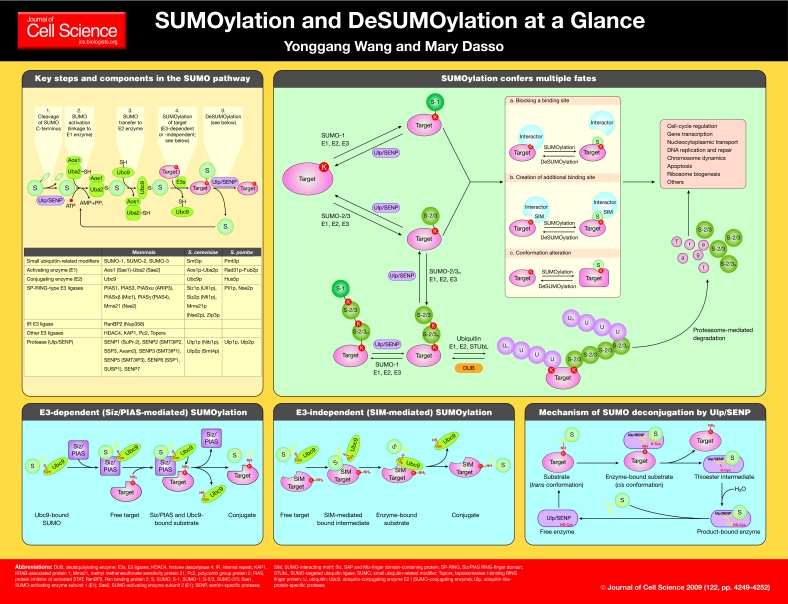
Small ubiquitin-related modifiers (SUMOs) are ubiquitin-like polypeptides that become covalently conjugated to cellular proteins in a manner similar to ubiquitylation (Johnson, 2004). More than 1000 proteins have been identified as potential SUMO-conjugation (SUMOylation) targets (Hochstrasser, 2009), and this pathway has been implicated in controlling many aspects of cell physiology, including cell-cycle regulation, transcription, nucleocytoplasmic transport, DNA replication and repair, chromosome dynamics, apoptosis, and ribosome biogenesis.
Both SUMOylation and deSUMOylation (SUMO deconjugation) are highly dynamic processes. This poster article discusses the enzymes that mediate SUMOylation and deSUMOylation, concentrating on their mechanisms of action. Although it is not possible to discuss individual SUMOylated targets in detail, we will mention emerging paradigms that explain how SUMOylation might direct the fate of target proteins.
SUMO paralogues
Yeast express a single SUMO paralogue, called Smt3p in Saccharomyces cerevisiae. Mammalian cells express three major SUMO paralogues, called SUMO-1, SUMO-2 and SUMO-3. SUMO-2 and SUMO-3 are ∼95% identical to each other. In most contexts, SUMO-2 and SUMO-3 cannot be distinguished, and here we collectively refer to them as SUMO-2/3. SUMO-2 and SUMO-3 are each ∼45% identical to SUMO-1, and all mammalian paralogues are ∼ 45% identical to Smt3p.
Figure 1.
There are several important differences between the mammalian SUMO paralogues. First, some SUMO targets are conjugated only to SUMO-1, others only to SUMO-2/3, and still others to all SUMO paralogues (Vertegaal et al., 2006). Second, the overall cellular concentration of SUMO-2/3 is greater than that of SUMO-1, as is the pool of free protein available for conjugation (Saitoh and Hinchey, 2000). It is thus likely that the bulk of SUMOylation involves SUMO-2/3. Third, SUMO-1 and SUMO-2/3 show different subcellular localization patterns (Ayaydin and Dasso, 2004; Zhang et al., 2008). Fourth, photobleaching experiments suggest that SUMO-1 is less dynamic than the other SUMO paralogues, and its pattern of conjugation responds differently to heat shock and stress (Ayaydin and Dasso, 2004; Saitoh and Hinchey, 2000).
