Summary
At least 17 members of the protein disulphide isomerase (PDI) family of oxidoreductases are present in the endoplasmic reticulum (ER) of mammalian cells. They are thought to catalyse disulphide formation to aid folding or to regulate protein function; however, little is known about their individual functions. Here, we show that some proteins that enter the ER are clients for single oxidoreductases, whereas others are clients for several PDI-like enzymes. We previously identified potential substrates for ERp57, and here identify substrates for ERp18 and ERp46. In addition, we analysed the specificity of substrates towards PDI, ERp72, ERp57, ERp46, ERp18 and P5. Strikingly, ERp18 shows specificity towards a component of the complement cascade, pentraxin-related protein PTX3, whereas ERp46 has specificity towards peroxiredoxin-4, a thioredoxin peroxidase. By contrast, most PDI family members react with Ero1α. Moreover, P5 forms a non-covalent complex with immunoglobulin heavy chain binding protein (BiP) and shows specificity towards BiP client proteins. These findings highlight cooperation between BiP and P5, and demonstrate that individual PDI family members recognise specific substrate proteins.
Keywords: BiP, ERp18, ERp46, P5, protein disulphide isomerase
Introduction
The ER is a highly specialised organelle allowing the oxidative folding and post-translational modification of proteins entering the secretory pathway (Hwang et al., 1992). The compartmentalisation of the ER away from the cytosol ensures that the correct redox conditions exist to enable a distinct set of folding catalysts to facilitate the formation of disulphide bonds (Sevier and Kaiser, 2002). The protein disulphide isomerase (PDI) family of ER oxidoreductases is thought to be responsible for catalysing the formation, isomerisation and reduction of these disulphides (Hatahet and Ruddock, 2007). There are at least 17 identified members of this family (Ellgaard and Ruddock, 2005), each of which is characterised by the presence of at least one domain that is homologous to thioredoxin. Many of these domains contain a pair of active site cysteine residues (CxxC) that shuttle between the disulphide and dithiol form (Ferrari and Soling, 1999). To perform disulphide exchange reactions, the individual active sites must be maintained in either the oxidised disulphide form to allow disulphide formation, or the reduced dithiol form for isomerisation or reduction of disulphide bonds (Freedman, 1995). How the active sites are maintained in either their reduced or oxidised state and how the ER maintains an environment conducive to concurrent disulphide bond formation, isomerisation and reduction has been the subject of intense speculation for over 40 years. The components of the oxidative pathway have been identified (Frand and Kaiser, 1998; Pollard et al., 1998), and the role of glutathione in the reductive pathway has recently been highlighted (Chakravarthi and Bulleid, 2004; Jessop and Bulleid, 2004; Molteni et al., 2004). However, several key questions, such as substrate specificity of Ero1 and the respective roles of each oxidoreductase remain unanswered.
The members of the PDI family of oxidoreductases are not minor components of the ER; indeed several are highly abundant and ubiquitously expressed, so it is likely that they have important functions. There is now extensive evidence from work carried out in vitro (Lyles and Gilbert, 1991), in yeast (Laboissiere et al., 1995) and in mammalian systems (Bulleid and Freedman, 1988) demonstrating that PDI is capable of both the formation and isomerisation of disulphide bonds within proteins. Although ERp57 is highly homologous to PDI and shares the same arrangement of thioredoxin-like domains, studies carried out in vitro (Zapun et al., 1998) and in vivo (Antoniou et al., 2002; Jessop et al., 2007) suggest that ERp57 is a glycoprotein-specific oxidoreductase that catalyses the reduction of non-native disulphides. In addition, PDI acts not only as a molecular chaperone (Wilson et al., 1998), but also as a non-catalytic component of the enzymes prolyl 4-hydroxylase (Koivu et al., 1987) and microsomal triglyceride transfer protein (Wetterau et al., 1991). Hence, with just these two oxidoreductases, we see similar, but distinct, functions in catalysis of protein folding for subsets of protein substrates and in the regulation of protein function and polypeptide binding. It is highly likely that each oxidoreductase has a defined role to play in protein maturation, which might be specific to cell or tissue type, or to specific stages of development.
To identify substrates for other PDI family members, we exploited the fact that during the reduction of disulphide bonds, a mixed disulphide must form between the protein substrate and the enzyme involved in catalysis. In this study, we made stable cell lines expressing substrate-trapping mutant oxidoreductases, where the second cysteine of the active site was mutated to alanine. Using this approach, we were able to trap mixed disulphides between oxidoreductase and substrate. This enabled us to identify some of the substrates for individual PDI family members and to assess the specificity of these enzyme-substrate complexes. Our results highlight the fact that some proteins that enter the secretory pathway react with distinct PDI-like oxidoreductases that facilitate their folding, whereas others interact with several oxidoreductases. In addition, we found that the PDI P5 (also known as PDIA6) binds non-covalently to BiP, an interaction that has been seen previously (Meunier et al., 2002). We now find that this binding is redox dependent and that P5 reacts with substrates that are known to associate with BiP, including those targeted for ER-associated degradation. These results reveal a level of interaction between P5 and BiP not previously envisaged and point to substrate specificity of some ER oxidoreductases that are linked to an interaction with a specific chaperone system.
Results
Identification of substrates for PDI family members
Our previous work with ERp57 demonstrated that substrate-trapping mutants of PDI-like oxidoreductases, where the second cysteine in each of the active sites was converted to alanine, can be used to isolate and identify substrates for these enzymes (Jessop et al., 2007). We therefore extended the work to five other PDI family members, namely PDI, P5, ERp72, ERp46 and ERp18. Each of these enzymes contains at least one thioredoxin domain with a very similar CGHC (CGAC for ERp18) active site. We made stable cell lines expressing either the wild-type protein or a substrate-trapping mutant of each oxidoreductase. For this work we used a HT1080 human fibrosarcoma cell line, which is the same cell type we used to construct the ERp57 cell lines in our previous work, so that the results would be comparable. Each cell line expressed the V5-tagged wild-type or mutant proteins to equivalent levels (supplementary material Fig. S1).
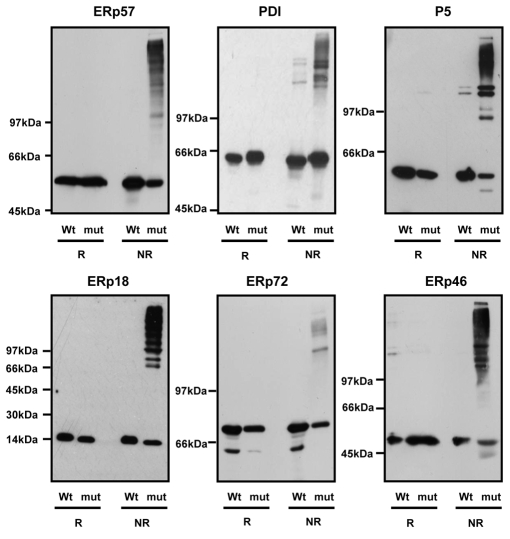
To assess the ability of the substrate-trapping mutants to form mixed disulphides with client proteins, we separated cell lysates under reducing and non-reducing conditions (Fig. 1). Each of the mutant oxidoreductases trapped substrates to varying degrees, as judged by a series of high molecular mass V5-reactive proteins, which were only present when the lysates were separated under non-reducing conditions (Fig. 1). The pattern of mixed disulphide-bonded complexes was different for each oxidoreductase, suggesting that they interacted with different client proteins. Very few mixed disulphides were seen with ERp72 and only a few distinct products were seen with PDI. These results suggest that either these proteins are not particularly active in reducing disulphides or they only react with client proteins under specific physiological conditions. Mixed disulphides were also seen with wild-type PDI and P5. It has been shown previously that PDI forms mixed disulphides with Ero1α (Mezghrani et al., 2001), and this was shown to be the case here (supplementary material Fig. S2). The mixed disulphides observed with wild-type P5 co-migrated with major mixed disulphides seen with the mutant protein.
