Abstract
This report provides a global view of how gene expression is affected by DNA replication. We analyzed synchronized cultures of Saccharomyces cerevisiae under conditions that prevent DNA replication initiation without delaying cell cycle progression. We use a higher-order singular value decomposition to integrate the global mRNA expression measured in the multiple time courses, detect and remove experimental artifacts and identify significant combinations of patterns of expression variation across the genes, time points and conditions. We find that, first, ∼88% of the global mRNA expression is independent of DNA replication. Second, the requirement of DNA replication for efficient histone gene expression is independent of conditions that elicit DNA damage checkpoint responses. Third, origin licensing decreases the expression of genes with origins near their 3′ ends, revealing that downstream origins can regulate the expression of upstream genes. This confirms previous predictions from mathematical modeling of a global causal coordination between DNA replication origin activity and mRNA expression, and shows that mathematical modeling of DNA microarray data can be used to correctly predict previously unknown biological modes of regulation.
Keywords: a higher-order singular value decomposition (HOSVD), cell cycle, DNA replication origin licensing and firing, DNA microarrays, mRNA expression
Introduction
DNA replication and transcription occur on a common template, and there are many ways in which these activities may influence each other. First, the passage of DNA replication forks offers an opportunity to change gene expression patterns. The transcription of capsid genes generally occurs late in most viral infections and is often dependent upon prior replication of the viral genome (Rosenthal and Brown, 1977; Thomas and Mathews, 1980; Toth et al, 1992). In bacteriophage T4, there is a direct coupling of replication and transcription because the sliding clamp processivity factor gp45 acts as a mobile transcriptional enhancer (Herendeen et al, 1989). Such coupling may serve a regulatory role: DNA replication differentially affects the transcription of the embryonic and somatic 5S rRNA genes in the frog Xenopus laevis (Wolffe and Brown, 1986). Second, juxtaposed genes and replication origins can influence each other's activity. The origin of the virus SV40 replication overlaps promoter and enhancer elements for both early and late gene expression (Cowan et al, 1973; Alwine et al, 1977), and there are many examples of transcription factors influencing replication origin function (DePamphilis, 1988). Moreover, induced transcription into a yeast Saccharomyces cerevisiae replication origin can inactivate it (Snyder et al, 1988). Finally, clashes between replication and transcription machinery are potentially important causes of genome instability. Machinery exists to minimize this instability (Liu and Alberts, 1995; Vilette et al, 1995; Wellinger et al, 2006) and the direction of replication and transcription can be coordinated to avoid clashes (Brewer, 1988).
We measured global gene expression in synchronized Saccharomyces cerevisiae cultures under two conditions that prevent DNA replication initiation without delaying cell-cycle progression. In the Cdc6− cells, depletion of the essential licensing factor Cdc6 prevents the replication origin licensing by preventing Mcm2–7, proteins necessary for the formation and maintenance of the prereplicative complex, from binding to origins during the cell cycle phase G1 (Piatti et al, 1995). In the Cdc45− cells, inactivation of the essential initiation factor Cdc45 prevents origin firing at a step after Mcm2–7 loading, and the Mcm2–7 proteins remain bound to origins even as the cells progress through the S, S/G2 and G2/M phases (Tercero et al, 2000). The duplicate time courses of a Cdc6 shutoff strain at the depleted condition of Cdc6− and its parental strain at the control condition of Cdc6+, and of a Cdc45 shutoff strain at the inactivated condition of Cdc45− and its parental strain at the control condition of Cdc45+, were sampled at approximately similar cell-cycle phases in both Cdc6+/45+ control conditions, starting at the exit from the pheromone-induced arrest and entry into the G1 phase through the S and S/G2 phases to the beginning of the G2/M phase just before nuclear division (Supplementary information Section 1, and Datasets 1 and 2). The experimental variation in sample batch, hybridization batch, DNA microarray platform and protocols (Gerke et al, 2006; Hu et al, 2007) was designed to be orthogonal to the biological variation in the condition of Mcm2–7 origin binding. This enables computational detection of experimental artifacts by using a higher-order SVD (Box 1 and Supplementary Figures 3–6, Sections 2 and 3, and Mathematica Notebook) as described earlier (Golub and Van Loan, 1996; Alter et al, 2000; Nielsen et al, 2002; Alter and Golub, 2004; Alter, 2006; Li and Klevecz, 2006; Omberg et al, 2007). The HOSVD reconstruction of the data cuboid is mathematically defined (Supplementary information Section 2.4), and used to computationally remove the experimental artifacts (Supplementary information Section 3.3). The two Cdc6− time courses are then averaged, and also separately the two Cdc45− and the four Cdc6+ and Cdc45+ control time courses (Supplementary information Dataset 3). The probabilistic significance of the enrichment of the time points in the averaged Cdc6+/45+ control in overexpressed or underexpressed cell cycle-regulated genes (Supplementary Figure 7 and Supplementary information Datasets 4 and 5), calculated (Supplementary information Section 2.5) as described (Tavazoie et al, 1999), is consistent with the flow cytometry measurements of cell synchrony in the Cdc6+ and Cdc45+ time courses (Supplementary Figures 1 and 2), as well as with previous analyses of α-factor synchronized cultures (Spellman et al, 1998).