Smt3p, SUMO-2 and SUMO-3 can form conjugated chains through a single conserved acceptor lysine (Bylebyl et al., 2003; Tatham et al., 2001). SUMO-1 does not have an equivalent lysine residue, and thus probably does not act as a link in elongating chains in vivo. However, SUMO-1 might terminate chains that are elongated through serial conjugation of SUMO-2/3 (Matic et al., 2008). Notably, although SUMOylation does not seem to rely upon the geometry of chain linkages to confer information, as ubiquitylation does (Pickart and Fushman, 2004), SUMO conjugates built from different paralogues or with different chain lengths can specify distinct target fates, as discussed below.
SUMO-interacting motifs
SUMO-interacting motifs (SIMs) play a central role in both the enzymology of the SUMO pathway and in the fate of conjugated species. The best-characterized class of SIM consists of a hydrophobic core ([V/I]-x-[V/I]-[V/I]) flanked by a cluster of negatively charged amino acids (Kerscher, 2007). The SIM hydrophobic core can bind to an interaction surface on SUMO proteins in a parallel or antiparallel orientation. The acidic residues adjacent to the core might contribute to the affinity, orientation or paralogue specificity of binding (Hecker et al., 2006; Meulmeester et al., 2008). A variant SIM was recently defined within the transcriptional repressor CoREST1, consisting of [I/V/L]-[D/E]-[I/V/L]-[D/E]-[I/V/L] with N-terminal acidic residues (Ouyang et al., 2009). This SIM is highly selective for SUMO-2/3 binding, and differs from previously identified SIMs because its core lacks a hydrophobic residue at position 4. Notably, the diversity of SIMs identified to date is much less than the 16 known ubiquitin-binding domains (Grabbe and Dikic, 2009); it is reasonable to speculate that additional SIMs remain to be discovered.
SUMO-processing, -activating and -conjugating enzymes
Newly translated SUMO proteins are cleaved to reveal C-terminal diglycine motifs. This processing is mediated by a family of proteases known as ubiquitin-like-protein-specific proteases (Ulps) in yeast and sentrin-specific proteases (SENPs) in mammals (Mukhopadhyay and Dasso, 2007). Ulps and SENPs also mediate deSUMOylation (see below).
All SUMO paralogues share the same activating (E1) and conjugating (E2) enzymes. These enzymes are structurally similar to E1 and E2 enzymes of ubiquitin, and they share many of the properties that have been demonstrated for those enzymes (Hochstrasser, 2009). The yeast SUMO E1 enzyme is a heterodimer consisting of Aos1p (also known as Sae1 in vertebrates) and Uba2p (also known as Sae2 in vertebrates), which show sequence similarity to the N-terminus and C-terminus of the monomeric ubiquitin E1 enzyme, respectively (Johnson et al., 1997). Aos1p-Uba2p catalyzes the formation of a high-energy thioester bond between Uba2p and the SUMO C-terminus, with ATP hydrolysis to AMP and pyrophosphate (Johnson et al., 1997; Lois and Lima, 2005). The activated SUMO is subsequently passed to a cysteine in the active site of the E2 enzyme, Ubc9, through an intermolecular thiol-transfer reaction.
Residues of Ubc9 that are directly involved in the transfer of SUMO act to orient the lysine of the target protein and to decrease its pKa, resulting in a higher occurrence of its activated, de-protonated state (Yunus and Lima, 2006). SUMO transfer from Ubc9 to some target proteins can occur through at least two ligase-independent mechanisms. First, many SUMOylated lysines lie within a consensus motif, Ψ-K-x-[D/E] (where Ψ is an aliphatic branched amino acid and x is any amino acid). Ubc9 can directly recognize this motif and conjugate the lysine residue within it (Bernier-Villamor et al., 2002). Second, some SUMO substrates contain SIMs that promote their own conjugation (Meulmeester et al., 2008; Zhu et al., 2008). These SIMs bind to the SUMO moiety to which Ubc9 is attached, thereby increasing its local concentration and facilitating SUMOylation. Because SIMs show paralogue preference, this mechanism allows targets to be modified in a paralogue-selective manner. Notably, mammalian Ubc9 can itself be SUMOylated on a nonconsensus lysine in its N-terminal helix (Knipscheer et al., 2008). This modification does not inhibit its activity per se, but alters its target preference, increasing the conjugation of substrates that contain SIMs, which bind to the SUMO that is conjugated to Ubc9.
