SUMMARY
Vpu proteins of pandemic HIV-1 M strains degrade the viral receptor CD4 and antagonize human tetherin to promote viral release and replication. We find that Vpus from SIVgsn, SIVmus and SIVmon infecting Cercopithecus primate species also degrade CD4 and antagonize tetherin. In contrast, SIVcpz, the immediate precursor of HIV-1, whose Vpu shares a common ancestry with SIVgsn/mus/mon Vpu, uses Nef rather than Vpu to counteract chimpanzee tetherin. Human tetherin, however, is resistant to Nef and thus poses a significant barrier to zoonotic transmission of SIVcpz to humans. Remarkably, Vpu from non-pandemic HIV-1 O strains are poor tetherin antagonists while those from the rare group N viruses do not degrade CD4. Thus, only HIV-1 M evolved a fully functional Vpu following the three independent cross-species transmissions that resulted in HIV-1 groups M, N, and O. This may explain why group M viruses are almost entirely responsible for the gobal HIV/AIDS pandemic.
INTRODUCTION
Primate lentiviruses such as HIV and SIV encode several accessory proteins. These include Vif, Vpr, Vpu, Vpx, and Nef, and are usually dispensable for viral growth in vitro. However, they are important for viral replication in vivo because they counteract host restriction factors, enhance viral replication and virion infectivity, or facilitate viral evasion of the adaptive immune response (reviewed in Malim and Emerman, 2008). Vif, vpr and nef are found in the genomes of all primate lentiviruses. In contrast, vpu genes were initially only found in HIV-1 and its precursor SIVcpz from chimpanzees (Pan troglodytes), but not in HIV-2 or other SIV strains (Cohen et al., 1988; Huet et al., 1990; Gao et al., 1999; Santiago et al., 2003). Subsequently, molecular characterization of SIVs from additional primate species revealed that the genomes of SIVgsn, SIVmon, SIVden and SIVmus from greater spot-nosed (Cercopithecus nictitans), mona (C. mona), Dent's mona (C. denti) and Mustached monkeys (C. cephus), as well as SIVgor from gorillas (Gorilla gorilla gorilla) also carry vpu genes (Barlow et al., 2003; Courgnaud et al., 2002; Courgnaud et al., 2003; Dazza et al., 2005; Takehisha et al., 2009). SIVcpz, which gave rise to pandemic (M, main) and non-pandemic (O, outlier and N, non-M, non-O) groups of HIV-1 and also to SIVgor (which is closely related to HIV-1 O; Van Heuverswyn et al., 2006), is the product of successive cross-species transmission and recombination events involving precursors of today’s SIVgsn/mus/mon/den and SIVrcm from red-capped mangabeys (Cercocebus torquatus) (Bailes et al., 2002). Thus, all vpu genes likely originated from a common ancestor of the SIVgsn/mus/mon/den lineage of primate lentiviruses (Kirchhoff, 2009).
Vpu is an ~80 amino acid integral class I membrane phosphoprotein (Cohen et al., 1988; Maldarelli et al., 1993). Studies performed with HIV-1 NL4-3 Vpu have established two main functions. First, Vpu induces the degradation of the primary viral receptor CD4 by a multi-step process that involves direct binding of Vpu to the cytoplasmic tail of CD4 in the endoplasmic reticulum (Bour et al., 1995, Margottin et al., 1998; Willey et al., 1992). Second, Vpu promotes the release of progeny virions from HIV-1-infected human cells (Strebel et al., 1988; Klimkait et al., 1990; Gottlinger et al., 1993) by antagonizing a recently identified restriction factor, termed tetherin (also known as CD317, BST2 or HM1.24), that is induced by interferon-alpha and results in the ‘tethering’ of nascent virions to the cellular plasma membrane (Neil et al., 2008; Van Damme et al., 2008). Tetherin has a broad antiviral activity and inhibits the release of various enveloped viruses (Jouvenet et al., 2009; Kaletsky et al., 2009; Sakuma et al., 2009). Furthermore, monkey and rodent tetherins block virion release but are not counteracted by HIV-1 Vpu (Goffinet et al., 2009; Gupta et al., 2009; McNatt et al., 2009; Wong et al., 2009).
To date, functional data have almost exclusively been derived from the Vpu protein of the T-cell line adapted HIV-1 NL4-3 molecular clone. It is thus unknown to what extent CD4 degradation and tetherin antagonism are conserved among the diverse Vpu proteins found in HIV-1 and SIVs and how Vpu function evolved following zoonotic transmissions of primate lentiviruses from monkeys to chimpanzees and, ultimately, to humans. To address these questions, we analyzed a panel of vpu constructs representing nearly the entire spectrum of primate lentiviruses known to encode this accessory gene, i.e. HIV-1 M, O and N, SIVcpz, SIVgor, SIVgsn, SIVmon and SIVmus. We show that all Vpu proteins, except those found in HIV-1 group N, degrade human CD4, whereas tetherin is antagonized in a more species-specific manner. Unexpectedly, we found that SIVcpz employs Nef rather than Vpu as a tetherin antagonist. Following transmission of SIVcpz to humans, HIV-1 group M Vpu and not Nef acquired an anti-tetherin activity in addition to its abilty to degrade CD4, most likely because the human tetherin has a deletion in its cytoplasmic domain that disrupts its susceptibility to Nef (Zhang et al., 2009). In contrast, Vpus of non-pandemic HIV-1 group O and N strains that resulted from independent zoonotic transmissions (Hahn et al., 2000; Van Heuverswyn and Peeters, 2007) either do not antagonize human tetherin (group O strains) or are unable to degrade CD4 (group N strains). Thus, the evolution of a fully functional Vpu protein may have been an important prerequisite for the efficient spread of HIV-1 group M in the human population.
RESULTS
Primate Lentiviral Vpus are Highly Variable but Exhibit some Conserved Features
To examine to what extent CD4 degradation and anti-tetherin activity are conserved among primate lentiviral Vpus, we analyzed a large panel of HIV-1 and SIV vpu alleles (Table S1). Our collection included Vpu constructs from 26 strains of HIV-1 group M, 11 strains of HIV-1 O, three strains of HIV-1 N, six strains of SIVcpzPtt from central chimpanzees (CPZ), four strains of SIVcpzPts from eastern chimpanzees, one strain of SIVgor from a Western lowland gorilla (GOR), two strains of SIVgsn from greater spot-nosed monkeys (GSN), one strain of SIVmon from a mona monkey (MON), and three strains of SIVmus from mustached monkeys (MUS). The HIV-1 group M Vpus included all nine subtypes, as well as ten vpu alleles (8 clade B and 2 clade C) from full-length infectious molecular clones of transmitted/founder viruses obtained by single genome amplification of plasma virion RNA (Salazar-Gonzales et al., 2009). Notably, all but two SIV vpu genes were amplified directly from the blood or fecal material of naturally infected monkeys or apes (Table S1).