Fig. 1.
Substrate-trapping mutants of PDI family members form mixed disulphides when expressed in HT1080 cells. Each panel represents results from a particular cell line expressing either wild-type (Wt) or a substrate-trapping mutant (mut) of the PDI family member indicated. Whole-cell lysates were separated by SDS-PAGE under either reducing (R) or non-reducing (NR) conditions. Ectopically expressed protein was visualised by western blot with antibody against the V5 tag. All gels were 7.5% acrylamide except for the ERp18 gel, which was a 7.5-12.5% gradient gel.
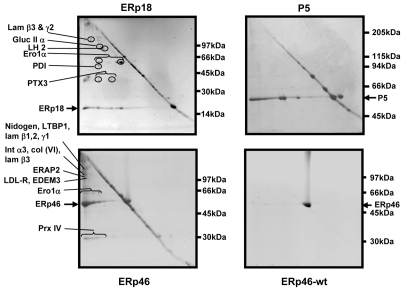
To identify the proteins forming mixed disulphides with the various PDI family members, we first captured the mixed disulphides by immunoisolation with agarose beads conjugated to an anti-V5 antibody. Mixed disulphides were eluted from the beads and separated by 2D SDS-PAGE. Non-reducing conditions were used for the first dimension, and reducing conditions for the second dimension to resolve the mixed disulphides and separate the substrate proteins according to their individual sizes. Proteins that had bound non-covalently migrated in a diagonal line, because these species were not part of a mixed disulphide and therefore migrated at the same rate under reducing and non-reducing conditions. The substrate proteins that formed part of a mixed disulphide resulted in spots or smears that migrated below the diagonal as they migrated further in the second dimension than the first. There were insufficient mixed disulphides to PDI or ERp72 to identify using this approach (results not shown). However, we were able to visualise client proteins from the ERp18 and the ERp46 cell lines that had formed mixed disulphides (Fig. 2). A control gel generated following lysis of HT1080 cells expressing wild-type ERp46 contained no proteins migrating faster in the second dimension than the first, demonstrating that the protein spots found with the mutant protein were due to substrate trapping. Resolved spots were excised from the gel and proteins identified by mass spectrometry (Table 1).
Fig. 2.
Resolution of substrates for PDI family members by 2D gel electrophoresis. HT1080 cells expressing either substrate-trapping mutant of ERp18, P5 or ERp46 or wild-type ERp46 were treated with NEM and lysed. Clarified lysates were immunoisolated with an anti-V5 antibody conjugated to agarose beads. Proteins were eluted by boiling in SDS and separated under non-reducing conditions. Gel lanes were excised and reduced with 50 mM DTT and separated in a second dimension. Proteins were visualised by silver staining. Proteins migrating faster in the second dimension than the first dimension were excised from the gel and identified by mass spectrometry. The identities of some of the excised protein spots are as indicated (see Table 1 for details). For P5, a 7.5% gel and for ERp18 and ERp46, a 7.5-12.5% gradient gel was used in both directions.
Table 1.
Characteristics of substrates forming mixed disulphides with PDI family members
| Substrate | SwissProt accession no. | Mixed disulphide partner(s) | Molecular mass (kDa) | Disulphides* | Glyc. |
|---|---|---|---|---|---|
| Ero1α | Q96HE7 | ERp57, ERp46, ERp18, PDI | 55 | 7 | Y |
| Lysyl hydroxylase 2 | O00469 | ERp57, ERp46, ERp18 | 85 | (13 Cys) | Y |
| Laminin β3 | Q13751 | ERp57, ERp46, ERp18 | 130 | ∼27 | Y |
| Laminins (β1, γ1) | P07942, P11047, | ERp57, ERp46 | 198, 178 | ∼54, 43 | Y |
| Integrins (α3, β2) | P26006, P05107, | ERp57, ERp46 | 119, 85 | 9, 28 | Y |
| Collagen α1(VI) | P12109 | ERp57, ERp46 | 109 | (20 Cys) | Y |
| LDL receptor | P01130 | ERp57, ERp46 | 95 | 30 | Y |
| Laminin γ2 | Q13753 | ERp57, ERp18 | 130 | 23 | Y |
| Integrins (α2, α6, β1, β5) | P17301, P23229, Q8WUM6, P18084 | ERp57 | 88, 88, 130, 127 | 12, 18, 42, 50 | Y |
| Plexin A1 | Q9UIW2 | ERp57 | 211 | (53 Cys) | Y |
| Agrin | O00468 | ERp57 | 215 | 15 | Y |
| Putative fibrillin | Q68CX6 | ERp57 | 235 | (34 Cys) | Y |
| Fibulin-like protein | Q12805 | ERp57 | 55 | 15 | Y |
| Discoidin | Q96PD2 | ERp57 | 85 | 4 | Y |
| Clusterin | P10909 | ERp57 | 52 | 5 | Y |
| Lysyl oxidase 2 | Q9Y4K0 | ERp57 | 87 | (42 Cys) | Y |
| Adam17 | P78536 | ERp57 | 93 | 7 | Y |
| Adam10 | O14672 | ERp57 | 84 | 3 | Y |
| Melanotransferrin | P08582 | ERp57 | 80 | 6 | Y |
| Glucosyltransferase family member | Q8NBJ5 | ERp57 | 72 | (13 Cys) | Y |
| Tapasin | O15533 | ERp57 | 48 | 2 | Y |
| Laminin β2 | P55268 | ERp46 | 196 | ∼50 | Y |
| Collagen α1(IV) | P02462 | ERp46 | 161 | (20 Cys) | Y |
| Collagen α2(IV) | P08572 | ERp46 | 168 | (21 Cys) | Y |
| ERAP1 | Q9BZQ6 | ERp46 | 107 | (10 Cys) | Y |
| ERAP2 | Q6P179 | ERp46 | 110 | (8 Cys) | Y |
| EDEM3 | Q9BZQ6 | ERp46 | 105 | (11 Cys) | Y |
| LTBP1 | P22064 | ERp46 | 153 | ∼66 | Y |
| Polycystic kidney disease 1 | Q7Z443 | ERp46 | 196 | (40 Cys) | Y |
| Aspartyl/asparagine β-hydroxylase | Q12797 | ERp46 | 86 | 7 | Y |
| Nidogen-1 | P14543 | ERp46 | 136 | 22 | Y |
| Peroxiredoxin IV | Q13162 | ERp46 | 31 | 4 | N |
| ER glucosidase II subunit α | Q14697 | ERp18 | 107 | (7 Cys) | Y |
| PTX3 | P26022 | ERp18 | 42 | 7 | Y |
Where it is not known how many disulphides are formed, the number of cysteines in a protein is given in parentheses
We were not able to identify any discernable client proteins using the 2D gel approach for P5, yet it formed complexes which were stabilised with disulphide bonds. This can clearly be seen on the 2D gel as a line of P5 migrating below the diagonal (Fig. 2). In addition, P5 itself formed intra-chain disulphides as evidenced by the appearance of a spot of P5 migrating above the diagonal. The identity of P5 in this position was verified by mass spectrometry. These results suggest that P5 either forms mixed disulphides with itself or with several client proteins, which cannot be visualised after resolution on 2D gel.