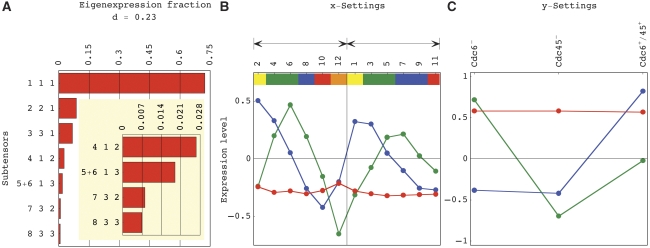
To uncover the cell cycle phase-dependent effects of Mcm2–7 origin binding, we use this HOSVD as described (Omberg et al, 2007) to separate the averaged data cuboid into a weighted sum of all possible combinations of three patterns of expression variation each: one across the 4270 genes, one across the 12 time points and one across the three conditions of Mcm2–7 origin binding (Figure 1, and Supplementary Figures 8–11 and Supplementary information Dataset 6). The significance of each combination of patterns, in terms of the fraction of the overall mRNA expression that this combination captures in the data cuboid, is proportional to its weight in this sum. We find that the seven most significant and unique combinations capture ∼94% of the mRNA expression of the 4270 genes (Table I).
Figure 1.
Significant and unique HOSVD combinations, that is, subtensors (Table I and Supplementary Figures 8–11). (A) Bar charts for the fractions of mRNA expression that the seven most significant combinations capture in the data cuboid. The fourth combination S(4,1,2), of the fourth pattern across the genes, the first across the time points and the second across the biological conditions, captures ∼2.7% of the expression of the 4270 genes. (B) Line-joined graphs of the first (red), second (blue) and third (green) expression patterns across the time points. The color bars indicate the cell cycle classifications of the time points in the averaged Cdc6+/45+ control (Supplementary Figure 7) as described (Spellman et al, 1998): M/G1 (yellow), G1 (green), S (blue), S/G2 (red) and G2/M (orange). The grid line separates the even and odd hybridization batches, indicated by black arrows. The first pattern is approximately time invariant. The second and third patterns describe oscillations consistent within the hybridization batches, that peak at the M/G1 and G1/S phases and trough at the S/G2 and G2/M phases, respectively. (C) Line-joined graphs of the first (red), second (blue) and third (green) patterns across the conditions. The first pattern is condition invariant. The second pattern correlates with underexpression in both conditions in which DNA replication is prevented, that is, in both Cdc6− and Cdc45− cells, relative to the averaged Cdc6+/45+ control. The third pattern correlates with overexpression in the Cdc6− cells and underexpression in the Cdc45− cells relative to the averaged control.
Table 1.
