versatile for selected biological assays than conventional fluorescent probes. These nanocrystals act as both redox-sensitive probes and central nanoscaffolds for constructing specific sensing assemblies.
Experimental
Peptide synthesis, labeling with metal complexes and assembly of the QD-complex conjugates
Polyhistidine-terminated peptides were synthesized manually using in-situ neutralization cycles for Boc-solid-phase synthesis following the procedures described in references 37,44. Sequences included: a generic peptide Ac-(His)6-Gly-Leu-Aib-Ala-Ala-Gly-Gly-His-Tyr-Gly-Cys-CONH2, a glutathione-modified polyprolyl peptide Glt-Aha-(Pro)9-Gly-Gly-(His)6-CONH2, a chymotrypsin substrate (Chym) Ac-(His)6-Gly-Leu-Aib-Ala-Ala-Gly-Gly-Trp-Gly-Cys-CONH2, and a thrombin substrate (Thr) (His)6-Gly-Leu-Ala-Aib-Ser-Gly-Phe-Pro-Arg-Gly-Arg-Cys-CONH2, where Ac is an acetyl group, CONH2 is an amide, Aib is alpha-amino isobutyric acid, Aha is aminohexanoic acid and Glt is glutathione (γ-glutamylcysteinylglycine). Each of the peptides used in this study includes a central short alpha helical segment, a terminal polyhistidine for interactions with the dot surface and an end function (e.g., Cys) for labeling with the metal complex. Maleimide-functionalized ruthenium phenanthroline (Ru-phen) complex was coupled to the cysteine-thiols of the peptides 45. For labeling, 1 mg of peptide was dissolved in 1 mL PBS (0.1 M sodium phosphate and 0.15 M NaCl pH 7.4) with 1 mg of Ru-phen-maleimide and incubated overnight at 4 °C with continuous agitation. Labeled peptide was purified from free Ru-phen-maleimide on Ni-NTA resin (Qiagen, Valencia CA), dialyzed against PBS and desalted using reverse phase 18 oligonucleotide purification cartridges (Applied Biosystems, Mountain View, CA). The purified peptide was quantitated using the Ru-phen-complex absorbance at 490 nm (5000 M-1cm-1), lyophilized and stored at -20°C. N-hydroxysuccinimide-(NHS) modified ferrocene was synthesized using a variation (using NHS instead of maleimide) of the procedure described in reference 46. The NHS group was reacted with the primary amine on the glutathione peptide in 10 mM NaTetraborate buffer pH 8.5, then purified as described above and quantitated using the Fc-complex absorbance at 264 nm (9500 M-1cm-1). Ruthenium chelate bis(bipyridine)-5-(isothiocyanatophenanthrolin)-Ru(PF6)2, Ru-bpy-phen-ITC (Sigma-Aldrich, St. Louis, MO), is a weak electroactive dye (with an absorption and emission maxima centered at 454 nm and 625 nm, respectively, see Figure S1) 47,48. Ru-bpy-phen-ITC was also reacted with the glutathione modified peptide and purified as described above. In order to reduce potential interference from FRET interactions, QDs that have the weakest spectral overlap with the Ru-bpy-phen absorption (551-nm emitting QDs) were used.
The QDs used in this study were CdSe-ZnS core-shell and CdSe core only nanocrystals with emission maxima centered at 520, 540, 551, 555, 565, 570, and 590 nm; absorption and emission spectra of two representative samples are shown in Figure 1. The CdSe cores were synthesized first by reacting organometallic precursors (for cadmium and selenium) in a hot coordinating solvent mixture 21, 49-51. The ZnS shell was also grown in hot coordinating solvent mixture on the CdSe cores using zinc and sulfur precursors at temperatures lower than those used for the cores 21, 49-51. The QDs were made hydrophilic by exchanging the native TOP/TOPO cap with either dihydrolipoic acid (DHLA) or polyethylene glycol-terminated DHLA (DHLA-PEG600/1000) ligands (see Figure 1b) 21,28. Self-assembled QD-peptide bioconjugates were prepared by mixing 30 picomoles of QDs with peptides at the desired molar ratio n in 10 mM Na Tetraborate buffer pH 8.5 or 10 mM Hepes buffer pH 8.0 and let incubate for 30 minutes at room temperature. Absorption spectra were collected from conjugate solutions using a UV-Vis HP 8453 diode array spectrophotometer (Agilent Technologies, Santa Clara, CA).
