Abstract
Plants are exposed to continual changes in the environment. The daily alternation between light and darkness results in massive recurring changes in the carbon budget, and leads to widespread changes in transcript levels. These diurnal changes are superimposed on slower changes in the environment. Quantitative molecular information about the numbers of ribosomes, of transcripts for 35 enzymes in central metabolism and their loading into polysomes is used to estimate translation rates in Arabidopsis rosettes, and explore the consequences for important sub-processes in plant growth. Translation rates for individual enzyme are compared with their abundance in the rosette to predict which enzymes are subject to rapid turnover every day, and which are synthesized at rates that would allow only slow adjustments to sustained changes of the environment, or resemble those needed to support the observed rate of growth. Global translation rates are used to estimate the energy costs of protein synthesis and relate them to the plant carbon budget, in particular the rates of starch degradation and respiration at night.
Keywords: Arabidopsis, polysomes, quantitative RT–PCR, ribosome, translation
Introduction
Plant growth is driven by photosynthetic assimilation of carbon (C). Nutrients like nitrate are absorbed by the roots and converted to amino acids in the leaves using light energy, or imported C in the roots. Sucrose and amino acids are transported to the shoot and root apex to support growth of more leaves and roots. Plants are unavoidably exposed to changes in the environment. One of the most striking changes is the daily alternation of light and darkness, which leads to an extreme and repeated alternation between two states, occurs every day in the natural environment, and can be precisely simulated in laboratory experiments. It results in a large positive balance of energy and C in the light period, and a deficit in the dark period. This is buffered by storing some of the newly fixed C as starch, and remobilizing it to support metabolism and growth during night (Geiger et al, 2000; Smith and Stitt, 2007). However, we do not know how energetically expensive processes like protein synthesis are regulated during these marked diurnal changes in the plant's energy budget.
Plants are also exposed to slower changes that occur in a time range of days or weeks, as a result of changing weather patterns and seasonal changes. A very large portion of the total leaf protein is invested in a single metabolic process. Owing to its very low rate of catalysis (Kcat=3 s−1), RubisCO represents over 30% of the total protein in a leaf (Farquhar et al, 2001; Zhu et al, 2007). Large amounts of protein are also invested in the synthesis of chlorophyll-binding proteins and other enzymes involved in the Calvin cycle and pathways for carbohydrate and amino acid synthesis. This raises the question about how plants integrate their response to the environment over a wide range of time spans to generate appropriate levels of proteins for photosynthesis and growth.
Thousands of genes undergo diurnal changes of their transcript level (Bläsing et al, 2005) driven by the circadian clock, light and sugars (Usadel et al, 2008). This includes many transcripts that encode enzymes in central metabolism. Gibon et al (2004b) developed a robotized platform to profile the maximum activities of over 20 enzymes, most of which show rather small diurnal changes (Smith et al, 2004; Gibon et al, 2004b). Although transcripts respond within hours, changes of enzyme abundances require days to adjust when plants are transferred to continuous darkness (Gibon et al, 2004b, 2006). Gibon et al (2004b) hypothesized that the rate of translation is so slow that several days are required to produce a major change in protein abundance. As a result, the rapid transient changes of transcripts would be integrated over a longer period of time to set the levels of enzymes and other proteins. This would buffer the enzymatic capacities in central metabolism against recurring changes caused by the light–dark cycle, while allowing them to adjust to sustained changes in the surroundings.
Protein synthesis occurs by the recruitment of transcripts to ribosomes, to form polysomes. Its synthesis represents a major portion of the total ATP consumption in animal and plant cells (Hachiya et al, 2007; Pace and Manahan, 2007; Proud, 2007). Energy is also required for the synthesis of amino acids. The conversion of nitrate to amino acids requires about five ATP molecules per amino acid (Penning de Vries, 1975; Hachiya et al, 2007). The synthesis of ribosomes requires energy, and diverts resources from other cellular components. Eukaryotic ribosomes typically contain one molecule of each of the four different ribosomal RNA (rRNA) species and one molecule of ca. 80 different ribosomal proteins (Perry, 2007). Ribosomal RNA and proteins represent >80 and 30–50% of the total RNA and protein, respectively, in a growing yeast cell (Warner, 1999; Perry, 2007). There is selective pressure to achieve a parsimonious use of the translational machinery (Beilharz and Preiss, 2007; Lackner et al, 2007). In budding yeast, up to 85% of the ribosomes are present in the polysomes. The ribosome density in polysomes is about one-fifth of the theoretical maximum (Arava et al, 2003; MacKay et al, 2004), which is consistent with the view that translation is generally regulated by the rate of initiation. A twofold decrease of transcript levels for some ribosomal proteins leads to a ribosome deficit and a minute growth reduction phenotype in Drosophila, indicating that the overall ribosomal number limits protein synthesis and growth (Perry, 2007).
The rate of translation of a given transcript species depends on transcript abundance, the proportion present in polysomes (often termed ‘ribosome occupancy'), the number of ribosomes present on the transcript (often termed ‘ribosome density'), and their speed of progression along the transcript (Arava et al, 2003; Beilharz and Preiss, 2004; Beyer et al, 2004; Brockmann et al, 2007). On average, about 70% of a given transcript species is occupied by ribosomes. Similar percentages of ribosomes (60%) and transcripts (59–82%) are loaded into polysomes in plants (Kawaguchi et al, 2004; Kawaguchi and Bailey-Serres, 2005).
In the following article, we describe the methods that allow quantitative analysis of ribosome and transcript concentrations, and polysome composition in Arabidopsis rosettes. These data allow us to predict translation rates, both globally, and for individual enzymes in central metabolism. We explore the consequences of these molecular events for important sub-processes in plant growth. First, we compare the rates of synthesis with the protein abundance to predict which enzymes are likely to be subject to rapid turnover. Second, we use these molecular data to estimate the costs of protein synthesis and relate to the C budget, in particular the rates of starch degradation and respiration at night.
Results
Experimental strategy
Figure 1 outlines our experimental strategy. Ribosome copy numbers are measured using quantitative real time RT–PCR (qRT–PCR) for rRNA, and polysome fractionation was used to estimate the proportion actively involved in translation. This information is used to estimate the overall rate of protein synthesis. qRT–PCR is combined with polysome fractionation to estimate the copy number of transcripts in polysomes, including 84 that encode enzymes involved in central plant metabolism. This information is used to estimate the rates of translation of individual transcripts. In parallel, quantitative proteomics and robotized measurements of maximum enzyme activities are used to provide two independent estimates of the amounts of these enzymes in the leaves. By comparing the estimated rates of synthesis with the measured abundance of total and individual proteins, and the rate of growth, it is possible to predict whether the global rate of protein synthesis and the rates of synthesis of individual proteins are of the same order as that requited for growth, or whether proteins are subject to rapid turnover.
Figure 1.
Quantitative analysis of translation in Arabidopsis rosette leaves.
Polysome fractionation
In preliminary experiments, we collected whole rosettes from 5-week-old wild-type Arabidopsis growing in a 12-h light–dark cycle at 6 times during the diurnal cycle, fractionated the material by centrifugation, and collected three fractions: the non-polysomal fraction (NPS) at the top of the gradient, the small polysomal fraction (SPS) with an estimated 2–4 ribosomes per polysome, and the large polysomal fraction (LPS) with an estimated ⩾5 ribosomes per polysome. The distribution of total RNA in polysome gradients was monitored by measuring A254 (Supplementary Figure 1). The proportion in the LPS decreased by about twofold during the night, and recovered within 2 h in the next light period (Supplementary Figure 2). In subsequent experiments, plants were collected at the end of the night (‘dark') period and after 2 h of illumination (‘light'), corresponding to the largest changes in polysome abundance.
Quantification of ribosomes
A254 does not distinguish between transcript RNA and rRNA, and does not distinguish between cytosolic, plastid and mitochondrial rRNA. Two complementary approaches were taken to quantify cytosolic and plastid ribosomes.
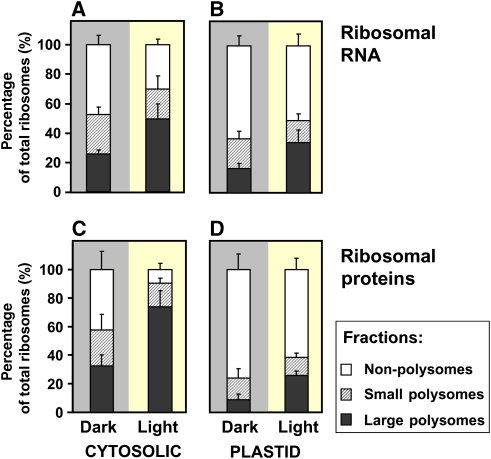
The first approach used qRT–PCR to determine the concentrations of 18S, 16S and 18S-like rRNAs, corresponding to the rRNA in the small subunit of the cytosolic, plastid and mitochondrial ribosomes, respectively. To allow precise quantification, the qRT–PCR data were normalized on four artificial control RNAs, which were added at a known concentration before purification of the rRNA from the gradient fractions. The total estimated concentration of ribosomes is about 0.10 nmol g−1 fresh weight (FW) (see, Table I and Calculations and assumptions section). Cytosolic ribosomes were threefold more abundant than plastid ribosomes, and 30-fold more abundant than mitochondrial ribosomes. This is in agreement with earlier reports that 26–36% of the total ribosomes in leaves are present in plastids (Dyer et al, 1971). The distribution of different rRNA species in the polysome gradient suggests that about twofold more ribosomes were present in the polysomes in the light period than in the dark period (Figure 2A and C). The proportion of cytosolic rRNA in the LPS fraction increased from 26 to 50%, whereas the proportion in SPS decreased slightly (from 27 to 20%), as expected in the case if the polysome population is shifting towards a higher number of ribosomes per transcript, and free cytosolic ribosomes decreased from 47 to 30%. A qualitatively similar picture was found for plastid rRNA, except that a smaller proportion was found in polysomes (Figure 2B). This conclusion is supported by the distribution of ribosomal proteins (Figure 2D). The distribution of mitochondrial rRNA was not investigated.
Table 1.
Ribosome content in the cytosol, plastids and mitochondria
| Ribosome content (mol g−1 FW) | ||
|---|---|---|
| Dark period | Light period | |
| Cytosol | 7.62E−11±1.56E−11 | 7.27E−11±1.79E−11 |
| Plastid | 2.64E−11±6.66E−12 | 2.57E−11±7.42E−12 |
| Mitochondrion | 2.25E−12±4.42E−13 | 2.21E−12±3.96E−13 |
The results are represented as mean±s.d. of three biological samples.
Estimation was done by qRT–PCR of SSU rRNA subunits of the cytosolic, plastidic and mitochondrial ribosomes.
Figure 2.
Distribution of ribosomes in different polysomal fractions in the dark and the light periods. (A, B) Ribosome number in each fraction was calculated by determining the amount of the SSU rRNA for cytosolic and plastid ribosomes by qRT–PCR, assuming each rRNA copy corresponds to one ribosome. (C, D) Ribosomal protein abundance in each fraction, calculated by normalizing the summed emPAI values for ribosomal proteins on total protein in the fraction.
The second approach used relative quantitative proteomics based on emPAI values (Ishihama et al, 2005) to estimate the abundance of ribosomal proteins in the polysome gradient fractions (Figure 2C and D). Briefly, the number of identified peptides per protein are corrected for the number of possible peptides from that protein, and taken as quantitative measure of the protein abundance (see Calculations and assumptions section). This approach confirmed that about twofold more ribosomes are present in polysomes in the light period than in the dark period, and that a larger proportion of cytosolic ribosomes than plastid ribosomes are present in polysomes. The measurements of protein abundance indicate a higher proportion of ribosomes in polysomes than the measurements of rRNA for cytosolic ribosomes. This might be for technical reasons. The NPS fraction is a complex matrix with a high proportion of proteins with other biological functions and emPAI may underestimate ribosomal proteins, especially in the light period, when they are strongly depleted in NPS.