Significant and unique HOSVD combinations, that is, subtensors
| Subtensor | Overexpression | Underexpression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fraction (%) | Annotation | J | j | P-value | Annotation | J | j | P-value | ||
| 1 | 1, 1, 1 | 72.3 | — | — | — | — | — | — | — | — |
| 2 | 2, 2, 1 | 8.7 | M/G1 | 84 | 41 | 1.5 × 10−33 | S/G2 | 88 | 27 | 6.3 × 10−16 |
| 3 | 3, 3, 1 | 6.7 | G1 and S | 263 | 103 | 1.1 × 10−77 | G2/M | 141 | 53 | 2.6 × 10−36 |
| 4 | 4, 1, 2 | 2.7 | 3′-end ARS | 153 | 14 | 1.1 × 10−2 | Histone genes | 9 | 9 | 9.1 × 10−13 |
| 5 | 5+6, 1, 3 | 1.9 | Histone genes | 9 | 7 | 1.5 × 10−8 | 3′-end ARS | 153 | 16 | 1.9 × 10−3 |
| 6 | 7, 3, 2 | 0.8 | Histone genes | 9 | 4 | 4.9 × 10−4 | — | — | — | — |
| 7 | 8, 3, 3 | 0.7 | — | — | — | — | 3′-end ARS | 153 | 17 | 6.9 × 10−4 |
The fractions of mRNA expression that the seven most significant combinations capture in the data cuboid (Figure 1), and the probabilistic significance of the enrichment of the corresponding patterns of expression variation across the genes in overexpressed or underexpressed cell cycle-regulated genes (Spellman et al, 1998), histone genes and genes with ARSs near their 3′ ends (Cherry et al, 1997; Nieduszynski et al, 2007). The P-value of each enrichment is calculated (Supplementary information Section 2.5) according to the annotations of the genes (Supplementary information Datasets 4 and 5), assuming hypergeometric distribution of the J annotations among the K=4270 genes, and of the subset of j⊆J annotations among the subset of k=200 genes with largest or smallest levels of expression in the corresponding pattern (Supplementary information Dataset 6), as described (Tavazoie et al, 1999).
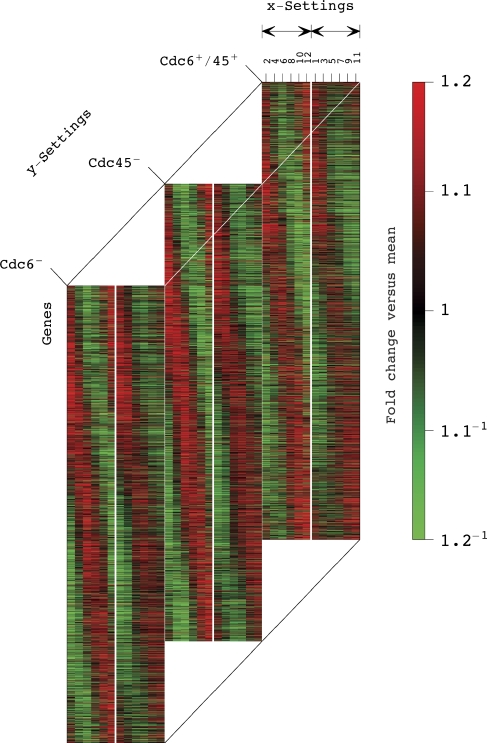
First, we find that ∼88% of the overall expression information in this data cuboid is independent of DNA replication and Mcm2–7 origin binding. This unperturbed expression is represented by the three most significant combinations, all of which correlate with condition-invariant overexpression. The first combination also correlates with time-invariant underexpression and represents steady-state expression. The second and third combinations describe oscillations consistent within the hybridization batches, which peak at the M/G1 and G1/S phases and trough at the S/G2 and G2/M phases in the averaged Cdc6+/45+ control time course, respectively. Consistently, the second and third combinations correlate with patterns of expression variation across the genes that are enriched in overexpressed M/G1 and, separately, G1 and S genes and underexpressed S/G2 and G2/M genes, respectively, with P-values <6.4 × 10−16. Taken together, the second and third combinations represent unperturbed cell cycle expression oscillations (Supplementary Figure 12). Upon sorting the genes by their levels of expression in the second and third HOSVD combinations (Supplementary Section 2.6), the picture that emerges is that of unperturbed global expression oscillations that are dominant in the Cdc6−, Cdc45− as well as in the Cdc6+/45+ time courses (Figure 2 and Supplementary information Dataset 3).
Figure 2.