SUMO ligases
SUMO ligases (E3 enzymes) facilitate the majority of SUMOylation under physiological conditions (Meulmeester et al., 2008). A number of SUMO ligases have been described, most of which seem to be specific to metazoans.
Siz/PIAS-family proteins
All eukaryotes express proteins with Siz/PIAS RING-finger-like domains (SP-RING domains), which are known as SAP and Miz-finger domain (Siz) proteins in yeast and protein inhibitor of activated STAT (PIAS) proteins in vertebrates (Hochstrasser, 2001). In budding yeast, Siz1p and Siz2p are required for most Smt3p conjugation (Johnson and Gupta, 2001; Takahashi et al., 2001). Other SP-RING proteins, Zip3p and Mms21p, promote assembly of the synaptonemal complex between homologous chromosomes during meiosis (Cheng et al., 2006) and DNA repair (Potts, 2009), respectively. The five vertebrate PIAS proteins (PIAS1, PIAS3, PIASxα, PIASxβ and PIASγ) have been implicated in many processes, including gene expression, signal transduction and genome maintenance (Palvimo, 2007).
Beyond their SP-RING domains, Siz/PIAS-family members share additional conserved motifs, including an N-terminal scaffold attachment factor (SAF)-A/B, acinus, PIAS (SAP) motif, a PINIT motif, a SIM, and a C-terminal domain that is rich in serine and acidic amino acids (S/DE domain) (Palvimo, 2007). The SAP domain directs the localization of Siz/PIAS proteins to chromatin within the nucleus (Azuma et al., 2005; Palvimo, 2007). Structural analysis of a Siz1 fragment that is sufficient for E3 activity in vitro shows that it has an elongated tripartite architecture, formed by its N-terminal PINIT domain, SP-RING domain and C-terminal domain, termed the SP-CTD (Yunus and Lima, 2009). The SP-RING and SP-CTD domains are required for activation of the Ubc9-SUMO thioester, whereas the PINIT domain directs SUMOylation to the correct target lysine.
RanBP2
RanBP2 is a nuclear-pore protein that localizes to the cytoplasmic face of the pore. RanBP2 possesses a domain called the internal repeat (IR) domain, which consists of two tandemly repeated sequences of around 50 residues (IR1 and IR2), separated by a 24-residue spacer (M). RanBP2 fragments containing the IR domain have SUMO ligase activity in vitro (Pichler et al., 2002). Structural analysis of a RanBP2 fragment containing the IR1 and M domains indicates that RanBP2 enhances Ubc9 activity without direct contacts to the target protein (Reverter and Lima, 2005). It has thus been proposed that RanBP2 promotes SUMOylation by aligning the Ubc9-SUMO thioester complex in an optimal configuration for substrate interaction with the active site of Ubc9 and for catalysis. Notably, the IR domain of RanBP2 binds extremely stably to Ubc9 and the SUMO-1-conjugated form of RanGAP1, the activating protein for the GTPase Ran (Matunis et al., 1998; Saitoh et al., 1998). Structural analysis yielded the puzzling result that this binding abolishes the ability of RanBP2 to promote multiple rounds of target SUMOylation (Reverter and Lima, 2005). It will clearly be important to establish how this inhibition is overcome for RanBP2 to act as an E3 enzyme in its physiological context.
Other SUMO ligases
Additional proteins that have been reported as potential SUMO ligases include histone deacetylase 4 (HDAC4), KRAB-associated protein 1 (KPA1), Pc2 and Topors.
HDAC4 is a histone deacetylase that is a SUMOylation target. HDAC4 expression enhances the SUMOylation of myocyte-specific enhancer factor 2 (MEF2), as well as other targets (Geiss-Friedlander and Melchior, 2007; Zhao et al., 2005). HDAC4 can bind to Ubc9, suggesting that it acts as an E3 enzyme. It has alternatively been proposed that HDAC4 enhances SUMOylation by other means, such as promoting the phosphorylation of target proteins at sites adjacent to conjugated lysine residues (Yang and Gregoire, 2006).