P5 forms a non-covalent interaction with BiP and ERp94
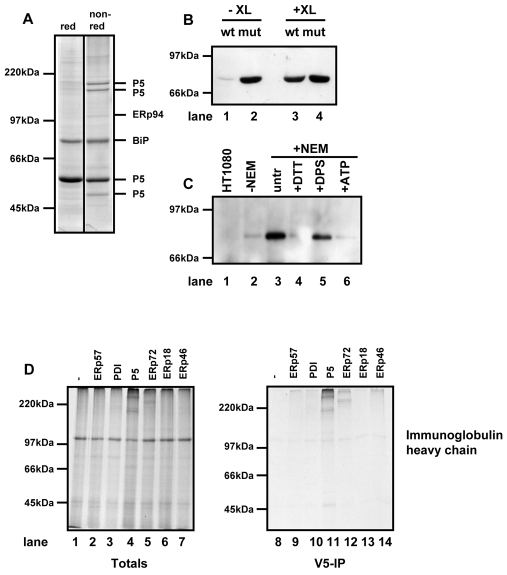
When the V5-immunoisolated material from the P5 substrate-trapping cell line was analysed by non-reducing SDS-PAGE followed by Coomassie blue staining, six prominent protein bands were observed (Fig. 3A). When these proteins were identified by mass spectrometry they were found to be P5, BiP and ERp94; four different bands containing P5 were seen, which migrated as a single band when separated under reducing conditions (Fig. 3A), suggesting that they differ only in redox form. The faster-migrating species probably represent reduced or intrachain disulphide-bonded forms, whereas the lower-mobility species are most likely interchain disulphide-bonded dimers. Note that the slower-migrating redox forms of P5 were also seen to a much lesser extent in the wild-type cell line (Fig. 1) and are therefore unlikely to arise as a result of the mutations introduced. Moreover, the association with BiP and ERp94 was non-covalent, because each migrated with an apparent molecular mass of the individual protein.
Fig. 3.
P5 forms a non-covalent interaction with BiP and associates with a BiP client protein. (A) HT1080 cells expressing substrate-trapping mutant of P5 was treated with NEM and lysed. Clarified lysates were immunoisolated with an anti-V5 antibody conjugated to agarose beads. Proteins were eluted by boiling in SDS and separated under reducing (red) or non-reducing (non-red) conditions. The protein bands were excised from the non-reducing gel and the indicated proteins identified by mass spectrometry. (B) HT1080 cells expressing either wild-type (wt) or the substrate-trapping mutant (mut) of P5 were either not treated (-XL) or treated (+XL) with a crosslinking agent. Cells were lysed and V5-tagged P5 immunoisolated using anti-V5 antibody immobilised on agarose beads. The immunoisolate was separated on by SDS-PAGE and western blotted with antibody against BiP. (C) Untransfected HT1080 cells (lane 1) or cells expressing the substrate-trapping mutant of P5 (lanes 2-6) were pretreated with 10 mM DTT (lane 4), 1 mM DPS (lane 5) or untreated (lanes 1, 2, 3, 6). Cells were either lysed in lysis buffer in the absence of NEM (lane 2) or lysed in the presence of NEM (lanes 1, 3-5) or in the presence of NEM and ATP (lane 6). V5-tagged P5 was immunoisolated with V5 agarose. The immunoisolate was separated by SDS-PAGE and western blotted with antibody against BiP. (D) Human immunoglobulin heavy chain was translated in the presence of SP cells prepared from either HT1080 cells (lanes 1 and 8) or cells expressing the substrate-trapping mutants of the PDI family members as indicated (lanes 2-7 and 9-14). Following translation for 2 hours, SP cells were isolated and products of translation were either separated by non-reducing SDS-PAGE immediately (totals; lanes 1-7) or following immunoisolation with V5-agarose (V5-IP; lanes 8-14). Radiolabelled proteins were visualised following autoradiography.
P5 and BiP have previously been shown to form a complex that can be stabilised by the addition of a crosslinking agent (Meunier et al., 2002). Using a similar crosslinking approach, we confirmed that such a complex exists within cells expressing wild-type P5 (Fig. 3B, lanes 3 and 4). V5-tagged P5 was first immunoisolated with the V5 antibody and BiP was detected in western blots of the resulting immunoisolated material. Even in the absence of crosslinker, BiP was isolated from the mutant cell line; a small amount was even isolated from the wild-type cell line (Fig. 3B, lanes 1 and 2). We also found that the interaction of BiP with the substrate-trapping mutant of P5 was ATP dependent (Fig. 3C, lanes 3 and 6). Moreover, the interaction between P5 and BiP was lost following treatment of cells with the reducing agent dithiothreitol (DTT), but stabilised with the oxidising agent dithiodipyridine (DPS) (Fig. 3C, lanes 4 and 5). The redox dependence of the interaction of P5 and BiP suggests a possible mechanism of regulation: the fact that we see a prolonged interaction of BiP with the substrate-trapping mutant of P5 could suggest that mutating the active site of P5 locks it in a redox state that favours its interaction with BiP.
To determine whether the interaction of P5 with BiP was a consequence of the reactivity of P5 towards BiP substrates we investigated the role of P5 in the folding and assembly of the BiP substrate immunoglobulin heavy chain using an in vitro translation system supplemented with semi-permeabilised cells (SP cells). In such a system, SP cells prepared from cells grown in culture are added to a reticulocyte lysate in the presence of an RNA transcript and radiolabelled amino acid (Wilson et al., 1995). The reticulocyte lysate does not contain added reducing agent; therefore, disulphide bond formation can occur co- and post-translationally within proteins translocated into the ER of the SP cells (Bulleid et al., 1996). When the immunoglobulin heavy chain was translated in such a system it was translocated into the ER lumen of the SP cells, formed disulphide bonds and assembled into an interchain disulphide-bonded dimer (supplementary material Fig. S3). When we prepared SP cells from cell lines expressing substrate-trapping PDI family members, we could determine whether any mixed disulphides were formed (Fig. 3D). When the translation products were separated under non-reducing conditions, we could clearly see that mixed disulphides formed when the immunoglobulin heavy chain was translated in the P5 cell line (Fig. 3D, lanes 1-7). In addition, some mixed disulphide formation was seen with ERp72 but no mixed disulphides were evident with ERp57, PDI, ERp18 or ERp46. The mixed disulphides were confirmed when the translation products were immunoisolated with the V5 antibody, which specifically isolates the ectopically expressed PDI family member (Fig. 3D, lanes 8-14). These results clearly show that P5 and ERp72 form mixed disulphides with the immunoglobulin heavy chain during its folding in the ER lumen and suggest that P5, and to a certain extent ERp72, react with BiP client proteins.
Assessing substrate specificity
When the identified client proteins for ERp18 and ERp46 are compared with our previous results with ERp57, it is clear that some proteins form mixed disulphides with all tested oxidoreductases whereas others specifically interact with one enzyme (Table 1). For example, the large heavily disulphide-bonded secreted proteins such as the laminins are clients for ERp57, ERp46 and ERp18, whereas other proteins show more restricted interactions. ERp18 also formed much fewer mixed disulphides than either ERp46 or ERp57, indicating a more restricted range of clients. These results could, however, be explained simply by our ability to actually isolate enough protein to allow identification by mass spectrometry. Therefore, an alternative approach was required to test the specificity of each substrate for individual oxidoreductases. Hence, we translated individual potential substrates into SP cells prepared from the cell lines expressing substrate-trapping PDI family members. The formation of a mixed disulphide between the translated protein and V5-tagged oxidoreductase can be evaluated by immunoisolation of translation products with V5 antibody. Table 2 summarises all the results obtained using this approach.