The averaged data cuboid of mRNA expression of the 4270 genes across the 12 time points and across the three biological conditions (Supplementary information Dataset 3). Raster display with overexpression (red), no change in expression (black) and underexpression (green), where the expression of each gene is centered at its time-invariant level. The genes are sorted by their angular distances θ:=arctan(U:3/U:2) between the second and third HOSVD combinations (Supplementary information Section 2.6), which represent the unperturbed cell cycle expression oscillations (Box 1 and Supplementary Figure 12). The white lines separate the even and odd hybridization batches, indicated by black arrows. The picture of global expression oscillations in the averaged Cdc6+/45+ control time course is consistent with previous genome-wide mRNA expression analyses of synchronized Saccharomyces cerevisiae cultures (Alter et al, 2000; Alter and Golub, 2004; Alter, 2006; Li and Klevecz, 2006; Klevecz et al, 2008). The picture that emerges is that of unperturbed global expression oscillations that are dominant in the Cdc6−, Cdc45− as well as in the Cdc6+/45+ time courses.
It was recently shown that the cell cycle phase of the peak expression of ∼70% of 1271 cell cycle-regulated genes is conserved in cells that do not express the S phase and mitotic cyclins-encoding genes CLB1-6 and are, therefore, unable to replicate DNA (Orlando et al, 2008). Our analysis of the 4270 genes that were selected on the basis of data quality alone significantly lowers this upper bound to replication-dependent mRNA expression. Our results are consistent with the idea that the program of cell cycle-regulated transcription may be largely independent of underlying cell cycle events, such as DNA replication (Simon et al, 2001).
Second, we find that ∼3.5% of the overall expression of the 4270 genes depends on DNA replication but is independent of the method of origin inactivation. These replication-dependent perturbations in mRNA expression are represented by the fourth and sixth combinations, which correlate with overexpression in the averaged control and underexpression in both Cdc45− and Cdc6− time courses. The fourth combination also correlates with time-invariant underexpression and with expression variation across the genes that is enriched in underexpressed histone genes with the P-value <9.2 × 10−13. The sixth combination correlates with overexpression of histone genes at the G1/S phase with the corresponding P-value <4.9 × 10−4. Taken together, the time-averaged and G1/S expression of histone genes is reduced in both situations where DNA replication is prevented, indicating that DNA replication is required for efficient histone gene expression.
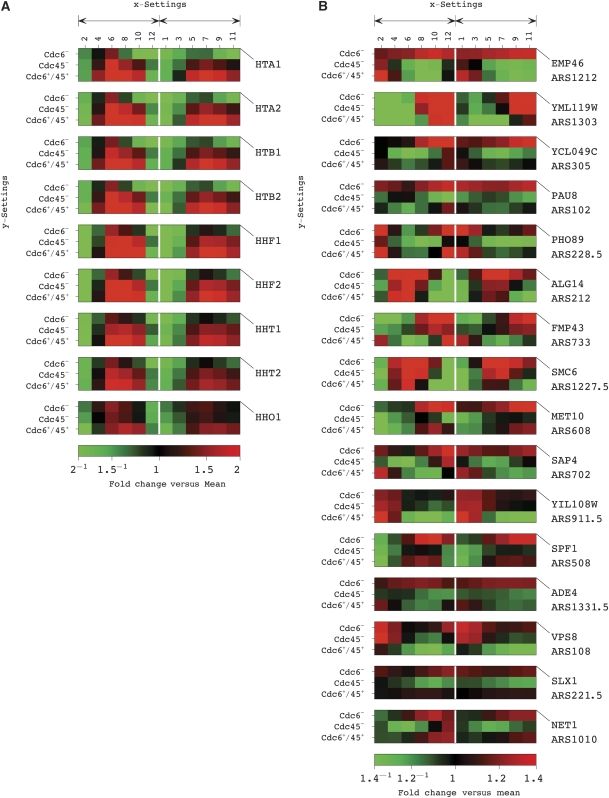
To examine the joint effects of DNA replication and origin binding on global mRNA expression, we classified the 4270 genes into intersections of the fourth through seventh combinations of expression patterns (Supplementary information Dataset 6). Enrichment in overexpressed histone genes was also observed for the fifth combination with the P-value <1.5 × 10−8. Among the 1294 genes that are underexpressed in the fourth and overexpressed in the fifth and sixth combinations, the four most significant genes, in terms of the fraction of mRNA expression that they capture in these combinations, are the histone genes HTA1, HTA2, HTB1 and HTB2. Six of the nine histone genes are among the ten most significant genes, an enrichment that corresponds to a P-value ∼2.1 × 10−15. Overall, the histone genes are overexpressed in the control relative to the Cdc6− condition, in which the Mcm2–7 licensing of origins and subsequent DNA replication are prevented, and to a lesser extent also relative to the Cdc45− condition, in which DNA replication is prevented but only after the origins are licensed (Figure 3a).