The human co-repressor KRAB-associated protein 1 (KAP1) possesses PHD-finger domains that catalyze intramolecular SUMOylation of an adjacent KAP1 bromodomain (Peng and Wysocka, 2008). SUMOylation stabilizes the association of the bromodomain with the chromatin modifiers, thus promoting the establishment of gene silencing. Structural analysis suggests that the PHD finger and the bromodomain cooperate as an integrated unit to recruit Ubc9 and facilitate SUMOylation (Zeng et al., 2008).
Mammalian Pc2 is a polycomb-group protein that can act as a SUMO ligase for the transcriptional co-repressor CtBP (Wotton and Merrill, 2007). Pc2 can bind to both Ubc9 and its conjugation targets, and it seems to have a rather limited spectrum of targets.
Topors is a RING-finger protein that binds DNA topoisomerase I and p53. It possesses both RING-finger-dependent ubiquitin ligase activity and RING-finger-independent SUMO ligase activity (Weger et al., 2005). Topors has a SIM (Hecker et al., 2006) and acts as a SUMO ligase in vitro (Hammer et al., 2007).
SUMO-deconjugating enzymes
Ulps/SENPs are responsible both for processing SUMO peptides and for deconjugating SUMOylated species (Hay, 2007; Mukhopadhyay and Dasso, 2007). Ulps/SENPs share a conserved ∼200-amino-acid catalytic domain that is typically found near their C-terminus.
There are two Ulps in budding yeast: Ulp1p and Ulp2p (Li and Hochstrasser, 1999; Li and Hochstrasser, 2000). Ulp1p localizes to the nuclear envelope and is encoded by an essential gene (Li and Hochstrasser, 1999). Overexpression of processed Smt3p weakly rescues Δulp1 cells, but unprocessed Smt3p does not (Li and Hochstrasser, 1999), suggesting that one essential function of Ulp1p is Smt3p maturation. Ulp2p localizes in the nucleoplasm (Li and Hochstrasser, 2000) and is particularly important for dismantling poly-Smt3p chains (Bylebyl et al., 2003). Although not essential for vegetative growth, Ulp2p has roles in chromosome segregation, meiotic development and recovery from cell-cycle checkpoint arrest (Li and Hochstrasser, 2000).
Mammals have six SENPs: SENP1, SENP2, SENP3, SENP5, SENP6 and SENP7. SENP1-3 and SENP5 are more similar to Ulp1p than to Ulp2p, whereas SENP6 and SENP7 are more Ulp2-like (Mukhopadhyay and Dasso, 2007). SENP1 and SENP2 localize to the nuclear envelope and have processing and deconjugation activity for both SUMO-1 and SUMO-2/3. By contrast, all other SENPs have a strong preference for SUMO-2/3. SENP3 and SENP5 localize in nucleoli and catalyze SUMO-2/3 processing and deconjugation (Di Bacco et al., 2006; Gong and Yeh, 2006). Similar to Ulp1p (Panse et al., 2006), SENP3 and SENP5 have important roles in ribosome biogenesis (Yun et al., 2008). SENP6 and SENP7 localize within the nucleoplasm and are implicated in the editing of poly-SUMO chains (Lima and Reverter, 2008; Mukhopadhyay et al., 2006; Shen et al., 2009).