Taken together, both approaches reveal marked changes in ribosomal loading into polysomes between the dark period and the light period, with 53–58% of the cytosolic ribosomes being loaded in the dark period, and 70–90% in the light period. The values in the light are similar to those in rapidly growing yeast (see Introduction).
Global rates of protein synthesis compared with growth requirements
Information about the numbers of ribosomes and the proportion found in polysomes was used to estimate the overall rate of protein synthesis. The summed concentration of cytosolic, plastid and mitochondrial ribosomes is about 0.1 nmol g−1 FW (see Table I). Assuming that a ribosome adds 3 amino acids per s (see Calculations and assumptions section), these could catalyze the addition of 26 μmol amino acids per g FW day−1, equivalent to the synthesis of about 3 mg protein g−1 FW day−1. The actual rate of protein synthesis is lower, because only 70% of the ribosomes are in polysomes in the light period, and 40% in the dark period (Figure 2), resulting in an estimated synthesis rate of 1.8 mg protein g−1 FW day−1. Under the conditions used in these experiments, Arabidopsis contains about 15 mg protein g−1 FW (Gibon et al, 2004a, 2009; Hannemann et al, 2009) and grows exponentially (see e.g. Tschoep et al, 2009), with a relative growth rate of 0.15–0.20 g FW g−1 FW day−1, which is equivalent to the synthesis of 2.2–3.0 mg protein g−1 FW day−1. The rate of protein synthesis estimated from ribosome copy number therefore resembles that required for the observed rate of growth.
Ribosomal occupancy of transcripts encoding enzymes in central metabolism
We next investigated ribosomal occupancy of transcripts for 98 genes (Supplementary Table I) including the major members of the gene families for 35 enzymes of central metabolism (84 transcripts). We also included AtCAB1 and AtCAB2 as representatives of photosynthetic and circadian-regulated genes whose expressions peak at the beginning of the day (Ernst et al, 1990), three circadian-regulated genes whose expressions peak at the end of the day (GER3, CAT3, GRP7) and 9 transcripts for ‘house-keeping' genes that are frequently used for normalization of transcriptional analyses (Czechowski et al, 2005). They are all nuclear encoded, except for the plastid-encoded RubisCO large subunit (RBCL).
We used qRT–PCR to investigate the distribution of these 98 transcripts in the polysome gradients. To allow precise quantification, the qRT–PCR data were normalized on four artificial control RNAs that were added at a known concentration before purification of RNA in the gradient fractions. This allowed copy numbers of each transcript species to be determined per fraction. In addition to being necessary for subsequent calculations (see below), this bypassed the problems associated with normalization of transcript levels between fractions in polysome gradients. Transcript levels are usually normalized to total RNA. However, the relative levels of transcript and rRNA probably change across a polysome gradient. ‘House-keeping' genes are often used for normalization of qRT–PCR data between different organs or environment treatments, but cannot be used for this purpose in polysome fractions, because it is not known whether they are subject to translational regulation. The proportion of transcript in each fraction of the polysome gradient can be easily calculated using qRT–PCR data in combination with internal standards, by simply comparing the numbers of transcripts against internal standard as a control across the whole gradient. The transcript concentrations in the rosette (unfractionated RNA) and polysome fractions, and the ribosomal occupancy (i.e. the proportion of a transcript in the SPS and LPS fractions) are provided in Supplementary Table II, along with the information about the length of the coding sequence, and length and molecular weight of the encoded peptide.
Ribosomal occupancy of the 98 transcripts analyzed, varied between 40–95% in the dark period, and 50–90% in the light period (Figure 3). Most transcripts show a marked increase in occupancy between the dark and the light periods, with an increase of between 5 and 55% in absolute terms, and between 10 and 100% relative to the value in the dark period. ‘House-keeping' genes showed a similar response to other transcripts. RBCL, RBCS-1B and RBCS-2B did not show any increase in ribosomal occupancy in the light (see below for further discussion). The increase in ribosomal occupancy in the light is smaller than the twofold increase of ribosome in polysomes (Figure 2), indicating that the average number of ribosomes per transcript probably increases in the light period.
Figure 3.
Scatter plot comparing the ribosome occupancy of 98 transcripts in the night and in the light period. Ribosome occupancy was calculated as (SPS+LPS)/(NPS+SPS+LPS). Green circles represent photosynthetic proteins, green filled circles indicate RBCS gene family and RBCL, and red circles indicate genes that are classified as ‘house-keeping' in expression studies. The plot is generated using data provided in Supplementary Table II.
Ribosomal occupancy was weakly but significantly dependent on transcript concentration, with a Pearson's R2 value of 0.065 (P=0.011) in the dark period, and 0.102 (P=0.001) in the light period (Figure 4). Transcript concentrations varied by three orders of magnitude. There was a large range of ribosomal occupancy (from 40 to >80%) for transcripts with the same concentration. Some of this was due to the effect of light, but there was still a large range when transcripts are considered in one condition. This indicates that ribosomal occupancy depends more on individual features of transcripts than their concentrations.
Figure 4.
Scatter plot for transcript abundance versus ribosome occupancy in the night and the light period. Ribosomal occupancy was calculated as (SPS+LPS)/(NPS+SPS+LPS). Blue and orange symbols denote plant material collected in the dark and light periods, respectively. Filled symbols denote the RBCS gene family (•) and RBCL (▪). Ribosomal occupancy is weakly, but significantly dependent on transcript concentration in the dark period (Pearson's R2=0.065, P-value=0.011) and light period (Pearson's R2=0.102, P-value=0.001). The plot is generated by using the data provided in Supplementary Table II.
For almost all transcripts except those encoding RubisCO subunits, there are 10–100 times lesser transcripts in the SPS fraction than in the LPS fraction. This is shown by the low log10 ratio of SPS/LPS in Figure 5. The proportion of transcript in SPS compared with LPS decreases as the proportion in the NPS fraction decreases, resulting in a near-linear relation in this semi-log plot. This empirical relationship indicates that initiation and ribosome progression are determined in a similar manner for all these transcripts. It is consistent with initiation being the limiting step; an increased probability of initiation will result in a decreased fraction of the transcript in the NPS fraction, and will also result in an increased average density of ribosomes per transcript, resulting in a decrease of the proportion found in the SPS compared with the LPS fraction. However, the amount of transcript in the NPS fraction is higher than would be expected from a simple binomial distribution (data not shown); indicating that the probability that ribosomes are recruited to a free transcript is lower than for a transcript that already has bound ribosomes.
Figure 5.
Relation between the fraction of transcript in the non-polysomal fraction (NPS) and the distribution of transcript between the small (SPS) and large polysomal (LPS) fractions. Blue and orange symbols denote plant material collected in the dark and light periods, respectively. Filled symbols denote the RBCS gene family (•) and RBCL (▪). The plot is generated using data provided in Supplementary Table II.
The five solid symbols in Figures 3, 4 and 5 depict the response of RBCS-1A, RBCS-1B, RBCS-2B, RBCS-3B (the small nuclear-encoded subunit of RubisCO) and RBCL (the large plastid-encoded subunit of RubisCO). These five transcripts deviate strongly from the general relationship between NPS and SPS/LPS. These five transcripts show a high occupancy in the dark as well as the light period, and a large proportion of the transcript is present in the SPS fraction (18–39% and 9–25% in the dark period and light period, respectively) (Figure 5). This could indicate that RBCL and RBCS transcripts are subject to complex translational control (see Discussion section).
Estimation of translation rates
The rate of synthesis of the proteins encoded by these 98 transcripts was estimated from the transcript abundance in the SPS and LPS fractions, multiplied by the ribosome density per translating transcript (see Calculations and assumptions section, and Supplementary Table II). The calculation assumes an elongation rate of 3 amino acids per ribosome per s, an average of 3 ribosomes per transcript in the SPS fraction, and a ribosome density of 6.6 ribosomes per kb coding sequence, (Brandt et al, 2009) in the LPS fraction.
The estimated rates of protein synthesis (mol protein g−1 FW h−1) ranged from 2.5E−15 to 2.9E−09 in the dark period, and 6.5E−15 to 4.3E−10 in the light period (Table II and Supplementary Table II). Among the enzymes involved in primary metabolism, RubisCO is the most rapidly synthesized enzyme, reflecting the high abundance of this enzyme in leaves (see Introduction section). Other rapidly synthesized enzymes include several Calvin cycle enzymes (e.g. aldolase, NADP–GAPDH, PGK and TPI), enzymes involved in nitrogen assimilation (e.g. AlaAT, NR and GS), and NAD–GAPDH and NAD–MDH. The relatively high rates of synthesis of PEP carboxylase, NADP–IDH, aconitase and PK compared with other glycolytic enzymes may reflect the fact that these enzymes are required to synthesize 2-oxoglutarate, which acts as the C acceptor during nitrate and ammonium assimilation. The relatively high rate of synthesis of glycerate kinase may be related to the fact that in leaves this enzyme is required for photorespiration. Fluxes through this pathway are roughly 15–20% of those through photosynthesis (Zhu et al, 2007), and much higher than in respiratory metabolism. Most enzymes showed an estimated 50–100% increase in the rate of synthesis in the light period compared to the dark period. To further interpret the biological significance of these rates of synthesis, we compared them with the estimated amount of each enzyme in the rosette.
Table 2.
Estimated rates of protein synthesis of the different enzymes in Arabidopsis rosette in the dark and light periods
| Enzyme | Estimated translation rate (mol h−1 g−1 FW) | |
|---|---|---|
| Dark period | Light period | |
| Ribulose-1,5-bisphosphate carboxylase (RubisCO) | 6.34E–10 | 8.60E–10 |
| Fructose-bisphosphate aldolase (aldolase) | 2.84E–11 | 5.97E–11 |
| NADP–glyceraldehyde 3-phosphate dehydrogenase (NADP–GAPDH) | 2.94E–11 | 5.13E–11 |
| Alanine aminotransferase (AlaAT) | 2.35E–11 | 3.40E–11 |
| NAD–glyceraldehyde 3-phosphate dehydrogenase (NAD–GAPDH) | 1.48E–11 | 2.60E–11 |
| NAD–malate dehydrogenase (NAD–MDH) | 1.60E–11 | 2.03E–11 |
| Nitrate reductase (NR) | 2.30E–11 | 9.20E–12 |
| Glutamine synthetase (GS) | 1.41E–11 | 1.42E–11 |
| Phosphoglycerokinase (PGK) | 1.33E–11 | 1.41E–11 |
| Triose phosphateisomerase (TPI) | 8.24E–12 | 1.18E–11 |
| ADP-glucose pyrophosphorylase (AGPase) | 3.54E–12 | 5.94E–12 |
| Phosphoenolpyruvate carboxylase (PEP carboxylase) | 3.90E–12 | 4.75E–12 |
| NADP–isocitrate dehydrogenase (NADP–IDH) | 5.11E–12 | 3.35E–12 |
| Transketolase (TK) | 4.01E–12 | 3.58E–12 |
| Aconitase | 2.45E–12 | 4.44E–12 |
| Pyruvate kinase (PK) | 2.27E–12 | 3.07E–12 |
| NADP–malate dehydrogenase (NADP–MDH) | 2.27E–12 | 3.05E–12 |
| Acid invertase (INV) | 2.29E–12 | 2.05E–12 |
| Glycerate kinase (GK) | 1.32E–12 | 1.56E–12 |
| Glucose-6-phosphate isomerase (PGI) | 1.09E–12 | 1.77E–12 |
| UDP-glucose pyrophosphorylase (UGPase) | 9.04E–13 | 1.63E–12 |
| Ferredoxin–glutamate synthase (Fd–GOGAT) | 1.39E–12 | 1.14E–12 |
| Phosphoglucomutase (PGM) | 8.23E–13 | 1.44E–12 |
| PPi-phosphofructokinase (PFP) | 7.25E–13 | 1.18E–12 |
| Fructose-1,6-bisphosphatase, cytosolic (cytFBPase) | 6.29E–13 | 1.25E–12 |
| Sucrose phosphate synthase (SPS) | 7.92E–13 | 9.80E–13 |
| Fructokinase (FK) | 7.56E–13 | 9.20E–13 |
| Fumarase (FUM) | 4.92E–13 | 9.92E–13 |
| Glucose-6-phosphate dehydrogenase (G6PDH) | 7.57E–13 | 6.36E–13 |
| Aspartate aminotransferase (AspAT) | 4.96E–13 | 6.22E–13 |
| NAD–isocitrate dehydrogenase (NAD–IDH) | 4.49E–13 | 5.69E–13 |
| NAD–glutamate dehydrogenase (NAD–GDH) | 5.46E–13 | 3.08E–13 |
| Glucokinase/hexokinase (HK) | 2.40E–13 | 4.57E–13 |
| ATP-phosphofructokinase (PFK) | 3.06E–13 | 3.83E–13 |
| Shikimate 5-dehydrogenase (Shikimate DH) | 2.34E–13 | 3.62E–13 |
The raw data and calculations are provided in Supplementary Table II.