Cyclic voltammetry
Indium Tin Oxide (ITO) coated glass slides (Rs = 10 Ω) were purchased from Delta Tech. Ltd (Stillwater, MN). Poly(allylamine hydrochloride) (PAH, Aldrich) was used as received. De-ionized (DI) water (18.2 MΩ) was obtained from a Milli-Q water purification system (Millipore, Burlington, MA). ITO slides were cleaned in a methanol base bath (10% (wt/vol) KOH in MeOH) at room temperature for 15 min, then rinsed copiously with DI water, sonicated for 5 minutes in DI water and dried under a N2 stream. ITO surfaces were modified with a PAH layer, by exposure to a 0.3% (wt/vol) solution of PAH in water for 30 min. at room temp. then rinsed with DI water and dried under N2. The slides were immersed in 500 nM QD solutions in a 10 mM borate buffer at pH 8.6 for 2 hours, to allow capture of a QD layer on the PAH-coated slide. A similar procedure for immobilizing a layer of metal complexes (free and bound to peptides) onto the ITO substrates was used. The slides were then rinsed with DI water and immediately placed in the electrochemical cell for analysis. All electrochemical measurements were performed under a Faraday cage in the 3-electrode geometry, using ITO as one working electrode, a Pt counter electrode (Bioanalytical Systems, Inc., BAS, West Lafayette, IN), and a Ag/AgCl, 3 M KCl reference electrode. A 3 mL glass cell (CH Instruments, Austin TX) was used and measurements were driven by an electrochemical workstation (Model 750, CH Instruments). A ~1 cm2 area of the ITO working electrode was exposed to the PBS electrolyte. All voltammograms were collected under ambient conditions. Before each measurement the electrolyte was purged with Ar and during each measurement a blanket of Ar gas was kept above the solution. Each voltammogram was initiated at the negative potential limit at a scan rate of 50 mV/s following a 2 s quiescence period. No signal was obtained in the same potential range on a PAH modified ITO electrode not containing QDs or metal complexes. Background subtracted voltammograms were generated using software provided by CH instruments.
Steady-state and time-resolved fluorescence
Ensemble PL spectra were collected from solutions of QD-peptide-metal complex conjugates and the corresponding control samples using a Tecan Safire Dual Monochromator Plate Reader (Tecan, Research Triangle Park, NC). Samples were measured in triplicate and error bars are shown where appropriate. The redox quenching efficiency η was determined by measuring the fluorescence intensity of a solution of unlabeled QD-peptide (PL0) and the intensity of a solution of QD-peptide-redox-complex conjugates (PLn) as: η = [PL0-PLn]/PL0 (Eq. 1). Time-resolved PL decays were collected on a time-correlated single photon counting (TCSPC) set-up. The 800 nm line was frequency-doubled using a barium borate crystal (Photop Technologies) to provide a pulsed excitation line at 400 nm used in all our experiments. The system is equipped with a mode locked tunable (920 to 710 nm) titanium sapphire laser source having a repetition rate of 80 MHz (Wideband Mai Tai, Newport Corp.) 52. Typical instrument response had a full width at half-maximum of ~45 picoseconds. Fluorescence decay curves were best fitted to a three exponential decay function using a least squares fit algorithm in SigmaPlot 6.0 (SPSS Inc.). An average lifetime and the corresponding FRET efficiency were extracted from the experimental decay data (using equation 2) for every sample. Fluorescence decay curves were fitted to a three exponential decay function 34,36,48, and average decay rates k0 and kn were measured in the absence and presence of n Ru-complex per QD. Quenching efficiencies η from time-resolved PL were then estimated using the relation: η = (kn - k0)/kn (Eq. 2) which can also be expressed in terms of the corresponding lifetimes as η = (τ0 - τn)/τ0.
Supplementary Material
Acknowledgements
The authors thank Pankaj Singhal and Philippe Guyot-Sionnest for stimulating discussions and Kim Sapsford (USFDA) for the control peptide. The authors acknowledge Stephen Lee and Ilya Elashvilli of the CB Directorate/Physical S&T Division (DTRA), ONR, NRL and the NRL-NSI for financial support. The fluorescent lifetime instrumentation for this work was provided by an NSF-CRIF award (CHE-0342973) to D.S.E. T.P. acknowledges a post-doctoral fellowship from the Fondation pour la Recherche Medicale (France).