The sites of SENP1, SENP2 and Ulp1p that are engaged during processing or deconjugation are shallow clefts lined with conserved amino acids (Mossessova and Lima, 2000; Reverter and Lima, 2004; Shen et al., 2006b). In both reactions, the C-terminus of SUMO lies within these clefts as an elongated strand, and conserved tryptophans and other adjacent residues of Ulp1p, SENP1 and SENP2 clamp the diglycine motif of SUMO in a hydrophobic `tunnel'. During deconjugation, such binding requires minimal structural distortion of the target protein, which explains how Ulps/SENPs can deconjugate many SUMOylated species with only modest target specificity. In the SUMO processing reaction, Ulps/SENPs induce the isomerization of the scissile peptide bond, resulting in a 90° kink in the SUMO C-terminal tail (Reverter and Lima, 2006; Shen et al., 2006a). In deconjugation reactions, Ulps/SENPs induce the scissile isopeptide bond between the C-terminus of SUMO and the ε-amine group of the lysine residue of the target protein to adopt a cis configuration, resulting in a similar 90° kink (Reverter and Lima, 2006; Shen et al., 2006a). For both peptide and amide bonds, the kinked cis conformations facilitate hydrolysis of the bond.
The fates of SUMOylated species
The consequences of SUMOylation are diverse, including alteration of the activity, localization and/or stability of the target protein (Geiss-Friedlander and Melchior, 2007). Frequently, these consequences result from recognition of conjugated species by SIM-containing proteins. SUMOylation might also cause the loss of binding partners, or cause conformational changes that alter the enzymatic activity of the target protein. Some SIM-mediated interactions prevent deconjugation by limiting the access of Ulps/SENPs to the conjugated proteins. In cases in which the interacting SIMs have intrinsic paralogue preference, they will selectively protect targets that are modified with the preferred SUMO paralogue (Zhu et al., 2009).
Few SUMOylation targets show quantitative modification. Notably, SUMOylation can play an important regulatory role even under these circumstances (Geiss-Friedlander and Melchior, 2007). This might be explained by the observation that SUMOylation can promote the assembly of protein complexes, such as in transcriptionally repressed chromatin, that remain stable despite subsequent deSUMOylation (Geiss-Friedlander and Melchior, 2007). Additionally, SUMOylation might function within the catalytic cycle of targets, facilitating enzymatic turnover (Hardeland et al., 2002). In both of these cases, SUMOylated species are transient intermediates that facilitate stable changes in target proteins.
Crosstalk between the SUMO and ubiquitin pathways
A particularly exciting development in this field has revealed an important point of crosstalk between the SUMO and ubiquitin pathways (Hunter and Sun, 2008): a subset of targets become conjugated with multiple SUMOs, and can be recognized by SUMO-targeted ubiquitin ligases (STUbLs), causing the proteasomal degradation of these targets. It is currently believed that STUbLs operate primarily through recognition of poly-SUMO chains, although it remains possible that they might recognize some targets that are mono-SUMOylated at numerous sites.
Rfp1p and Rfp2p are fission-yeast RING-finger proteins that possess N-terminal SIMs (Sun et al., 2007). They are genetically redundant and there is no visible phenotype for loss of either gene encoding these proteins. However, cells must possess at least one of these proteins for growth and genome stability. Both Rfp1p and Rfp2p heterodimerize with the Slx8p protein, a RING-finger ubiquitin ligase. Together, they mediate the ubiquitylation of poly-SUMOylated targets, resulting in their proteasomal destruction. The Slx5p-Slx8p heterodimer acts similarly in budding yeast (Hunter and Sun, 2008). The functions of Rfp1 and/or Rfp2 and Slx8 are performed by a single protein in human cells, RING-finger protein 4 (RNF4), which is the only confirmed mammalian STUbL (Sun et al., 2007).
Perspectives
Findings during the last 5 or 6 years have provided a much more sophisticated understanding of the SUMOylation pathway, revealing some aspects that are unique to this pathway and others that are probably common to pathways involving all ubiquitin-like proteins. However, the picture of SUMOylation is not yet complete, and we expect that studies of the SUMO pathway may yet hold more surprises. In particular, we look forward to future findings regarding the mechanisms of SUMO ligases and the possibility that additional SIMs remain to be discovered. Finally, we still have much to learn regarding the biological functions of this pathway and its interaction with ubiquitin and other regulatory pathways.
This work was supported through Eunice Kennedy Shriver National Institute of Child Health and Human Development Intramural funds (Z01 HD001902). Deposited in PMC for release after 12 months.