Table 2.
Mixed disulphide formation following in vitro translation of potential substrates for PDI family members*
| ERp57 | PDI | P5 | ERp72 | ERp18 | ERp46 | |
|---|---|---|---|---|---|---|
| IgG heavy chain | – | – | +++ | ++ | – | – |
| Ero1α | ++ | ++ | + | – | +++ | ++ |
| β1 integrin | +++ | – | ++ | – | – | ++ |
| β1 integrin + C | – | – | +++ | ++ | – | +++ |
| LDLR | +++ | – | +++ | + | – | +++ |
| LDLR + C | + | – | +++ | ++ | – | +++ |
| Clusterin | +++ | + | – | – | – | – |
| Clusterin + C | + | – | ++ | ++ | – | + |
| PTX3 | – | + | + | – | +++ | + |
| Prx-IV | – | – | +++ | – | – | +++ |
| C-propeptide | – | – | + | – | +++ | + |
| α1-antitrypsin | – | – | – | – | – | – |
| α1-antitrypsin-NHK | + | – | +++ | – | – | – |
Table shows level of mixed disulphide formed. –, no mixed disulphide; +, low level of mixed disulphide formed; ++, medium level of mixed disulphide formed; +++, high level of mixed disulphide formed. C, castanospermine
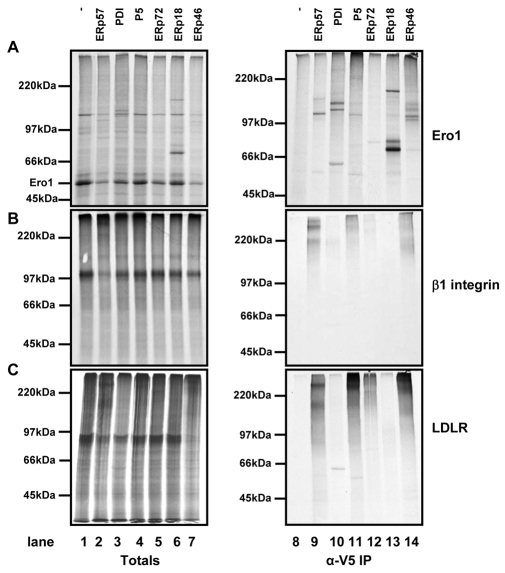
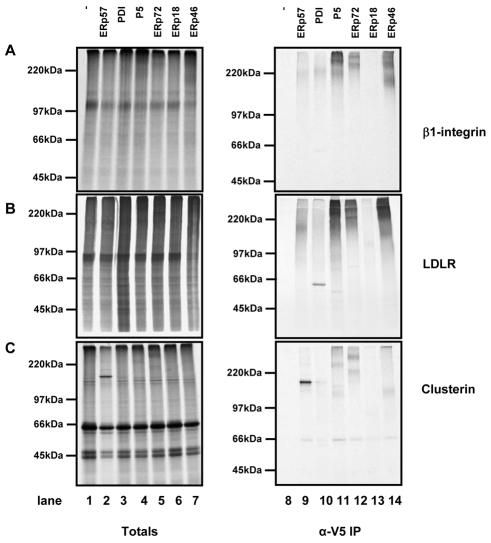
We first looked at three proteins, Ero1α, β1 integrin and low density lipoprotein receptor (LDLR), which had been shown from our mass spectrometry data to form mixed disulphides with several PDI family members. We separated translation products by non-reducing SDS-PAGE either before or after immunoisolation with the V5 antibody (Fig. 4). All PDI family members tested, apart from ERp72, formed mixed disulphides with Ero1α (Fig. 4A). Mixed disulphides can be seen by the immunoisolation of radiolabelled species with a mass equal to the sum of Ero1 and the V5-tagged enzyme. No mixed disulphides were seen with the untransfected cell line (Fig. 4A, lane 8). Some radiolabelled bands were seen at the correct size for the PDI family members (Fig. 4A, PDI, lane 10) which indicates that there is some translation of endogenous mRNA during the reaction. Several of the substrate-trapping enzymes formed more than one mixed disulphide, as judged by several banding patterns, which might indicate mixed disulphides between different cysteines within Ero1α or different active sites of the substrate-trapping mutants.
Fig. 4.
Assessing substrate specificity by in vitro translation of Ero1α, β1 integrin or LDLR. mRNA encoding Ero1α (A), β1 integrin (B), or LDLR (C) was translated in the presence of SP cells prepared from either HT1080 cells (lanes 1 and 8) or cells expressing the substrate-trapping mutants of the PDI family members as indicated (lanes 2-7 and 9-14). Following translation for 2 hours, SP cells were isolated and products of translation were either separated by non-reducing SDS-PAGE carried out immediately (totals; lanes 1-7) or following immunoisolation with V5-agarose (V5-IP; lanes 8-14). Radiolabelled proteins were visualised following autoradiography.
For β1 integrin, the results matched those obtained using the mass spectrometry approach. Mixed disulphides were seen with ERp57, ERp46 and to a certain extent P5 (Fig. 4B, lanes 9, 11, 14). A similar pattern was seen with LDLR, but with a stronger interaction with P5 and some mixed disulphides with ERp72. Both of these complex glycoproteins have several disulphides and appear to form mixed disulphides with multiple PDI family members, possibly because the substrate interacts both with the calnexin cycle and with other chaperone systems, such as that involving BiP. It is striking that neither of these proteins formed mixed disulphides with ERp18 or PDI, underscoring the specificity of substrate interaction with the PDI family members.
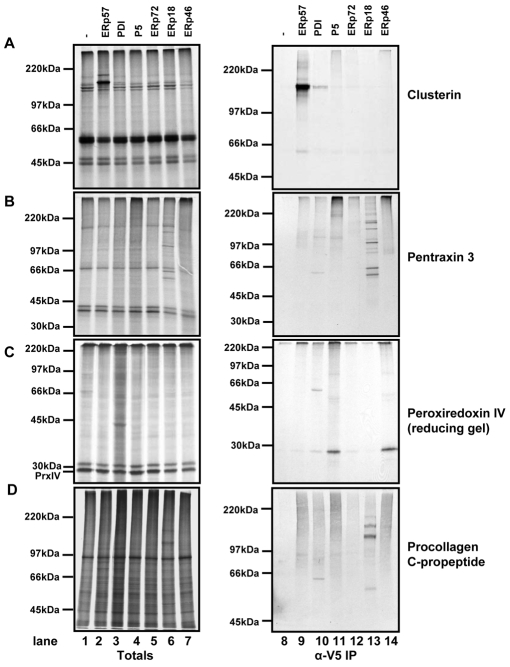
We next looked at three substrates that seemed to have a much narrower specificity towards particular PDI family members. From our mass spectrometry data, clusterin was only seen to form a mixed disulphide with ERp57, pentraxin-related protein PTX3 with ERp18, and peroxiredoxin-4 (Prx-IV) with ERp46. These results were generally mirrored when we translated the individual proteins into SP cells derived from our substrate-trapping cell lines (Fig. 5). Clusterin mainly formed a mixed disulphide with ERp57, although there was a faint interaction with PDI (Fig. 5A, lanes 9, 10). By contrast, PTX3 only formed distinct mixed disulphides with ERp18 and in addition formed multiple species (Fig. 5B, lane 13). When a folding time course of PTX3 was carried out, we found that the protein formed intra-chain disulphides at early time points as judged by the appearance of species that migrated faster than the fully reduced protein (supplementary material Fig. S4). At later time points, slower-migrating species were formed, indicative of the formation of oligomers stabilised by interchain disulphide bonds. Hence, the numerous mixed disulphides seen with ERp18 indicate that this enzyme forms mixed disulphides with several multimeric forms of PTX3 during its assembly to the secreted decameric form (Bottazzi et al., 1997).