Estimation of protein abundance
Protein abundance of metabolic enzymes in rosette leaves was estimated by two independent methods. In one approach, protein abundance was estimated from the maximum enzyme activity (original data provided in Supplementary Table III), corrected by literature values for the specific activity (Supplementary Table IV). In case of disagreement, the highest specific activity was chosen because an underestimate of the specific activity can easily occur due to loss of activity or incomplete purification. Enzyme activities were measured using a robotized enzyme determination platform, in which products are quantitatively determined in highly sensitive cycling assays that allow high dilution ratios and minimize the interference form other components in the extracts (Gibon et al, 2004b). For all assays, it was checked that the substrate concentration was saturating, and that the activity was linear with the amount of extract added. The results and calculations are summarized in Supplementary Table V.
In the second approach, relative protein amounts were estimated using the emPAI index (Supplementary Table VI). The emPAI index is calculated from the fraction of the number of experimentally identified tryptic peptides out of all detectable tryptic peptides within the mass range of the mass spectrometer (Ishihama et al, 2005). This method yields abundances that are linear over three orders of magnitude in protein concentration (10 nmol l−1–10 μmol l−1), although it does underestimate very highly abundant single proteins (Ishihama et al, 2005). In our data set, RubisCO subunits summed to >10% of all proteins identified, and are probably underestimated (Supplementary Table VI). The experimentally determined total protein content (15 mg g−1 FW) was used to convert the mol% values to concentrations (Supplementary Table VI). On a molar basis, proteins with photosynthetic functions contributed 30% of all identified proteins. Protein synthesis made up the second largest functional protein group (11%). Proteins with functions in amino acid metabolism, redox regulation and TCA cycle/organic acid transformation contributed with 5–6% each (Supplementary Table VI).
Both approaches are subject to experimental error and involve assumptions. Nevertheless, comparison of the resulting estimates for protein abundance (Figure 6 and Table III) revealed a highly significant agreement (Pearson's R2 =0.590, P=0.727E−06). The slope of the regression (0.73) deviated from 1, with a smaller dynamic range for the proteomics-based than for the enzyme activity-based estimates. This may reflect a slight attenuation inherent in emPAI. Four enzymes (PGI, PEP carboxylase, NADP–MDH, glycerate kinase) showed a >5-fold discrepancy between the two approaches, eight (TPI, PK, cytFBPase, NAD–MDH, aldolase, PGK, UGPase and fumarase) showed a 3–5-fold discrepancy, and the others showed <3-fold discrepancy. For four enzymes, cross-validation was not possible because no gene family members were detected in the proteomics analysis (ATP–PFK, G6PDH, HK, NAD–GDH) (see Supplementary Table VII).
Figure 6.
Relation between protein concentrations of metabolic enzymes calculated from specific enzyme activities or from the emPAI protein abundance index determined by mass spectrometric analysis. Filled symbol denotes RubisCO, which is not included in the Pearson's regression analysis. Protein abundances estimated using the two methods are highly significantly correlated (R2=0.592, P-value=0.681E−06). The plot is generated by using the data provided in Supplementary Tables V and VII.
Table 3.
Comparison of the estimated enzyme abundance in the rosette and the corresponding estimated time to synthesize the entire enzyme in the rosette in days (TP)
| Enzyme | Protein abundance (mol g−1 FW) | Days to synthesize all the protein in the rosette (TP) | |||
|---|---|---|---|---|---|
| Enzyme activities | Quantitative proteomics | Enzyme activities | Quantitative proteomics | Average | |
| Nitrate reductase (NR) | 4.38E–11 | 3.60E–11 | 0.14 | 0.12 | 0.13 |
| ATP-phosphofructokinase (PFK) | 5.96E–12 | NA | 0.73 | NA | 0.73 |
| Acid Invertase (INV) | 3.58E–11 | 4.29E–11 | 0.69 | 0.78 | 0.74 |
| Glucose-6-phosphate dehydrogenase (G6PDH) | 2.60E–11 | NA | 1.57 | NA | 1.57 |
| Glucokinase/hexokinase (HK) | 1.40E–11 | NA | 1.86 | NA | 1.86 |
| Alanine aminotransferase (AlaAT) | 9.04E–10 | 1.72E–09 | 1.35 | 2.51 | 1.93 |
| ADP-glucose pyrophosphorylase (AGPase) | 1.94E–10 | 2.13E–10 | 1.82 | 2.12 | 1.97 |
| Pyrophosphate-phosphofructokinase (PFP) | 5.24E–11 | 4.67E–11 | 2.43 | 1.65 | 2.04 |
| NADP–glyceraldehyde 3-phosphate dehydrogenase (NADP–GAPDH) | 1.12E–09 | 2.51E–09 | 1.25 | 2.98 | 2.11 |
| Ribulose-1,5-bisphosphate carboxylase (RuBisCO) | 5.17E–08 | 2.56E–08 | 2.95 | 1.28 | 2.12 |
| Glucose-6-phosphate isomerase (PGI)** | 2.27E–11 | 1.67E–10 | 0.70 | 4.78 | 2.74 |
| Phosphoenolpyruvate carboxylase (PEP carboxylase)** | 1.17E–10 | 5.61E–10 | 1.14 | 4.36 | 2.75 |
| Triose phosphate isomerase (TPI)* | 3.85E–10 | 1.02E–09 | 1.65 | 4.42 | 3.03 |
| Glutamine synthetase (GS) | 7.65E–10 | 1.35E–09 | 2.25 | 3.97 | 3.11 |
| Pyruvate kinase (PK)* | 7.75E–11 | 3.32E–10 | 1.24 | 5.13 | 3.18 |
| Fructose-1,6-bisphosphatase, cytosolic (cytFBPase)* | 2.43E–11 | 1.02E–10 | 1.21 | 6.77 | 3.99 |
| Sucrose phosphate synthase (SPS) | 1.1E–10 | 8.80E–11 | 5.37 | 3.90 | 4.64 |
| NAD–isocitrate dehydrogenase (NAD–IDH) | 5.09E–11 | 7.47E–11 | 4.23 | 5.47 | 4.85 |
| NAD–glyceraldehyde 3-phosphate dehydrogenase (NAD–GAPDH) | 2.97E–09 | 1.83E–09 | 6.55 | 3.88 | 5.21 |
| NADP–isocitrate dehydrogenase (NADP–IDH) | 4.88E–10 | 6.03E–10 | 5.03 | 6.06 | 5.54 |
| Shikimate 5-dehydrogenase (Shikimate DH) | 4.51E–11 | 4.67E–11 | 6.61 | 5.38 | 5.99 |
| NAD–malate dehydrogenase (NAD–MDH)* | 1.06E–09 | 4.17E–09 | 2.48 | 9.63 | 6.06 |
| Fructose-bisphosphate aldolase (aldolase)* | 9.96E–09 | 3.19E–09 | 10.78 | 3.41 | 7.10 |
| NAD–glutamate dehydrogenase (NAD–GDH) | 7.39E–11 | NA | 7.82 | NA | 7.82 |
| Phosphoglycerokinase (PGK)* | 1.25E–09 | 4.05E–09 | 3.81 | 12.30 | 8.06 |
| NADP–malate dehydrogenase (NADP–MDH)** | 1.06E–09 | 1.34E–10 | 16.96 | 2.31 | 9.63 |
| UDP–glucose pyrophosphorylase (UGPase)* | 1.54E–10 | 5.15E–10 | 5.52 | 14.21 | 9.86 |
| Fumarase (FUM)* | 2.70E–10 | 7.40E–11 | 17.10 | 4.64 | 10.87 |
| Fructokinase (FK) | 2.94E–10 | 2.02E–10 | 14.73 | 7.65 | 11.19 |
| Aconitase | 9.76E–10 | 7.44E–10 | 12.89 | 9.99 | 11.44 |
| Glycerate kinase (GK)** | 8.59E–10 | 9.60E–11 | 25.04 | 3.03 | 14.03 |
| Ferredoxin-dependent glutamate synthase (Fd–GOGAT) | 2.61E–10 | 5.97E–10 | 8.68 | 19.96 | 14.32 |
| Phosphoglucomutase (PGM) | 4.57E–10 | 5.40E–10 | 18.18 | 17.95 | 18.06 |
| Transketolase (TK) | 2.20E–09 | 1.25E–09 | 24.20 | 13.67 | 18.93 |
| Aspartate aminotransferase (AspAT) | 2.17E–10 | 4.59E–10 | 16.34 | 30.61 | 23.47 |
The raw data and calculations are given in Supplementary Tables V and VII. * and ** mark the enzymes that show a discrepancy bigger than 3 and 5-fold, respectively, between protein abundance calculated by the two methods. NA indicates the protein abundance column in which no peptides were quantified for the enzyme, and the TP means could not be calculated because the translation rate was not estimated for the protein quantified and/or the protein was not quantified in the samples by proteomics analysis. Calculations were based on enzymes activities and specific activities or on quantitative proteomics.
Bold type indicates values for TP that are supported by protein quantification using both enzyme activities and emPAI values. Italics indicate enzymes for which there was a large (>4-fold) discrepancy between the protein quantification provided by these two approaches. Normal face indicates where no emPAI estimates of protein abundance were available.
The protein abundances were inspected to retrospectively validate that the genes that were chosen for the qRT–PCR analysis represent major members of the gene families for these enzymes. The corresponding protein was detected by mass spectrometry for 54 of the 85 genes included in the qRT–PCR platform. No peptides were detected for any other member of the gene families, showing that the selected genes are the major family members (details are provided in Supplementary Table VII).
Comparison of protein synthesis rates with protein abundance
The estimated rates of protein synthesis were compared with the estimated protein concentration to calculate how many days it would require to synthesize the respective proteins in an Arabidopsis rosette (Table III). This will be referred to as TP. This term is used instead of T0.5, because part of the newly synthesized protein represents the flux to growth, rather than turnover of existing protein (see Discussion section). Interpretation will concentrate on cases in which similar estimates of the protein abundance were obtained from enzyme activities and emPAI.