References
- Ayaydin, F. and Dasso, M. (2004). Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15, 5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma, Y., Arnaoutov, A., Anan, T. and Dasso, M. (2005). PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor, V., Sampson, D. A., Matunis, M. J. and Lima, C. D. (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345-356. [DOI] [PubMed] [Google Scholar]
- Bylebyl, G. R., Belichenko, I. and Johnson, E. S. (2003). The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278, 44113-44120. [DOI] [PubMed] [Google Scholar]
- Cheng, C. H., Lo, Y. H., Liang, S. S., Ti, S. C., Lin, F. M., Yeh, C. H., Huang, H. Y. and Wang, T. F. (2006). SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20, 2067-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco, A., Ouyang, J., Lee, H. Y., Catic, A., Ploegh, H. and Gill, G. (2006). The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 26, 4489-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander, R. and Melchior, F. (2007). Concepts in sumoylation: a decade on. Nat. Rev Mol. Cell. Biol. 8, 947-956. [DOI] [PubMed] [Google Scholar]
- Gong, L. and Yeh, E. T. (2006). Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 281, 15869-15877. [DOI] [PubMed] [Google Scholar]
- Grabbe, C. and Dikic, I. (2009). Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 109, 1481-1494. [DOI] [PubMed] [Google Scholar]
- Hammer, E., Heilbronn, R. and Weger, S. (2007). The E3 ligase Topors induces the accumulation of polysumoylated forms of DNA topoisomerase I in vitro and in vivo. FEBS Lett. 581, 5418-5424. [DOI] [PubMed] [Google Scholar]
- Hardeland, U., Steinacher, R., Jiricny, J. and Schar, P. (2002). Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21, 1456-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, R. T. (2007). SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17, 370-376. [DOI] [PubMed] [Google Scholar]
- Hecker, C. M., Rabiller, M., Haglund, K., Bayer, P. and Dikic, I. (2006). Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117-16127. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2001). SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107, 5-8. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2009). Origin and function of ubiquitin-like proteins. Nature 458, 422-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, T. and Sun, H. (2008). Crosstalk between the SUMO and ubiquitin pathways. Ernst Schering Found. Symp. Proc. 1-16. [DOI] [PubMed]
- Johnson, E. S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73, 355-382. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S. and Gupta, A. A. (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735-744. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., Schwienhorst, I., Dohmen, R. J. and Blobel, G. (1997). The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O. (2007). SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer, P., Flotho, A., Klug, H., Olsen, J. V., van Dijk, W. J., Fish, A., Johnson, E. S., Mann, M., Sixma, T. K. and Pichler, A. (2008). Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell 31, 371-382. [DOI] [PubMed] [Google Scholar]
- Li, S. J. and Hochstrasser, M. (1999). A new protease required for cell-cycle progression in yeast. Nature 398, 246-251. [DOI] [PubMed] [Google Scholar]
- Li, S. J. and Hochstrasser, M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20, 2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, C. D. and Reverter, D. (2008). Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J. Biol. Chem. 283, 32045-32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois, L. M. and Lima, C. D. (2005). Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic, I., van Hagen, M., Schimmel, J., Macek, B., Ogg, S. C., Tatham, M. H., Hay, R. T., Lamond, A. I., Mann, M. and Vertegaal, A. C. (2008). In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell Proteomics 7, 132-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M. J., Wu, J. and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester, E., Kunze, M., Hsiao, H. H., Urlaub, H. and Melchior, F. (2008). Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610-669. [DOI] [PubMed] [Google Scholar]
- Mossessova, E. and Lima, C. D. (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865-876. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, D. and Dasso, M. (2007). Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286-295. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, D., Ayaydin, F., Kolli, N., Tan, S. H., Anan, T., Kametaka, A., Azuma, Y., Wilkinson, K. D. and Dasso, M. (2006). SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 174, 939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, J., Shi, Y., Valin, A., Xuan, Y. and Gill, G. (2009). Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell 34, 145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palvimo, J. J. (2007). PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 35, 1405-1408. [DOI] [PubMed] [Google Scholar]