Fig. 5.
Mixed disulphides are formed between clusterin, PTX3, Prx-IV and procollagen C-propeptide and specific PDI family members. The translation of clusterin (A), PTX3 (B), Prx-IV (C) and procollagen C-propeptide (D) were carried out as in Fig. 4 with the following modifications: PTX3, Prx-IV and procollagen C-propeptide samples were separated through 7.5-12.5% gradient gels, and Prx-IV samples were separated under reducing conditions.
We have previously shown that Prx-IV also forms multiple redox species (Tavender et al., 2008), which compromised our ability to see distinct mixed disulphides when the samples were separated under non-reducing conditions (results not shown). In addition, the protein does not contain any methionine residues, so only the cysteine residues were radiolabelled, which reduced the overall signal. To overcome this problem, we carried out immunoisolation of the translation products with V5 antibody and then separated the resulting immunoisolates under reducing conditions. The presence of Prx-IV indicates that an interaction had occurred between PDI family member and Prx-IV. The results clearly show that Prx-IV formed an interaction specifically with ERp46 and P5 (Fig. 5C, lanes 11 and 14). This supports the mass spectrometry data for ERp46 and indicates a clear specificity for mixed disulphide formation within the PDI family members.
Finally, we also investigated the interaction of a protein that is not normally expressed by HT1080 cells: the C-propeptide from type III procollagen (Bottomley et al., 2001). The ability to identify mixed disulphides when translating proteins into the various SP cells expressing substrate-trapping mutants would demonstrate the utility of this approach to screen proteins for their specificity towards particular PDI family members. Following translation into SP cells, the C-propeptide formed intra-chain disulphides, followed by trimer formation stabilised by interchain disulphides (supplementary material Fig. S5). Remarkably, when this protein was translated into the substrate-trapping cell lines, only ERp18 formed distinct mixed disulphides (Fig. 5D, lane 13).
Modulating substrate specificity
The results from mass spectrometry and in vitro translation demonstrate that some substrates interact with several PDI family members, whereas others substrates have much more defined specificities. The basis of this specificity could reside with molecular recognition between enzyme and substrate or could be defined by the targeting of substrates to specific chaperone systems to which a PDI family member is associated (Jessop et al., 2009). A classic example of the latter is the targeting of glycoproteins to the calnexin cycle. To determine whether the profile of mixed disulphides interacting with each PDI family member changes if targeting to the calnexin cycle is disrupted, we translated the glycoproteins β1 integrin, LDLR and clusterin in the presence of castanospermine, a drug that blocks glycan trimming and therefore reduces targeting to the calnexin cycle (Hebert et al., 1996). Such treatment diminished the formation of mixed disulphides of all three substrates with ERp57 (Fig. 6). Both β1 integrin and LDLR now formed stronger mixed disulphides with ERp72 (Fig. 6, lane 12), in addition to the previously observed interaction with P5 and ERp46. Moreover, clusterin, which almost exclusively interacted with ERp57 in the absence of castanospermine, now formed mixed disulphides with P5, ERp72 and ERp46. These results support previous work (Jessop et al., 2009; Solda et al., 2006), clearly demonstrating that, at least in the case of the calnexin cycle and ERp57, the specificity of substrates for a particular PDI family member is due to targeting of the protein to a chaperone complex.
Fig. 6.
Consequence of blocking the entry of substrates into the calnexin cycle on substrate specificity. The translation of β1 integrin (A), LDLR (B) and clusterin (C) was carried out as in Fig. 4 except that SP cells were pre-treated with castanospermine, which was also present during the translation reaction. Gels were run exactly as described in Figs 4 and 5.
Specificity during ER-associated degradation
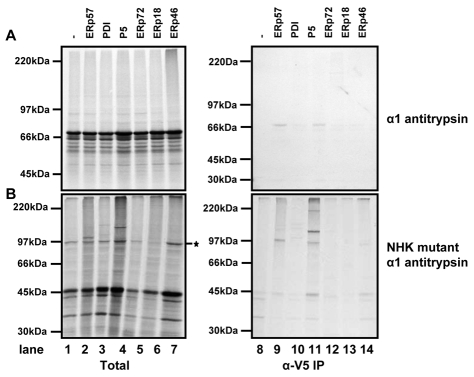
Most of the work on the specificity of PDI family members has focused on the role of these enzymes during the folding and assembly of proteins. Recently, it has become apparent that these enzymes might also have a role in the reduction of proteins that are misfolded and targeted for degradation in the cytosol via a process termed ER-associated degradation (ERAD) (Antoniou et al., 2002; Dong et al., 2008; Ushioda et al., 2008). These studies suggest that ERp57 or ERdj5 are involved in the reduction of disulphides bonds before retrotranslocation of proteins from the ER to the cytosol. To investigate whether any of the substrate-trapping PDI family members could form mixed disulphides with an ERAD substrate, we translated either the wild type or a mutant form of the glycoprotein α1-antitrypsin. The NHK mutant of α1-antitrypsin is known to misfold and be retained in the ER and is eventually degraded via ERAD. During this process, the single cysteine in NHK mutant of α1-antitrypsin forms an aberrant interchain disulphide bond that needs to be reduced before it can be retrotranslocated (Hosokawa et al., 2006). When wild-type α1-antitrypsin was translated in the presence of SP cells derived from the various cell lines expressing substrate-trapping PDI family members, no mixed disulphides were seen with any of the cell lines (Fig. 7A). This result was to be expected, because α1-antitrypsin would not normally form any disulphide bonds. However, when the NHK mutant was translated in the identical cell lines, clear mixed disulphides were seen with ERp57 and P5. The mixed disulphide with ERp57 is probably not surprising because this protein is glycosylated and would be expected to be targeted to calnexin. However, an interaction with P5 suggests that targeting to BiP or ERp94 occurs (Christianson et al., 2008), which might result from protein misfolding.
Fig. 7.
Substrate targeted for ERAD forms mixed disulphides with P5. Translation of either wild-type α1-antitrypsin (A) or the NHK mutant of α1-antitrypsin (B) was carried out as described in Fig. 4. The band labelled with an asterisk is the aberrantly interchain disulphide-bonded α1-antitrypsin.
Discussion
The presence of a large family of oxidoreductase enzymes characterised by their similarity to PDI and localisation to the ER has raised questions as to whether each enzyme catalyses specific reactions or has a distinct set of substrate proteins (Ellgaard and Ruddock, 2005; Hatahet and Ruddock, 2007). Our results demonstrate that although some redundancy does exist, there is clear specificity amongst these enzymes for protein substrates. The requirement for individual oxidoreductases to catalyse the folding of individual substrates has been recently elegantly highlighted. The removal of one such enzyme, AGR2, resulted in a complete block in the secretion of intestinal mucus (Park et al., 2009). The substrate-trapping approach used here has identified some of the substrates for individual PDI family members. As with AGR2, it is highly likely that there will be an absolute requirement for each of the PDI family members for the correct folding and assembly of some client proteins.