The estimated values for TP vary by about 200-fold. By far the lowest value is for NR (0.13 days; that is, all the protein in the rosette could be synthesized in ∼3 h). Acid invertase is predicted to have a TP in the order of 18 h, AlaAT and AGPase of under 2 days, PFP, NADP–GAPDH, TPI, GS and PK in the order of 2–4 days, and cytFBPase, SPS, NAD–IDH, NAD–GAPDH, NADP–IDH, shikimate DH, and NAD–MDH of 4–7 days. For aldolase, NAD–GDH, PGK, UGPase, FUM, FK, aconitase, Fd-GOGAT, PGM, TK and AspAT, the estimated TP was even higher. Most TP values are within a factor of 2 of the rate of synthesis that would actually be required for the flux to growth (approximately 15–20% per day, see above, which would require a TP of 5–7). As discussed later, the estimated TP of 2.12 for RubisCO is probably an underestimate.
Analogous calculations were carried out for the individual protein isoforms detected by emPAI (see Supplementary Table VII). In eleven cases, TP values for different isoforms of the same enzyme were calculated, and in 60% of the cases the estimated values for individual members of a given gene families are quite similar (less than 50% r.m.s.d.), including family members for PGI (4.2 and 5.6 days), AlaAT (2.0 and 2.6 days), NADP–GAPDH (2.3–3.6 days) and PGM (11.2 and 11.1 days). Genes for the cytosolic and plastid isoforms of TPI had estimated TP values of 2.7 and over 9 days. Our proteomics analysis detected some proteins that are encoded by some of the transcripts that were included as standards for the qRT–PCR analyses of polysome gradients (Supplementary Table VII). This allowed estimates of TP values for catalase (2.4 days), RubisCO activase (2.8 days), 60S ribosomal protein L13 (2.9 days) and GER3 (5.4 days).
Diurnal responses of selected enzymes
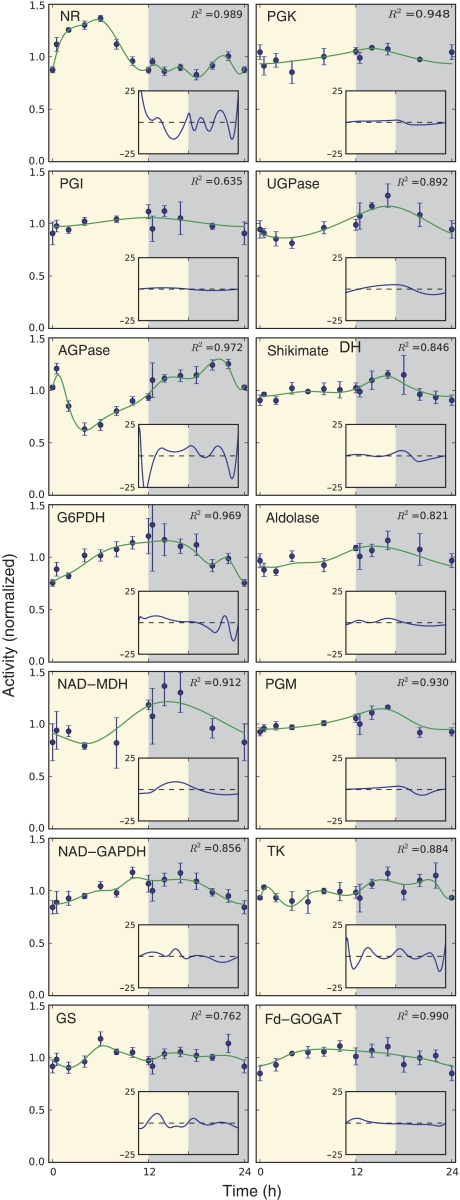
Data for the diurnal changes of enzyme activity from Gibon et al (2004b), and measurements on material collected in this study (see Supplementary Table III) were normalized and combined (for details see Material and Methods). For 14 enzymes, the data quality allowed interpolation of a smoothing cubic spline with an R2 value ⩾0.8 (Figure 7).
Figure 7.
Examples for diurnal changes in maximum enzyme activities. Enzyme activities from two independent experiments, containing 3 and 5 biological replicates, were averaged and normalized. The main plot shows the mean normalized activity and the error bars indicate s.e.m. values. Solid lines indicate an interpolating smoothing cubic spline fit to the data. The inserted plot shows the first derivative of the spline (the rate of change of the enzyme, percentage mean activity h−1).
NR was predicted to have a TP value of 0.13 days. In agreement, it shows a very rapid increase of activity during the first 1–2 h of the light period (see also Scheible et al, 1997; Matt et al, 2001; Kaiser and Huber, 2001). AGPase was predicted to have a TP value of about 2 days. Its activity decreases at the start of the light period, and recovers during the remainder of the diurnal cycle. G6PDH was estimated to have a relatively low TP value (1.6 days), and again showed marked diurnal changes of activity. PGI was predicted to have a relatively low TP, but did not show diurnal changes. NAD–GAPDH, NAD–MDH and UGPase show smaller diurnal changes of activity with amplitude of about 20%. The former two were predicted to have an intermediate TP (5.2 and 6.1 days, respectively). UGPase was predicted to have higher TP (9.9), but there was a large discrepancy between the two estimates of protein abundance. The other six enzymes (GS, shikimate-DH, PGK, Fd-GOGAT, PGM and TK) showed very small diurnal changes. Of these, five were predicted to have a high TP (3.1, 6.0, 8.1, 14.3, 18 and 18.9 days, respectively). Thus, the rapid turnover of NR and significant turnover of AGPase and G6PDH is predicted by our calculations, and good qualitative agreement achieved for 9 of the other 11 enzymes. Further, with the exception of NR and AGPase, most enzymes showed a maximum at the end of the day or in the early part of the night. This matches the prediction from the light-dependent increase in polysome loading that synthesis of most proteins will be faster in the light period than in the dark period (see Figure 3).
Codon usage
The rate of elongation is affected by codon usage (Wright, 1990, Brockmann et al, 2007). We estimated, for each gene, two measures of codon usage, the codon adaptation index (CAI; Sharp and Li, 1986) and the effective number of codons (Nc, Wright, 1990) (Supplementary Table X). As the scores generated by these two approaches are reciprocal (a high CAI signifies a biased codon usage, which resembles that of the most abundant transcripts in that species, whereas a low Nc signifies biased use of a small number of codons) there was a strong negative correlation between CAI and Nc (−0.7; Supplementary Table XI; Supplementary Figure S3). Codon usage also correlated with GC content and transcript length. Many earlier studies using Caenorhabditis elegans, budding yeast and Arabidopsis (Duret and Mouchiroud, 1999; Wright et al, 2004; Brockmann et al, 2007) found that highly abundant transcripts tended to be shorter and to have a biased codon usage. In our set of genes, the regression was driven by some highly abundant genes from the photosynthetic machinery and some ‘house-keeping' genes, which have high CAI, low Nc (Supplementary Table XI and Supplementary Figure S3) and a high GC content (see also Chiapello et al, 1998). CAI and Nc correlated with transcript length and transcript abundance (Spearman's r for CAI=0.48 and −0.36, respectively), but not with ribosome occupancy (Supplementary Table XI, Supplementary Figure S3). Genes encoding proteins with a low TP had an intermediate to low CAI, and intermediate to high Nc (Supplementary Table X), with scores resembling those of genes that encode enzymes with high TP values. CAI and Nc were unrelated to TP, irrespective of whether the latter were calculated by averaging across the entire enzyme family or for each protein (Spearman's r<0.01; Supplementary Table XI; Supplementary Figure S3).
Discussion
Quantitative analysis of ribosome and transcript numbers
Most expression profiling and proteomic studies provide static measurements of transcript and protein levels, using largely arbitrary units. They also usually lack information about the extent to which the transcripts are being translated. This makes it impossible to quantitatively compare and integrate data from these two functional levels. We present a toolbox that combines qRT–PCR with quantitative proteomics, and provides quantitative data about the concentrations of transcripts and ribosomes, and the recruitment of individual transcripts to polysomes. By combining this with information about protein abundance, we are able to predict rates of protein synthesis, relate them to the rate of growth and calculate the resulting energy costs. This provides a quantitative framework to integrate molecular information about transcript concentrations, ribosome numbers and protein synthesis with emergent properties like the energy balance and cellular growth.
Our calculations require several assumptions. We assume that translation is primarily regulated by the rate of initiation, and that all of the ribosomes in the polysomes are involved in active protein synthesis. Evidence for the general validity of the assumption is provided in Figure 5, with the exception of the RBCS gene family and RBCL (see below for further discussion). We also assume that literature values for ribosomal progression in yeast and animals (Mathews et al, 2007) are applicable to higher plants. An error here would lead to a systematic increase or decrease in all of the estimates of protein synthesis rates. Another source of error is that we do not know the precise number of ribosomes associated with each individual species of transcript. For the SPS fraction, this was estimated from polysome profiles (Arava et al, 2003; MacKay et al, 2004). For the LPS fraction, we used a ribosome density of 6.6 kb−1 on the basis of ultramicroscopy of Escherichia coli polysomes (Brandt et al, 2009). This is about threefold lower than the maximum theoretical packing, based on the structure of the ribosome, and is slightly higher than the packing estimated from detailed analyses of budding yeast polysomes (Arava et al, 2003; MacKay et al, 2004). We also assume that ribosome density is similar for all transcripts. This is an oversimplification, as transcripts with differing representations in the LPS fraction are likely to have different rates of initiation and, hence, differing ribosome densities.
The rate of elongation is increased by optimizing codon usage (Wright, 1990, Brockmann et al, 2007). We also found a trend of biased codon usage in genes with very abundant transcripts. However, codon usage was not integrated into the calculations of translation rates, for two reasons. First, we do not know how it affects the elongation rates in Arabidopsis. Second, if initiation is the limiting step, optimized codon usage may not increase the rate of translation of that particular transcript. Optimized codon usage in highly abundant transcripts might instead serve to increase the overall efficiency of translation, by recycling ribosomes from abundant transcripts into a common pool that is available to translate all transcripts, as proposed by Duret and Mouchiroud (1999).
Our calculations are obviously limited by the precision of our measurements. Precise quantification of changes of <twofold is difficult using qRT–PCR. We carried out all determinations on three biological replicates, and included technical replicates for polysome gradients and qRT–PCR determinations. This nevertheless affects the precision of the measurements of ribosome numbers and transcript concentrations, and calculations that are based on them. Similar limitations apply to emPAI in which, like other currently available general proteomics methods, highly abundant proteins tend to be underestimated (Ishihama et al, 2005) and rare proteins tend to be not detected. For this reason, we took advantage of the fact that proteins that we studied can also be quantified by measuring enzymatic activity. Enzyme activities can be measured with great precision (Gibon et al, 2004b). However, activity measurements may not always precisely reflect the changes in protein levels, and are often the sum of two or more individual proteins. Furthermore, the calculation of protein amounts requires reliable literature values for specific activities. Despite these shortcomings, there was a good quantitative agreement between the proteins levels estimated from emPAI and enzyme activities, with only 4 out of 31 showing >5-fold discrepancy, and 8 >3–5-fold discrepancy.
Dynamics of turnover of enzymes in central metabolism
There are >1000-fold differences in the estimated rates of synthesis of individual proteins from central C and N metabolism (Table II), and >200-fold differences in the estimated time to synthesize the protein in the rosette (TP) (Table III). Most enzymes have estimated rates of synthesis that are quite low compared with the amount of protein in the leaf, resulting in estimates for TP of four or more days (Figure 3). As Arabidopsis has a relative growth rate of 0.15–0.20 under the conditions used in our study (Gibon et al, 2009; Tschoep et al, 2009), a full complement of new enzymes must be synthesized every 5–7 days. The estimated rates of synthesis of most enzymes are similar to those needed for growth. These enzymes also show only small diurnal changes of their maximum activity (Figure 7). Although these diurnal changes may underestimate the rate of synthesis, if degradation is occurring at the same time, they provide independent evidence that many enzymes do not undergo rapid turnover.