Figure 3.
DNA replication-dependent and Mcm2–7 origin binding-dependent gene expression. Raster display, in which the expression of each gene is centered at its time-invariant level. (A) DNA replication is required for efficient histone gene expression. Raster display of histone gene expression shows that histone genes are overexpressed in the Cdc6+/45+ control, relative to the Cdc6− condition, and to a lesser extent also relative to the Cdc45− condition, in a highly correlated manner. (B) Origin licensing decreases the expression of genes with origins near their 3′ ends. Raster display of the expression of the 16 most significant genes in this class shows that these genes are overexpressed in the Cdc6− relative to the Cdc45− time courses, and to a lesser extent also relative to the control, in a manner less correlated than that of the histone genes.
Previous work has shown that the coupling of histone mRNA levels to DNA replication is primarily due to transcriptional regulatory mechanisms (Lycan et al, 1987). Because in our study the Rad53 checkpoint kinase is not activated in either the Cdc6− or Cdc45− conditions as previously described (Piatti et al, 1995; Tercero et al, 2000), and because we did not observe any significant enrichment in DNA damage-induced genes (Jelinsky and Samson, 1999) in the fourth through seventh combinations of expression patterns, in which the corresponding P-values >9.4 × 10−2, we suggest that these effects on histone gene expression are directly dependent on DNA replication status, independent of DNA damage checkpoint responses.
Third, we find that ∼2.6% of the mRNA expression is affected by Mcm2–7 origin binding. The origin binding-dependent perturbations are represented by the fifth and seventh combinations, which correlate with overexpression in the Cdc6− and underexpression in the Cdc45− cultures. The fifth combination correlates with time-invariant underexpression that is enriched in underexpressed genes with autonomously replicating sequences (ARSs) adjacent to their 3′ ends, defined as genes with at least one confirmed ARS at a distance of less than 100 nucleotides from their respective 3′ ends (Cherry et al, 1997; Nieduszynski et al, 2007) with the P-value <1.9 × 10−3. The seventh combination correlates with G2/M overexpression of genes with ARSs near their 3′ ends, with the P-value <6.9 × 10−4. Taken together, origin licensing decreases time-averaged and G2/M expression of genes with origins near their 3′ ends. We did not observe any significant enrichment in genes with ARSs near their 5′ ends nor did we observe any significant enrichment in genes that overlap ARSs, where all the corresponding P-values were >1.2 × 10−1. We suggest that origin licensing may affect the expression of adjacent genes by interfering with transcription elongation and/or pre-mRNA 3′-end processing (Proudfoot, 2004; Gilmartin, 2005; Weiner, 2005; Rosonina et al, 2006), thus destabilizing the mRNA transcripts. The accumulation of mRNA transcripts of this class of genes throughout the Cdc6− relative to the Cdc45− time courses is consistent with the observed peak in their differential expression late in the time courses at the G2/M phase (Figure 3b).
Of the 153 genes with ARSs near their 3′ ends, 16 are among the 100 most significant of the 1412 genes that are overexpressed in the fourth and underexpressed in the fifth and seventh combinations, an enrichment that corresponds to a P-value <3.4 × 10−7. No other significant enrichment was observed among these 100 genes, nor was an enrichment in gene ontology (GO) (Cherry et al, 1997) annotations observed among the 16 genes with ARSs near their 3′ ends. Of these 153 and 16 genes, only 24 and 5, respectively, are cell cycle regulated. These 16 genes are overexpressed in the Cdc6− condition, in which the origins are unlicensed, relative to the Cdc45− condition, in which Mcm2–7 bind origins throughout the time course, and to a lesser extent also relative to the control, in which Mcm2–7 bind origins only during G1. The expression of these genes is not as highly correlated as that of the nine genes for histones, consistent with this class of genes being co-degraded but not necessarily co-transcribed. Previous studies have shown that transcription can interfere with the function of downstream origins (Snyder et al, 1988; Nieduszynski et al, 2005; Donato et al, 2006). Our results reveal that downstream origins can also interfere with the expression of upstream genes. This interference requires origin licensing but does not require origin firing. We suggest that cells may exploit this complex reciprocal arrangement in different contexts to regulate gene expression or origin activation.