- Panse, V. G., Kressler, D., Pauli, A., Petfalski, E., Gnadig, M., Tollervey, D. and Hurt, E. (2006). Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7, 1311-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J. and Wysocka, J. (2008). It takes a PHD to SUMO. Trends Biochem. Sci. 33, 191-194. [DOI] [PubMed] [Google Scholar]
- Pichler, A., Gast, A., Seeler, J. S., Dejean, A. and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109-120. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M. and Fushman, D. (2004). Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610-616. [DOI] [PubMed] [Google Scholar]
- Potts, P. R. (2009). The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 8, 499-506. [DOI] [PubMed] [Google Scholar]
- Reverter, D. and Lima, C. D. (2004). A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12, 1519-1531. [DOI] [PubMed] [Google Scholar]
- Reverter, D. and Lima, C. D. (2005). Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter, D. and Lima, C. D. (2006). Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat. Struct. Mol. Biol. 13, 1060-1068. [DOI] [PubMed] [Google Scholar]
- Saitoh, H. and Hinchey, J. (2000). Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252-6258. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., Sparrow, D. B., Shiomi, T., Pu, R. T., Nishimoto, T., Mohun, T. J. and Dasso, M. (1998). Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8, 121-124. [DOI] [PubMed] [Google Scholar]
- Shen, L., Tatham, M. H., Dong, C., Zagorska, A., Naismith, J. H. and Hay, R. T. (2006a). SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat. Struct. Mol. Biol. 13, 1069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. N., Dong, C., Liu, H., Naismith, J. H. and Hay, R. T. (2006b). The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem. J. 397, 279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. N., Geoffroy, M. C., Jaffray, E. G. and Hay, R. T. (2009). Characterization of SENP7, a SUMO-2/-3 specific isopeptidase. Biochem. J. 2, 223-230. [DOI] [PubMed] [Google Scholar]
- Sun, H., Leverson, J. D. and Hunter, T. (2007). Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Kahyo, T., Toh, E. A., Yasuda, H. and Kikuchi, Y. (2001). Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276, 48973-48977. [DOI] [PubMed] [Google Scholar]
- Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H. and Hay, R. T. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368-35374. [DOI] [PubMed] [Google Scholar]
- Vertegaal, A. C., Andersen, J. S., Ogg, S. C., Hay, R. T., Mann, M. and Lamond, A. I. (2006). Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics 5, 2298-2310. [DOI] [PubMed] [Google Scholar]
- Weger, S., Hammer, E. and Heilbronn, R. (2005). Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 579, 5007-5012. [DOI] [PubMed] [Google Scholar]
- Wotton, D. and Merrill, J. C. (2007). Pc2 and SUMOylation. Biochem. Soc. Trans. 35, 1401-1404. [DOI] [PubMed] [Google Scholar]
- Yang, X. J. and Gregoire, S. (2006). A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell 23, 779-786. [DOI] [PubMed] [Google Scholar]
- Yun, C., Wang, Y., Mukhopadhyay, D., Backlund, P., Kolli, N., Yergey, A., Wilkinson, K. D. and Dasso, M. (2008). Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 183, 589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus, A. A. and Lima, C. D. (2006). Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491-499. [DOI] [PubMed] [Google Scholar]
- Yunus, A. A. and Lima, C. D. (2009). Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L., Yap, K. L., Ivanov, A. V., Wang, X., Mujtaba, S., Plotnikova, O., Rauscher, F. J., 3rd. and Zhou, M. M. (2008). Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 15, 626-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. D., Goeres, J., Zhang, H., Yen, T. J., Porter, A. C. and Matunis, M. J. (2008). SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29, 729-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Sternsdorf, T., Bolger, T. A., Evans, R. M. and Yao, T. P. (2005). Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell. Biol. 25, 8456-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Zhu, S., Guzzo, C. M., Ellis, N. A., Sung, K. S., Choi, C. Y. and Matunis, M. J. (2008). Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283, 29405-29415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S., Goeres, J., Sixt, K. M., Bekes, M., Zhang, X. D., Salvesen, G. S. and Matunis, M. J. (2009). Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol. Cell 33, 570-580. [DOI] [PMC free article] [PubMed] [Google Scholar]