As already mentioned, our calculation of translation rates depends on the assumption that translation is primarily regulated by the rate of initiation. An unusually high proportion of the RBCL and RBCS transcripts was present in the SPS fraction (Figure 5), indicating that their translation may be regulated in a more complex manner. RBCL, like many other plastid-encoded genes, is subject to sophisticated translation regulation (Marín-Navarro et al, 2007). The accumulation of RBCL leads to autoregulation of the translation of the RBCL transcript (Wostrikoff and Stern, 2007), allowing RBCL production to be adjusted to changes in the supply of the nuclear-encoded RBCS subunit (Rodermel et al, 1996). Complex translational regulation of RBCS may be needed to coordinate the provision of RBCS with RBCL synthesis in the plastid. An alternative explanation for the high proportion of RBCS transcripts in the SPS fraction can probably be discounted. Most of the investigated transcripts had a length of >250 amino acids, with most being >500 amino acids (see Supplementary Table I). However, RBCS is a rather small protein, with a length of about 180 amino acids. Given that a eukaryotic ribosome physically occupies about 12 codons (Wolin and Walter, 1988), and that the maximum packing of ribosomes on polysomes may be about 4–6 times lower than the maximal packing density (MacKay et al, 2004; Lackner et al, 2007; Qin et al, 2007), RBCS transcripts may be too short to accommodate more than 4–5 ribosomes. RBCS also has a highly biased codon usage that might allow rapid elongation, and further decrease the average number of ribosomes per transcript. However, ATGRP7, At ELF5a and ATGER3 also have short coding sequences (176, 160 and 211 bp, respectively) and AtGER3 and AtELF5a have a similarly biased codon usage, but did not have a high proportion of transcripts in the SPS fraction (Supplementary Tables I and II). This indicates that a short open reading frame and biased codon usage are not the main reasons for the high proportion of RBCS transcripts in the SPS fraction. Complex translational regulation may explain the unexpectedly low estimated value of TP for RBCL and RBCS-1A (<1 day). RubisCO represents about 30% of the total leaf protein (see Introduction) and is not though to be subject to rapid turnover. Simpson et al (1981) reported half-lives of RubisCO of about 7 days in mature maize leaves, whereas Esquivel et al (1998) found that RubisCO turned over at a similar rate as total protein in mature maize, sorghum and wheat leaves. This discrepancy may be due to specific problems in using our data to estimate translation rates for RBCS and RBCL.
Examples of enzymes with rapid synthesis times
For some enzymes, the estimated rates of synthesis are high, relative to their abundance. Several of these also show marked changes in their maximum enzyme activities (Figure 7) and protein abundance during diurnal cycles, in particular NR and AGPase. These reflect changes in corresponding protein amounts (Scheible et al, 1997; Weiner and Kaiser, 1999; Gibon et al, 2004a). These two enzymes catalyze key reactions in nitrogen and C metabolism, which need to be regulated during the diurnal cycle. Both are subject to multilevel transcriptional, translational and posttranslational regulation (Stitt et al, 2007; Lillo, 2008). NR catalyzes the first step in nitrate reduction, which is the major source of inorganic nitrogen for most plants (Foyer et al, 2000). NR expression and activity respond rapidly to the availability of nitrate and C, and the accumulation of downstream products of nitrogen assimilation (Lillo, 2008). As NR catalyzes a side reaction with nitrite to produce NO (Rockel et al, 2002; Lea et al, 2004), it is also essential to inactivate NR in the dark when nitrite reductase is restricted by the availability of reduced ferredoxin. AGPase is the key regulatory enzyme for starch synthesis, and is, therefore, intimately involved in the regulatory network that determines how much C is stored to support respiration and growth at night (Smith and Stitt, 2007; Stitt et al, 2007).
Relation between protein synthesis and the diurnal C and energy budget
Central metabolism represents a major part of the total protein in an Arabidopsis rosette (see Introduction). The finding that most enzymes have a TP >4 days indicates that there is little protein turnover. An independent and more direct estimate of the overall rate of protein synthesis can be made based on the ribosome numbers. Arabidopsis rosettes contain about 0.1 nmol ribosomes g−1 FW. This would support a maximum rate of protein synthesis of approximately 26 μmol amino acids g−1 FW day−1 or 3 mg protein g−1 FW day−1, or synthesis of the protein in a rosette (15 mg g−1 FW) in about 5 days. Ribosomal loading in polysomes indicates that the actual rate is about 40% lower. This resembles the estimated TP values for many individual enzymes. It also resembles the rate of protein synthesis that is needed (2.2–3.0 mg protein g−1 FW day−1) to account for the rate of growth (0.15–0.2 g FW g−1 FW day−1).
These comparisons are dependent on the assumed rate of ribosome progression (3 amino acids s−1). If the rate of ribosome progression were faster, the estimated rates of protein synthesis would rise, allowing a surplus for protein turnover. Protein degradation is of great importance for signalling (Bassham, 2007; Vierstra, 2009). However, this is unlikely to require large-scale protein turnover. Large-scale protein turnover in plants occurs in a time frame of days as they adjust to changes in the environment (see Introduction), and as nitrogen is remobilized from senescing leaves to support growth of seeds and tubers.
There are considerable energetic costs associated with protein synthesis. The conversion of one molecule of nitrate or ammonium to amino acids requires about 5 ATP (Penning de Vries, 1975; Hachiya et al, 2007). The addition of an amino acid to a growing peptide chain requires two ATP molecules for amino acid activation and another two ATP for peptide bond formation and ribosome translation, plus additional costs of about another ATP, for error correction and the synthesis of sequences that are removed during protein maturation (Noguchi et al, 2001). Thus, if they were active, 0.1 nmol ribosomes g−1 FW would consume about 260 μmol ATP g−1 FW day−1. ATP is supplied by photophosphorylation in the light period, and by respiration of carbohydrates and other metabolites in the dark period. This will be thermodynamically less efficient, and also requires that enough respiratory substrate can be stored to sustain this demand. In our study, Arabidopsis was grown in a 12-h light–dark cycle. If protein synthesis were to continue at maximal rates during the night about 65 μmol hexose g−1 FW night−1 would be required to supply the amino acids (assuming an average of three Cs per amino acid), and a respiration rate of about 3.6 μmol CO2 g−1 FW (equivalent to another 7 μmol hexose g−1 FW of starch) would be required to deliver the ATP (assuming an ideal phosphorylation potential of 3). In our growth conditions, Arabidopsis contains about 40 μmol glucose g−1 FW as starch at the end of the light period, and has a rate of respiration of about 8 μmol CO2 g−1 FW h−1 (Gibon et al, 2004a; Hannemann et al, 2009). This is insufficient to provide C and energy that are required to support high rates of protein synthesis during the night.
This potential C and energy deficit is avoided in two ways. First, plants accumulate large amounts of amino acids during the light period (Matt et al, 2001; Fritz et al, 2006). Up to 30 μmol amino acids g−1 FW are accumulated in Arabidopsis rosettes in the light period, and used at night (Gibon et al, 2004a, 2006, 2009). Second, the rate of protein synthesis is decreased in the dark period. The distribution of ribosomes between the free and polysomal fractions (Figure 2) indicates that protein synthesis is about twofold lower in the dark period than in the light period. Changes in the ribosomal occupancy of transcripts (Figure 3) indicate a smaller decrease, but this is probably an underestimate because it does not take into account that the number of ribosomes per transcript probably decreases in the dark period. These diurnal adjustments in amino acid metabolism and protein synthesis mean that the nocturnal requirement of C skeletons for amino acid synthesis is effectively abolished, whereas the respiratory cost is decreased by about 75%.
In conclusion, quantitative measurements of transcripts, ribosomes, polysomes and protein levels provide information that can be used to estimate protein synthesis and turnover rates. This provides a framework in which the metabolic and energetic costs of protein synthesis and turnover can be related to basic molecular components that affect cellular growth. It allows proteins to be identified that are subject to rapid turnover. In this study, this approach was used to study the dynamics of enzymes that are involved in C and nitrogen assimilation. However, by extending the range of transcripts to include signaling components, it may be possible to use an analogous approach to transcriptional signaling responses, and identify transcripts that are likely to have a pivotal role because changes in their concentration are likely to lead to a rapid change in the concentration of the encoded protein. It will also be possible to investigate whether changes of ribosome concentration and loading can be used to predict how protein content and growth are modified in genotypes with altered regulation of ribosomal biogenesis.
Calculations and assumptions
Overall rate of protein synthesis
Literature values for the rate of ribosome progression in eukaryotic cells vary from 1–8 amino acids per second per translating ribosome, depending on the cellular conditions and the mRNA (for reviews, see Ryazanov et al, 1991; Mathews et al, 2007), with typical values of 4–5 for animal cells at 25–26°C (e.g. Lodish and Jacobsen, 1972; Palmiter, 1974) and 7–8 for with yeast cells at 30°C (e.g. Arava et al, 2003). The plants used in our study were grown at 20°C. On the basis of the published values for animal cells and assuming a Q10 of about 2.5, we used an elongation rate of 3 amino acids s−1 in our calculations. A measured ribosome concentration of 0.1 nmol g−1 FW (summed cytosolic, plastid and mitochondrial ribosomes; see Table I) to allow for the addition of about 26 μmol amino acids g−1 FW day−1, or (assuming an average molecular weight of an amino acid of 118.9 g mol−1, Hachiya et al, 2007) the synthesis of about 3 mg protein day−1 g−1 FW.
Rate of translation of individual transcripts
The rate of synthesis of the individual proteins (copies of protein per hour per g FW) were estimated from the transcript abundance (copies of transcript per g FW) in the SPS and LPS fractions multiplied by the ribosome density per translating transcript (see below for more details) and the rate of ribosome progression along the transcript. The rates of protein synthesis for transcripts in the SPS and LPS fractions were calculated separately, and then summed (Supplementary Table II). The general formula used was: rate of protein synthesis (mol protein g−1 FW h−1)=transcript copy number in the fraction (mol g−1 FW) × ribosome density on transcripts in that fraction (ribosome per transcript) × rate of ribosome progression (amino acids added per s) × 3600. SPS and LPS transcript levels were calculated as copy number g−1 FW/Avogadro constant. The calculations used an elongation rate of 3 amino acids per ribosome per sec (see above). Ribosome density for the SPS fraction was empirically estimated as 3 per transcript, by plotting defined peaks from the polysome profile against the relative distance from the start of the profile and fitting an exponential curve (see MacKay et al, 2004). Ribosome density for the LPS fraction was estimated using a ribosome density of 6.6 ribosome per kb of open reading frame (see Brandt et al, 2009) multiplied by the known length of the open reading frame in a given transcript.
Estimation of abundance of enzyme proteins in the rosette
All the 1002 emPAI index values (see Materials and Methods) were normalized against the total sum of emPAI values of all 1002 proteins identified in the leaf extract to calculate the molar fraction of each protein in the sample (mol%). The molar fractions were converted to concentrations (mol g−1 FW) taking into account an experimentally determined average protein molecular weight of 53.5 kDa, and a measured total protein concentration of 15 mg g−1 FW rosette (Supplementary Table VI). In general, as only a small proportion of all proteins of the leaf were identified, abundance values calculated from emPAI will result in overestimation of the true amounts. emPAI values and calculated protein concentrations are listed in Supplementary Table VI and the subset corresponding to the metabolic enzymes investigated in this study are listed in Supplementary Table VII. The full list of identified peptides is available in Supplementary Table X.
Estimation of ribosomal protein abundance in polysome fractions
For each polysomal fraction (LPS, SPS and NPS), the identified ribosomal proteins were classified to specific families of cytosolic or plastid ribosomes based on MapMan (Thimm et al, 2004). Usually, assignment of peptides to a protein requires at least one proteotypic peptide (i.e. a peptide that is uniquely attributable to a single protein). However, high sequence similarity between members of gene families (like ribosomal proteins that are encode by gene families consisting of 2–8 genes in higher plants, TAIR8) can result in no proteotypic peptides being found. In our data set, 30% of all identified peptides for ribosomal proteins matched more than one ribosomal protein, and on the protein level for about 20% of the ribosomal proteins no clear assignment of protein species could be made. In these cases, all possible protein names are listed. emPAI values were scored only once, even if multiple protein matches were possible, assuming that peptide abundances would be higher (more spectra) if several proteins contribute to the pool of peptides. A full list of identified peptides and proteins in the polysome gradients including their emPAI values is provided in Supplementary Table VIII and IX.