Although individual substrates were identified for ERp57, ERp46 and ERp18, we were unable to identify substrates for PDI, ERp72 or P5 using the 2D gel analysis followed by mass spectrometry. It was clear that, with the exception of Ero1α, the PDI substrate-trapping mutant does not form many mixed disulphides. This observation was confirmed when we carried out in vitro translation of individual substrates into SP cells. The fact that a mixed disulphide is observed between PDI and Ero1α might be explained by the reduction of one of the regulatory or active site disulphides within Ero1α by PDI (Appenzeller-Herzog et al., 2008; Baker et al., 2008). The absence of other mixed disulphides could be explained if the main cellular function of PDI is to catalyse disulphide formation. The substrate-trapping mutant cannot act as a donor of a disulphide bond, so a mixed disulphide would only be formed during the reduction of a substrate. In yeast, the essential function of PDI is to catalyse disulphide exchange, although a role for the isomerase activity of PDI cannot be ruled out (Xiao et al., 2004). The ability of PDI to reduce substrates in vitro is well established, therefore it seems unlikely that such a reaction does not take place in cells. It might be that the mixed disulphides formed between PDI and substrates are very unstable, even with the substrate-trapping mutant, and are resolved before the free thiols can be alkylated with N-ethylmaleimide (NEM). These intermediates could be resolved either by glutathione present within the ER lumen or by free thiols within the substrate protein.
For ERp72, only a low level of mixed disulphides was observed and this could not be identified by mass spectrometry. The main substrates that formed mixed disulphides with this enzyme following in vitro translation were glycoproteins, which were particularly apparent when the interaction with calnexin was blocked. A recent structural analysis of the bb′ domain of ERp72 shows strong similarity to ERp57, although ERp72 does not interact with calnexin (Kozlov et al., 2009). It might be that when substrates that are normally acted upon by ERp57 are prevented from entering the calnexin cycle they can then become substrates for ERp72. It has also been shown that the folding of substrates that enter the calnexin cycle is impaired in ERp57-knockout cells. Preventing these substrates from interacting with calnexin then allows their efficient folding (Jessop et al., 2009), presumably by allowing an interaction with enzymes such as ERp72 (Solda et al., 2006). It is equally possible that the main substrates for ERp72 might not be expressed in HT1080 cells; indeed, it has been shown previously that ERp72 associates with mutant thyroglobulin, a protein that is expressed specifically in thyrocytes (Menon et al., 2007).
We showed that P5 forms a non-covalent interaction with BiP; an interaction that has been demonstrated previously (Meunier et al., 2002). In the cell line expressing wild-type P5, this interaction was stabilised by the addition of a crosslinking agent, demonstrating that the association is not dependent upon the active site mutations. The presence of both intrachain and interchain disulphide-bonded forms of the substrate-trapping mutant of P5 is noteworthy. It raises the possibility that mixed disulphides between one active site in P5 and its substrate could be resolved by a second active site, either in the same protein or in an adjacent molecule. The interaction between P5 and BiP is destabilised when P5 is reduced, hinting at a possible mechanism of regulation. In this scenario, P5 could be recruited to BiP under conditions of oxidative load in the ER, thereby assisting in the resolution of non-native disulphides in substrates that become misfolded and targeted to BiP. It has been shown previously that hyperoxidising conditions lead to aberrant disulphide formation in proteins that become associated with BiP in large complexes (Marciniak et al., 2004). The presence of P5 in these complexes would facilitate refolding of these proteins.
There was a clear contrast in the ability to identify substrates forming mixed disulphides with P5 by mass spectrometry analysis and by proteins translated in vitro. In addition, the classical substrate for BiP, immunoglobulin heavy chain, formed mixed disulphides with P5, as did other substrates that are likely to require BiP for their folding. The simplest explanation for these results is that proteins that are targeted to BiP for their folding can become substrates for P5. Such an association of a PDI family member with a chaperone system would mirror the situation with ERp57 and the calnexin cycle. The consequence of a protein requiring an interaction with a chaperone, such as calnexin or BiP, to assist their retention in the ER or their folding, would be that a specialised oxidoreductase becomes associated with the chaperone. As the specificity of BiP is such that it will interact with most secreted proteins, our inability to identify specific substrates for P5 by mass spectrometry would simply reflect the large number of interactions that take place. As well as forming mixed disulphides with folding proteins, P5 also formed a mixed disulphide with a substrate for ERAD. In this case, the misfolded protein is likely to bind to BiP, thereby targeting the protein to P5 to act as a reductase. Any aberrant disulphides would then be removed before retrotranslocation of the protein from the ER to the cytosol for degradation. A similar role has been suggested for ERdj5 (Ushioda et al., 2008), which has been shown to accelerate the reduction and degradation of the NHK variant of α1-antitrypsin.
The ability of ERp46 to form a mixed disulphide with Prx-IV highlights the fact that some oxidoreductases are likely to catalyse disulphide-exchange reactions that are part of the normal redox regulation of enzymatic activity. Prx-IV is an ER-localised thioredoxin peroxidase, which is able to catalyse the conversion of hydrogen peroxide to water (Tavender et al., 2008; Wood et al., 2003). During its enzymatic cycle, the active site cysteine reacts with H2O2 to form H2O, and in the process the thiol group is oxidised to form sulphenic acid. The sulphenic group then reacts with a second thiol group in a neighbouring Prx-IV molecule to form an interchain disulphide. This disulphide must be reduced to complete the catalytic cycle: a reaction that is probably catalysed by a disulphide reductase. The formation of a mixed disulphide between ERp46 and Prx-IV indicates that this oxidoreductase could fulfil the role of disulphide reductase during the recycling of Prx-IV. Such a role for a PDI family member in the regulation of protein activity has already been suggested for ERp57, which might regulate the ER calcium pump SERCA2b by modulating the redox state of a key intrachain disulphide bond (Li and Camacho, 2004).
The ability of ERp18 to specifically form mixed disulphides with PTX3 during its assembly into a decamer and with the C-propeptide of procollagen during its assembly into a trimer is intriguing. Structural analysis of ERp18 has revealed that it contains a loop insertion that positions a number of hydrophobic residues close to the active site (Rowe, 2009). This loop insertion is only present in ERp18, AGR2 and AGR3, which makes it a prime candidate for interacting directly with substrates and conferring specificity to the enzyme. Further mutagenesis needs to be carried out to determine whether this is indeed the region of substrate binding.
In summary, we have begun to clarify why there are so many members of the PDI family of oxidoreductases in the ER of mammalian cells. Although there might be some demarcation of oxidation and reductase function, there is clear specificity in the types of client proteins with which each enzyme reacts. This specificity might lie with the targeting of substrates to particular chaperones (ERp57, P5) but might also be due to distinct protein-protein interactions (ERp46, ERp18). Once more substrates for individual enzymes are identified, it should be possible to determine the general features that characterise specificity.