References
- 1.Efros AL, Rosen M. Ann. Rev. Mater. Sci. 2000;30:475–521. [Google Scholar]
- 2.Murray CB, Kagan CR, Bawendi MG. Ann. Rev. Mater. Sci. 2000;30:545–610. [Google Scholar]
- 3.Kippeny T, Swafford LA, Rosenthal SJ. J. Chem. Ed. 2002;79:1094–1100. [Google Scholar]
- 4.Klimov VI. J. Phys. Chem. B. 2006;110:16827–16845. doi: 10.1021/jp0615959. [DOI] [PubMed] [Google Scholar]
- 5.Mattoussi H, Cumming AW, Murray CB, Bawendi MG, Ober R. Phys. Review B. 1998;58:7850–7863. [Google Scholar]
- 6.Pons T, Uyeda HT, Medintz IL, Mattoussi H. J. Phys. Chem. B. 2006;110:20308–20316. doi: 10.1021/jp065041h. [DOI] [PubMed] [Google Scholar]
- 7.Klimov VI, McBranch DW, Leatherdale CA, Bawendi MG. Phys. Review B. 1999;60:13740–13749. [Google Scholar]
- 8.Klimov VI, Mikhailovsky AA, McBranch DW, Leatherdale CA, Bawendi MG. Science. 2000;287:1011–1013. doi: 10.1126/science.287.5455.1011. [DOI] [PubMed] [Google Scholar]
- 9.Shim M, Wang CJ, Guyot-Sionnest P. J. Phys. Chem. B. 2001;105:2369–2373. [Google Scholar]
- 10.Wang CJ, Shim M, Guyot-Sionnest P. Science. 2001;291:2390–2392. doi: 10.1126/science.291.5512.2390. [DOI] [PubMed] [Google Scholar]
- 11.Yildiz I, Tomasulo M, Raymo FM. P. N. A. S. (U. S. A) 2006;103:11457–11460. doi: 10.1073/pnas.0602384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson NA, Lian TQ. Annu. Rev. Phys. Chem. 2005;56:491–519. doi: 10.1146/annurev.physchem.55.091602.094347. [DOI] [PubMed] [Google Scholar]
- 13.Klimov VI, Mikhailovsky AA, McBranch DW, Leatherdale CA, Bawendi MG. Phys. Review B. 2000;61:R13349–R13352. doi: 10.1126/science.287.5455.1011. [DOI] [PubMed] [Google Scholar]
- 14.Schlamp MC, Peng XG, Alivisatos AP. J. App. Phys. 1997;82:5837–5842. [Google Scholar]
- 15.Mattoussi H, Radzilowski LH, Dabbousi BO, Thomas EL, Bawendi MG, Rubner MF. J. App. Phys. 1998;83:7965–7974. [Google Scholar]
- 16.Coe S, Woo W-K, Bawendi M, Bulovic V. Nature. 2002;420:800–803. doi: 10.1038/nature01217. [DOI] [PubMed] [Google Scholar]
- 17.Gur I, Fromer NA, Geier ML, Alivisatos AP. Science. 2005;310:462–465. doi: 10.1126/science.1117908. [DOI] [PubMed] [Google Scholar]
- 18.Klimov VI, Ivanov SA, Nanda J, Achermann M, Bezel I, McGuire JA, Piryatinski A. Nature. 2007;447:441–446. doi: 10.1038/nature05839. [DOI] [PubMed] [Google Scholar]
- 19.Bruchez M, Jr., Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 20.Chan WCW, Nie S. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 21.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J. Am. Chem. Soc. 2000;122:12142–12150. [Google Scholar]
- 22.Katz E, Willner I. Angew. Chem. Int. Ed. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 23.Medintz I, Uyeda H, Goldman E, Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 24.Ipe BI, Niemeyer CM. Angew. Chem. Int. Ed. 2006;45:504–507. doi: 10.1002/anie.200503084. [DOI] [PubMed] [Google Scholar]
- 25.Clarke JC, Annette Hollmann CA, Zhang Z, Suffern D, Bradforth SE, Dimitrijevic NM, Minarik WG, Nadeau JL. Nat. Mater. 2006;5:409–417. doi: 10.1038/nmat1631. [DOI] [PubMed] [Google Scholar]
- 26.Sandros MG, Gao D, Benson DE. J. Am. Chem. Soc. 2005;127:12198–12199. doi: 10.1021/ja054166h. [DOI] [PubMed] [Google Scholar]
- 27.Aryal BP, Benson DE. J. Am. Chem. Soc. 2006;128:15986–15987. doi: 10.1021/ja066658f. [DOI] [PubMed] [Google Scholar]