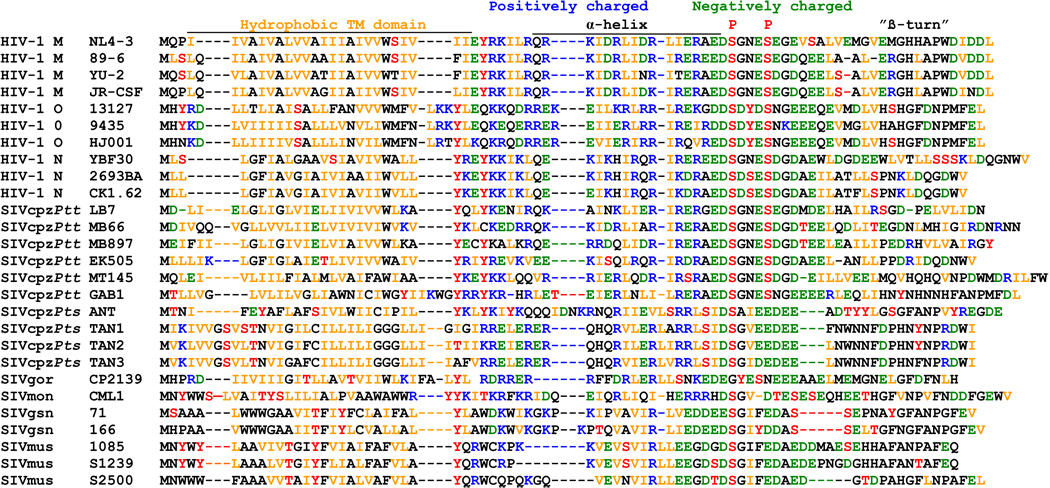
An amino acid alignment of representative primate lentiviral Vpus shows that they are highly variable. Nonetheless, some features, including a hydrophobic N-terminal membrane anchor domain, a putative central α-helical region with hydrophobic, basic and acidic residues and an acidic C-proximal region, are usually conserved (Figure 1). Two serine residues (S52 and S56), known to be phosphorylated by casein kinase II and required for CD4 degradation by NL4-3 Vpu (Schubert and Strebel, 1994; Paul and Jabbar, 1997), were present in all HIV-1 and SIVcpzPtt Vpus. In the remaining Vpus, the number and location of serine residues varied, although at least one potential serine phosphorylation site was always preserved (Figure 1). Thus, despite their exceedingly high variability, primate lentiviral Vpus exhibit common sequence signatures.
Figure 1.
Alignment of HIV-1 and SIV Vpu amino acid sequences. The NL4-3 Vpu sequence is shown on top for comparison. The hydrophobic transmembrane (TM) domain, the central charged region, the position of two serine phosphorylation sites and a β-turn motif in the NL4-3 Vpu protein are indicated. Dashes indicate gaps introduced to optimize the alignment. Acidic residues (E, D) are highlighted in green, basic residues (K, R) in blue, hydrophobic residues (I, V, L) in orange and residues that can potentially be phosphorylated (S, T, Y) in red.
All Vpu proteins - except those from HIV-1 group N - induce CD4 degradation
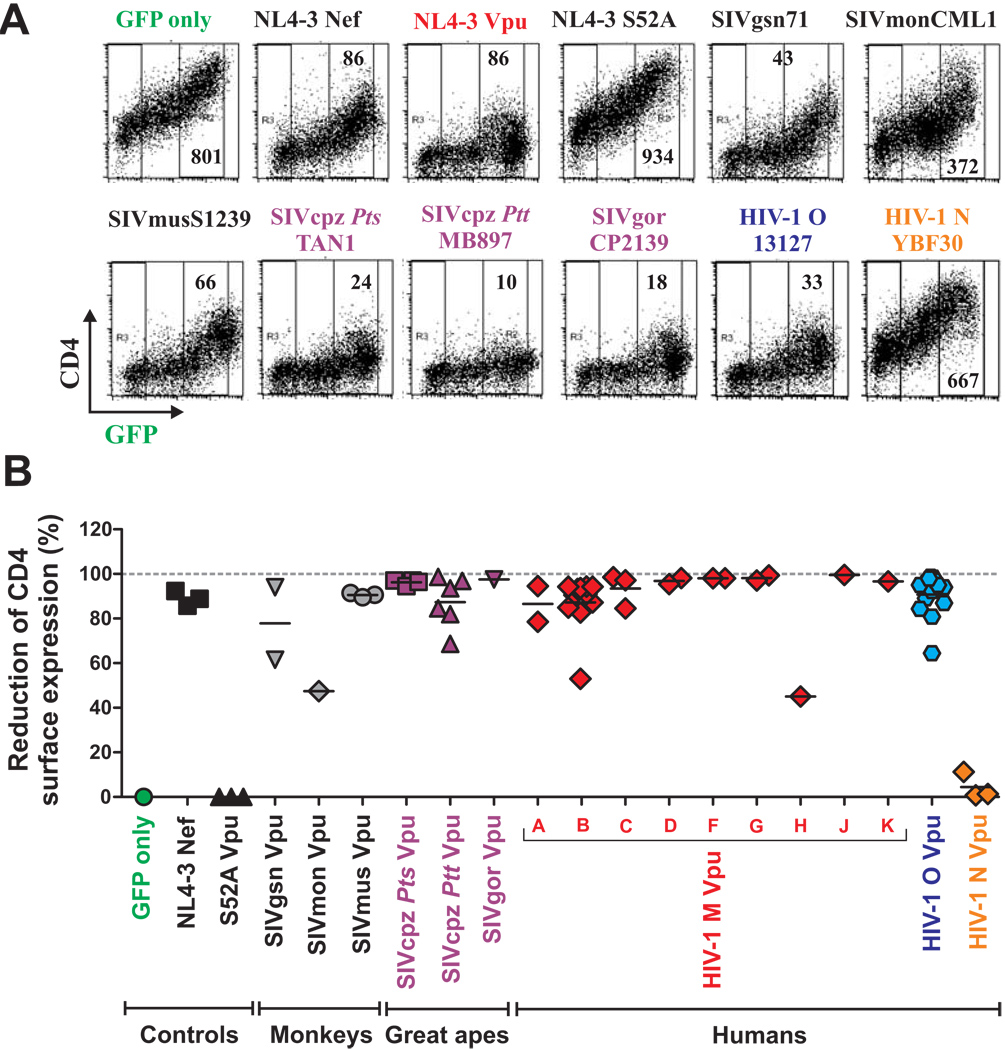
To determine which primate lentiviral Vpus prevent cell surface expression of CD4, we cotransfected 293T cells with vectors coexpressing Vpu and GFP (or GFP alone for control) together with a human (HU) CD4 expression construct. In the absence of Vpu, cells expressed high levels of CD4 and GFP (Figure 2A). However, upon coexpression of most Vpus, CD4 surface expression was markedly decreased (examples are shown in Figure 2A). This effect was Vpu specific since it was abrogated by a single S52A mutation in the cytosolic domain of the HIV-1 NL4-3 Vpu, previously shown to be critical for CD4 degradation (Schubert and Strebel, 1994; Paul and Jabbar, 1997). Quantitative analysis of CD4 surface levels (Fig. 2B) showed that most SIVgsn, SIVmon, SIVmus, SIVcpz and SIVgor, as well as HIV-1 M and O Vpus reduced CD4 cell surface expression by more than 80% (Figure 2B). In striking contrast, each of three HIV-1 group N Vpu proteins lacked this function (Figure 2B). These functional differences were statistically highly significant (p<0.0001). Analysis of Vpus containing a C-terminal AU-1-tag confirmed these results and showed that all but one construct expressed proteins of the expected size (Figure S1 and data not shown). These results show that the ability of blocking CD4 cell surface expression is highly conserved among primate lentiviral Vpus - with the notable exception of HIV-1 N Vpu, which lacks this function (Figure 2B).
Figure 2. Suppression of CD4 surface expression by HIV-1 and SIV Vpus.
(A) FACS analysis of 293T cells cotransfected with a CD4 expression vector and pCGCG plasmids expressing GFP alone (GFP only) or together with the indicated vpu alleles. Constructs expressing the NL4-3 Nef and the mutant NL4-3 S52A Vpu were used as positive and negative controls, respectively.
(B) Reduction of Vpu-mediated CD4 expression in 293T cells. Shown is the reduction in the levels of CD4 cell surface expression relative to those measured in cells transfected with the GFP only control vector. The range of GFP expression used for the calculation is indicated in panel A. Each symbol represents one of the 57 vpu alleles examined (Table S1) or the indicated controls. Shown are average values derived from three experiments. The GFP control is color coded green, vpu alleles derived from SIVgsn, SIVmon and SIVmus grey, SIVcpz and SIVgor magenta, HIV-1 M red, O blue and N orange.