Materials and Methods
DNA constructs
The ERp57 expression constructs were as published previously (Jessop et al., 2007). The expression constructs for PDI, P5, ERp72, ERp18 and for wild-type ERp46 were all constructed in pcDNA3.1(+) (Invitrogen) by amplification of the coding sequence to include a V5 tag followed by a KDEL ER retrieval sequence and a stop codon at the 3′ end. Mutagenesis of the C-terminal active site cysteine to alanine in PDI, P5, ERp72 and ERp18 was performed by PCR. The mutant ERp46 with mutations in each of the active sites along with the V5 tag and KDEL sequence was synthesised de novo (Genescript). The β1 integrin, clusterin, peroxiredoxin-4 and procollagen C-propeptide constructs were as published previously (Bottomley et al., 2001; Jessop et al., 2007; Tavender et al., 2008). The Ero1α construct was a gift from Roberto Sitia (DIBIT, Milan, Italy); the α1-antitrypsin wild-type and NHK mutant constructs were a gift from Lisa Swanton (University of Manchester, Manchester, UK) and the LDLR a gift from Ineke Braakman (University of Utrecht, Utrecht, The Netherlands). The PTX3 construct was purchased from Origene. The mouse monoclonal anti-V5 antibody was purchased from Invitrogen and the V5-agarose beads from Sigma.
Construction of stable cell lines
Plasmids were linearised with PvuI before transfecting into subconfluent HT1080 human fibroblasts with Fugene8 (Roche) according to the manufacturer's instructions. Stable cell-lines were selected with G418 (Sigma) for 14 days before colonies were isolated and screened by western blotting for expression of the respective PDI family member using a mouse monoclonal anti-V5 antibody (Invitrogen).
2D gel electrophoresis
HT1080 cells expressing substrate-trapping mutants were treated with 25 mM NEM to preserve mixed disulphides. Cells (1×108) were lysed in 1% (v/v) Triton X-100, 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM ethylenediamine tetra-acetic acid (EDTA) and 0.5 mM phenylmethylsulphonyl fluoride (PMSF) (lysis buffer). Clarified lysates were pre-incubated with protein-A Sepharose for 30 minutes to limit non-specific binding, before incubation with anti-V5 antibody conjugated to agarose beads (Sigma) for 16 hours at 4°C. Beads were washed four times with 100× bed volume 1% (w/v) deoxycholic acid, 0.1% (w/v) SDS, 1% (v/v) Triton X-100, 50 mM Tris-HCl, pH 8.0, 500 mM NaCl and 0.5 mM PMSF (RIPA buffer). Proteins were eluted in non-reducing SDS-PAGE sample buffer [0.25 mM Tris-HCl, pH 6.8, 2% (w/v) SDS, 20% (v/v) glycerol, 0.004% (w/v) bromophenol blue] and boiled before being separated by SDS-PAGE. Gel lanes were excised and incubated in buffer containing 50 mM DTT for 10 minutes before separation through a second SDS-PAGE gel. Proteins were visualised by silver staining. Spots corresponding to proteins that had formed mixed disulphides were excised from the gel, digested with trypsin and peptides identified by tandem mass spectrometry (LC/MS/MS) on a quadrupole-time-of-flight (QTOF) instrument. Peptides were matched against predicted peptides in the current version of Mass Spectrometry Database (MSDB) using Mascot software.
Transcription and translation in vitro
DNA was transcribed and proteins translated essentially as described previously (Jessop et al., 2007; Wilson et al., 1995). RNA transcripts were translated in a rabbit reticulocyte lysate (Flexilysate, Promega) and where specified in the presence of SP cells. Where appropriate SP cells were preincubated with castanospermine (1 mM) for 5 minutes. For the substrate-trapping experiments, translation was carried out for 2 hours, and reactions stopped by the addition of NEM to a final concentration of 25 mM. SP cells were isolated by centrifugation, resuspended in 50 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl, 1% v/v Triton X-100 and a third of the sample removed. The remaining two thirds of the sample was immunoisolated with the V5 antibody and protein-A Sepharose. Both the total translation product aliquot and the immunoisolated material were added to SDS-PAGE sample buffer in preparation for electrophoresis. For the time course experiments, initiation of protein synthesis was allowed to proceed for 5 minutes at 30°C before inhibition with 1 mM aurintricarboxylic acid (ATCA; Sigma), followed by further incubation at 30°C to allow elongation and post-translational modification. At specified times, 25 mM NEM was added at 4°C to prevent disulphide exchange and halt further folding. SP cells were isolated and resuspended in SDS-PAGE sample buffer. Samples were separated by SDS-PAGE under non-reducing conditions (unless stated otherwise), dried and exposed to Fuji BioMax MR film (GRI, Essex, UK).
P5 and BiP interactions
To stabilise interactions between BiP and P5, cells were treated with the thiol-cleavable crosslinking agent dithiobis(succinimidylpropionate) (DSP; 1 mM) for 1 hour and then incubated with 100 mM glycine and 40 mM NEM for an additional 15 minutes to quench the crosslinker. The cells were lysed in lysis buffer and clarified by centrifugation at 16,100 g for 10 minutes. P5 was immunoisolated from the resulting clarified lysates and separated by SDS-PAGE under reducing conditions. Associated BiP was visualised by western blotting using an anti-BiP antibody (a gift from Richard Zimmerman, Saarland University, Saarbruecken, Germany).
For isolating P5-BiP complexes in the absence of crosslinking agent, cells were first treated with 20 mM NEM for 5 minutes and lysed in 50 mM Tris-HCl buffer, pH 7.4, containing, 150 mM NaCl, 2.5 mM EDTA, 5 mM EGTA, 1% Triton X-100, protease inhibitor cocktail, 1 U/ml apyrase, with 20 mM NEM, unless stated otherwise. To determine the ATP dependence of the interaction, cells were lysed in 50 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl, 2 mM ATP, 2 mM MgCl2, 1% Triton X-100, protease inhibitor cocktail with 20 mM NEM. Lysates were clarified and P5 isolated and associated BiP visualised as described above. Where appropriate, cells were pretreated with 10 mM DTT or 1 mM DPS before addition of NEM and cell lysis.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/23/4287/DC1
We would like to acknowledge Roberto Sitia, Lisa Swanton, Richard Zimmerman and Ineke Braakman, for reagents. The work was support by grants from the BBSRC (D00764) and the Wellcome Trust (074081). Deposited in PMC for release after 6 months.