The emPAI values (Ishihama et al, 2005) for each protein in a given polysomal fraction were divided by the total sum of emPAI values for all proteins in the same fraction to calculate the mol% abundance of each ribosomal protein. Subsequently, the abundance fraction (mol%) of each protein was summed across all cytosolic and plastid ribosomal proteins, and used as a basis for comparison between the polysomal fractions. Total RNA was used to normalize between different gradients. rRNA was found to be the major component of the cellular RNA and the ratio between rRNA and transcript RNA did not depend on the light regime (Supplementary Table II), making total RNA a justified measure for normalizing gradients with respect to ribosome numbers between replicates and different treatments.
Estimation of protein abundance from enzyme activity measurements
Protein abundance was estimated from the enzyme activity measured in the leaves (original data provided in Supplementary Table III), corrected by literature values for the specific activity (literature information concerning the enzyme structures and the specific activities is supplied in Supplementary Table IV). Specific activities and enzyme structures were identified from BRENDA database (http://www.brenda-enzymes.info/) and literature (e.g. Lea, 1990). In case of disagreement, the highest specific activity was chosen because an underestimate of the specific activity can easily occur due to loss of activity during purification, or incomplete purification of the enzyme. The literature value for the specific activity (nmol min−1 mg−1 protein) was multiplied by the molecular weight of one catalytic site to give the specific activity per mol protein (nmol min−1 mol−1 protein, see column ‘Maximun Specific Activity at 25°C' of Supplementary Table V). In most cases there is one active site per protein, but in some cases the enzyme is a heterodimer consisting of one catalytic and one non-catalytic peptide. The average enzyme activity during a diurnal cycle (nmol min−1 g−1 FW) is then divided by the specific activity (nmol min−1 mol−1 protein) to estimate the protein abundance (mol g−1 FW, see column ‘Estimated Protein Abundance during the Diurnal Cycle' of Supplementary Table V).
TP, the time in days needed to synthesize all of a given protein in the rosette
The estimated rates of protein synthesis were compared with the estimated protein concentration in Arabidopsis rosettes to estimate how long it would require to synthesize all the protein in an Arabidopsis rosette (TP). TP (days to synthesize all the protein in the rosette) was estimated as (protein abundance in the leaf (mol g−1 FW))/(translation rate (mol h−1 g−1 FW) × 24). Two independent estimates were obtained, first (Supplementary Table V), by dividing the summed rates of synthesis for all family members covered by the qRT–PCR platform (see column ‘Estimated Translation Rate' of Supplementary Table V) by the protein abundance estimated from enzyme activities (column ‘Estimated Protein Abundance during the Diurnal Cycle' of Supplementary Table V). A second set (see columns X-AC of Supplementary Table VII) was obtained by comparing the rate of protein synthesis on a gene-by-gene basis (see columns G–J of Supplementary Table VII) with protein abundance estimated from emPAI (see columns R–W of Supplementary Table VII) for all family members that were quantifiable by emPAI and were covered by the qRT–PCR platform, and summing the resulting estimates. The results are summarized in Table III.
Normalization and spline fitting to enzyme activities
In most cases, enzyme activities were measured during the diurnal cycle in two separate experiments. In each experiment, 3–5 replicate biological samples (each containing at least 5 rosettes) were processed. To combine the data, activities of each enzyme were first normalized to the mean activity observed over the complete diurnal cycle. Next, the mean normalized activity for each time point in the diurnal cycle as well as its s.e.m. value were calculated. Using the SciPy library of scientific software (Jones et al, 2001), a periodic interpolating smoothing B-spline of order 3 was fitted through these data, using the s.e.m. values as weights. The s.e.m. values, and not s.d. values, were used as weights because different numbers of data points were available for different time points—some time points only being measured in one experiment—leading to some data being known with greater certainty than others. The data were assumed periodic with identical values at 0 and 24 h. A smoothing condition of Σ(w(y−g))2⩽6 (where g(x) is the smoothed interpolation of (x,y) and values for w are the weights) was applied to the spline; this was found empirically to give the optimal trade-off between closeness and smoothness of fit.
Codon usage
CAI values (Sharp and Li, 1986) for the genes quantified were calculated as follows. First, the dataset atge100 was downloaded from CSBDB (Steinhauser et al, 2004). This comprises one array per plant part and developmental stage from the series AtgenExpress but does not include mutants. The data set was RMA normalized and the probe set-wise average abundance calculated. The hundred most highly expressed probe sets were extracted and from this set reference and ambiguous probe sets were removed. The coding sequences corresponding to these probe sets were obtained by using the TAIR online tool (www.arabidopsis.org). These sequences were used to construct codon usage tables for highly expressed Arabidopsis genes using the cusp program of the EMBOSS suite (Rice et al, 2000). Subsequently, these tables were used to calculate the CAI using the tool ‘cai' from the EMBOSS suite. Using the CAI server (Puigbo et al, 2008), we could also calculate the effective number of codon index (Nc, Wright, 1990) and the total as well as the GC content at the first, second and third position.
Materials and methods
Reagents
All chemicals were purchased from Sigma-Aldrich (Schnelldorf, Germany), except for NADH (Roche; Mannheim, Germany). All enzymes used for analysis were purchased from Roche except invertase (Sigma-Aldrich) and UMP-kinase, which was overexpressed in E. coli and purified as described by Serina et al (1995).
Plant growth
Arabidopsis thaliana ecotype Col-0 was grown in GS90 soil under 12-h light–dark cycle at a light intensity of 150 μmol m−2 s−1 and 20°C for 5 weeks (Thimm et al, 2004), at which time flowering had not commenced. The samples were immediately frozen under ambient irradiance in liquid nitrogen. The samples were powdered using Labman Automation (Stocksley, UK) and stored at −80°C until use.
Replication
Samples were collected for three independently grown sets of plants, each containing typically, at least, 15 rosettes. For each biological replicate two polysome gradients were run, fractionated and separately analyzed. For each unfractionated and gradient fraction RNA, two and three technical replicates were carried out for qRT–PCR analysis of transcript and rRNA species, respectively, and one for emPAI. Enzyme activities were separately measured once in the three biological samples.
Isolation of total RNA, polysomes, polysomal RNA and DNase I digestion
Total unfractionated RNA was isolated from 100 mg of homogenized leaf tissue using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Polysomes were fractionated from crude leaf extracts as described previously (Kawaguchi et al, 2003) with modifications. Briefly, 100 mg of pulverized leaf tissue was hydrated in 0.5 ml of extraction buffer (200 mM Tris (pH 9.0), 200 mM KCl, 25 mM MgCl2, 25 mM ethylene glycol-bis (β-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) (pH 8.3), 1 mg ml−1 heparin, 5 mM dithiothreitol (DTT), 50 μg ml−1 cycloheximide, 50 μg ml−1 chloramphenicol, 1% (v/v) Triton X-100, 1% (v/v) Brij-35, 1% (v/v) Tween-20, 1% (v/v) IGEPAL CA-630, 2% (v/v) polyoxyethylene-10-tridecyl-ether and 1% (v/v) sodium deoxycolate). Hydrated tissue was placed at 4°C on a QIAshredder column of the RNeasy Plant Mini kit (Qiagen, Hilden, Germany) and centrifuged at 16 000 g for 1 min to remove cell debris. The supernatant was separated in a 5 ml (20–60% w/v) sucrose gradient by ultracentrifugation at 27 500 g for 90 min at 4°C (SW 55Ti rotor, Beckman Coulter, Krefeld, Germany). Afterwards the gradients were fractionated in 14 fractions of approximately 350 μl by using Programmable Density Gradient Fractionation System (Teledyne Isco, Lincoln, NE, USA), which continuously recorded the ribosome absorbance at 254 nm (ribosome profile). The A254 value of polysome profiles varied by less than one-half fraction among six gradients for each condition (three biological and two technical replicates).
Polysome levels were determined by calculating the area under the polysome profile after subtracting the gradient baseline absorbance (absorbance of a gradient loaded with extraction buffer). The area of each polysome profile was normalized to an equal value to account for differences in sample loading. Levels of non-polysomes (NPS: gradient region containing mRNP complexes, 40/60S and 30/50S ribosome subunits, 70/80S ribosomes and one ribosome per transcript), small polysomes, (SPS: gradient region containing two to 4 ribosomes per mRNA), and large polysomes, (LPS: gradient region containing five or more ribosomes per transcript) were determined by calculating corresponding peak areas of the gradient regions. The areas corresponding to the NPS, SPS and large LPS fractions were reported as a percentage of the total area under the profile. These calculations were carried out using a script written in R (CoreTeam RD, 2008). The lower boundary was represented by the 30/40S subunit peak and the upper boundary by the bottom of the gradient.
Fractions corresponding to NPS (1–6), SPS (7–9) and LPS (10–14) were combined. RNA was precipitated with an equal volume of 8 M guanidine hydrochloride and two volumes of ethanol at −20°C overnight. After centrifugation at 12 000 g for 30 min (JA-25.50 rotor, Beckman Coulter, Krefeld, Germany), the RNA pellet was re-suspended in 450 μl of RLT buffer from the Plant RNeasy Mini kit (Qiagen, Hilden, Germany), and the RNA was recovered following the manufacturer's protocol. To confirm that the observed sedimentation pattern is the result of polysome association, control gradients were carried out in which extracts were incubated with 50 mM EDTA (pH 8.0) and the gradient contained EDTA (100 mM) instead of MgCl2. During the extraction, the first on-column DNase I (Qiagen, Hilden, Germany) digestion was carried out according to the manufacturer′s protocol. The unfractionated total RNA was digested for the second time using Turbo DNA-free DNase I (Applied Biosystems/Ambion, Darmstadt, Germany) following manufacturer's instructions. RNA concentration and integrity were measured before and after DNase I digestion with a NanoDrop ND-1000 UV–Vis spectrophotometer (NanoDrop Technologies, Wilmington, USA) and an Agilent-2100 Bioanalyzer using RNA 6000 NanoChip (Agilent Technologies, Böblingen, Germany). Samples were reproducibly of high quality and integrity. Absence of genomic DNA contamination in DNase I-treated samples was subsequently confirmed by qRT–PCR using primers designed to amplify a 155-bp and a 633-bp fragment of the ACT2 (At3g18780) transcript and gene, respectively, and primers designed to amplify an intron sequence of the MAF5 gene (At5g65080; primer specifications are given in Supplementary Table I).