SIVcpz, SIVgor and HIV-1 group O Vpus do not Efficiently Antagonize Tetherin
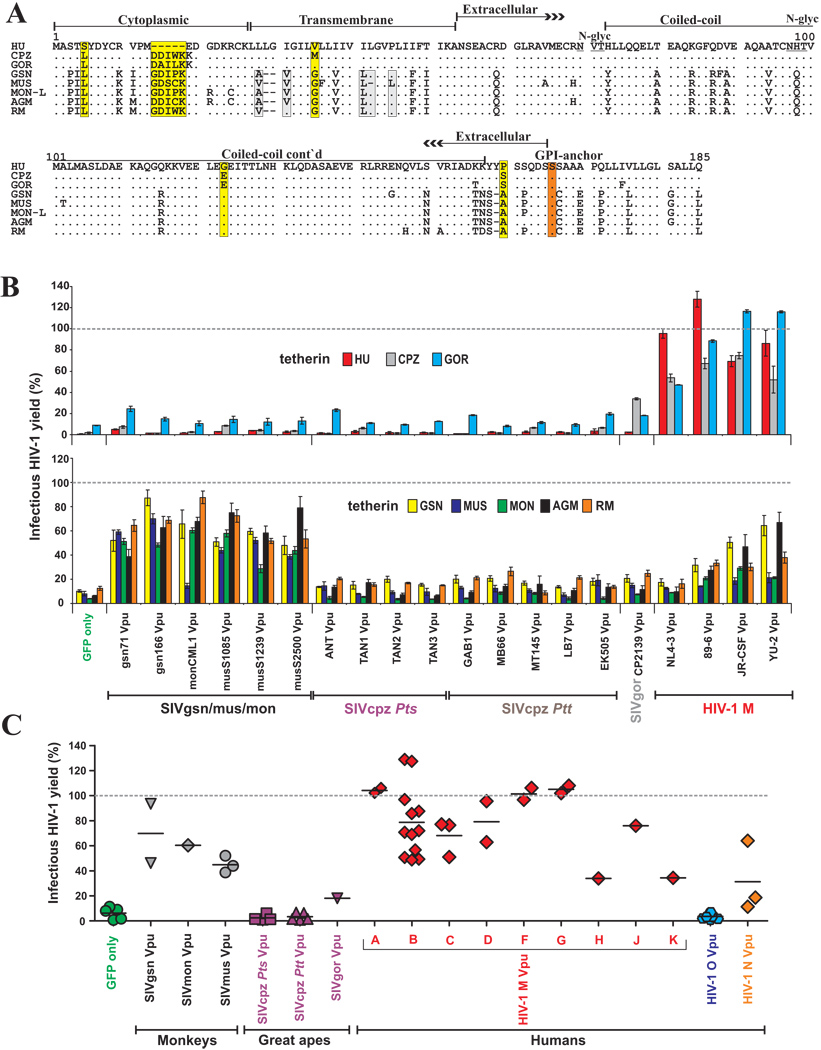
It has recently been shown that the HIV-1 NL4-3 Vpu antagonizes tetherin in a species-specific manner, i.e. it exhibited activity against human but not monkey (i.e. rhesus macaque, RM and African green monkey, AGM) or rodent tetherins (Goffinet et al., 2009; Gupta et al., 2009; McNatt et al., 2009). To determine the anti-tetherin activity of other primate lentiviral Vpus we generated vectors that expressed GSN, MON, MUS, CPZ, GOR, HU, AGM and RM tetherin molecules (Table S2). As shown in Figure 3A, the sequence of the transmembrane (TM) domain of tetherin, which has been evolving under positive selection and determines its susceptibility to HIV-1 NL4-3 Vpu (McNatt et al., 2009; Gupta et al., 2009), varies substantially between higher primates and monkeys, but is conserved between humans, gorillas and chimpanzees (except for a V35M variation between HU and CPZ tetherins; Figure 3A). In comparison, the TM domains of the six monkey tetherin sequences differed at a total of four amino acid positions. All tetherin proteins were expressed and suppressed HIV-1 release in the absence of Vpu (Figure S2).
Figure 3. Tetherin antagonism by primate lentiviral Vpu proteins.
(A) Alignment of tetherin amino acid sequences from humans (HU), chimpanzees (CPZ), gorillas (GOR), Greater spot-nosed monkeys (GSN), Mustached monkeys (MUS), Mona monkeys (MON-L, C. mona lowei), African green monkeys (AGM) and rhesus macaques (RM). Amino acid identity is indicated by dots and gaps by dashes. Differences between HU and CPZ tetherins are highlighted by yellow boxes and variations in the TM domain of monkey tetherins by grey boxes. Known domains, the serine GPI anchor site (red box) and two potential glycosylation sites (underlined), are indicated.
(B) Effect of various Vpus on infectious virus release in the presence of the indicated tetherin molecules. 293T cells were cotransfected with HIV-1 ΔVpu NL4-3 (2 µg), pCGCG vectors expressing GFP alone or together with Vpu (500 ng), and tetherin expression constructs (50 ng). Viral supernatants were obtained 2 days later and used to measure the quantity of infectious HIV-1 in the culture supernatants by infecting TZM-bl indicator cells. Shown are average values ±SD (n=3) of infectious virion yield relative to those obtained in the absence of tetherin expression vector (100%). The results were confirmed in at least two independent experiments.
(C) Infectious virus release from 293T cells expressing vpu alleles from the indicated primate lentiviruses and tetherins from their respective host species. The experiments were performed as described in panel B. Each symbol represents the average infectious virus release (n=3) obtained in the presence of one of the 55 individual vpu alleles analyzed.