References
- Antoniou, A. N., Ford, S., Alphey, M., Osborne, A., Elliott, T. and Powis, S. J. (2002). The oxidoreductase ERp57 efficiently reduces partially folded in preference to fully folded MHC class I molecules. EMBO J. 21, 2655-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog, C., Riemer, J., Christensen, B., Sorensen, E. S. and Ellgaard, L. (2008). A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J. 3, 2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, K. M., Chakravarthi, S., Langton, K. P., Sheppard, A. M., Lu, H. and Bulleid, N. J. (2008). Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 27, 2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottazzi, B., Vouret-Craviari, V., Bastone, A., De Gioia, L., Matteucci, C., Peri, G., Spreafico, F., Pausa, M., D'Ettorre, C., Gianazza, E. et al. (1997). Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J. Biol. Chem. 272, 32817-32823. [DOI] [PubMed] [Google Scholar]
- Bottomley, M. J., Batten, M. R., Lumb, R. A. and Bulleid, N. J. (2001). Quality control in the endoplasmic reticulum: PDI mediates the ER retention of unassembled procollagen C-propeptides. Curr. Biol. 11, 1114-1118. [DOI] [PubMed] [Google Scholar]
- Bulleid, N. J. and Freedman, R. B. (1988). Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature 335, 649-651. [DOI] [PubMed] [Google Scholar]
- Bulleid, N. J., Wilson, R. and Lees, J. F. (1996). Type-III procollagen assembly in semi-intact cells: chain association, nucleation and triple-helix folding do not require formation of inter-chain disulphide bonds but triple-helix nucleation does require hydroxylation. Biochem. J. 317, 195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi, S. and Bulleid, N. J. (2004). Glutathione is required to regulate the formation of native disulphide bonds within proteins entering the secretory pathway. J. Biol. Chem. 279, 39872-39879. [DOI] [PubMed] [Google Scholar]
- Christianson, J. C., Shaler, T. A., Tyler, R. E. and Kopito, R. R. (2008). OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, M., Bridges, J. P., Apsley, K., Xu, Y. and Weaver, T. E. (2008). ERdj4 and ERdj5 Are Required for Endoplasmic Reticulum-associated Protein Degradation of Misfolded Surfactant Protein C. Mol. Biol. Cell 19, 2620-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L. and Ruddock, L. W. (2005). The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6, 28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, D. M. and Soling, H. D. (1999). The protein disulphide-isomerase family: unravelling a string of folds. Biochem. J. 339, 1-10. [PMC free article] [PubMed] [Google Scholar]
- Frand, A. R. and Kaiser, C. A. (1998). The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1, 161-170. [DOI] [PubMed] [Google Scholar]
- Freedman, R. B. (1995). The formation of protein disulphide bonds. Curr. Opin. Struct. Biol. 5, 85-91. [DOI] [PubMed] [Google Scholar]
- Hatahet, F. and Ruddock, L. W. (2007). Substrate recognition by the protein disulfide isomerases. FEBS J. 274, 5223-5234. [DOI] [PubMed] [Google Scholar]
- Hebert, D. N., Foellmer, B. and Helenius, A. (1996). Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 15, 2961-2968. [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, N., Wada, I., Natsuka, Y. and Nagata, K. (2006). EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells 11, 465-476. [DOI] [PubMed] [Google Scholar]
- Hwang, C., Sinskey, A. J. and Lodish, H. F. (1992). Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496-1502. [DOI] [PubMed] [Google Scholar]
- Jessop, C. E. and Bulleid, N. J. (2004). Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 279, 55341-55347. [DOI] [PubMed] [Google Scholar]
- Jessop, C. E., Chakravarthi, S., Garbi, N., Hammerling, G. J., Lovell, S. and Bulleid, N. J. (2007). ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 26, 28-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop, C. E., Tavender, T. J., Watkins, R. H., Chambers, J. E. and Bulleid, N. J. (2009). Substrate specificity of the oxidoreductase ERp57 is determined primarily by its interaction with calnexin and calreticulin. J. Biol. Chem. 284, 2194-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivu, J., Myllyla, R., Helaakoski, T., Pihlajaniemi, T., Tasanen, K. and Kivirikko, K. I. (1987). A single polypeptide acts both as the beta subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase. J. Biol. Chem. 262, 6447-6449. [PubMed] [Google Scholar]
- Kozlov, G., Maattanen, P., Schrag, J. D., Hura, G. L., Gabrielli, L., Cygler, M., Thomas, D. Y. and Gehring, K. (2009). Structure of the noncatalytic domains and global fold of the protein disulfide isomerase ERp72. Structure 17, 651-659. [DOI] [PubMed] [Google Scholar]
- Laboissiere, M. C., Sturley, S. L. and Raines, R. T. (1995). The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J. Biol. Chem. 270, 28006-28009. [DOI] [PubMed] [Google Scholar]
- Li, Y. and Camacho, P. (2004). Ca2+-dependent redox modulation of SERCA 2b by ERp57. J. Cell Biol. 164, 35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles, M. M. and Gilbert, H. F. (1991). Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry 30, 613-619. [DOI] [PubMed] [Google Scholar]
- Marciniak, S. J., Yun, C. Y., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., Nagata, K., Harding, H. P. and Ron, D. (2004). CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, S., Lee, J., Abplanalp, W. A., Yoo, S. E., Agui, T., Furudate, S., Kim, P. S. and Arvan, P. (2007). Oxidoreductase interactions include a role for ERp72 engagement with mutant thyroglobulin from the rdw/rdw rat dwarf. J. Biol. Chem. 282, 6183-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier, L., Usherwood, Y. K., Chung, K. T. and Hendershot, L. M. (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezghrani, A., Fassio, A., Benham, A., Simmen, T., Braakman, I. and Sitia, R. (2001). Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 20, 6288-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni, S. N., Fassio, A., Ciriolo, M. R., Filomeni, G., Pasqualetto, E., Fagioli, C. and Sitia, R. (2004). Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J. Biol. Chem. 279, 32667-32673. [DOI] [PubMed] [Google Scholar]
- Park, S. W., Zhen, G., Verhaeghe, C., Nakagami, Y., Nguyenvu, L. T., Barczak, A. J., Killeen, N. and Erle, D. J. (2009). The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 106, 6950-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, M. G., Travers, K. J. and Weissman, J. S. (1998). Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1, 171-182. [DOI] [PubMed] [Google Scholar]
- Rowe, M. L., Ruddock, L. W., Kelly, G., Schmidt, J. M., Williamson, R. A. and Howard, M. J. (2009). Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase. Biochemistry 48, 4596-4606. [DOI] [PubMed] [Google Scholar]
- Sevier, C. S. and Kaiser, C. A. (2002). Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell. Biol. 3, 836-847. [DOI] [PubMed] [Google Scholar]
- Solda, T., Garbi, N., Hammerling, G. J. and Molinari, M. (2006). Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J. Biol. Chem. 281, 6219-6226. [DOI] [PubMed] [Google Scholar]
- Tavender, T. J., Sheppard, A. M. and Bulleid, N. J. (2008). Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem. J. 411, 191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda, R., Hoseki, J., Araki, K., Jansen, G., Thomas, D. Y. and Nagata, K. (2008). ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569-572. [DOI] [PubMed] [Google Scholar]
- Wetterau, J. R., Combs, K. A., McLean, L. R., Spinner, S. N. and Aggerbeck, L. P. (1991). Protein disulfide isomerase appears necessary to maintain the catalytically active structure of the microsomal triglyceride transfer protein. Biochemistry 30, 9728-9735. [DOI] [PubMed] [Google Scholar]
- Wilson, R., Allen, A. J., Oliver, J., Brookman, J. L., High, S. and Bulleid, N. J. (1995). The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem. J. 307, 679-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R., Lees, J. F. and Bulleid, N. J. (1998). Protein disulfide isomerase acts as a molecular chaperone during the assembly of procollagen. J. Biol. Chem. 273, 9637-9643. [DOI] [PubMed] [Google Scholar]
- Wood, Z. A., Poole, L. B. and Karplus, P. A. (2003). Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650-653. [DOI] [PubMed] [Google Scholar]
- Xiao, R., Wilkinson, B., Solovyov, A., Winther, J. R., Holmgren, A., Lundstrom-Ljung, J. and Gilbert, H. F. (2004). The contributions of protein disulfide isomerase and its homologues to oxidative protein folding in the yeast endoplasmic reticulum. J. Biol. Chem. 279, 49780-49786. [DOI] [PubMed] [Google Scholar]
- Zapun, A., Darby, N. J., Tessier, D. C., Michalak, M., Bergeron, J. J. and Thomas, D. Y. (1998). Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J. Biol. Chem. 273, 6009-6012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.