Normalization of the transcript and translation profiles
The commonly used normalization methods used to normalize expression data, including qRT–PCR, are based on the assumption that there is no net difference in overall mRNA levels between different samples. However, this assumption is not applicable for polysome profiling because the overall mRNA level, as well as the mRNA levels of specific genes, is expected to differ a lot between different fractions of the gradient. Owing to this and to control for variable losses of mRNA resulting from fractions during purification, and for variation from differences in cDNA synthesis processivity and PCR efficiency, a normalization method was used by doping a ‘normalization mix' of four different artificial mRNAs with known concentrations into the samples as internal standard for a quantitative normalization. The ‘normalization mix' contained four different commercially available artificial mRNAs, utility spike controls from Lucidea Univeral ScoreCard spikes (GE Healthcare, Buckinghamshire, UK) and Alien qRT–PCR Inhibitor Alert (Stratagene-Agilent Technologies, Waldbronn, Germany). The spike-in controls were added at a concentration of 2.0E10, 2.0E09, 2.0E08 and 2.0E06 copies of Utility1, Utility2, Utility3 and Alien, respectively per 200 and 100 mg of fresh weight (FW) tissue to the unfractionated leaf extract and to the NPS, SPS and LPS pooled fractions before proceeding with the RNA isolation protocols.
cDNA synthesis and quality control
The cDNA was synthesized using 1 μg of total DNase I-treated RNA using SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen, Karlsruhe, Germany), according to the manufacturer's instructions, using a mixture of polyT primers, and separately for rRNA and RBCL analysis using a mixture of polyT and random primers (von Ahlfen et al, 2007). RNA integrity and processivity of the reverse transcriptase in each sample were estimated by determining the 5′/3′ ratio of GAPDH transcript (AT1G13440; primer specifications are given in Supplementary Table I) qRT–PCR amplification with two primer pairs from either the 5′ (GAPDH5 primers) or 3′ (GAPDH3 primers) region. All samples presented good 5′/3′ GAPDH amplification ratios, between 1 and 2.
RT–PCR primer design and test
The set of 105 genes included the genes that encode the major gene family isoforms of enzymes from primary C and N metabolism analyzed in Gibon et al (2004b), a set of TPS-like genes expressed in leaves, reference genes, and exogenous spike transcripts. The primer list and its specifications are given in Supplementary Table I. The gene sequences and models for A. thaliana genes were retrieved from TAIR (The Arabidopsis Information Resource; http://www.arabidopsis.org/) database (TAIR6 Genome Release) and used to design primers by Eurofins MWG GmbH (Ebersberg, Germany). A standard set of reaction conditions and a set of stringent criteria were used as follows: Tm of 60±2°C, PCR amplicon length of 60–150 bp, primer length of 20–30 bp and a GC content of 35–55%. If gene structure allowed, at least one primer was designed to cover an exon–exon junction. The specificity of the primer pair sequence was checked against Arabidopsis transcripts (TAIR6_CDS) from TAIR database using the BLAST program for short, nearly exact matches (NCBI, The National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/blast). The specificity of the amplicons was checked by qRT–PCR dissociation curve analysis. The PCR efficiencies of the polymerase chain reactions were estimated using the LinRegPCR software (Ramakers et al, 2003) and ranged between 1.7 and 2.1 with R2⩾0.995 (Supplementary Table I).
Quantitative RT–PCR conditions and analysis
PCR reactions were carried out in an optical 384-well plate with an ABI PRISM 7900 HT sequence detection system (Applied Biosystems Deutschland, Darmstadt, Germany). A 5-μl reaction containing 0.5 μl of cDNA (cDNA was diluted to 1/10 for all genes except for the rRNA genes that were diluted to 1/600), 200 nM of each gene-specific primer and 2.5 μl of Power SYBR Green PCR Master Mix (Applied Biosystems Deutschland), was used to monitor double-strand DNA synthesis. A master mix of sufficient cDNA and 2 × Power SYBR Green PCR Master Mix was prepared before dispensing into individual well, to reduce pipetting errors and to ensure that each reaction contained an equal amount of cDNA. The 384-well plates were prepared using PerkinElmer Evolution P3 Precision Pipetting Platform (PerkinElmer Life Sciences, Rodgau–Jügesheim, Germany). The qRT–PCR reactions were carried out following the recommended thermal profile: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After 40 cycles, the specificity of the amplifications was tested by heating from 60 to 95°C with a ramp speed of 1.9°C min−1, resulting in melting curves. Data analysis was carried out using SDS 2.3 software (Applied Biosystems Deutschland). To generate a baseline-subtracted plot of the logarithmic increase in fluorescence signal (Rn) versus cycle number, baseline data were collected between cycles 3 and 15. All amplification plots were analyzed with an Rn threshold of 0.2 to obtain threshold cycle (Ct) values. The concentration of a target gene in the samples was calculated using the spike-in controls. Spike-in controls were added to each sample before RNA extraction, and the efficiency and Ct values were used to generate a standard curve (see above ‘Normalization of the transcript and translation profiles'). One standard curve was used for each sample. All standard curves had R2 value higher than 0.98. Each data point for a specific gene in a sample was plotted against the standard curve to calculate the concentration of mRNA in the unit of copy per gram fresh weight tissue (copy g−1 FW).
Quantitative analysis of ribosomal small subunit RNAs
The ribosome number within each polysomal fraction was calculated by determining the amounts of cytosolic, plastid and mitochondrial rRNA small subunits by qRT–PCR on the basis that each of these ribosomal RNAs corresponds to one ribosome. The primers were designed to amplify the genes for cytosolic (AT2G01010 and AT3G41768), plastid (ATCG00920 and ATCG01210) and mitochondrial (ATMG01390) rRNA small subunits (primer specifications are given in Supplementary Table I).
Protein identification by tandem mass spectrometry
Tryptic peptide mixtures were analyzed by LC/MS/MS using nanoflow HPLC (Proxeon Biosystems, Denmark) and an Orbitrap hybrid mass spectrometer (LTQ-Orbitrap, Thermo Electron, USA) as mass analyzer. Peptides were eluted from a 75-μm analytical column (Reprosil C18, Dr Maisch GmbH, Germany) on a linear gradient running from 4 to 64% (v/v) acetonitrile in 90 min and sprayed directly into the LTQ-Orbitrap mass spectrometer. Proteins were identified by tandem mass spectrometry (MS/MS) by information-dependent acquisition of fragmentation spectra of multiple-charged peptides. Up to five data-dependent MS/MS spectra were acquired in the linear ion trap for each FTMS full scan spectrum acquired at 30 000 FWHM resolution settings with an overall cycle time of approximately 1 s. Fragment MS/MS spectra from raw files were extracted as DTA files and then merged to peak lists using default settings of DTASuperCharge version 1.18 (http://www.msquant.sourceforge.net) with a tolerance for precursor ion detection of 50 ppm. Fragmentation spectra were searched against a non-redundant database consisting of the complete Arabidopsis protein database (TAIR8 Genome Release; www.arabidopsis.org) to which commonly observed contaminants (human keratin, trypsin and lysyl endopeptidase) have been added. Mascot algorithm (version 2.2.0; Matrix Science, UK, www.matrixscience.com) was used for database matching. Following search parameters were applied: trypsin was used as the cleaving enzyme, peptide mass tolerance of 10 ppm, MS/MS tolerance of 0.8 Da, one missed cleavage allowed. Carbamidomethylation of cysteine was set as a fixed modification and methionine oxidation was chosen as variable modification. Only peptides with a length of more than five amino acids were considered. In general, peptides were accepted without manual interpretation if they had a Mascot score greater than 32 (as defined by Mascot P<0.01 significance threshold). Peptides with a score greater than 24 (as defined by Mascot P<0.05) were manually inspected for requiring a series of three consecutive y or b ions and assignment of major peaks in the spectrum to be accepted. Using a decoy database, the estimated false-discovery rate of peptide identification was 0.81% at the chosen Mascot confidence cutoff score of 95%.
The primary MS data has been deposited at PRIDE (http://www.ebi.ac.uk/pride) under the accession numbers 9886–9893 inclusive.
Quantitative analysis of ribosomal proteins in polysome gradient fractions
Polysomal fractions were collected as described and protein was precipitated using acetone. Protein pellets were re-suspended in 6 M urea/2 M thiourea and subsequently in-solution digested using trypsin after reduction and alkylation with iodoacetamide. Tryptic peptide mixtures from each polysome fraction were analyzed by LC/MS/MS to identify peptides (listed in Supplementary Table VIII). emPAI protein abundance index values (Ishihama et al, 2005) from Mascot (see Supplementary Tables IX) were used for quantification. Conversion to ribosomal protein concentrations is described in Calculations and assumptions section.
Quantification of whole leaf protein abundances
Total protein was extracted from 20 mg leaf material, and 50 μg of protein was separated on a gradient (4–12%) SDS–polyacrylamide gel. The whole gel lane was cut into ten slices of roughly equal protein content and protein was in-gel digested (Olsen et al, 2004). Tryptic peptide mixtures were analyzed by LC/MS/MS as described above. All extracted peak lists were combined into one single file for database search by Mascot to obtain emPAI protein abundance index values (Ishihama et al, 2005). In total, emPAI values were obtained for 1002 proteins (Supplementary Table VI). Conversion to protein concentrations is described in Calculations and assumptions section.
Assay of enzyme activities
Enzyme extraction was carried out as described by Nunes-Nesi et al (2007). INV, AGPase, AlaAT, AspAT, cytFBPase, FK, FUM, G6PDH, HK, NAD–GAPDH, NADP–GAPDH, NAD–GDH, GS, Fd–GOGAT, NADP–IDH, NR, PEP carboxylase, PFP, PK, TK, SDH and SPS were assayed as described by Gibon et al (2004b); NAD–IDH as described by Nunes-Nesi et al (2007); PGK and GK as described by Huege et al (2007); RubisCO as described by Sulpice et al (2007); TPI as described by Burrell et al (1994); NAD–MDH as described by Jenner et al (2001); NADP–MDH as described by Scheibe and Stitt (1988); PGI as described by Cross et al (2006); PGM as described by Manjunath et al (1998); and PFK and UGPase as described by Keurentjes et al (2008). The assay for aconitase was adapted from Jenner et al (2001) by assaying the NADPH through a cycling assay as carried out in the study by Gibon et al (2004b). Aldolase was assayed by incubating crude extract or dihydroxyacetone phosphate standards for 20 min in fresh medium containing 0 or 5 mM fructose-1,6-bisphosphate, 1 U ml−1 triose-phosphate isomerase, 2 U ml−1 glycerol-3- phosphate dehydrogenase, 0.3 mM NAD+, 5 mM MgCl2, 1 mM EDTA, 0.05% (v/v) Triton X100 and 100 mM Tricine buffer (pH 8.5). The reaction was stopped with an equal volume of 0.5 M HCl. After incubation for 10 min at room temperature and neutralization with 0.5 M NaOH, glycerol-3-phosphate was determined as described by Gibon et al (2004b). Protein concentration was measured as described by Gibon et al (2004b).
Supplementary Material
Supplemental Figures 1-3
Information about genes
Transcript levels in unfractionated extracts and polysome gradient fractions, and estimation of rates of translation
Enzyme activities
Literature information about enzymes
Protein abundance and Tp estimated from enzyme activities
Protein abundance estimated from quantitative proteomics
Protein abundance and Tp for enzymes of primary metabolism estimated from quantitative proteomics
Empai values for all proteins identified in the different ribosomal fractions (NPS, SPS, LPS) in dark and 2h light for three biological replica sets
List of all unique peptides with score above the 95% significance cutoff (score >32) identified in the different ribosomal fractions (NPS; SPS; LPS) from three biological replicates and the two time points (dark=0 and 2h light=2)
This table contains for each gene the estimated CAI and Nc,the length of the open reading frame, the GC content of the entire open reading frame and of the codon positon 3, the mean copy number of the transcript in the dark and in the light, and the ribosomal occupancy [(SPS+LPS)/(NPS+SPS+LPS)
Spearmens regression coefficient for correlations between CAI, Nc, the average transcript abundance in the dark and in the light, polysome loading in the dark and the light, the open reading frame length, and GC content of the entire open reading frame, and at positons 1, 2 and 3 of codons
Acknowledgments
We are grateful to Dr John Lunn for overexpression and purification of UMP-kinase. JH was supported by the Humboldt Foundation. We acknowledge the Max Planck Society, the European Commission (FP6 Integrated project ‘Agronomics' LSHG-CT-2006-037704) and the German Ministry of Education and Research (GoFORSYS, 0313924) for financial support.
Footnotes
AJB is or has served as a scientific advisor and/or consultant to NuMedii, Genstruct, Prevendia, Tercica, Eli Lilly and Company, and Johnson and Johnson.