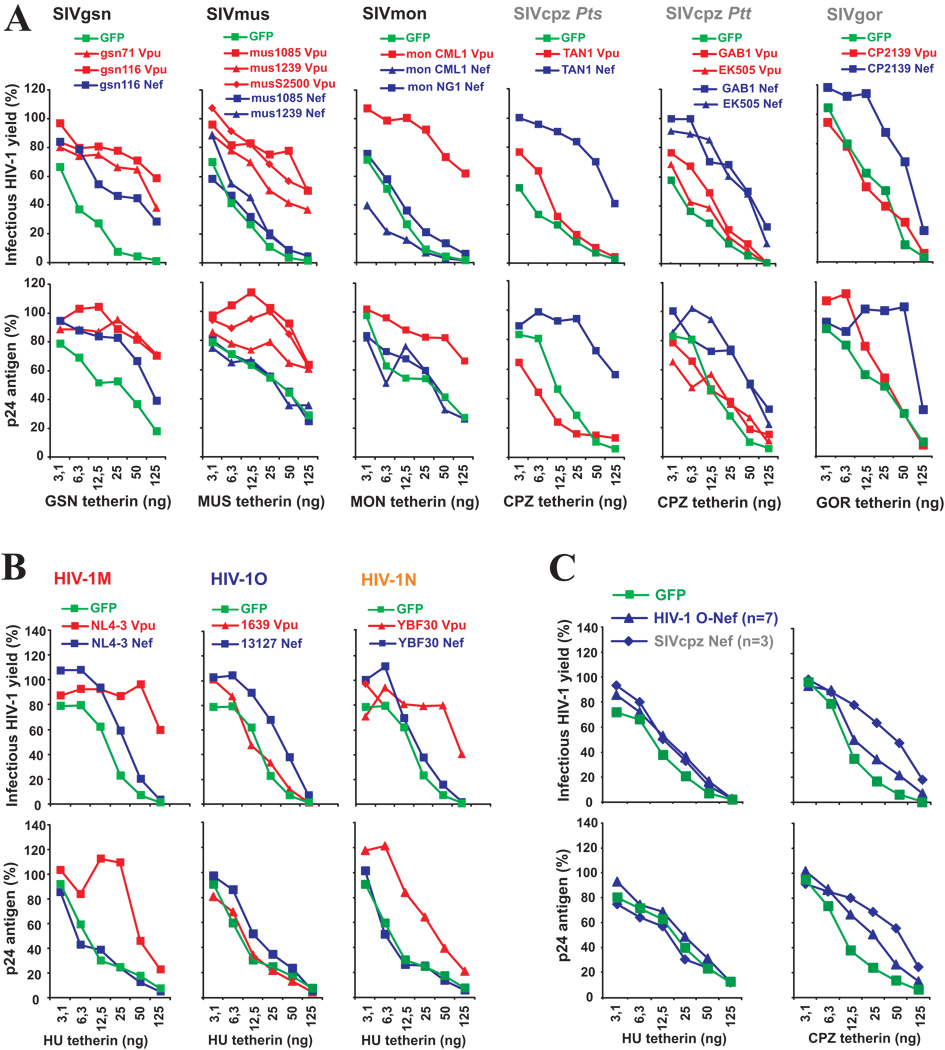
To test whether the various HIV-1 and SIV Vpus are capable of counteracting tetherins from their respective hosts, as well as those from other species, we measured infectious virus yields from 293T cells following cotransfection of a vpu-deleted (ΔVpu) HIV-1 proviral construct (Rucker et al., 2004) with Vpu and tetherin expression plasmids. Our data showed that only HIV-1 Vpus were effective antagonists of HU, CPZ and GOR tetherins (Figure 3B). In comparison, SIVgsn, SIVmon and SIVmus Vpus counteracted the tetherins of their respective host, as well as other monkey species (Figure 3B). The exception was SIVmon CML1 Vpu, which was inactive against the MUS tetherin, which differs from the other monkey tetherins by a L36F substitution and a single amino acid deletion (ΔL42) in the TM domain (Figure 3A, B). Thus, HIV-1 group M Vpus potently antagonized tetherins of apes and humans but were less effective against the monkey restriction factors, whereas SIVgsn, SIVmon and SIVmus Vpus showed the opposite phenotype. Interestingly, the HIV-1 JR-CSF and YU-2 Vpus exhibited some activity against GSN and AGM tetherins (Figure 3B), indicating that certain HIV-1 Vpus have a broader anti-tetherin activity than the NL4-3 Vpu (Goffinet et al., 2009; Gupta et al., 2009; Jia et al., 2009; McNatt et al., 2009). In agreement with previous studies (McNatt et al., 2009), an exchange of the TM domain in HU tetherin with that of AGM or RM rendered it resistant to HIV-1 Vpus but conferred sensitivity to SIVmon and SIVmus Vpus (Figure S3).
While the functional profiles of HIV-1, SIVgsn, SIVmon and SIVmus Vpus were as expected, the observed anti tetherin activities of ape virus Vpus came as a surprise: all SIVcpz Vpus, including those derived from the closest relatives of the HIV-1 group M (i.e. LB7, MB897, MB66) (Keele et al., 2006), failed to antagonize all eight different tetherins including that derived from their cognate CPZ host (Figure 3B). Similarly, the Vpu protein of SIVgor had only poor anti-tetherin activity. These results were confirmed by expressing the various Vpu proteins in cis in the context of infectious HIV-1 molecular clones, using both tetherin transfected 239T cells as well as HeLa cells expressing endogenous tetherin (see Supplemental Results and Figure S4, Figure S5 and Figure S6). To control for intracellular viral gene expression, we also examined the anti-tetherin activity of primate lentiviral Vpus by Western blot analysis. The results confirmed that HIV-1 Vpus efficiently counteracted HU and CPZ tetherins, but were less effective against those of AGM or RM, whereas Vpu proteins from SIVgsn and SIVmus showed the opposite phenotype. They also confirmed that SIVcpz Vpus lack significant anti-tetherin activity (Figure S7). Importantly, the Western blots also demonstrated that the various Vpus acted specifically at the level of virion release into the culture supernatants, and did not influence cell-associated levels of p24 antigen expression.
Our finding that SIVcpz Vpus are very poor tetherin antagonists suggested that HIV-1 group M Vpus gained anti-tetherin activity during adaptation to the new human host. Thus, we next examined whether Vpus from non-pandemic HIV-1 groups O and N strains (which represent independent cross-species transmissions; Hahn et al., 2000; Van Heuverswyn and Peeters, 2007) also evolved the capability of antagonizing HU tetherin. Strikingly, we found that (similarly to SIVcpz and SIVgor Vpus) HIV-1 group O Vpus generally displayed little if any activity against the human tetherin (Figure 3C). In contrast, one HIV-1 group N Vpu (YBF30) efficiently counteracted HU tetherin, while two others had more modest effects (Figure 3C). These results were confirmed at various levels of HU tetherin expression (Figure S8). Thus, only HIV-1 group M and N, but not HIV-1 group O Vpus, have acquired significant anti-tetherin activity during viral adaptation to the human host.
Nef proteins from SIVcpz and SIVgor, but not from HIV-1 group O, antagonize Tetherin
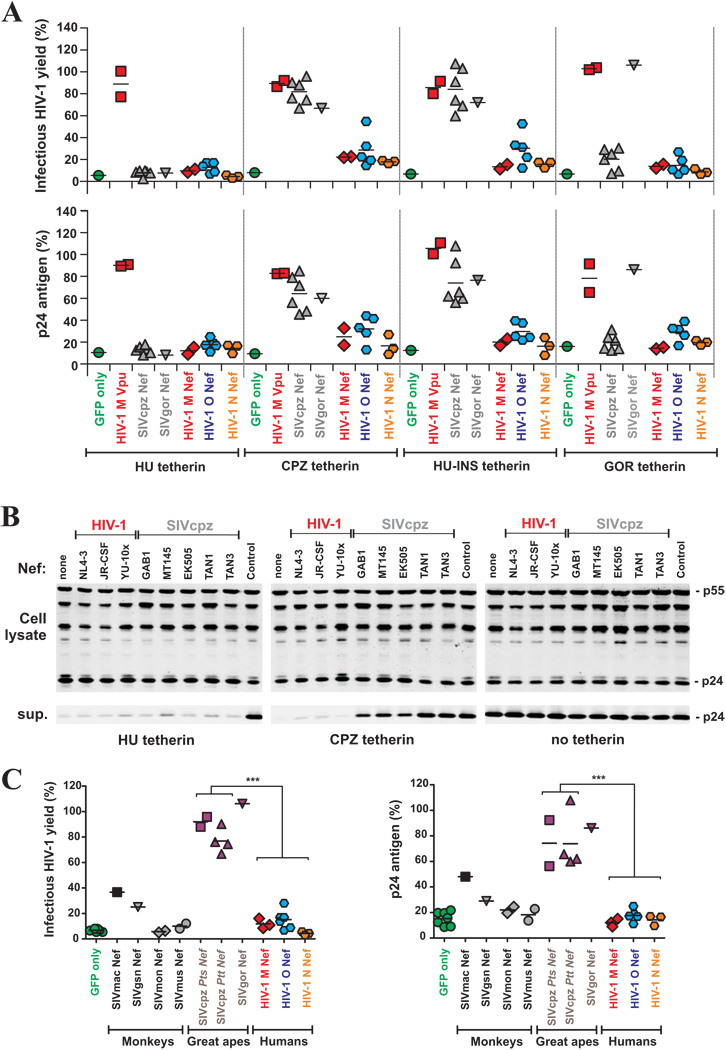
Recently, it has been shown that the Nef proteins of some SIVs that lack a vpu gene (i.e. those of SIVsmm/mac, SIVagm and SIVblu) counteract tetherin (Jia et al., 2009; Zhang et al., 2009). We therefore examined whether SIVcpz, SIVgor and HIV-1 group O employ Nef rather than Vpu to antagonize this restriction factor. Since primate lentiviral Nef proteins enhance virion infectivity (Münch et al., 2007), it was expected that expression of Nef would enhance the infectious virus yield but not p24 antigen release in the absence of tetherin (examples shown in Figure S9). To account for this confounding variable, we normalized all infectious virus yields to that obtained in the presence of Nef but the absence of tetherin (100%). After this adjustment the relative levels of virus release measured by infectivity assays and by p24 antigen ELISA correlated extremely well (R2 =0.93; Figure S9). We also measured the levels of cell-associated p24 expression by ELISA and Western blot and found that neither Nef nor tetherin affected these values.