A global pattern of correlation between DNA binding of Mcm2–7 and reduced expression of adjacent genes, most of which are not cell cycle regulated, during the cell cycle phase G1 was discovered from previous mathematical modeling of DNA microarray data (Alter and Golub, 2004; Omberg et al, 2007), in which the mathematical variables and operations were shown to represent biological reality (Alter, 2006). The mathematical variables, patterns uncovered in the data, were shown to correlate with activities of cellular elements, such as regulators or transcription factors. The operations, such as classification, rotation or reconstruction in subspaces of these patterns, were shown to simulate experimental observation of the correlations and possibly even the causal coordination of these activities (Supplementary information Section 2). Of the 153 genes with ARSs near their 3′ ends, the ARSs near 151 were identified in Mcm2–7 high-throughput binding assays, and for the ARSs near 139 of those, including all of the 16 significant genes, consensus sequence elements were identified (Wyrick et al, 2001; Xu et al, 2006). Our results, therefore, suggest that a causal relation underlies this correlation, that is, the binding of Mcm2–7 is responsible for diminished expression of adjacent genes, and show that mathematical modeling of DNA microarray data can be used to correctly predict previously unknown biological modes of regulation.
Higher-order singular value decomposition (HOSVD).
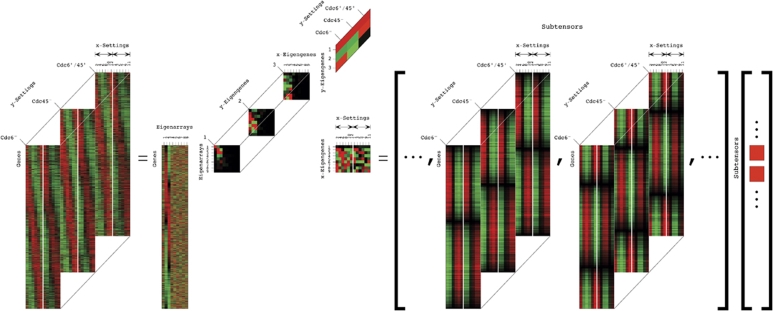
The structure of the data in this study is of an order higher than that of a matrix. Genes, time points and conditions of Mcm2–7 origin binding, each represent a degree of freedom in a cuboid, that is, a third-order tensor. Unfolded into a matrix, these degrees of freedom are lost and much of the information in the data tensor might also be lost. We integrate these data by using a tensor HOSVD (Supplementary information Sections 2 and 3, and Mathematica Notebook). This HOSVD uncovers in the data tensor (Supplementary information Dataset 3) patterns of mRNA expression variation across the genes, time points and conditions, as depicted in the raster display of Supplementary information Equation (1) (left) with overexpression (red), no change in expression (black) and underexpression (green). This HOSVD was recently reformulated in analogy with the matrix singular value decomposition (SVD) (Golub and Van Loan, 1996; Alter et al, 2000; Nielsen et al, 2002; Alter and Golub, 2004; Alter, 2006; Li and Klevecz, 2006) such that it separates the data tensor into a weighted sum of combinations of three patterns each, that is, ‘subtensors', as depicted in the raster display of Supplementary information Equation (7) (right), where the second and third HOSVD combinations and their corresponding weights are shown explicitly. In these raster displays, each expression pattern across the time points is centered at its time-invariant level. The genes are sorted by their ‘angular distances' between the second and third HOSVD combinations (Supplementary information Section 2.6), which represent the unperturbed cell cycle expression oscillations (Figure 2 and Supplementary Figure 12). The white lines separate the even and odd hybridization batches, indicated by black arrows. This reformulation of the HOSVD was shown to enable its interpretation in terms of the cellular states, biological processes and experimental artifacts that compose the data tensor by defining the significance of each combination of patterns, and the operation of rotation in a subspace of these combinations (Omberg et al, 2007). In this study, we extend this analogy to mathematically define the operation of reconstruction in a subspace of combinations (Supplementary information Section 2.4), and use it to computationally remove experimental artifacts from the global mRNA expression measured in multiple time courses (Supplementary information Section 3.3).