References
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC (2007) Plant autophagy—more than a starvation response. Curr Opinion Plant Biol 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T (2004) Translational profiling: the genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic 3: 103–111 [DOI] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T (2007) Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A, Hollunder J, Nasheuer H-P, Wilhelm T (2004) Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics 3: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W (2009) The native 3D organization of bacterial polysomes. Cell 136: 261–271 [DOI] [PubMed] [Google Scholar]
- BRENDA. The Comprehensive Enzyme Information System (http://www.brenda-enzymes.info/)
- Brockmann R, Beyer A, Heinisch JJ, Wilhelm T (2007) Posttranscriptional expression regulation: what determines translation rates? PLoS Comp Biol 3: 0531–0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell MM, Mooney PJ, Blundy M, Carter D, Wilson F, Green J, Blundy KS, Aprees T (1994) Genetic manipulation of 6-phosphofructokinase in potato-tubers. Planta 194: 95–101 [Google Scholar]
- Chiapello H, Lisacek F, Caboche M, Henaut A (1998) Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209: GC1–GC38 [DOI] [PubMed] [Google Scholar]
- CoreTeam RD (2008) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M (2006) Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol 142: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D (1999) Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila and Arabidopsis. Proc Natl Acad Sci USA 96: 4482–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer TA, Miller RH, Greenwood AD (1971) Leaf nucleic acids: I. Characteristics and role in the differentiation of plastids. J Exp Bot 22: 125–136 [Google Scholar]
- Ernst D, Apfelbock A, Bergmann A, Weyrauch C (1990) Rhythmic regulation of the light-harvesting chlorophyll a/b protein and the small subunit of ribulose-1,5-bisphosphate carboxylase mRNA in rye seedlings. Photochem Photobiol 52: 22–33 [DOI] [PubMed] [Google Scholar]
- Esquivel MG, Ferreira RB, Teixeira AR (1998) Protein degradation in C-3 and C-4 plants with particular reference to ribulose bisphosphate carboxylase and glycolate oxidase. J Exp Bot 49: 807–816 [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA (2001) Models of photosynthesis. Plant Physiol 125: 42–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Ferrario-Mery S, Huber SC (2000) Regulation of carbon fluxes in the cytosol: coordination of sucrose synthesis, nitrate reduction and organic acid and amino acid biosynthesis. In Photosynthesis: Physiology and metabolism, Leegood RC, Sharkey TD, von Caemmerer S (eds), pp 177–203. Dordrecht, Netherlands: Kluwer Academic Publishers [Google Scholar]
- Fritz C, Mueller C, Matt P, Feil R, Stitt M (2006) Impact of the C–N status on the amino acid profile in tobacco source leaves. Plant Cell Environ 29: 2055–2076 [DOI] [PubMed] [Google Scholar]
- Geiger DR, Servaites JC, Fuchs MA (2000) Role of starch in carbon translocation and partitioning at the plant level. Funct Plant Biol 27: 571–582 [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004b) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M (2004a) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Pyl E-T, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing O, Kamlage B, Hoehne M, Trethewey R, Stitt M (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Terashima I, Noguchi K (2007) Increase in respiratory cost at high growth temperature is attributed to high protein turnover cost in Petunia × hybrida petals. Plant Cell Environ 30: 1269–1283 [DOI] [PubMed] [Google Scholar]
- Hannemann J, Poorter H, Usadel B, Bläsing OE, Finck A, Tardieu F, Atkin OK, Pons T, Stitt M, Gibon Y (2009) Xeml Lab: a tool that supports the design of experiments at a graphical interface and generates computer-readable meta-data files, which capture information about genotypes, growth conditions, environmental perturbations and sampling strategy. Plant Cell Environ 32: 1185–1200 [DOI] [PubMed] [Google Scholar]
- Huege J, Sulpice R, Gibon Y, Lisec J, Koehl K, Kopka J (2007) GC-EI-TOF-MS analysis of in vivo carbon-partitioning into soluble metabolite pools of higher plants by monitoring isotope dilution after (CO2)-C-13 labelling. Phytochem Rev 68: 2258–2272 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E, Oilphant T, Peterson P (2001) SciPy: open source scientific tools for Python. http://www.scipy.org
- Kaiser WM, Huber SC (2001) Post-translational regulation of nitrate reductase: mechanism, physiological relevance and environmental triggers. J Exp Bot 52: 1981–1989 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J (2005) mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res 33: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Williams AJ, Bray EA, Bailey-Serres J (2003) Water-deficit-induced translational control in Nicotiana tabacum. Plant Cell Environ 26: 221–229 [Google Scholar]
- Keurentjes J, Sulpice R, Gibon Y, Steinhauser M-C, Fu J, Koornneef M, Stitt M, Vreugdenhil D (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea US, Tem Hoopen F, Provan F, Kaiser WM, Meyer C, Lillo C (2004) Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in high nitrite excretion and NO emission from leaf and root tissue. Planta 219: 59–65 [DOI] [PubMed] [Google Scholar]
- Lillo C (2008) Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J 415: 11–19 [DOI] [PubMed] [Google Scholar]
- Lodish HF, Jacobsen M (1972) Regulation of hemoglobin synthesis. Equal rates of translation and termination of α- and β-globin chains. J Biol Chem 247: 3622–3629 [PubMed] [Google Scholar]
- MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR (2004) Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics 3: 478–489 [DOI] [PubMed] [Google Scholar]
- Manjunath S, Lee CH, VanWinkle P, Bailey-Serres J (1998) Molecular and biochemical characterization of cytosolic phosphoglucomutase in maize. Expression during development and in response to oxygen deprivation. Plant Physiol 117: 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Navarro J, Manuell A, Wu J, P Mayfield S (2007) Chloroplast translation regulation. Photosynthesis Res 94: 359–374 [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB (2007) Origins and principles of translational control. In Translational Control in Biology and Medicine, Mathews MB, Sonenberg N, Hershey JWB (eds), pp 1–40. New York, USA: Cold Spring Harbor laboratory press [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24: 1119–1137 [Google Scholar]
- NCBI. The National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast)
- Noguchi K, Nakajima N, Terashima I (2001) Acclimation of leaf respiratory properties in Alocasia odora following reciprocal transfers of plants between high- and low-light environments. Plant Cell Environ 24: 831–839 [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Ong S-E, Mann M (2004) Trypsin cleaves exclusively c-terminal to arginine and lysine residues. Mol Cell Proteomics 3: 608–614 [DOI] [PubMed] [Google Scholar]
- Pace DA, Manahan DT (2007) Cost of protein synthesis and energy allocation during development of Antarctic sea urchin embryos and larvae. Biol Bull 212: 115–129 [DOI] [PubMed] [Google Scholar]
- Palmiter DR (1974) Differential rates of initiation on conalbumin and ovalbumin messenger ribonucleic acid in reticulocyte lysates. J Biol Chem 249: 6779–6787 [PubMed] [Google Scholar]
- Penning de Vries FWT (1975) The cost of maintenance processes in plant cells. Ann Bot 39: 77–92 [Google Scholar]
- Perry RP (2007) Balanced production of ribosomal proteins. Gene 401: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG (2007) Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J 403: 217–234 [DOI] [PubMed] [Google Scholar]
- Puigbo P, Bravo IG, Garcia-Vallve S (2008) E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI). BMC Bioinformatics 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Ahn S, Speed T, Rubin G (2007) Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol 8: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trend Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53: 102–110 [PubMed] [Google Scholar]
- Rodermel S, Haley J, Jiang C-Z, Tsai C-H, Bogorad L (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93: 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazanov AG, Rudkin BB, Spirin AS (1991) Regulation of protein synthesis at the elongation stage: new insights into the control of gene expression in eukaryotes. FEBS Lett 285: 170–175 [DOI] [PubMed] [Google Scholar]
- Scheibe R, Stitt M (1988) Comparison of NADP-malate dehydrogenase activation, QA reduction and O2 evolution in spinach leaves. Plant Physiol Biochem 26: 473–481 [Google Scholar]
- Scheible W-R, González-Fontes A, Morcuende R, Lauerer M, Geiger M, Glaab J, Gojon A, Schulze E-D, Stitt M (1997) Tobacco mutants with a decreased number of functional nia genes compensate by modifying the diurnal regulation of transcription, post-translational modification and turnover of nitrate reductase. Planta 203: 304–319 [DOI] [PubMed] [Google Scholar]
- Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles AM, Barzu O (1995) Escherichia coli Ump-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine-nucleotides and UTP. Biochemistry (Mosc) 34: 5066–5074 [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH (1986) An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol 24: 28–38 [DOI] [PubMed] [Google Scholar]
- Simpson E, Cooke RJ, Davies DD (1981) Measurement of protein degradation in leaves of Zea mays using [3H]acetic anhydride and tritiated water. Plant Physiol 67: 1214–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser D, Usadel B, Lüdemann A, Thimm O, Kopka J (2004) CSBDB: A comprehensive systems biology database. Bioinformatics 20: 3647–3651 [DOI] [PubMed] [Google Scholar]
- Stitt M, Gibon Y, Lunn JE, Piques M (2007) Multilevel genomics analysis of carbon signalling during low carbon availability: coordinating the supply and utilisation of carbon in a fluctuating environment. Funct Plant Biol 34: 526–549 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Tschoep H, von Korff M, Bussis D, Usadel B, Hohne M, Witucka-Wall H, Altmann T, Stitt M, Gibon Y (2007) Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ 30: 1163–1175 [DOI] [PubMed] [Google Scholar]
- TAIR. The Arabidopsis Information Resource (http://www.arabidopsis.org/)
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Usadel B, Blasing OE, Gibon Y, Retzlaff K, Hohne M, Gunther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- von Ahlfen S, Missel A, Bendrat K, Schlumpberger M (2007) Determinants of RNA quality from FFPE samples. PLoS ONE 2: e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Weiner H, Kaiser WM (1999) 14-3-3 proteins control proteolysis of nitrate reductase in spinach leaves. FEBS Lett 455: 75–78 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Walter P (1988) Ribosome pausing and stacking during translation of a eukaryotic messenger-RNA. EMBO J 7: 3559–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl Acad Sci USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F (1990) The effective number of codons used in a gene. Gene 87: 23–29 [DOI] [PubMed] [Google Scholar]
- Wright SI, Yau CBK, Loosely M, Meyers BC (2004) Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol Biol Evol 21: 1719–1726 [DOI] [PubMed] [Google Scholar]
- Zhu X-G, de Sturler E, Long SP (2007) Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: a numerical simulation using an evolutionary algorithm. Plant Physiol 145: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures 1-3
Information about genes
Transcript levels in unfractionated extracts and polysome gradient fractions, and estimation of rates of translation
Enzyme activities
Literature information about enzymes
Protein abundance and Tp estimated from enzyme activities
Protein abundance estimated from quantitative proteomics
Protein abundance and Tp for enzymes of primary metabolism estimated from quantitative proteomics
Empai values for all proteins identified in the different ribosomal fractions (NPS, SPS, LPS) in dark and 2h light for three biological replica sets
List of all unique peptides with score above the 95% significance cutoff (score >32) identified in the different ribosomal fractions (NPS; SPS; LPS) from three biological replicates and the two time points (dark=0 and 2h light=2)
This table contains for each gene the estimated CAI and Nc,the length of the open reading frame, the GC content of the entire open reading frame and of the codon positon 3, the mean copy number of the transcript in the dark and in the light, and the ribosomal occupancy [(SPS+LPS)/(NPS+SPS+LPS)
Spearmens regression coefficient for correlations between CAI, Nc, the average transcript abundance in the dark and in the light, polysome loading in the dark and the light, the open reading frame length, and GC content of the entire open reading frame, and at positons 1, 2 and 3 of codons