First, we examined whether HIV-1, SIVcpz and SIVgor Nefs (Table S3) antagonize tetherin from apes and humans by cotransfecting 293T cells with an HIV-1 proviral construct lacking vpu and nef, tetherin expression vectors and constructs expressing various Nef proteins. The results demonstrated that SIVcpz and SIVgor Nefs antagonize the tetherin molecules from their cognate hosts but not the tetherin variant found in humans (Figure 4A). Insertion of five amino acids (DDIWK) that are missing in the cytoplasmic domain of HU tetherin (Figure 3A) and are known to confer sensitivity to SIVmac and SIVagm Nef (Jia et al., 2009; Zhang et al., 2009) fully restored its susceptibility to SIVcpz and SIVgor Nef antagonism (HU-INS, Figure 4A). The disruptive effect of the N-terminal deletion in HU tetherin was highly significant (p<0.0001). Some HIV-1 Nefs displayed modest activities against the CPZ, GOR and HU-INS tetherins, but did not antagonize HU tetherin. The SIVgor Nef was more effective against the GOR tetherin than any of six SIVcpz Nefs. Notably, the GOR and CPZ tetherin proteins differ at only two amino acid positions in the cytoplasmic domain (D15A, W17L) at the same location that governs sensitivity to other Nef proteins (Figure 4A). This strongly suggests that SIVgor Nef has specifically adapted to antagonize tetherin in gorillas. To confirm these findings we generated vpu-defective mutants of replication-competent HIV-1 NL4-3 proviral constructs that coexpress GFP and different HIV-1 and SIVcpz nef alleles from bicistronic RNAs (Schindler et al., 2006) and analyzed viral protein expression and release using Western blot analyses. Our data confirmed that all SIVcpz Nefs antagonize CPZ (but not HU) tetherin (Figure 4B). Notably, Nef proteins from other SIVs that encode Vpu proteins, i.e. SIVgsn, SIVmon and SIVmus, did usually not counteract tetherins from their respective hosts (Figure 4C). In agreement with published data (Jia et al., 2009; Zhang et al., 2009), we found that the nef alleles from SIVmac and SIVagm antagonize RM and AGM tetherins, albeit less efficiently than SIVcpz Nefs (Figure 4C, and data not shown).
Figure 4. Tetherin antagonism by primate lentiviral Nef proteins.
(A) Infectious virus and p24 antigen yield from 293T cells cotransfected with the proviral HIV-1 NL4-3 ΔVpu ΔNef construct (Rucker et al., 2004) containing disrupted vpu and nef genes (2 µg), pCGCG vectors expressing the indicated nef alleles or the NL4-3 Vpu protein (500 ng), in combination with plasmids expressing the HU, CPZ or GOR tetherins or a Hu tetherin variant in which the N-terminal deletion was restored (HU-INS) (50 ng). The upper panel shows the average values derived from triplicate infections of TZM-bl indicator cells and the lower panel the quantity of p24 antigen in the culture supernatant. All values are shown relative to those obtained in the absence of tetherin expression vector (100%) the levels of cellular p24 expression did not differ significantly (data not shown).
(B) Western blot analysis of cell and virion lysates following cotransfection of 293T cells with vpu-defective proviral Vpu/Nef/GFP HIV-1 NL4-3 constructs expressing the indicated nef alleles and 50 ng of empty vector (control) or HU or CPZ tetherin-HA expression plasmids. Cell and virion lysates were probed with an anti HIV-1 capsid p24 monoclonal antibody. A proviral HIV-1 NL4-3 construct containing defects in both vpu and nef genes was used in the “none” and “control” lanes. Sup., cell culture supernatant.
(C) Infectious virus release from 293T cells expressing nef alleles from the indicated primate lentiviruses and tetherins from their respective host species. The experiments were performed as described in panel A. The symbols represent the average infectious virus release (n=3) obtained in the presence of one of the 25 nef alleles analyzed. ***, p<0.0001.
To detect possible weak anti-tetherin activities, we next compared the effects of various primate lentiviral vpu and nef alleles on virus release at different doses of plasmids expressing the tetherins from their respective hosts. These titration experiments confirmed that SIVgsn, SIVmus and SIVmon use mainly Vpu to antagonize tetherin (Figure 5A). The exception was SIVgsn116 Nef which also displayed significant anti-tetherin activity, suggesting that this primate lentivirus may use both accessory proteins to counteract this restriction factor. The titration analyses also confirmed that SIVcpz and SIVgor use Nef and HIV-1 groups M and N use Vpu to antagonize tetherin (Figure 5A, B). We also assessed whether HIV-1 O Nefs counteract HU tetherin at lower concentrations, since a weak enhancement of infectious virus release in the presence of HU tetherin was observed in some experiments (Figure 5B). These titration analyses showed that HIV-1 group O Nefs are just as inactive against HU tetherin as those of SIVcpz (Figure 5C). Moreover, HIV-1 group O Nefs were less potent antagonists of CPZ tetherin than SIVcpz Nefs (Figure 4A, Figure 5C). Thus, HIV-1 group O strains appear to have lost some of their Nef-associated anti-CPZ-tetherin activity following transmission to humans. Taken together, our results demonstrate that SIVcpz and SIVgor (but not HIV-1 groups M, N and O) use Nef rather than Vpu to antagonize tetherin.
Figure 5. Titration of anti-tetherin antagonism.
(A, B) Virus release from 293T cells following transfection with 2 µg of a ΔVpu/ Nef proviral NL4-3 construct, 500 ng of SIV (A) or HIV-1 (B) Vpu (color coded red) or Nef (color coded blue) expression constructs and varying amounts of plasmids expressing the tetherin molecules from the respective host species. Infectious virus was determined by infection of TZM-bl indicator cells (upper panels) and p24 ELISA (lower panels) is shown as a percentage of that detected in the absence of tetherin (100%). All infections shown in panels A–C were performed in triplicate and the results were confirmed in an independent experiment.
(C) HIV-1 O Nefs are inactive against HU tetherin. The assays were performed as described in panels A and B. Shown are average values obtained using seven group O (HJ428, HJ036, HJ736, HJ100, HJ256, 13127 and 8161) and three SIVcpz Ptt (Gab1, MT145, MB897) nef alleles.