Supplementary Material
Supplementary Text; Supplementary Figures 1–12; Supplementary Tables I–II
A Mathematica 5.2 code file, executable by Mathematica 5.2 and readable by Mathematica Player, freely available at http://www.wolfram.com/products/player/.
A PDF format file, readable by Adobe Acrobat Reader.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating relative mRNA expression levels of 4,771 probes of the UT DNA microarrays that correspond to the K=4,270 genes across 24 samples.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating relative mRNA expression levels of 8,540 probes of the WU DNA microarrays that correspond to the K=4,270 genes across 72 samples.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating the averaged log2 of the relative mRNA expression of the K=4,270 genes across the L=12 time points and across the M=3 conditions of Mcm2-7 origin binding. The genes are sorted by their angular distances between the second and third HOSVD combinations (SI Sec. 2.6 and Dataset 6), which represent the unperturbed cell cycle expression oscillations (Fig. 2 and SI Fig. 12). The angular distance of each gene is also listed.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, reproducing descriptions and genomic coordinates [32] of the 325 confirmed ARSs in Saccharomyces cerevisiae.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, reproducing cell cycle annotations [26], DNA damage responses [31], descriptions and genomic coordinates [33] and of the 4,270 Saccharomyces cerevisiae genes.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating the eigenarrays and superpositions of eigenarrays that define the global gene expression patterns of the seven significant and unique subtensors of the averaged data cuboid. The expression levels of the genes in the intersections of the fourth through seventh HOSVD combinations, as computed by using the corresponding eigenarrays, are also tabulated. The ten significant among the 1,294 genes that are underexpressed in the fourth and overexpressed in the fifth and sixth combinations are enriched in histone genes. The 100 significant among the 1,412 genes that are overexpressed in the fourth and underexpressed in the fifth and seventh combinations are enriched in genes with ARSs near their 3′ ends.
Acknowledgments
We thank BA Cohen and VR Iyer and his laboratory members, Z Hu, PJ Killion and S Shivaswamy, for technical assistance with DNA microarrays from the Washington University Microarray Core and at the University of Texas, respectively, and AW Murray for insightful comments. We also thank GH Golub for introducing us to matrix and tensor computations, and the American Institute of Mathematics in Palo Alto for hosting the 2004 Workshop on Tensor Decompositions where some of this study was carried out. This study was supported by Cancer Research UK (JFXD) and National Human Genome Research Institute R01 Grant HG004302 and National Science Foundation CAREER Award DMS-0847173 (OA).
Footnotes
The authors declare that they have no conflict of interest.
References
- Alter O (2006) Discovery of principles of nature from mathematical modeling of DNA microarray data. Proc Natl Acad Sci USA 103: 16063–16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D (2000) Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA 97: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O, Golub GH (2004) Integrative analysis of genome-scale data by using pseudoinverse projection predicts novel correlation between DNA replication and RNA transcription. Proc Natl Acad Sci USA 103: 16577–16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine JC, Reed SI, Stark GR (1977) Characterization of the autoregulation of simian virus 40 gene A. J Virol 24: 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53: 679–686 [DOI] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, Botstein D (1997) Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67–73 [PMC free article] [PubMed] [Google Scholar]
- Cowan K, Tegtmeyer P, Anthony DD (1973) Relationship of replication and transcription of Simian Virus 40 DNA. Proc Natl Acad Sci USA 70: 1927–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML (1988) Transcriptional elements as components of eukaryotic origins of DNA replication. Cell 52: 635–638 [DOI] [PubMed] [Google Scholar]
- Donato JJ, Chung SC, Tye BK (2006) Genome-wide hierarchy of replication origin usage in Saccharomyces cerevisiae. PLoS Genet 2:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke JP, Chen CT, Cohen BA (2006) Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174: 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM (2005) Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev 19: 2517–2521 [DOI] [PubMed] [Google Scholar]
- Golub GH, Van Loan CF (1996) Matrix Computations, 3rd edn. Baltimore, Maryland, USA: Johns Hopkins University Press [Google Scholar]
- Herendeen DR, Kassavetis GA, Barry J, Alberts BM, Geiduschek EP (1989) Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science 245: 952–958 [DOI] [PubMed] [Google Scholar]