- 28.Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. J. Am. Chem. Soc. 2005;127:3870–3878. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- 29.Sapsford KE, Pons T, Medintz IL, Higashiya S, Brunel FM, Dawson PE, Mattoussi H. J. Phys. Chem. C. 2007;111:11528–11538. [Google Scholar]
- 30.Pons T, Medintz IL, Wang X, English DS, Mattoussi H. J. Am. Chem. Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- 31.Kucur E, Riegler J, Urban GA, Nann T. J. Chem. Phys. 2003;119:2333–2337. [Google Scholar]; Kucüur E, Bucking W, Giernoth R, Nann T. J. Phys. Chem. B. 2005;109:20355–20360. doi: 10.1021/jp053891b. [DOI] [PubMed] [Google Scholar]
- 32.Kucur E, Bucking W, Arenz S, Giernoth R, Nann T. ChemPhysChem. 2006;7:77–81. doi: 10.1002/cphc.200500284. [DOI] [PubMed] [Google Scholar]
- 33.Lakowicz JR. Principles of Fluorescence Spectroscopy. Third Edition Springer; New York: 2006. [Google Scholar]
- 34.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. J. Am. Chem. Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 35.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat. Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 36.Pons T, Medintz IL, Sykora M, Mattoussi H. Phys. Rev. B. 2006;73:245302. [Google Scholar]
- 37.Medintz IL, Dawson PE, Clapp AR, Uyeda HT, Chang EL, Deschamps JR, Mattoussi H. Nat. Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 38.Vinayakan R, Shanmugapriya T, Nair PV, Ramamurthy P, Thomas KG. J. Phys. Chem.C. 2007;111:10146–10149. [Google Scholar]
- 39.Schlag EW, Sheu S-Y, Yang D-Y, Selzle HL, Lin SH. Angew. Chem. Int. Ed. 2007;46:3196–3210. doi: 10.1002/anie.200601623. [DOI] [PubMed] [Google Scholar]
- 40.Gray HB, Winkler JR. Annu. Rev. Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 41.Abdel Malak R, Gao Z, Wishart JF, Isied SS. J. Am. Chem. Soc. 2004;126:13888–13889. doi: 10.1021/ja0401040. [DOI] [PubMed] [Google Scholar]
- 42.Sykora M, Petruska MA, Alstrum-Acevedo J, Bezel I, Meyer TJ, Klimov VI. J. Am. Chem. Soc. 2006;128:9984–9985. doi: 10.1021/ja061556a. [DOI] [PubMed] [Google Scholar]
- 43.Tucker JHR, Collinson SR. Chem. Soc. Rev. 2002;31:147–156. doi: 10.1039/a804251i. [DOI] [PubMed] [Google Scholar]
- 44.Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB. Int. J. Pept. Protein. Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 45.Trammell SA, Goldston HM, Jr., Tran PT, Tender LM, Conrad DW, Benson DE, Hellinga HW. Bioconj. Chem. 2001;12:643–647. doi: 10.1021/bc010022q. [DOI] [PubMed] [Google Scholar]
- 46.DiGleria K, Hill HAO, Wong LL. FEBS Letters. 1996;390:142–144. doi: 10.1016/0014-5793(96)00648-5. [DOI] [PubMed] [Google Scholar]
- 47.Szmacinski H, Terpetshnig E, Lakowicz JR. Biophys. Chem. 1996;62:109–120. doi: 10.1016/s0301-4622(96)02199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clapp AR, Medintz IL, Fisher BR, Anderson GP, Mattoussi H. J. Am. Chem. Soc. 2005;127:1242–1250. doi: 10.1021/ja045676z. [DOI] [PubMed] [Google Scholar]
- 49.Hines MA, Guyot-Sionnest P. J. Phys. Chem. 1996;100:468–471. [Google Scholar]
- 50.Peng ZA, Peng X. J. Am. Chem. Soc. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 51.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. J. Phys. Chem. B. 1997;101:9463–9475. [Google Scholar]
- 52.Grimes AF, Call SE, Vicente DA, English DS, Harbron EJ. J. Phys. Chem. B. 2006;110:19183–19190. doi: 10.1021/jp0613236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.