DISCUSSION
CD4 Degradation and Tetherin Antagonism are Conserved Among Primate Lentiviruses
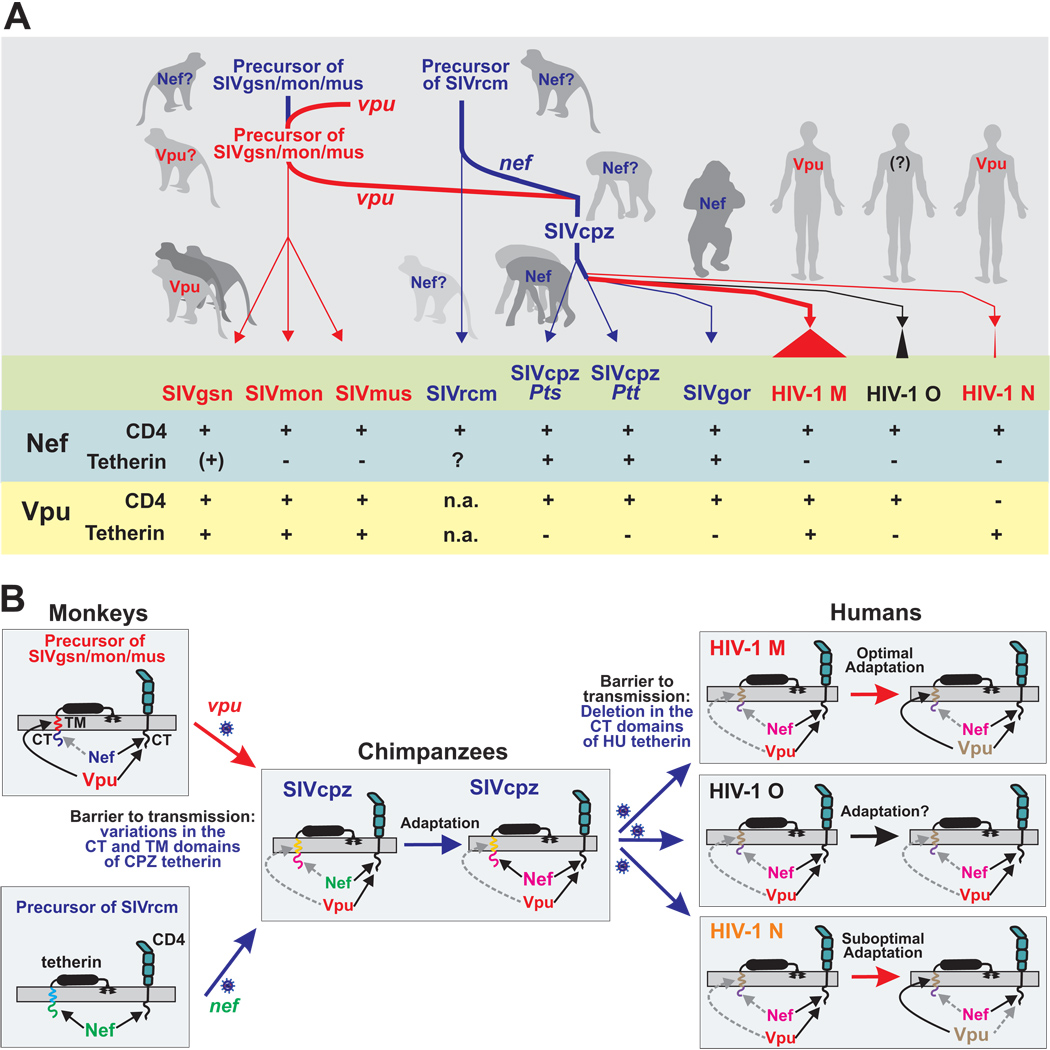
In this study, we examined the evolution of Vpu function in the primate lentiviruses whose ancestors gave rise to HIV-1. We show that the two major functions of the HIV-1 NL4-3 Vpu protein, i.e. CD4 degradation and enhancement of virion release by counteracting tetherin, are conserved in other vpu-containing non-human primate lentiviruses (summarized in Figure 6A). Nonetheless, important differences were noted. First, Vpu appears to require some host-specific adaptation in order to be an effective tetherin antagonist since Vpus from SIVgsn, SIVmus and SIVmon were only active against monkey tetherins and those from HIV-1 group M were only effective against ape and human tetherins (Figure 3B). In comparison, host-specific adaptation seems less important for CD4 downmodulation since the great majority of primate lentiviral Vpu and Nef proteins is active against human CD4 (Figure 2; Schindler et al., 2006). Second, we found that SIVcpz employs Nef rather than Vpu to antagonize tetherin (Figure 3C and 4C). This finding was unexpected given that the vpu genes of SIVcpz, SIVgsn, SIVmus and SIVmon share a common ancestor (Schindler et al., 2006) and because previous studies suggested that only SIVs that do not contain a vpu gene use Nef to antagonize tetherin (Jia et al., 2009; Zhang et al., 2009). Third, we found that SIV Nefs generally fail to counteract HU tetherin because a deletion in the TM domain of the human restriction factor disrupts its susceptibility to Nef (Figure 4A) (Jia et al., 2009; Zhang et al., 2009). Together, these findings suggest that tetherin poses a significant - albeit not insurmountable - barrier to transmission of non-human primate lentiviruses to humans. Moreover, it seems that only the pandemic HIV-1 M group has entirely cleared this hurdle because its Vpu protein evolved to become an effective tetherin antagonist, yet maintained its ability to degrade CD4. We therefore speculate that the acquisition of a fully functional Vpu protein by the main group of HIV-1 contributed to the global spread of the AIDS pandemic.
Figure 6. Hypothetical model of tetherin driven Vpu and Nef evolution.
(A) Schematic of the acquisition of vpu and the subsequent transfer of vpu and nef genes from monkeys to chimpanzees and to humans by zoonotic primate lentiviral transmissions. The events that led to the emergence of pandemic HIV-1 group M are indicated by thick lines. Nef-mediated tetherin antagonism is indicated by blue and Vpu-mediated tetherin antagonism by red lines, respectively. The lower part summarized the results of the present study and previous data on Nef function. Note that the upper part is hypothetical as the ancient vpu and nef genes are not available for analysis.
(B) Tetherin-driven evolution of Vpu and Nef function. Nef is present in all primate lentiviruses and interacts with the cytoplasmic tails (CTs) of tetherin and CD4. Vpu was initially acquired by a precursor of SIVgsn/mus/mon and also targets the CT of CD4 but antagonizes tetherin by interacting with its transmembrane (TM) domain. The vpu-containing precursor of SIVgsn/mon/mus (upper left) recombined with that of SIVrcm (lower left) in chimpanzees and the resulting recombinant virus, i.e. SIVcpz, was subsequently transmitted to humans. Since Nef and Vpu inhibit tetherin in a species-specific manner they were most likely initially poorly active against CPZ tetherin. Subsequently, Nef evolved to become an effective tetherin antagonist in chimpanzees (middle). Upon cross-species transmission of SIVcpz to humans, however, this Nef function was most likely disrupted by a unique deletion in the CT of the human restriction factor. Subsequently, Vpu evolved to counteract HU tetherin during the emergence of pandemic HIV-1 M strains (upper right). In comparison, HIV-1 O Vpus remained poor tetherin antagonists (middle right) and those from HIV-1 N lost the CD4 degradation activity (lower right). Matching colors indicate that Vpu and Nef recognize their TM and CT interaction sites in the tetherin molecules. Broken grey arrows indicate disrupted or impaired activities.