- Hu Z, Killion PJ, Iyer VR (2007) Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39: 683–687 [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD (1999) Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA 96: 1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevecz RR, Li CM, Marcus I, Frankel PH (2008) Collective behavior in gene regulation: the cell is an oscillator, the cell cycle a developmental process. FEBS J 275: 2372–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Klevecz RR (2006) A rapid genome-scale response of the transcriptional oscillator to perturbation reveals a period-doubling path to phenotypic change. Proc Natl Acad Sci USA 103: 16254–16259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Alberts BM (1995) Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Lycan DE, Osley MA, Hereford LM (1987) Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol 7: 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Blow JJ, Donaldson AD (2005) The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res 33: 2410–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD (2007) OriDB: a DNA replication origin database. Nucleic Acids Res 35: D40–D46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O'Connell JX, Ferro M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M (2002) Molecular characterisation of soft tissue tumours: a gene expression study. Lancet 359: 1301–1307 [DOI] [PubMed] [Google Scholar]
- Omberg L, Golub GH, Alter O (2007) Tensor higher-order singular value decomposition for integrative analysis of DNA microarray data from different studies. Proc Natl Acad Sci USA 104: 18371–18376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB (2008) Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J 14: 3788–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N (2004) New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Rosenthal LJ, Brown M (1977) The control of SV40 transcription during a lytic infection: late RNA synthesis in the presence of inhibitors of DNA replication. Nucleic Acids Res 4: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Kaneko S, Manley JL (2006) Terminating the transcript: breaking up is hard to do. Genes Dev 20: 1050–1056 [DOI] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, Young RA (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697–708 [DOI] [PubMed] [Google Scholar]
- Snyder M, Sapolsky RJ, Davis RW (1988) Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol 8: 2184–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nat Genet 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JFX (2000) DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J 19: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GP, Mathews MB (1980) DNA replication and the early to late transition in adenovirus infection. Cell 22: 523–533 [DOI] [PubMed] [Google Scholar]
- Toth M, Doerfler W, Shenk T (1992) Adenovirus DNA replication facilitates binding of the MLTF/USF transcription factor to the viral major late promoter within infected cells. Nucleic Acids Res 20: 5143–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D, Ehrlich SD, Michel B (1995) Transcription-induced deletions in Escherichia coli plasmids. Mol Microbiol 17: 493–504 [DOI] [PubMed] [Google Scholar]
- Weiner AM (2005) E Pluribus Unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell 20: 168–170 [DOI] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A (2006) Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Brown DD (1986) DNA replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell 47: 217–227 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavaré S (2006) Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text; Supplementary Figures 1–12; Supplementary Tables I–II
A Mathematica 5.2 code file, executable by Mathematica 5.2 and readable by Mathematica Player, freely available at http://www.wolfram.com/products/player/.
A PDF format file, readable by Adobe Acrobat Reader.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating relative mRNA expression levels of 4,771 probes of the UT DNA microarrays that correspond to the K=4,270 genes across 24 samples.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating relative mRNA expression levels of 8,540 probes of the WU DNA microarrays that correspond to the K=4,270 genes across 72 samples.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating the averaged log2 of the relative mRNA expression of the K=4,270 genes across the L=12 time points and across the M=3 conditions of Mcm2-7 origin binding. The genes are sorted by their angular distances between the second and third HOSVD combinations (SI Sec. 2.6 and Dataset 6), which represent the unperturbed cell cycle expression oscillations (Fig. 2 and SI Fig. 12). The angular distance of each gene is also listed.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, reproducing descriptions and genomic coordinates [32] of the 325 confirmed ARSs in Saccharomyces cerevisiae.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, reproducing cell cycle annotations [26], DNA damage responses [31], descriptions and genomic coordinates [33] and of the 4,270 Saccharomyces cerevisiae genes.
A tab-delimited text format file, readable by both Mathematica and Microsoft Excel, tabulating the eigenarrays and superpositions of eigenarrays that define the global gene expression patterns of the seven significant and unique subtensors of the averaged data cuboid. The expression levels of the genes in the intersections of the fourth through seventh HOSVD combinations, as computed by using the corresponding eigenarrays, are also tabulated. The ten significant among the 1,294 genes that are underexpressed in the fourth and overexpressed in the fifth and sixth combinations are enriched in histone genes. The 100 significant among the 1,412 genes that are overexpressed in the fourth and underexpressed in the fifth and seventh combinations are enriched in genes with ARSs near their 3′ ends.