Switching between Nef- and Vpu-mediated antagonism of Tetherin may have Facilitated Primate Lentiviral Zoonoses
It may seem surprising that SIVcpz uses Nef to antagonize tetherin but switched to use Vpu following transmission to humans. Although further studies are necessary to fully elucidate the evolution of Vpu and Nef functions, a possible scenario is outlined in Figure 6. Recent data show that some primate lentiviruses that do not encode a vpu gene use Nef to antagonize tetherin (Jia et al., 2009; Zhang et al., 2009). Our observation that Vpus from SIVgsn, SIVmus and SIVmon are all capable of counteracting their cognate tetherin suggests that this function was already present in the vpu-containing common ancestor of these viruses (Figure 6B, upper left). Evolutionary analyses indicate that SIVcpz received vpu from the precursor of the SIVgsn/mus/mon lineage, while its nef gene is derived from the SIVrcm lineage (Bailes et al., 2003; Schindler et al., 2006). Thus, during the adaptation of SIVcpz to its chimpanzee host, either Nef or Vpu could have served as the tetherin antagonist. Because both Nef and Vpu antagonize tetherin in a host-specific manner, it is likely that initially they were both only poorly active against the CPZ tetherin (Figure 6B, middle). Our data show that Nef subsequently evolved to become an effective antagonist of CPZ tetherin, possibly because the cytoplasmic tail (CT) of the chimpanzee tetherin differs less from that of monkey tetherins than the TM domain which governs susceptibility to Vpu (Figure 3A). Furthermore, the recombinant virus that comprises SIVcpz (Bailes et al., 2003) may have had a selective advantage over its non-mosaic precursors because its Vpu was capable of degrading CD4 while its Nef was capable of exerting an anti-tetherin activity. Upon cross-species transmission of SIVcpz to humans, however, the Nef-mediated tetherin antagonism was lost, most likely because a deletion in the cytoplasmic domain of the human tetherin abrogated its sensitivity to Nef. This deletion, which may have evolved in humans following encounters with other pathogens, made it difficult, if not impossible, for Nef to restore its anti-tetherin function. Hence, none of the 14 HIV-1 group M, N and O Nefs that we tested had an appreciable activity against HU tetherin. In contrast, the SIVgor Nef was able to adapt to antagonize the GOR tetherin, most likely because that restriction factor differs from that of CPZ only by two amino acid substitutions in the CT domain. Consistent with this hypothesis are recent findings showing that SIVsmm and SIVmac Nefs are antagonists of monkey but not human tetherin (Jia et al., 2009; Zhang et al., 2009). Furthermore, HIV-2, which originated from SIVsmm, appears to use Env to facilitate the release of progeny virions from human cells (Bour and Strebel, 1996), although it remains to be elucidated whether this effect is due to Env-mediated tetherin antagonism. Our data show that the HIV-1 group M Vpu evolved to become an effective antagonist of HU tetherin (Figure 6B, upper right). Thus, the flexibility to switch between Nef and Vpu (which is lacking in HIV-2) to counteract tetherin may have facilitated two cross-species transmission events which may otherwise have been less successful: (i) the transmission of SIVgsn/mus/mon to chimpanzees and (ii) the transmission of SIVcpz to humans (Figure 6).
Multi-functionality of Accessory Proteins May Increase the Potential for Cross-Species Transmission of Primate Lentiviruses
In contrast to their effect on tetherin, the ability of SIV Vpus to degrade CD4 has been well preserved, and this is also true for most SIV Nef proteins (Schindler et al., 2006). Thus, at least two primate lentiviral accessory genes, i.e. nef and vpu, that antagonize host restriction factors in a species-specific manner also have activities that seem largely independent of the host species. It thus seems beneficial for primate lentiviruses to target antiviral factors by proteins that exhibit functional plasticity. A viral gene encoding a product that becomes entirely non-functional, and cannot readily adapt to a new host environment after cross-species transmission is likely prone to elimination by the rapid accumulation of mutations. However, genes encoding proteins capable of performing multiple functions, including at least one that is not species-specific, will be maintained and may evolve to regain lost activities, sometimes perhaps even at the cost of losing another activity. The acquisition of anti-tetherin activity by HIV-1 group M Vpus, and the loss of anti-CD4 activity by HIV-1 group N Vpus may represent examples of this.
Tetherin May Represent a Barrier for Cross-specicies Transmissions of SIVs to Humans
While the specificity of the Vpu/tetherin interaction resembles that of other anti-retroviral host restriction factors and their viral antagonist, such as APOBEC3G and Vif (Gaddis et al., 2004; Sawyer et al., 2004; Schröfelbauer et al., 2004; Zennou and Bieniasz, 2006), there is one important difference: other restriction factors such as APOBEC3G and TRIM5α are unlikely to pose a barrier to cross-species transmission of SIVcpz from chimpanzees to humans. Specifically, the SIVcpz Vif protein is fully capable of eliminating the antiviral activity of human APOBEC3G (Gaddis et al., 2004), while the SIVcpz capsid is not susceptible to restriction by human TRIM5α (Kratovac et al., 2008). Altogether, these previous studies suggested that SIVcpz did not have to overcome a major species barrier to transmission to humans because chimpanzees and humans are genetically closely related. Our finding that SIVcpz Vpus and Nefs were generally poorly active in antagonizing HU tetherin challenges this dogma and strongly suggests that tetherin constitutes a significant barrier to transmission. Our data also provide compelling evidence that host-specific adapation allowed pandemic HIV-1 M strains to overcome this restriction because its Vpu protein gained anti-tetherin activity while maintaining its ability to degrade CD4. In the absence of such adaptation, SIVcpz strains that have crossed the species barrier may not spread efficiently. This may explain why just one of three independent chimpanzee-to-human transmissions of SIVcpz has led to a pandemic. Our finding that Vpus from all group M clades are capable of antagonizing tetherin suggests that this function was acquired early in the evolution of HIV-1 group M. Moreover, the ten vpu alleles from transmitted/founder HIV-1 strains that are responsible for productive infection in humans all antagonized tetherin. In contrast, Vpu proteins of HIV-1 group O are poor tetherin antagonists and this is also true for this group’s Nef proteins. It will thus be interesting to investigate whether HIV-1 O has evolved other mechanisms to counteract this restriction factor (similar to HIV-2). Finally, Vpu alleles from some HIV-1 group N strains exhibit Vpu mediated anti-tetherin activity (Figure 3C) but appear to have lost the CD4 degradation function (Figure 2B). Thus, imperfect adaptation of HIV-1 O and N to the human host may explain why they have spread substantially less efficiently in the human population than HIV-1 M.
Conclusions
In summary, our data suggest that evolutionary switches between Nef- and Vpu-mediated tetherin antagonism facilitated the cross-species transmission that ultimately led to the emergence of pandemic HIV-1 strains. The differences in HIV-1 group M, O and N Vpu function identified here should provide important insights into the role of Vpu and tetherin antagonism in virus transmission and pathogenesis. It has been previously shown that the primate lentiviral lineage that gave rise to HIV-1 lost its ability to suppress T-cell activation (Schindler et al., 2006). Thus, HIV-1 may need a particularly effective tetherin antagonist for effective virus release because it induces higher levels of inflammation (and thus possibly tetherin) than non-pathogenic natural SIV infections (Kirchhoff, 2009). Finally, the present report shows that - with respect to Nef and Vpu function - SIVgsn, SIVmon and SIVmus more closely resemble HIV-1 M, than SIVcpz or other known SIVs. Thus, studies of these natural SIV infections may provide relevant insights into the pathogenesis of AIDS in humans.
MATERIALS AND METHODS
Vpu, Nef and Tetherin Expression Vectors
Constructs expressing N-terminally HA-tagged tetherins have been described previously (Neil et al., 2008; McNatt et al., 2009). Vpu alleles from the different subtypes of HIV-1 M and some tetherin variants were chemically synthesized (GenScript) (Table S1, Table S2). Cloning of vpu, nef and tetherin alleles into the bi-cistronic CMV-based pCGCG expression vector coexpressing the green fluorescent protein (GFP) was essentially performed as described previously (Greenberg et al., 1998; Schindler et al., 2003). See the Supplemental Experimental Procedures for details.
Proviral HIV-1 Constructs
Generation of HIV-1 (NL4-3-based) proviral constructs containing a deletion of nucleotides 3 to 120 of the vpu coding frame (ΔVpu) or functional nef genes followed by an IRES and the eGFP gene (Nef/eGFP) has been described previously (Rucker et al., 2004; Schindler et al., 2003; Schindler et al., 2006). To generate vpu-defective derivatives of the HIV-1 Nef/eGFP constructs (ΔVpu/Nef/eGFP), restriction fragments encompassing the nef-IRES-eGFP region were cloned into the ΔVpu proviral HIV-1 construct using the unique StuI and NotI restriction sites in the env gene and the flanking vector sequence, respectively. Proviral HIV-1 NL4-3 vectors expressing various vpu alleles independently of overlapping env gene sequences were generated using splice-overlap extension PCR and standard cloning techniques as outlined in the Supplemental Experimental Procedures. All PCR-derived inserts were sequenced to confirm their accuracy.
Amplification of HIV-1 N and O nef and vpu alleles
Blood samples were obtained from HIV-1 infected people living in Cameroon. Genomic DNA was extracted from buffy-coat and fragments spanning the nef and vpu genes were amplified by nested or semi-nested PCR. For details, see the Supplemental Experimental Procedures.
Cell culture and transfections
Cells were cultured and transfected as described in the Supplemental Experimental Procedures.
CD4 cell surface expression
To determine the effect of Vpu on CD4 cell surface expression, 293T cells were transfected by the calcium phosphate method in triplicate with 1 µg of a CD4 expression vector and 5 µg of pCGCG GFP/Vpu or GFP/Vpu-AU1 constructs expressing GFP alone or together with untagged or tagged Vpus. Two days post-transfection CD4 expression was examined by FACS analysis as described previously (Schindler et al., 2003). See Supplemental Experimental Procedures for details.
Virus yield and infectivity assays
Virus yield and infectivity assay were conducted in principle as described (Munch et al., 2007; Neil et al., 2008; McNatt et al., 2009; Zhang et al., 2009). For details, see the Supplemental Experimental Procedures.
Western blot analysis
The effects of Vpu and Nef on viral particle release were monitored as described previously (Neil et al., 2008; NcNatt et al., 2009; Zhang et al., 2009). Expression of Vpu and tetherin, as well as of viral genes by proviral NL4-3 UIE constructs was examined as described in the Supplemental Experimental Procedures.
Sequence analyses and accession numbers
Tetherin and Vpu amino acid sequences were aligned using Multalign and ClustalW2, respectively, followed by some manual editing. Newly derived vpu, nef and tetherin sequences will be submitted to the GenBank sequence database and accession numbers will be provided upon acceptance of the manuscript.
Statistical analysis
All statistical calculations were performed with a two-tailed unpaired Students-t-test using Graph Pad Prism Version 5.0. P values <0.05 were considered significant. Correlations were calculated with the linear regression module.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas Mertens for support, Daniela Krnavek, Kerstin Regensburger, Martha Mayer and Nicola Schrott for excellent technical assistance, Guido Silvestri, Ingrid Bennett and Nathalie Arhel for critical reading of the manuscript, and Welkin Johnson for the SM tetherin. The molecular p89.6, pYU-2 and pYK-JRCSF clones and TZM-bl cells were obtained through the NIH AIDS Research and Reference Reagent Program. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) and grants from National Institutes of Health to FK (RO1 AI06757), BHH (R37 AI50529, R01 AI58715, P30 AI27767) and TH (R01 AI078788).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Barlow KL, Ajao AO, Clewley JP. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona) J. Virol. 2003;77:6879–6888. doi: 10.1128/JVI.77.12.6879-6888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 1996;70:8285–8300. doi: 10.1128/jvi.70.12.8285-8300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Schubert U, Strebel K. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J. Virol. 1995;69:1510–1520. doi: 10.1128/jvi.69.3.1510-1520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler IF, Pandream I, Marxm PA, Apetrei C. HIV genetic diversity: biological and public health consequences. Curr. HIV Res. 2007;5:23–45. doi: 10.2174/157016207779316297. [DOI] [PubMed] [Google Scholar]
- Cohen EA, Terwilliger EF, Sodroski JG, Haseltine WA. Identification of a protein encoded by the vpu gene of HIV-1. Nature. 1988;334:532–534. doi: 10.1038/334532a0. [DOI] [PubMed] [Google Scholar]
- Courgnaud V, Abela B, Pourrut X, Mpoudi-Ngole E, Loul S, Delaporte E, Peeters M. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 2003;77:12523–12534. doi: 10.1128/JVI.77.23.12523-12534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Salemi M, Pourrut X, Mpoudi-Ngole E, Abela B, Auzel P, Bibollet-Ruche F, Hahn BH, Vandamme AM, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nicititans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 2002;76:8298–8309. doi: 10.1128/JVI.76.16.8298-8309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazza MC, Ekwalanga M, Nende M, Shamamba KB, Bitshi P, Paraskevis D, Saragosti S. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's Mona monkey (Cercopithecus mona denti) J. Virol. 2005;79:8560–8571. doi: 10.1128/JVI.79.13.8560-8571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis NC, Sheehy AM, Ahmad KM, Swanson CM, Bishop KN, Beer BE, Marx PA, Gao F, Bibollet-Ruche F, Hahn BH, Malim MH. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 2004;78:12041–12046. doi: 10.1128/JVI.78.21.12041-12046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Gomez LM, Pacyniak E, Flick M, Hout DR, Gomez ML, Nerrienet E, Ayouba A, Santiago ML, Hahn BH, Stephens EB. Vpu-mediated CD4 downregulation and degradation is conserved among highly divergent SIV(cpz) strains. Virology. 2005;335:46–60. doi: 10.1016/j.virol.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Cohen EA, Haseltine WA. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. USA. 1993;90:7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Iafrate AJ, Skowronski J. The SH3 domain binding surface and an acidic motif in HIV 1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Hué S, Schaller T, Verschoor E, Pillay D, Towers GJ. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 2009;5:e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLOS Pathogens. doi: 10.1371/journal.ppat.1000429. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci. USA. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 2009;7:467–476. doi: 10.1038/nrmicro2111. [DOI] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J. Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratovac Z, Virgen CA, Bibollet-Ruche F, Hahn BH, Bieniasz PD, Hatziioannou T. Primate lentivirus capsid sensitivity to TRIM5 proteins. J. Virol. 2008;82:6772–6777. doi: 10.1128/JVI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Chen MY, Willey RL, Strebel K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type 1 integral membrane protein. J. Virol. 1993;67:5056–5061. doi: 10.1128/jvi.67.8.5056-5061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-TrCP, that interacts with HIV-1 Vpu, connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- Münch J, Rajan D, Schindler M, Specht A, Rücker E, Novembre FJ, Nerrienet E, Müller-Trutwin MC, Peeters M, Hahn BH, Kirchhoff F. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol. 2007;81:13852–13864. doi: 10.1128/JVI.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnsonm WE, Neil SJ, Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Paul M, Jabbar MA. Phosphorylation of both phosphoacceptor sites in the HIV-1 Vpu cytoplasmic domain is essential for Vpu-mediated ER degradation of CD4. Virology. 1997;232:207–216. doi: 10.1006/viro.1997.8541. [DOI] [PubMed] [Google Scholar]
- Rucker E, Grivel JC, Münch J, Kirchhoff F, Margolis L. Vpr and Vpu are important for efficient human immunodeficiency virus type 1 replication and CD4+ T-cell depletion in human lymphoid tissue ex vivo. J Virol. 2004;78:12689–12693. doi: 10.1128/JVI.78.22.12689-12693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. Inhibition of Lassa and Marburg Virus Production by Tetherin. J. Virol. 2009;83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, Muller MN, Emery M, Goldenberg DA, Lwanga JS. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J Virol. 2003;77:2233–2242. doi: 10.1128/JVI.77.3.2233-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Würfl S, Benaroch P, Greenough TC, Daniels R, Easterbrook P, Brenner M, Münch J, Kirchhoff F. Downmodulation of Mature MHC Class II and Up-regulation of Invariant Chain Cell Surface Expression are Well Conserved Functions of Human and Simian Immunodeficiency Virus nef alleles. J. Virol. 2003;77:10548–10556. doi: 10.1128/JVI.77.19.10548-10556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Münch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Müller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Schröfelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl. Acad. Sci. USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Strebel K. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Klimkait T, Martin MA. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 2009;83:1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Peeters M. The Origins of HIV and Implications for the Global Epidemic. Curr Infect Dis Rep. 2007;9:338–346. doi: 10.1007/s11908-007-0052-x. [DOI] [PubMed] [Google Scholar]
- Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SK, Connole M, Sullivan JS, Choe H, Carville A, Farzan M. A New World primate deficient in tetherin-mediated restriction of human immunodeficiency virus type 1. J. Virol. 2009;83:8771–8780. doi: 10.1128/JVI.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. Nef Proteins from Simian Immunodeficiency Viruses Are Tetherin Antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.